Abstract
The secreted phospholipase A2 (sPLA2) isoform, sPLA2-IIA, has been implicated in a variety of diseases and conditions, including bacteremia, cardiovascular disease, COVID-19, sepsis, adult respiratory distress syndrome, and certain cancers. Given its significant role in these conditions, understanding the regulatory mechanisms impacting its levels is crucial. Genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs), including rs11573156, that are associated with circulating levels of sPLA2-IIA. The work in the manuscript leveraged 4 publicly available datasets to investigate the mechanism by which rs11573156 influences sPLA2-IIA levels via bioinformatics and modeling analysis. Through genotype-tissue expression (GTEx), 234 expression quantitative trait loci (eQTLs) were identified for the gene that encodes for sPLA2-IIA, PLA2G2A. SNP2TFBS was used to ascertain the binding affinities between transcription factors (TFs) to both the reference and alternative alleles of identified eQTL SNPs. Subsequently, candidate TF-SNP interactions were cross-referenced with the ChIP-seq results in matched tissues from ENCODE. SP1-rs11573156 emerged as the significant TF-SNP pair in the liver. Further analysis revealed that the upregulation of PLA2G2A transcript levels through the rs11573156 variant was likely affected by tissue SP1 protein levels. Using an ordinary differential equation based on Michaelis-Menten kinetic assumptions, we modeled the dependence of PLA2G2A transcription on SP1 protein levels, incorporating the SNP influence. Collectively, our analysis strongly suggests that the difference in the binding dynamics of SP1 to different rs11573156 alleles may underlie the allele-specific PLA2G2A expression in different tissues, a mechanistic model that awaits future direct experimental validation. This mechanism likely contributes to the variation in circulating sPLA2-IIA protein levels in the human population, with implications for a wide range of human diseases.
Keywords: sPLA2-IIA, genetic polymorphism, SNP, eQTL, transcription factor, transcription factor binding affinity
Introduction
Members of the secreted phospholipase A2 (sPLA2) family of enzymes play important roles in numerous biological processes, chiefly by cleaving the sn-2 position of glycerophospholipids. This hydrolysis results in an unsaturated fatty acid (UFA) and a 2-lyso-phospholipid. In humans, the 11 isoforms (PLA2-IB, PLA2-IIA, PLA2-IIC, PLA2-IID, PLA2-IIE, PLA2-IIF, PLA2-III, PLA2-V, PLA2-X, PLA2-XIIA, and PLA2-XIIB) have distinct cellular distributions, enzymatic specificities, and preferences for different polar head groups and UFAs at the sn-3 and sn-2 positions, respectively.1-4 Collectively, sPLA2 isoforms participate in multiple physiological and pathophysiological activities, generating proinflammatory and anti-inflammatory signaling molecules. Their varied functions also encompass remodeling of membrane phospholipids, food phospholipid degradation, and the hydrolysis of bacterial membranes. Notably, the sPLA2-IIA isoform, encoded by the PLA2G2A gene, efficiently hydrolyzes bacterial membrane phospholipids. This is especially the case for Gram-positive bacteria, where sPLA2-IIA, even at minimal concentrations, plays an essential antimicrobial role in innate immunity.5-8
Furthermore, circulating sPLA2-IIA levels can be reliably measured and have been identified as key biomarkers indicative of the severity or susceptibility linked with cardiovascular disease, 9 rheumatoid arthritis,10,11 cancer,12-15 sepsis, 16 and viral infections including human hepatitis virus B 17 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). 18 Notably, experimental and clinical evidence show that sPLA2-IIA plays a critical role in hypotension and multiple organ failure observed in sepsis, COVID-19, and respiratory distress syndrome.19-21 While numerous genetic variants influencing mRNA levels or blood sPLA2-IIA protein levels have been identified, the precise molecular mechanism by which these variants impact sPLA2-IIA levels remains to be elucidated. Therefore, gaining insight into the genetic and molecular mechanisms that regulate circulating and tissue sPLA2-IIA levels is crucial, as this knowledge can identify populations and individuals vulnerable to either reduced or excessive production of this pivotal enzyme.
The single nucleotide polymorphism (SNP) rs11573156, where G is defined as the reference allele, and C is defined to be an alternative allele in the coding strand sequence, has been strongly associated with circulating levels of sPLA2-IIA. This association is supported by 5 studies that reveal a dose-dependent allele association between circulating levels and activity. 22 For instance, sPLA2-IIA levels increased by 57% to 62% in those with the homozygous alternative allele (CC) compared to the homozygous reference (GG).
Recognizing the significance of SNPs like rs11573156, our study aimed to understand the genetic and molecular foundations of PLA2G2A expression. This research developed an analysis pipeline (outlined in Supplementary Figure 1), by using data from 4 publicly available resources (genotype-tissue expression [GTEx], SNP2TFBS, ENCODE, and Human Protein Atlas [HPA]) and mathematical modeling to explore how rs11573156 modulates sPLA2-IIA levels. More specifically, we used these datasets to examine transcription factor (TF) binding sites where PLA2G2A-eQTLs are located and how different alleles at rs11573156 influence TF-binding affinity. A simple ODE model was constructed, assuming Michaelis-Menten kinetics, to simulate how varying SP1-rs11573156 binding affects allele-specific PLA2G2A transcription. This model was tested with data of SP1 protein level and PLA2G2A expression across various tissues, as obtained from HPA and GTEx, respectively. Our findings suggest that rs11573156 affects PLA2G2A transcription via the differential binding affinities of the 2 alleles to SP1, which likely constitutes an important genetic/molecular mechanism affecting circulating sPLA2-IIA levels.
