Abstract
Previous analyses using in situ hybridization alone or together with PCR have yielded conflicting results regarding the cell type in which latent varicella-zoster virus (VZV) resides. We separated human trigeminal ganglia (TG) into neuronal and nonneuronal fractions, followed by primary and nested PCR to quantitate VZV DNA at the single cell level. Both TG from each of eight cadavers were dissociated and separated into neuronal and nonneuronal cell suspensions by differential filtration. Analysis of the neuron fraction (5,000 neurons per sample) revealed VZV DNA in 9 of 16 samples, with copy numbers ranging from 1 to 12, whereas only 2 of 16 nonneuronal cell samples were positive for VZV DNA, with 1 copy each. Further analysis of 10 samples of 100 neurons and the corresponding nonneuronal cell fractions from each TG of a single subject revealed VZV DNA in 3 of 10 samples of the left TG (range, 2 to 5 copies) and in 1 of 10 samples of the right TG (2 copies) but in none of the 20 nonneuronal cell fractions. These data indicate that latent VZV DNA is present primarily, if not exclusively, in neurons, at a frequency of two to five copies per latently infected neuron.
Varicella-zoster virus (VZV), a ubiquitous neurotropic alphaherpesvirus, causes chickenpox (varicella), becomes latent in dorsal root ganglia at all levels of the neuraxis, and may reactivate decades later to produce shingles (zoster). Because of the morbidity associated with zoster and its attendant neurological complications, prevention of virus reactivation is an issue of paramount importance. Thus, the initial detection of latent VZV in human ganglia by nucleic acid hybridization techniques (8) led to efforts in several laboratories to determine the physical state of virus in latently infected human ganglia, and to identify the cell type in which VZV becomes latent. VZV was first detected exclusively in neurons (9, 10), then in perineuronal satellite cells (3, 14), and later in both neurons and various nonneuronal cells of latently infected human ganglia (14). Although two further studies corroborated the initial finding of VZV latency exclusively in neurons (6, 11), the exact location of latent VZV remains controversial. Because all of these studies were carried out by using in situ hybridization (ISH) or a combination of ISH and PCR, we examined the site of VZV latency by using a different approach. Immediately after death of the subject, we removed ganglia, separated the cells by size, identified them as neurons or nonneuronal cells, and quantitated VZV DNA by PCR in our different cell-type populations.
MATERIALS AND METHODS
Human tissue.
Both trigeminal ganglia (TG) were removed from eight subjects within 24 h of death (range, 12 to 24 h). Table 1 summarizes the clinical information on the subjects. At autopsy, there were no skin lesions attributable to VZV, and no subject was immunocompromised before death.
TABLE 1.
Clinical features of subjects
| Subject | Age (yr)/sexa | Cause of death |
|---|---|---|
| 1 | 44/M | Cirrhosis/liver failure |
| 2 | 66/M | Pancreatic cancer |
| 3 | 67/M | Metastatic melanoma |
| 4 | 50/M | Prostate cancer |
| 5 | 51/M | Perforated bowel |
| 6 | 55/F | Gastric surgery postoperative complications |
| 7 | 67/M | Ruptured abdominal aortic aneurysm |
| 8 | 66/F | Myocardial infarction |
M, male; F, female.
We adapted the cell separation protocol of Sawtell (16) to human TG removed at autopsy. Our study consisted of two parts. In part A, samples of ganglia containing 5,000 neurons and associated nonneuronal cells (satellite and Schwann cells, fibroblasts, endothelial, and blood cells) were separated by size into neuronal (20 to 70 μm in diameter) and nonneuronal (<20 μm in diameter) fractions by passage through nylon filters. The VZV DNA content was quantitated in each fraction. In part B, two ganglia found in part A to contain the most abundant VZV DNA were further examined for VZV DNA copy number by using samples of 100 neurons and associated nonneuronal cells.
Fixation, dissociation, and cell separation of TG.
The TG were placed into 40 ml of Streck tissue fixative (STF; Streck Laboratories, Inc., Omaha, Nebr.). After 1 h, the TG were removed, diced finely with a scalpel, returned to fresh STF for 12 to 16 h, centrifuged at 800 × g for 10 min, and washed with 50 ml of nuclease-free water. After two further rounds of centrifugation and washing, ganglia pieces were placed into 10 ml of 0.25% collagenase (CLS I; Worthington Biochemical Corp., Lakewood, N.J.) in Hanks balanced salt solution containing 1% penicillin-streptomycin-amphotericin B (Gibco BRL, Gaithersburg, Md.) adjusted to pH 7.4, followed by incubation, with shaking, at 37°C for 24 to 48 h. Aliquots (10 μl) were removed and examined microscopically to monitor tissue dissociation. If the tissue was not completely dissociated, the tube was centrifuged as described above, and the supernatant fluid was removed and stored at 4°C. Any remaining undissociated tissue was retreated with fresh collagenase. Complete dissociation often required several treatments.