Materials and Methods
Collection of PLA2G2A eQTLs in different tissues
The GTEx Portal V8 (https://gtexportal.org) 23 was used to determine PLA2G2A expression quantitative trait loci (eQTLs) across 18 tissues including thyroid, testis, stomach, skin-sun exposed (lower leg), skin-not sun exposed (suprapubic), prostate, ovary, nerve-tibial, muscle-skeletal, lung, liver, heart-left ventricle, heart-atrial appendage, esophagus-muscularis, esophagus-gastroesophageal junction, artery-aorta, adipose-visceral (omentum), and adipose-subcutaneous. The GTEx includes data measured in tissues collected within 24 hours of death for 948 individuals (32.9% female in total; 84.6% white with 34.3% of them female, 12.9% African American with 30.5% female, 1.3% Asian with 41.7% female, and 1.1% unknown with 16.7% female). The genotype data based on whole genome sequencing from 838 donors were applied to eQTL analyses in the GTEx V8, where only variants with MAF ⩾1% across all 838 samples were included. A significant eQTL was determined by a false discovery rate (FDR) threshold of ⩽0.05. The normalized effect size (NES) of the eQTLs was defined as the slope of the linear regression model between genotype and expression. The NES was calculated as the effect of the alternative allele relative to the reference allele in the human genome reference GRCh38/ hg38. A more detailed description is on the GTEs Portal site (https://gtexportal.org/home/methods). A positive NES indicates upregulation, while a negative NES indicates downregulation of the affected gene.
TF binding to eQTL alleles
The SNP2TFBS tool (http://ccg.vital-it.ch/snp2tfbs/) was used to predict TF-eQTL SNP binding variations. SNP2TFBS is a comprehensive database of regulatory SNPs affecting predicted transcription factor binding site (TFBS) affinity. 24 An SNP’s effect on TF binding is estimated based on a position weight matrix (PWM) model for the binding specificity of the corresponding factor, which are fixed length TFBS models represented by a matrix of probabilities reflecting the occurrence frequencies of bases at binding site positions. SNP2TFBS uses the JASPAR database (http://jaspar.genereg.net) as a source of the TF-binding profiles, which provides PWMs as base frequency matrices. SNP2TFBS reports raw binding scores, which are computed as the sum of the position-specific weights over all bases of the binding site. It uses SNP data from the 1000 Genomes Project (minor allele frequency > 0.001), human genome assembly GRCh37/hg19, and RefSeq gene annotations provided by ANNOVAR. 25 In this study, eQTLs affecting PLA2G2A gene expression were queried with SNP2TFBS to obtain a list of predicted TFs that differentially bind to those SNPs.
Verification of the predicted TF-eQTL binding by ChIP-seq dataset
ChIP-seq Peaks (340 TFs in 129 cell and tissue types) from ENCODE 3, downloaded from the UCSC Genome Browser (https://genome.ucsc.edu/) alignment to GRCh38/hg38, were used to examine whether the candidate TFs bind to the predicted eQTL regions. This dataset represents a large-scale TF-binding profile based on ChIP-seq experiments generated by the ENCODE Consortium between February 2011 and November 2018.26,27 If candidate TFs had SNP2TFBS-predicted differential binding to PLA2G2A SNPs, we checked whether the predicted TF binding occurred with the eQTL-corresponding regions with the PLA2G2A gene locus (chromosome 1; 18990581-20971970) in the matched tissue.
Gene expression and genotyping data
The RNA-sequencing (RNA-seq) data (TPMs) were obtained from the GTEx portal open access database (https://gtexportal.org) on April 21, 2022. The genotyping data were obtained per dbGaP approval and downloaded as dbGaP accession number phg001219.v1 on October 02, 2022.
log TPM values used in Figure 4 were computed by (1) applying a log (TPM + 1) transformation to all raw TPM values retrieved from GTEx, (2) computing the mean of transformed values from step 1, grouping on both tissue and allele, and (3) subtracting the GG/GC from the CC values from step 2, grouping on tissue.
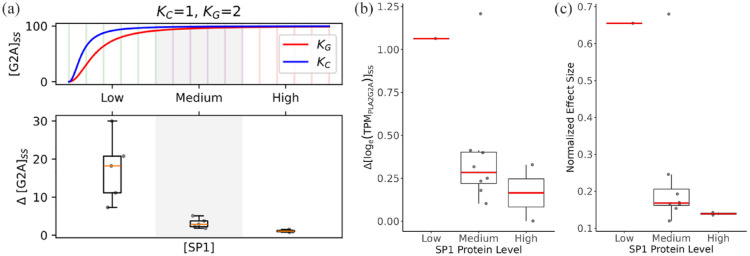
Figure 4.
The impact of SP1 protein levels on allele-specific PLA2G2A gene expression across tissues. (a) Modeled PLA2G2A allele expression. In the upper panel, the ODE across an arbitrary range of SP1 level ([SP1]) (x-axis) was simulated to get the steady-state PLA2G2A expression level ([G2A]ss) corresponding to the C and G alleles of rs11573156 (Kc and KG, respectively). In the lower panel, (y-axis) indicates the difference in the C and G allele PLA2G2A expression levels, which is box-plotted against [SP1] (x-axis) spanning 3 categories (with 5 bins in each category, as indicated in the top panel). The model parameters are (b) Empirical delta PLA2G2A allele expression. The differences (delta logTPM) of PLA2G2A transcript levels by genotype (C vs G allele at rs11573156) were calculated in each tissue based on the GTEx dataset and box-plotted against the 3 categories of SP1 protein levels (low, medium, and high) as in a. (c) Normalized effect size of rs11573156 in each tissue box-plotted against the 3 categories of SP1 protein levels as in a.
SP1 protein level in different tissues
SP1 protein expression data by tissue are publicly available from the HPA (https://www.proteinatlas.org/about/download). Expression values are semiquantitative, derived from immunohistochemistry of tissue microarrays, and take on 4 possible values: “Not Detected,” “Low,” “Medium,” and “High.” Tissues not present in GTEx were omitted. When a tissue had multiple recorded cell types, the protein expression profile in the cell type that exhibits the largest cellular population within the tissue was selected.
Modeling SP1 level-dependent allele-specific PLA2G2A gene expression in tissues
A simple ordinary differential equation (ODE) model was developed to simulate and test the correlation between SP1-eQTL binding and PLA2G2A expression in different tissues:
| (1) |
The structure of the equation was based on standard Hill function kinetics. describes the rate of the change of PLA2G2A transcript concentration over time. is a rate constant describing SP1-dependent PLA2G2A transcription. with the specific instances of KC and KG for CC and GG/GC alleles, respectively, is a Hill constant inversely correlated with the eQTL allele-specific binding affinity to SP1. is a Hill coefficient. is the degradation rate constant of PLA2G2A transcripts. The steady-state concentration of PLA2G2A expression is given by the following equation:
| (2) |
To explore qualitative insights into the system’s general behavior, simple, generic parameter values were selected as follows: = 1, 0.01, = 2, = 1. With this base parameter set, and with KG > KC, reflecting the higher binding affinity of SP1 to the CC versus GG/GC allele of the PLA2G2A promoter, consistent qualitative patterns are observed (Supplementary Figure 2). PLA2G2A steady-state concentrations were simulated over a range of SP1 protein concentrations from 0 to 20 arbitrary units.
Results
eQTLs of PLA2G2A in human tissues
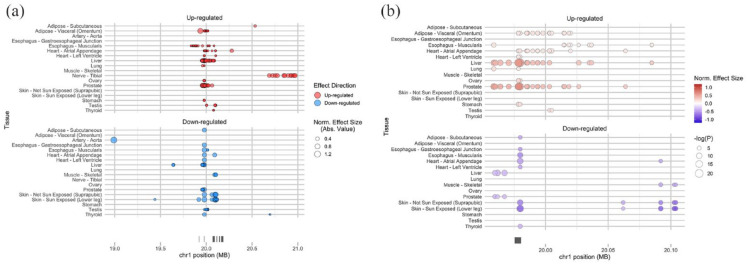
To investigate the potential genetic basis for variations in PLA2G2A expression in humans, the relationship between the SNPs in the human genome and PLA2G2A expression levels were examined. Specifically, we leveraged data from the GTEx project, which correlates genome-wide SNPs with variations in gene expression across multiple human tissues. Our analysis identified 234 eQTLs significantly associated with PLA2G2A expression (with an FDR ⩽ 0.05) in GTEx (V8) (Figure 1a). Located on chromosome 1 (hg38; chr1: 19975431-19980416), these eQTLs primarily occupy noncoding regions proximal to and flanking the PLA2G2A gene locus. Of the 234 eQTLs, 171 are associated with increased PLA2G2A mRNA expression, while 63 are linked with its downregulation. Interestingly, 37 of these eQTLs were observed in 2 or more tissues, with each displaying a consistent effect (upregulating or downregulating) on PLA2G2A expression across these tissues (Figure 1b, Supplementary Table 1). This consistency pointed to the recurring roles these 37 eQTLs play in modulating PLA2G2A expression in the human body. Consequently, our subsequent analysis was centered on these 37 eQTLs.
Figure 1.
Genome-wide SNPs that Impact PLA2G2A Gene Expression in Multiple Tissues. (a) The location of 234 eQTLs for PLA2G2A. X-axis shows the chromosomal position, and Y-axis shows tissues. The marker size indicates the magnitude of the effect size. The marker color indicates the effect direction in alternative alleles compared to reference alleles. (b) The location of 37 eQTLs shared among more than one tissue. X-axis shows the chromosomal position, and Y-axis shows tissues. The marker size indicates the magnitude of the negative logarithm of the P value. The color intensity indicates the magnitude of normalized effect size. The color indicates the effect direction in alternative alleles compared to reference alleles. Red: up-regulated, Blue: down-regulated. The genome-wide SNPs were examined using data from the GTEx project. The normalized effect size (NES) of the eQTLs is defined as the slope of the linear regression and is computed as the effect of the alternative allele (ALT) relative to the reference allele (REF) in the human genome reference GRCh38/hg38, as described in GTEx Portal (https://gtexportal.org/home/methods).