To estimate the number of neurons, 10-μl aliquots of dissociated ganglia were removed and examined microscopically. Large cells (>20 μm in diameter) morphologically consistent with neurons were counted. Volumes containing a mixed population of 5,000 neurons together with their associated nonneuronal cells were sorted by repeated filtration through nylon mesh. Suspended cells were filtered through a 70-μm-pore-size SpectraMesh (Spectrum, Laguna Hills, Calif.) to remove most large tissue remnants, and the flowthrough was centrifuged at 800 × g for 10 min, resuspended in 10 ml of STF, and heated to 70°C for 15 min. After this postfixation, some mixed cells were stored at 4°C and could be used days to weeks later for separation into neurons and nonneuronal cells. Another portion of the mixed cells was immediately separated into neurons and nonneuronal cells by filtration through either 20-μm-pore-size SpectraMesh or 20-μm-pore-size CellTrics nylon mesh cartridges (BioErgonomics, Inc., White Bear Lake, Minn.), which retained the larger neurons. While still in the cartridge, large neurons were washed with 10 ml of STF. The mesh was cut from the cartridge with a scalpel, placed into 2 ml of STF, and agitated on a Vortex mixer to remove most neurons from the mesh. Neuronal and nonneuronal fractions were centrifuged at 800 × g for 10 min and washed with 2 ml of nuclease-free water. After two further rounds of centrifugation and washing, 10-μl aliquots of the nonneuronal fraction were examined microscopically for neurons; fractions revealing neurons were further filtered through 10-μm-pore-size SpectraMesh, and only the flowthrough was retained.
For part B, populations of neurons and nonneuronal cells were similarly prepared from aliquots containing approximately 100 neurons.
Immunocytochemistry.
Samples of each cell suspension population (mixed, neuronal, and nonneuronal) and a suspension of liver cells were air dried onto Superfrost/Plus glass microscope slides (Fisher Scientific, Pittsburgh, Pa.). Slides were incubated overnight at room temperature, washed for 5 min in phosphate-buffered saline (pH 7.4) (PBS), and incubated for 4 h in blocking buffer (PBS, 1% fraction V bovine serum albumin and 0.5% Igepal CA-630) (Sigma, St. Louis, Mo.). Mouse monoclonal anti-neurofilament-200 antibody (clone N52, N-0142; Sigma) diluted 1:2,500 in blocking buffer was applied to the slides within an area drawn with a Pap Pen (Research Products International Corp., Mount Prospect, Ill.). Slides were incubated in a humid chamber at room temperature for 4 h, washed three times with PBS, followed by incubation at room temperature for 2 h with alkaline phosphatase-conjugated horse anti-mouse immunoglobulin G (IgG) antibody (Vector Laboratories, Inc., Burlingame, Calif.) diluted 1:300 in blocking buffer. Slides were then washed three times with PBS and placed in buffer 1 (100 mM Tris-HCl, pH 7.5; 150 mM NaCl) for 5 min, followed by treatment with buffer 2 (100 mM Tris-HCl, pH 9.5; 100 mM NaCl; 50 mM MgCl2) for 10 min. Color was developed by using buffer 2 containing levamisole (0.24 mg/ml; Sigma), nitroblue tetrazolium (0.45 mg/ml), and BCIP (5-bromo-4-chloro-1-indolylphosphate toluidinium; 0.175 mg/ml; Boehringer Mannheim, Indianapolis, Ind.) for 4 h and a washing for 5 min in distilled water. Coverslips were applied over Immu-Mount (Shandon, Pittsburgh, Pa.).
The nonneuronal population was similarly processed and incubated with a 1:1,000 dilution of rabbit anti-actin antibody (A-2668; Sigma), followed by 1:300 dilution of alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (Vector). Additional mixed-cell populations were stained with either acridine orange for RNA or Luxol-fast blue for myelin.
DNase treatment.
PCR performed on 5 μl of the liquid phase obtained after centrifugation (800 × g for 10 min) of the cell populations described above confirmed the presence of extracellular β-actin DNA, most likely from burst cells (data not shown). To eliminate the possibility that extracellular VZV DNA might also be present, all cell populations were treated as described previously (16) with DNase immobilized on polyvinyl beads (F7m; Bio 101, Inc., Vista, Calif.) suspended in reaction buffer (20 mM Tris-HCl, pH 7.6; 5 mM MgCl2; 5 mM CaCl2) at a concentration of 1,000 beads per μl.
For part A, samples initially containing approximately 5,000 neurons and their associated nonneuronal cells were used. Samples were either separated by filtration into neuronal and nonneuronal populations or not separated. All samples were centrifuged in 2-ml microcentrifuge tubes at 800 × g for 10 min and resuspended in 500 μl of reaction buffer containing DNase as described above. Based on our microscopic observations indicating approximately 100 nonneuronal cells per neuron, approximately 500,000 beads were present for 500,000 cells in the mixed population. After cell separation, the nonneuronal cell-to-bead ratio was slightly less than 1:1 and was approximately 1:100 for neurons. Samples were placed on a mixing apparatus at 37°C for at least 16 h; we were careful to ensure that the tubes were completely inverted so that the DNase beads were always in contact with the liquid containing the cells. After 16 h, tubes were heated to 95°C for 5 min to inactivate the DNase and centrifuged at 800 × g for 10 min, and then all but approximately 5 μl of the reaction buffer was carefully removed.
For part B, cell populations based on 100 neurons were centrifuged in PCR tubes, resuspended in 50 μl of reaction buffer, and treated as described above.
DNA extraction.