Identification of eQTL-bound TFs
Expression quantitative trait loci have the potential to modulate gene expression by various mechanisms, including transcription, RNA processing, RNA stability, and translation.28-30 One of the predominant mechanisms involves the alteration of TF-binding sites on the targeted gene promoters.31-33 To identify the TF-binding sites that may be affected by PLA2G2A eQTLs, we examined the 37 eQTLs (Figure 1b, Supplementary Table 1) using SNP2TFBS, a computer program that predicts how an SNP may affect TF-binding site sequence and binding affinity. 24 From the SNP2TFBS analysis, 9 of the 37 eQTLs were associated with known TF-binding sites. Notably, 7 of these 9 eQTLs had the potential to influence TF-binding affinities due to sequence variations between the reference and alternative SNP alleles (Table 1). In particular, 7 eQTLs were predicted to affect the binding of 8 TFs: EBF1, ZNF263, SP1, PPARG_RXRA, SREBF1, SREBF2, Gata1, and Foxq1 (Table 1). Normalized effect sizes of significant tissue-eQTLs were obtained for these 7 eQTLs from the GTEx Portal (Table 1).
Table 1.
Candidate TFs associated with PLA2G2A gene expression determined by SNP2TFBS.
| SNP (eQTL) rsID | Ref | Alt | Effect | eQTL tissues | Effect size | Matched TFs |
|---|---|---|---|---|---|---|
| rs12733637 | A | G | down | Liver | −0.39 | EBF1 |
| Prostate | −0.34 | |||||
| rs1891320 | C | T | up | Liver | 0.45 | ZNF263 |
| Prostate | 0.50 | |||||
| rs11573156 | G | C | up | Liver | 0.66 | SP1 |
| Prostate | 0.68 | |||||
| Ovary | 0.25 | |||||
| Adipose-Visceral (Omentum) | 0.17 | |||||
| Stomach | 0.19 | |||||
| Heart-Atrial Appendage | 0.15 | |||||
| Heart-Left Ventricle | 0.17 | |||||
| rs1796923 | T | G | down | Skin-Sun Exposed (Lower leg) | −0.80 | PPARG_RXRA |
| Heart-Atrial Appendage | −0.51 | |||||
| Skin-Not Sun Exposed (Suprapubic) | −0.65 | |||||
| Esophagus-Muscularis | −0.62 | |||||
| Thyroid | −0.40 | |||||
| Heart-Left Ventricle | −0.48 | |||||
| Adipose-Subcutaneous | −0.48 | |||||
| Esophagus-Gastroesophageal Junction | −0.54 | |||||
| rs114156017 | C | T | up | Liver | 0.42 | SREBF1 SREBF2 |
| Prostate | 0.41 | |||||
| Adipose-Visceral (Omentum) | 0.18 | |||||
| Heart-Atrial Appendage | 0.16 | |||||
| rs149238894 | G | A | up | Liver | 0.40 | GATA1 |
| Prostate | 0.42 | |||||
| Adipose-Visceral (Omentum) | 0.18 | |||||
| Heart-Atrial Appendage | 0.15 | |||||
| rs139466428 | TG | T | up | Prostate | 0.38 | FOXQ1 |
| Adipose-Visceral (Omentum) | 0.15 | |||||
| Esophagus-Muscularis | 0.13 |
Ref , reference allele; Alt , alternative allele; Effect , the direction of the eQTL’s effect on PLA2G2A gene expression; “up” means the alternative allele upregulates PLA2G2A gene expression compared to the reference allele, and “down” means the alternative allele downregulates PLA2G2A gene expression compared to the reference allele.; eQTL tissues , tissue distribution of each eQTL; Effect size , the normalized effect size of each significant tissue-eQTL obtained from GTEx Portal, which was computed as the effect of the alternative allele relative to the reference allele in the human genome reference GRCh38/hg38; TF , transcription factor.
Verification of TF-eQTL binding
The relationships between eQTLs and TFs in the tissues they influence were next examined. We posited that if an eQTL modulates gene expression (in this case PLA2G2A) by affecting TF-binding affinity, there would be TF binding to the eQTL site in the tissues where the eQTL has an effect. To address this question, we used the ENCODE chromatin immunoprecipitation (ChIP)-seq dataset, which maps the chromatin-binding patterns of 340 TFs across 129 cell lines or tissue types. From the 8 TFs hypothesized to be influenced by PLA2G2A eQTLs, 6 TFs were evaluated in the ENCODE dataset (Table 2). However, only one TF, SP1, had been studied in an eQTL-matched tissue, specifically, the liver.
Table 2.
The list of candidate TFs examined in the ENCODE ChIP-seq dataset.
| TF | Cell line | Origin of cell line |
|---|---|---|
| EBF1 | GM12878 | B-lymphocyte |
| ZNF263 | HEK293 | Kidney |
| SP1 | A549 H1-hESC HEK293T HepG2 K562 Liver MCF-7 |
Lung Embryo Kidney Liver Bone marrow Liver Breast |
| GATA1 | Erythroblast K562 |
Erythroblast Bone marrow |
| SREBF1 | A549 K562 MCF-7 |
Lung Bone marrow Breast |
| SREBF2 | A549 HeLa-S3 |
Lung Uterus cervix |
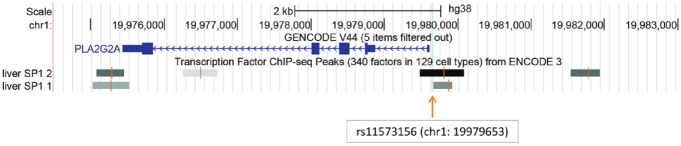
The ENCODE data revealed that SP1 indeed binds to the chromatin region of the eQTL rs11573156 in the liver (Figure 2). Consistently, among all tissues examined in GTEx, the liver displayed the most robust eQTL effect (P < 5 × 10−21, NES = 0.66) of rs11573156 on PLA2G2A expression (Supplementary Table 2). Therefore, the subsequent analysis focused on the SP1-rs11573156 binding in modulating PLA2G2A expression.
Figure 2.
Display of confirmed SP1 binding regions in the liver. The chromatin regions of SP1 binding in the liver are shown around the rs11573156 eQTL site in the UCSC Genome Browser. The SP1 binding was determined by ChIP-seq (ENCODE 3 dataset). The gray and black rectangles indicate the respective SP1-binding regions (ChIP-seq peaks) in liver tissues derived from separate individuals. A vertical light blue line indicates the location of rs11573156.
SP1 bound to rs11573156 as a transcriptional activator of PLA2G2A
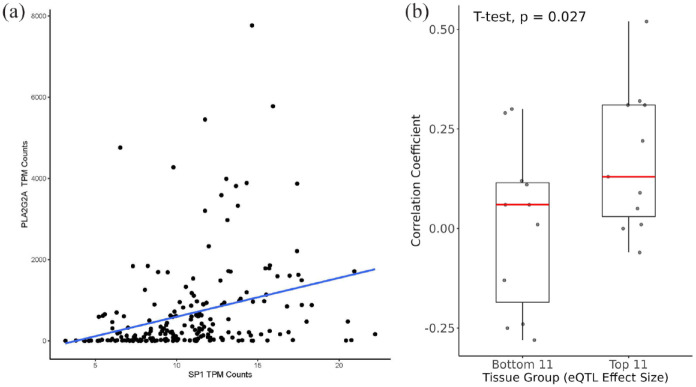
If SP1 binds to the eQTL rs11573156 in the liver and thereby influences PLA2G2A expression, a discernible correlation between liver SP1 and PLA2G2A would be anticipated. Specifically, considering the predictive higher binding affinity of SP1 to the alternative allele (C) of rs11573156 over the reference allele (G) (as shown in Table 1) coupled with the positive effect of the alternative allele on PLA2G2A expression, we postulated that there would be a positive correlation between SP1 and PLA2G2A levels in the liver. In the GTEx dataset containing 208 liver donor samples, SP1 was positively associated with PLA2G2A expression (correlation coefficient, r = 0.31; Figure 3a). This result supported the role of SP1 as a transcriptional activator of the PLA2G2A gene in the liver.
Figure 3.
Associations between PLA2G2A and SP1 gene expression in GTEx. (a) Correlation analysis of the gene expression between PLA2G2A and SP1 in the liver (r = 0.31). X-axis shows transcripts per million (TPM) of SP1 and Y-axis shows TPM of PLA2G2A. (b) Box plots of the correlation coefficients between SP1 and PLA2G2A TPMs across tissues. Top 11: Top 11 tissues by multitissue posterior probability of effect (m-value) of the eQTL rs11573156. Bottom 11: bottom 11 tissues by m-value.
To examine how the correlation between the gene expression of SP1 and PLA2G2A changes in various tissues with different effect sizes of rs11573156, we next focused on the top 11 and bottom 11 tissues or tissue subtypes by probability assigned by the GTEx project (out of a total of 48) where the effect of eQTL rs11573156 on PLA2G2A expression was most and least pronounced. The top 11 tissues were defined by a single-tissue P value <.001 and the multitissue posterior probability m-value = 1 (Supplementary Table 2), indicating the highest level of confidence in the eQTL effect within those tissues. We hypothesized that if SP1 binds to the eQTL rs11573156 and elevates PLA2G2A expression, then the correlation between SP1 with PLA2G2A would be stronger in these 11 tissues compared to others. This expectation was validated in our analysis. As illustrated in Figure 3b, the correlation coefficients between SP1 and PLA2G2A in these top 11 tissues (with an m-value of 1) were significantly higher (P = .03) than in the control tissues (bottom 11 with m-value <.21; Supplementary Table 2). This result suggested that tissues with larger allele effects of rs11573156 exhibited stronger correlations between SP1 and PLA2G2A, consistent with rs11573156 exerting its impact on PLA2G2A expression through SP1.