For part A, DNA was extracted by using a QIAamp affinity chromatography kit (Qiagen, Inc., Chatsworth, Calif.) according to the manufacturer’s instructions for tissue-based DNA extraction, and then eluted with 50 μl of nuclease-free water.
For part B, the small number of cells precluded DNA extraction, and crude cell lysates were prepared instead (16). The total volume in the PCR tubes, including cells, DNase beads (inactivated), and DNase buffer remnant, was brought to 50 μl by the addition of 45 μl of conversion buffer (20 mM Tris-HCl, pH 8.4; 3 mM MgCl2; 50 mM KCl). After the addition of 2 μl of a 10-μg/μl proteinase K solution (Gibco BRL) tubes were kept at 50°C for several hours. When examination of the contents of the PCR tubes under an inverted microscope revealed no intact cells, tubes were heated to 95°C for 10 min to inactivate the proteinase K.
To ensure complete disruption of cells, some samples in both parts A and B were repeatedly quick-frozen in liquid nitrogen and thawed at 37°C (average, three to five times), or mixed with 425- to 600-μm-diameter acid-washed glass beads (Sigma) and agitated on a Vortex mixer. When necessary, samples were treated with additional proteinase K as described above.
Plasmid construction for PCR quantitation.
We used plasmid pBSal-C (1, 2) into which unique HindIII and SpeI sites had been introduced outside the VZV gene 21 open reading frame (ORF). Plasmid DNA was extracted (Qiagen), linearized by HindIII digestion, quantitated by spectrophotometry, and diluted to yield 104, 103, 102, 101, 100, and 10−1 copies of VZV ORF 21 in 10 μl of nuclease-free water.
A plasmid containing 476 bp of human actin cDNA was constructed by PCR amplification of human cDNA with actin-specific primers A1 and A2 (Table 2). The amplified actin fragment was inserted into PCR-II (Invitrogen, Carlsbad, Calif.), and plasmid DNA was extracted, digested with EcoRI, quantitated by spectrophotometry, and diluted to yield 107, 106, 105, 104, 103, 102, 101, 100, and 10−1 copies of β-actin DNA in 10 μl of nuclease-free water.
TABLE 2.
Oligonucleotides used in this study
| Gene and name | Sequence (5′ to 3′) | Location | Orientationa |
|---|---|---|---|
| VZV ORF21 | |||
| V1 | CCGAATCTATTGCAACCGTG | 33475b | Sense |
| V2 | TTGTCCTATTATCAGCGTCGG | 33791 | Antisense |
| V3 | GCCCCAACCTCATATATGAG | 33546 | Sense |
| V4 | GAAATTACGTCTCTGACCGTTG | 33763 | Antisense |
| VP | ACAAGGCAGCAGTTTCATTCG | 33588 | Sense |
| β-Actin | |||
| A1 | GATGCATTGTTACAGGAAGT | 3260c | Sense |
| A2 | TCATACATCTCAAGTTGGGGG | 3500 | Antisense |
| AP | CAAGTCCACACAGGGGAGGTG | 3339 | Sense |
PCR.
For part A, 5 μl of DNA extracted from the three cell populations of each of 16 ganglia was amplified by using β-actin primers A1 and A2. β-Actin plasmid standards in a background of 100 ng of pGEM3z (Promega) DNA were similarly amplified. The remaining 45 μl of extracted DNA was amplified by using VZV gene 21 primers V1 and V2 (Table 2). VZV gene 21 plasmid standards in a background of 100 ng of BSC-1 DNA were similarly amplified. We chose VZV gene 21 because we have been able to detect as few as 10 copies of latent VZV DNA with gene 21 primers (1, 2). The 100-μl PCR reaction mixture consisted of 10 μl of PCR Buffer II (Perkin-Elmer, Branchburg, N.J.) (final concentration: 10 mM Tris-HCl, pH 8.3; 50 mM KCl), 6 μl of 25 mM MgCl2 (final concentration, 1.5 mM), 16 μl of 1.25 mM deoxynucleoside triphosphate (dNTP) mix (final concentration, 200 μM) (Boehringer Mannheim, Indianapolis, Ind.), 5 μl of 20 μM upstream and downstream primers (final concentration, 1 μM) (Genosys Biotechnologies, Inc., Woodlands, Tex.), and 0.5 μl of a 5-U/μl Taq DNA polymerase mixture (final concentration, 2.5 U/100 μl). Tubes were placed in a GeneAmp 2400 (Perkin-Elmer Cetus, Norwalk, Conn.), heated to 95°C for 5 min, and cycled 40 times through the following sequence: 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s, with no ramp time delays, followed by a final cycle of 95°C for 1 min, 55°C for 2 min, and 72°C for 7 min. After primary PCR for VZV gene 21, 5 μl was removed and reamplified (by nested PCR) in the above reaction mixture for various cycles by using VZV gene 21 primers V3 and V4 (Table 2).