Model differential SP1-rs11573156 binding-mediated PLA2G2A expression
Next, we asked why the allele effect of rs11573156 varied depending on the tissue type. In the top 11 tissues in Figure 3b, there was a striking variation in the effect size of the eQTL rs11573156, spanning over a 5-fold range from 0.12 to 0.68 (Supplementary Table 2). To explore the potential molecular mechanism explaining how SP1-rs11573156 binding may account for the varying effect size of rs11573156 on PLA2G2A expression, we constructed a simple ODE model based on the premise of Hill function kinetics:
| (3) |
In this model, the change of PLA2G2A transcript level over time ( ) is a function of SP1 (TF) protein level (with and being constants, see Method for details). The difference between the alternative allele (C) and the reference allele (G) of rs11573156 is reflected in the value of the Hill constant that is inversely correlated with SP1 binding affinity representing a higher SP1 binding affinity to the alternative allele C than the reference allele G).
This minimalist model suggests that because of the difference in SP1- rs11573156 binding affinity a general trend regarding the steady-state level of PLA2G2A transcripts, [G2A]ss, can be observed (Figure 4a and Supplementary Figure 2): (1) [G2A]ss is higher with the alternative allele (C) than with the reference allele (G), (2) the discrepancy in [G2A]ss between the 2 alleles, represented as Δ[G2A]ss, becomes more pronounced at lower concentrations of SP1 protein levels, and (3) this divergence, Δ[G2A]ss, gradually diminishes as SP1 concentrations rise. This result can largely be explained by the “saturating” effect at high SP1 levels, where the excess amount of SP1 protein has the capacity to fully transactivate PLA2G2A even to the lower binding affinity allele of rs11573156.
Validation of model predictions across tissues
To test the model predictions, we obtained the SP1 protein levels across diverse tissues from the HPA. SP1 protein levels were emphasized for a couple of reasons: (1) It is the protein level (rather than the mRNA level) of the TF SP1 that impacts PLA2G2A expression and (2) the SP1 mRNA levels across different tissues may not always correlate well with SP1 protein levels, due to tissue-specific differences in posttranscriptional modifications, translation, and protein stability, and so on.
Human Protein Atlas estimates protein levels in different human tissues in 3 categories, low, medium, and high. The categorical SP1 protein levels in the top 11 tissues are shown in Suppl. Table 2. When the PLA2G2A transcript level differential (ΔlogTPM) between the C and G alleles in each of the 11 tissues was plotted against their corresponding SP1 protein categories, a clear trend emerged. This trend was consistent with the model’s predictions: the difference in PLA2G2A transcript levels between alleles C and G was most significant at the lowest SP1 protein level, tapering off as SP1 levels ramped up to medium and then high (Figure 4b). Similarly, the effect size of the eQTL rs11573156, which is positively correlated with the PLA2G2A level difference between the C and G alleles, also exhibits a similar trend that is inversely correlated with SP1 protein level in the tissue (Figure 4c). Cumulative, these results strongly support the hypothesis that variations in PLA2G2A gene expression associated with the eQTL rs11573156 alleles are remarkably, and inversely, influenced by the tissue-specific SP1 protein levels.
Discussion
Much of the early work with the sPLA2-IIA isoform centered on its bactericidal role where it was shown to increase 100 to 1000-fold (up to 5 μg/ml) in serum/plasma during infectious sepsis. 34 However, studies beginning in the mid-1980s, also identified its capacity at persistently high concentrations to induce lethal multiple organ failure associated with sepsis.18,20,21 In fact, Nyman and colleagues demonstrated that sPLA2-IIA levels could effectively predicted lethal multiple organ failure when measured at later time points (>3 days in the intensive care unit [ICU]) in sepsis patients. 20
More recently, our group discovered that deceased COVID-19 patients, primarily those succumbing to lethal multiple organ failure, also exhibited considerably elevated levels of circulating, catalytically active sPLA2-IIA. 18 Elevated sPLA2-IIA levels paralleled several indices of COVID-19 disease severity such as kidney dysfunction, hypoxia, and multiple organ dysfunction. Moreover, 3 machine learning models identified sPLA2-IIA levels as the pivotal factor in differentiating patients who died from sepsis due to COVID-19. A pressing question emerging from these studies and other clinical conditions, such as cardiovascular disease, 9 rheumatoid arthritis, 10 and cancer,12-15 is: Why do only certain patients produce and release high concentrations of sPLA2-IIA from tissues, especially the liver?
Several GWASs have demonstrated the impact of genetic variation on sPLA2-IIA levels and further revealed that rs11573156 G > C polymorphism is highly associated with circulating level of sPLA2-IIA.35-38 The mean allele frequencies across all global populations are 77% for the G reference allele and 23% for the C alternative allele. Frequencies of the C allele varying from 5% (Asian) to 36% (Estonian) in different populations.