For part B, tubes containing 50 μl of crude cell lysates, DNase beads and proteinase K (both inactivated) were converted to a PCR-enabling environment by the addition of 21 μl of 100 mM KCl (final concentration, 50 mM), 16 μl of 1.25 mM dNTP mix (final concentration, 200 μM), 5 μl of 20 μM upstream and downstream primers (final concentration, 1 μM), and 0.5 μl of a 5-U/μl mixture of Taq DNA polymerase (final concentration, 2.5 U/100 μl). The final volume was 100 μl and final concentrations of Tris-HCl and MgCl2 were 10 and 1.5 mM, respectively. The PCR primers, cycling conditions, and standard plasmid preparations were identical to those in part A. After primary PCR for VZV gene 21, 5 μl was removed and nested as described above.
PCR product detection.
PCR products were resolved by electrophoresis through 2% agarose gels containing 0.5 μg of ethidium bromide per ml, electrophoretically transferred to nylon membranes (Zeta-Probe; Bio-Rad Laboratories, Hercules, Calif.), and hybridized overnight at 42°C to 32P-end-labeled oligonucleotides internal to the appropriate PCR primers (VP and AP; Table 2). Blots were washed with oligonucleotide wash buffer, followed by two washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2% sodium dodecyl sulfate for 15 min each at 42°C. Images were developed by exposure to a PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, Calif.) and analyzed with the corresponding ImageQuant software. Graphical representations of the data were generated by using SigmaPlot version 4.01 (SPSS, Inc., Chicago, Ill.).
RESULTS
Dissociation and cell-type separation of human TG.
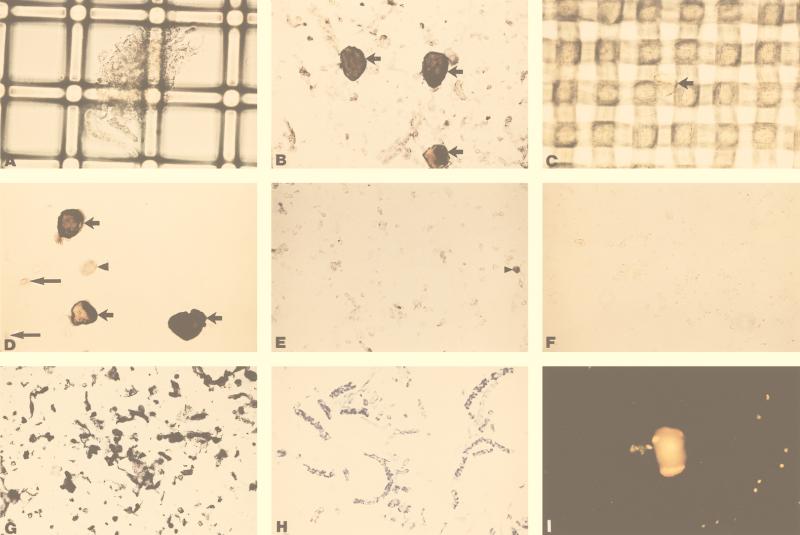
Figure 1 shows the separation of dissociated human ganglia into neuronal and nonneuronal cell populations. After collagenase treatment, tissue debris was retained on 70-μm filters (Fig. 1A), while neuronal and nonneuronal cells were collected in the filtrate (B). Neuronal and nonneuronal cells were further separated by retention (C) and liberation (D) of neurons on the 20-μm mesh, while most nonneuronal cells were collected in the filtrate (E). Immunocytochemistry revealed neurofilament-positive staining in neurons (B and D) and in neuronal fragments (E) but not in nonneuronal cells (E) or in liver cells (F). The small, clear nonneuronal cells in panel E retained their cellular contents, as indicated by the uniform positive staining for actin (G). The Luxol-fast blue-positive thin, filamentous material often seen in suspensions were identified as myelinated axon fragments (H). The diffuse cytoplasmic fluorescence seen after acridine orange staining (I) indicated that cells retain RNA after dissociation from intact ganglia.
FIG. 1.
Cell-type separation of dissociated human TG. (A) Large debris from a trigeminal ganglion captured by 70-μm mesh. (B) Flowthrough from 70-μm mesh contains neuronal and nonneuronal cells, including small debris. Arrows depict three large neurofilament-positive cells stained as described in Materials and Methods. (C) Neuron (arrow) retained by 20-μm mesh. (D) Three neurofilament-positive cells retained by 20-μm mesh (short arrows), together with two remaining nonneuronal cells (long arrows) and a cell fragment (arrowhead). (E) Small neurofilament-positive fragments (arrowhead) but no neurofilament-positive cells in the nonneuronal cell population after 20-μm mesh filtration. (F) Absence of neurofilament staining in control dissociated liver cells. (G) Positive actin staining in 20-μm filtrate (nonneuronal cells in E). (H) Luxol-fast blue staining of filamentous debris often seen in suspensions, revealing myelinated axon fragments. (I) Acridine orange staining of mixed-cell populations: diffuse orange cytoplasm represents retained cell RNA.
Quantitation of VZV DNA by PCR.
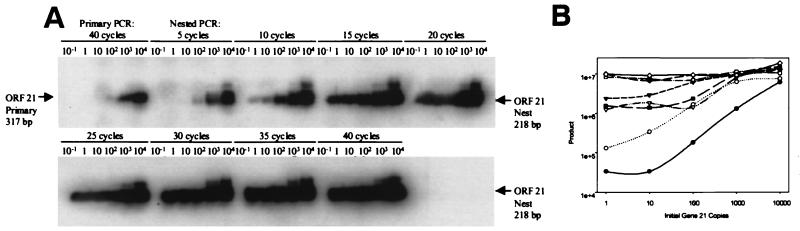
After 40 PCR cycles, 102 copies of target DNA were detected by hybridization (Fig. 2A). To increase the limit of detection, nested PCR was performed with multiple cycles. After 10 cycles, a single-copy target (Fig. 2A) and a linear relationship (r2 = 0.96) between log10 PCR product and log10 initial target copy number was seen (Fig. 2B), thus allowing quantitation.
FIG. 2.
Quantitation of VZV DNA. (A) PCR of dilutions from 10−1 to 104 copies of VZV gene 21 for various nested PCR cycles. (B) Graph of quantitation from part A, showing a linear relation between product and initial copy number for 10 cycles of nested PCR. Number of nested cycles: ●, 5; ○, 10; ▾, 15; ▿, 20; ■, 25; □, 30; ⧫, 35; ◊, 40.
Cell-type distribution of VZV DNA in dissociated TG from eight subjects (part A).
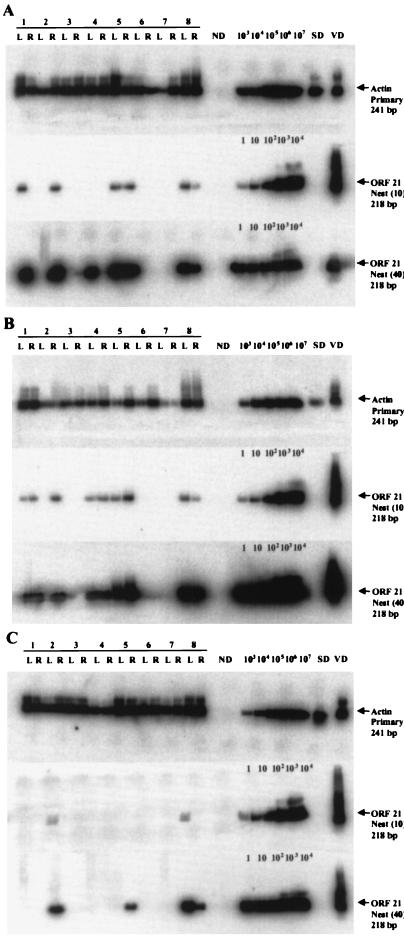
The top blots in each panel of Fig. 3 show actin DNA in mixed (A), neuronal (B), and nonneuronal (C) cells from all 16 ganglia. The middle blots in each panel reveal VZV gene 21 DNA in 6 of 16 mixed cell populations (A), 9 of 16 neuronal populations (B), and in 2 of 16 nonneuronal populations. Quantitative analysis (40 cycles of primary PCR, followed by 10 cycles of nesting) indicated the presence of 1 to 12 copies of latent viral DNA per sample (Table 3). Exhaustively nested PCR (40 cycles of primary PCR, followed by 40 cycles of nesting, bottom blots in each panel) revealed VZV DNA in 7 of 16 mixed cell populations (A), in 9 of 16 neuronal cell populations (B), and in 4 of 16 nonneuronal cell populations (C). When DNA was omitted from the PCR reaction (ND), no amplification product was detected. No VZV DNA was found after amplification with VZV-specific primers by using a suspension of mixed cells from a human TG, previously shown not to contain VZV DNA, spiked with 1,000 copies of VZV gene 21 plasmid DNA and treated with DNase attached to beads prior to proteinase K. Thus, no extracellular VZV DNA was present (SD, middle and bottom blots in each panel). In contrast, PCR with β-actin primers was positive (SD, top blots in each panel), indicating that intracellular DNA (actin) remained intact. β-Actin and VZV gene 21 DNA in DNA extracted from VZV-infected cells (VD) served as positive controls.
FIG. 3.
Cell-type distribution of VZV DNA in dissociated TG from eight subjects. Left (L) and right (R) TG from eight subjects were dissociated and processed into mixed-cell (A), neuronal (B), and nonneuronal (C) populations based on a 5,000-neuron count. The top blot in each panel shows the PCR amplification for β-actin in all samples. The middle blot in each panel shows VZV gene 21 DNA after 40 cycles of primary PCR, followed by 10 cycles of nested PCR. Positive signals were seen in six mixed-cell samples, nine neuronal-cell samples, and two nonneuronal-cell samples. Quantitative analysis revealed 1 to 12 copies of VZV DNA in five mixed-cell samples, seven neuronal-cell samples, and two nonneuronal-cell samples. VZV DNA corresponding to less than 0.5 copies was present in one mixed-cell sample and two neuronal-cell samples. The bottom blot in each panel shows VZV gene 21 DNA after 40 cycles of primary PCR, followed by 40 cycles of nested PCR. Intense signals were seen in seven mixed-cell samples, nine neuronal-cell samples, and four nonneuronal-cell samples. ND, no added DNA; SD, dissociated non-latently-infected human TG cells spiked with 1,000 copies of VZV gene 21 plasmid, digested with DNase attached to beads before proteinase K treatment, which did not amplify VZV gene 21 DNA (extracellular DNA contamination control); VD, DNA extracted from VZV-infected BSC-1 cells. Quantitation of 1 to 104 copies of gene 21 was performed as described in Fig. 2.