This study demonstrated that among the 234 eQTLs for PLA2G2A, rs11573156 in the liver showed the most robust effect. This SNP exclusively affects PLA2G2A gene expression, and it is not identified as an eQTL with any of the other 11 sPLA2 genes, including the 6 isoforms (PLA2G2E, PLA2GA, PLA2G5, PLA2G2D, PLA2GF, and PLA2G2C) that occur as a gene cluster (hg38 chr1:19 920 009-20 186 518) on chromosome 1 (data not shown). Furthermore, the pQTL (protein quantitative trait locus) analysis also indicated that rs11573156 is strongly associated with circulating protein levels of sPLA2-IIA.36-39
The rs11573156 SNP is positioned immediately upstream of the 5′ untranslated region (5′-UTR) in the consensus transcript of the PLA2G2A gene, as designated by the MANE Select project, 40 acting at a TF-binding site that regulates PLA2G2A expression. This study, through our developed analysis pipeline (as outlined in Supplementary Figure 1), suggests that this region containing rs11573156 is a crucial SP1 binding site that modulates PLA2G2A transcript levels. Specifically, in tissues such as the liver where the SP1 protein levels are low, the difference in the binding affinity of SP1 to the G versus C allele leads to a significant disparity in SP1 binding dynamics and occupancy at the rs11573156 site, resulting in a notable difference in allele-specific PLA2G2A expression. In contrast, in tissues with high SP1 protein levels (e.g. skin and lung), SP1 saturates both the high and low affinity binding sites (i.e, both the C and G allele), thereby diminishing the allele-specific difference in PLA2G2A expression. That is, as summarized in Supplementary Figure 3, our study suggests that the PLA2G2A transcription depended on both (1) the SP1 binding affinity affected by the alleles of rs11573156 and (2) the concentration of SP1 protein in a given tissue, which could explain tissue-specific rs11573156 effect size. Moreover, there appears to be a direct relationship between PLA2G2A transcription and sPLA2-IIA protein level, as indicated by the fact that rs11573156 also serves as a pQTL for sPLA2-IIA. Notably, the liver is believed to be the primary organ to produce sPLA2-IIA released into circulation.41-43 This aligns with the observation that rs11573156 alleles via their differential binding affinity to SP1 modulate allele-specific PLA2G2A expression in the liver, and correspondingly, sPLA2-IIA protein level variations in circulation.
This study has important limitations. First, the ENCODE ChIP-seq dataset does not include all tissues where the eQTL rs11573156 exhibits significant effect on PLA2G2A gene expression, including the liver, prostate, ovary, stomach, adipose-visceral, heart-left ventricle, and heart-atrial appendage. Among these tissues, only the liver has the ChIP-seq data of SP1. It remains unclear whether SP1-mediated allele-specific PLA2G2A expression also occurs in the other tissues. In addition, there were several other TF-eQTL combinations possibly showing allele-dependent binding affinity influencing the expression of other sPLA2 isoforms. These include RFX1-rs71647137 for PLA2G5, CTCF-rs2070873 for PLA2G1B, NRF1-rs762521 and CTCF-rs11407099 for PLA2G10, ZNF143-rs9621185 and NFYB-rs5997969 for PLA2G3. Further analysis of such associations is currently limited by the unavailability of ChIP-seq datasets in the matched tissue types. Second, this study uses datasets from diverse publicly available resources, including GTEx, SNP2TFBS, ENCODE, and HPA. Comparing data from different sources presents inherent challenges and uncertainties. To mitigate these concerns, we employed complementary measures, by (1) focusing our main analysis on the liver, using multiple datasets from the same tissue to ensure context consistency, (2) adopting a categorical analysis approach, grouping tissues based on SP1 protein levels, enabling identification of broader trends and robust patterns across datasets (GTEx and HPA) and tissues (Figure 4b and c) independently validating our findings through a simple ODE model that successfully accounts for allele-specific PLA2G2A expression across 11 tissue types (Figure 4a). Third, this study focused on investigating the effect of TF-SNP binding dynamics on PLA2G2A expression. It is worth noting that some SNPs could also impact gene expression via a variety of other mechanisms, including mRNA stability, subcellular localization, micro RNAs (miRNAs) modulation, and epigenetic modulation.44,45 Future studies with a wider range of experimental datasets and more sophisticated models are needed to expand our understanding of allele-specific PLA2G2A expression. Finally, it is important to note that our findings in this study, based on computational and modeling analysis, need to be carefully tested and validated in future experimental investigations involving cell models derived from various tissues and genotypes.
Given the diverse functions and diseases where sPLA 2 -IIA plays a modulatory role, it is imperative to determine how levels of this pivotal enzyme are controlled and whether its synthesis can be inhibited. This article describes a TF-binding interaction between SP1 and rs11573156 (the SNP that has been most associated sPLA2-IIA levels), which likely accounts for the allele-specific PLA2G2A expression in the liver. Unraveling a genetic molecular mechanism that controls sPLA2-IIA levels in the primary organ of its generation represents an exciting finding in the field. Studies like this pave the way for future precision medicine approaches that may modulate PLA2G2A expression by altering the dynamics of SP1 binding to SNPs, such as rs11573156, especially in potentially fatal diseases like infectious sepsis.
Supplemental Material
Supplemental material, sj-docx-1-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-2-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-3-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-4-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-5-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Center for Complementary and Integrative Health (grant no. R01 AT008621; FHC), U.S. Department of Agriculture (grant no. ARZT-1361680-H23-157; FHC), and TRIF Improving Health Initiative (Univ. of Arizona; GY and FHC).
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: FHC is a cofounder of Tyrian Omega, Inc, and Resonance Pharma, Inc. These companies use microorganisms to generate omaga-3 fatty acids and develop diagnostic and therapeutics for lipid targets, respectively. These commercial relationships are managed by the Office for Responsible Outside Interests at the University of Arizona. The remaining authors had nothing to disclose.