TABLE 3.
PCR results for 5,000-neuron samples from eight subjects
| Subject no./sidea | PCR results for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mixed cellsb
|
Neuronal cellsb
|
Nonneuronal cellsb
|
|||||||
| Actin | ORF21/Q | ORF21/N | Actin | ORF21/Q | ORF21/N | Actin | ORF21/Q | ORF21/N | |
| 1/L | + | 4 | + | + | 0 | + | + | 0 | − |
| 1/R | + | 0 | − | + | 1 | + | + | 0 | − |
| 2/L | + | 0 | − | + | 0 | − | + | 0 | − |
| 2/R | + | 3 | + | + | 2 | + | + | 1 | + |
| 3/L | + | 0 | − | + | 0 | − | + | 0 | − |
| 3/R | + | 0 | − | + | 0 | − | + | 0 | − |
| 4/L | + | 0 | + | + | 1 | + | + | 0 | − |
| 4/R | + | 0 | − | + | 1 | + | + | 0 | − |
| 5/L | + | 2 | + | + | 1 | + | + | 0 | − |
| 5/R | + | 3 | + | + | 12 | + | + | 0 | + |
| 6/L | + | 0 | − | + | 0 | − | + | 0 | − |
| 6/R | + | 0 | − | + | 0 | − | + | 0 | − |
| 7/L | + | 0 | − | + | 0 | − | + | 0 | − |
| 7/R | + | 0 | − | + | 0 | − | + | 0 | − |
| 8/L | + | 4 | + | + | 2 | + | + | 1 | + |
| 8/R | + | 0 | + | + | 0 | + | + | 0 | + |
L, left TG; R, right TG.
Actin, primary PCR (40 cycles). The presence (+) or absence (−) of the β-actin gene in the sample is indicated. ORF 21Q, primary PCR (40 cycles), followed by nested PCR (10 cycles). The number of VZV gene 21 copies after quantitation against standards is indicated. ORF 21N, primary PCR (40 cycles), followed by nested PCR (40 cycles). The presence (+) or absence (−) of VZV gene 21 at the highest possible sensitivity is indicated.
Presence of VZV DNA in dissociated TG from a single subject: copy number per cell (Part B).
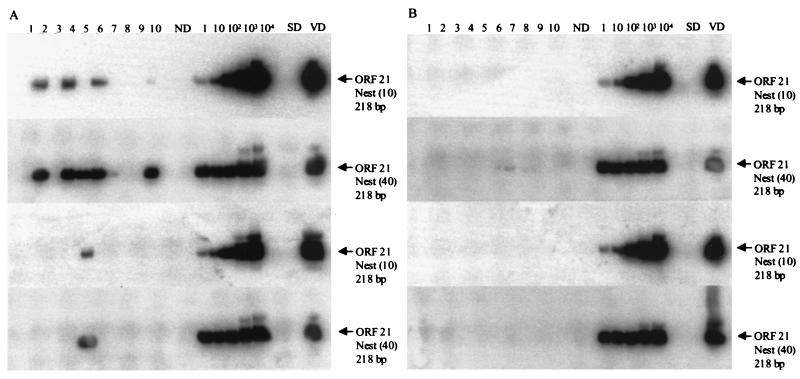
Figure 4 shows the abundance of latent VZV DNA in the left and right TG of subject 8. Ten samples from each ganglion, each containing approximately 100 neurons and associated nonneuronal cells, were separated into neuronal and nonneuronal populations and analyzed for VZV DNA. The samples were not analyzed for β-actin DNA since the crude cell lysates could only be used with one set of PCR primers. VZV DNA was detected in 3 of 10 samples from neurons of the left TG (Fig. 4A, top blot), and in 1 of 10 samples of neurons from the right TG (Fig. 4A, third blot). Quantitative analysis (40 cycles of primary PCR, followed by 10 cycles of nesting) revealed two to five copies of latent VZV DNA per sample (Table 4). No VZV DNA was detected in nonneuronal cells (Fig. 4B). Exhaustively nested PCR (40 cycles of primary PCR, followed by 40 cycles of nesting), revealed VZV DNA in 5 of 10 samples from the neuronal populations of the left TG (Fig. 4A, second blot) and in 1 of 10 samples from the right TG (Fig. 4A, bottom blot) but not in the nonneuronal cell populations (Fig. 4B). Control lanes containing no DNA showed no amplification product (ND lanes), whereas VZV-infected BSC-1 cell DNA amplified with VZV gene 21 primers (VD lanes). Further, dissociated non-latently-infected human TG cells spiked with 1,000 copies of VZV gene 21 plasmid, digested with DNase attached to beads before proteinase K treatment, did not amplify VZV gene 21 DNA (SD lanes).
FIG. 4.
VZV DNA copy number in dissociated TG. Ten aliquots each from left (upper two blots in panels A and B) and right (lower two blots in panels A and B) TG from subject 8 were dissociated and processed into neuronal (A) and nonneuronal (B) cells based on 100-neuron counts. The top blot in each panel shows two to five copies of VZV DNA in three neuronal-cell samples and no VZV DNA in any nonneuronal-cell samples, respectively, after 40 cycles of primary PCR for VZV gene 21 followed by 10 cycles of nested PCR. The second blot in each panel shows a strong VZV DNA signal in five neuronal-cell samples and no VZV DNA in any of ten nonneuronal-cell samples, respectively, after PCR for 40 cycles of primary PCR for VZV gene 21 followed by 40 cycles of nested PCR. The third blot in each panel shows two copies of VZV DNA in a single neuronal-cell sample and no VZV DNA in any of 10 nonneuronal-cell samples, respectively, after 40 cycles of primary PCR for VZV gene 21, followed by 10 cycles of nested PCR. The bottom blot in each panel shows VZV DNA in one neuronal-cell sample and no VZV DNA in any of ten nonneuronal-cell samples, respectively, after 40 cycles of primary PCR for VZV gene 21 followed by 40 cycles of nested PCR. ND, SD, VD, and quantitation were as described in Fig. 2 and 3.
TABLE 4.
PCR results from 100-neuron samples from subject 8
| Subject (side/sample no.)a | PCR results for:
|
|||
|---|---|---|---|---|
| Neuronal cellsb
|
Nonneuronal cellsb
|
|||
| ORF21/Q | ORF21/N | ORF21/Q | ORF21/N | |
| L/1 | 0 | − | 0 | − |
| L/2 | 2 | + | 0 | − |
| L/3 | 0 | − | 0 | − |
| L/4 | 5 | + | 0 | − |
| L/5 | 0 | + | 0 | − |
| L/6 | 2 | + | 0 | − |
| L/7 | 0 | − | 0 | − |
| L/8 | 0 | − | 0 | − |
| L/9 | 0 | − | 0 | − |
| L/10 | 0 | + | 0 | − |
| R/1 | 0 | − | 0 | − |
| R/2 | 0 | − | 0 | − |
| R/3 | 0 | − | 0 | − |
| R/4 | 0 | − | 0 | − |
| R/5 | 2 | + | 0 | − |
| R/6 | 0 | − | 0 | − |
| R/7 | 0 | − | 0 | − |
| R/8 | 0 | − | 0 | − |
| R/9 | 0 | − | 0 | − |
| R/10 | 0 | − | 0 | − |
Side: L, left TG; R, right TG. Sample number: successive 100-neuron samples.
ORF21/Q, primary PCR (40 cycles), followed by nested PCR (10 cycles). The number of VZV gene 21 copies after quantitation against standards is indicated. ORF21/N, primary PCR (40 cycles), followed by nested PCR (40 cycles). The presence (+) or absence (−) of VZV gene 21 at the highest possible sensitivity is indicated.
DISCUSSION
After primary infection, both herpes simplex virus (HSV) and VZV become latent in human ganglia. Studies with ISH have identified neurons as the exclusive site of HSV latency, and most ISH studies, including PCR-ISH, have also identified neurons as the predominant, if not the exclusive site of VZV latency (6, 9–11). Dueland et al. (6) examined seven TG by PCR-ISH and found VZV gene 40 exclusively in neurons of five subjects. Other investigators, however, have found VZV in nonneuronal satellite cells (3, 14). Multiple problems are associated with PCR-ISH. We have found that the PCR product diffuses from the site of amplification, a fact which may explain the detection of VZV DNA only in the cytoplasm of latently infected neurons by Dueland et al. (6). Another problem is that the nuclei of nonneuronal cells in ganglia, especially satellite cells, are small, and signals produced by PCR-ISH are difficult to discern. To determine whether the discrepant findings might reflect problems inherent in the techniques of ISH and PCR-ISH, we used a completely different procedure to identify the cell type infected by VZV during latency.
We adapted the procedures of Sawtell (16) to human TG recovered at autopsy. By immediately placing the finely minced ganglia into STF, the nucleic acid and protein content of cells was preserved, after which cells were separated by size, observed morphologically, stained immunocytochemically, and examined for VZV DNA by PCR. Filtration of the dissociated TG through a 70-μm mesh removed the larger debris. Most neurons are 20 to 50 μm in diameter and were separated from nonneuronal cells by size. Neurons were retained on the 20-μm mesh, and the flowthrough contained nonneuronal cells with few contaminating neurons. Further filtering through a 10-μm mesh removed all visible neurons. To avoid any extracellular DNA contamination, all dissociated cell samples were treated with DNase immobilized on beads.
When DNA was extracted from samples of mixed, neuronal, and nonneuronal cells based on 5,000 neurons and their associated nonneuronal cells (part A), the VZV gene 21 DNA was found in most mixed and neuronal cell population samples and in a few nonneuronal cell populations. One positive sample from the mixed-cell population was negative in the corresponding neuronal cell population, and three positive samples from the neuronal cell population were negative in the mixed-cell populations. Since the nested PCR reactions were repeated multiple times on the primary PCR product and produced identical results (Tables 3 and 4), those findings are probably due to random variation of VZV DNA content in relatively small sample sizes. The nonneuronal populations that were positive for VZV gene 21 may represent nonneuronal cells harboring latent virus, contaminating extracellular DNA, or the presence of residual neurons incompletely removed by filtration (16). Further analysis of both TG from a single subject found to be VZV DNA positive in both neuronal and nonneuronal populations revealed VZV DNA only in the neuronal population when PCR was carried out on separated aliquots of 100 neurons and their associated nonneuronal cells. Thus, residual neurons were present in the nonneuronal cell populations separated in part A. Since all samples originated from the same aliquot and since only a few samples were positive, the positive samples most likely contained DNA contributed by a single cell. Overall, there were two to five copies of VZV DNA per latently infected neuron.
Some samples in both parts A and B were negative during quantitation but were consistently positive after exhaustive nested PCR (6/48 in part A and 2/20 in part B). It is possible that some VZV DNA was tightly bound, perhaps to histones, and only released after a few cycles of primary PCR. Although the ability of VZV to bind to nucleosomes during latency has not been studied, latent HSV-1 is known to be tightly bound to histones (5).
From microscopic observations of small aliquots, we estimated that there were 100 nonneuronal cells per neuron in our mixed cell suspensions. Thus, our findings of two to five copies of VZV DNA per 100 neurons (or 10,000 total cells) compares favorably with our previous demonstration that showed a low VZV DNA burden during latency in ganglia of 6 to 31 copies per 100,000 cells (13). This is significantly lower than the HSV DNA burden in latently infected human ganglia (7).
In summary, analysis of human ganglia separated into neuronal and nonneuronal cell fractions revealed that VZV is primarily, if not exclusively, in neurons. Quantitative analysis of VZV DNA in sorted cells confirmed the low VZV DNA burden previously described in latently infected human ganglia.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants AG 06127 and NS 32623 from the National Institutes of Health. James LaGuardia is supported by Public Health Service grant NS 07321 from the National Institutes of Health.
We thank John Smith for tissue collection and Ravi Mahalingam and Mary Wellish for technical advice. We thank Marina Hoffman for editorial review and Cathy Allen for preparing the manuscript.
REFERENCES
- 1.Cohrs R, Mahalingam R, Dueland A N, Wolf W, Wellish M, Gilden D H. Restricted transcription of varicella-zoster virus in latently infected human trigeminal and thoracic ganglia. J Infect Dis. 1992;166(Suppl. 1):24–29. doi: 10.1093/infdis/166.supplement_1.s24. [DOI] [PubMed] [Google Scholar]
- 2.Cohrs R J, Barbour M, Gilden D H. Varicella-zoster virus gene 21: transcriptional start site and promoter region. J Virol. 1998;72:42–47. doi: 10.1128/jvi.72.1.42-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croen K D, Ostrove J M, Dragovic L J, Straus S E. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc Natl Acad Sci USA. 1988;85:9773–9777. doi: 10.1073/pnas.85.24.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davison A J, Scott J E. The complete DNA sequence of the varicella-zoster virus. J Gen Virol. 1986;67:1759–1786. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 5.Deshmane S L, Fraser N W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989;63:943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dueland A N, Ranneberg-Nilsen T, Degre M. Detection of latent varicella zoster virus DNA and human gene sequences in human trigeminal ganglia by in situ amplification combined with in situ hybridization. Arch Virol. 1995;140:2055–2066. doi: 10.1007/BF01322692. [DOI] [PubMed] [Google Scholar]
- 7.Efstathiou S E, Minson A C, Field H J, Anderson J R, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and humans. J Virol. 1986;57:446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilden D H, Vafai A, Shtram Y, Becker Y, Devlin M, Wellish M. Varicella-zoster virus DNA in human sensory ganglia. Nature. 1983;306:478–480. doi: 10.1038/306478a0. [DOI] [PubMed] [Google Scholar]
- 9.Gilden D H, Rozenman Y, Murray R, Devlin M, Vafai A. Detection of varicella-zoster virus nucleic acid in neurons of normal human thoracic ganglia. Ann Neurol. 1987;22:377–380. doi: 10.1002/ana.410220315. [DOI] [PubMed] [Google Scholar]
- 10.Hyman R W, Ecker J R, Tenser R B. Varicella-zoster virus RNA in human trigeminal ganglia. Lancet. 1983;ii:814–816. doi: 10.1016/s0140-6736(83)90736-5. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy P G, Grinfeld E, Gow J W. Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc Natl Acad Sci USA. 1998;95:4658–4662. doi: 10.1073/pnas.95.8.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lungu O, Annunziato P W, Gershon A, Staugaitis S M, Josefson D, LaRussa P, Silverstein S J. Reactivated and latent varicella-zoster virus in human dorsal root ganglia. Proc Natl Acad Sci USA. 1995;92:10980–10984. doi: 10.1073/pnas.92.24.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahalingam R, Wellish M, Lederer D, Forghani B, Cohrs R, Gilden D H. Quantitation of latent varicella-zoster virus DNA in human trigeminal ganglia by polymerase chain reaction. J Virol. 1993;67:2381–2384. doi: 10.1128/jvi.67.4.2381-2384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier J L, Straus S E. Varicella-zoster virus DNA polymerase and major DNA-binding protein genes have overlapping divergent promoters. J Virol. 1993;67:7573–7581. doi: 10.1128/jvi.67.12.7573-7581.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T. Molecular structure of the human cytoplasmic β-actin gene: interspecies homology of sequences in the introns. Proc Natl Acad Sci USA. 1985;82:6133–6137. doi: 10.1073/pnas.82.18.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawtell N M. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J Virol. 1997;71:5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]