Author Contributions: Investigation, AH and GY; Methodology, EL and GY; Formal analysis, EL and LJ; Writing—original draft, AH; Visualization, AH, EL, LJ, MW, and SS; Data curation, LJ; Validation, BH; Writing—review & editing, BH, JCW, HHZ, GY, and FHC; Conceptualization, GY and FHC; Supervision, FHC and GY; Funding acquisition, FHC and GY. All authors have read and agreed to the published version of the manuscript.
ORCID iD: Aki Hara  https://orcid.org/0009-0003-7548-648X
https://orcid.org/0009-0003-7548-648X
Data Availability Statement: Publicly available datasets were analyzed in this study. PLA2G2A-eQTL data can be found here: (https://gtexportal.org/home/gene/PLA2G2A/GTEx Analysis Release V8 [dbGaP Accession phs000424.v8.p2]). Transcription Factor ChIP-seq peaks data can be found here: [https://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=1825404726_a88kgTdbGWEtaDzgNNYYcmotx69C&db=hg38&g=encTfChipPk/Sourcedataversion:ENCODE3Nov2018]. The genotyping data are not publicly available due to protected access and requires the approved dbGaP application.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50(suppl):S237-S242. doi: 10.1194/jlr.R800033-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cupillard L, Koumanov K, Mattéi MG, Lazdunski M, Lambeau G. Cloning, chromosomal mapping, and expression of a novel human secretory phospholipase A2. J Biol Chem. 1997;272:15745-15752. doi: 10.1074/jbc.272.25.15745 [DOI] [PubMed] [Google Scholar]
- 3. Murakami M, Sato H, Miki Y, Yamamoto K, Taketomi Y. A new era of secreted phospholipase A2. J Lipid Res. 2015;56:1248-1261. doi: 10.1194/jlr.R058123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Six DA, Dennis EA. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488:1-19. doi: 10.1016/s1388-1981(00)00105-0 [DOI] [PubMed] [Google Scholar]
- 5. Dore E, Boilard E. Roles of secreted phospholipase A(2) group IIA in inflammation and host defense. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:789-802. doi: 10.1016/j.bbalip.2018.08.017 [DOI] [PubMed] [Google Scholar]
- 6. Grönroos JO, Laine VJO, Nevalainen TJ. Bactericidal group IIA phospholipase A2 in serum of patients with bacterial infections. J Infect Dis. 2002;185:1767-1772. doi: 10.1086/340821 [DOI] [PubMed] [Google Scholar]
- 7. Movert E, Wu Y, Lambeau G, Kahn F, Touqui L, Areschoug T. Secreted group IIA phospholipase A2 protects humans against the group B streptococcus: experimental and clinical evidence. J Infect Dis. 2013;208:2025-2035. doi: 10.1093/infdis/jit359 [DOI] [PubMed] [Google Scholar]
- 8. van Hensbergen VP, Wu Y, van Sorge NM, Touqui L. Type IIA secreted phospholipase A2 in host defense against bacterial infections. Trends Immunol. 2020;41:313-326. doi: 10.1016/j.it.2020.02.003 [DOI] [PubMed] [Google Scholar]
- 9. Kugiyama K, Ota Y, Takazoe K, et al. Circulating levels of secretory type II phospholipase A(2) predict coronary events in patients with coronary artery disease. Circulation. 1999;100:1280-1284. doi: 10.1161/01.cir.100.12.1280 [DOI] [PubMed] [Google Scholar]
- 10. Liu NJ, Chapman R, Lin Y, et al. Point of care testing of phospholipase A2 group IIA for serological diagnosis of rheumatoid arthritis. Nanoscale. 2016;8:4482-4485. doi: 10.1039/c5nr08423g [DOI] [PubMed] [Google Scholar]
- 11. Seilhamer JJ, Pruzanski W, Vadas P, et al. Cloning and recombinant expression of phospholipase A2 present in rheumatoid arthritic synovial fluid. J Biol Chem. 1989;264:5335-5338. doi: 10.1016/S0021-9258(18)83549-9 [DOI] [PubMed] [Google Scholar]
- 12. Dong Z, Liu Y, Scott KF, et al. Secretory phospholipase A2-IIa is involved in prostate cancer progression and may potentially serve as a biomarker for prostate cancer. Carcinogenesis. 2010;31:1948-1955. doi: 10.1093/carcin/bgq188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kupert E, Anderson M, Liu Y, et al. Plasma secretory phospholipase A2-IIa as a potential biomarker for lung cancer in patients with solitary pulmonary nodules. BMC Cancer. 2011;11:513. doi: 10.1186/1471-2407-11-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oleksowicz L, Liu Y, Bracken RB, et al. Secretory phospholipase A2-IIa is a target gene of the HER/HER2-elicited pathway and a potential plasma biomarker for poor prognosis of prostate cancer. Prostate. 2012;72:1140-1149. doi: 10.1002/pros.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Yu H, Xu H, Yang L. Expression of secreted phospholipase A2-Group IIA correlates with prognosis of gastric adenocarcinoma. Oncol Lett. 2015;10:3050-3058. doi: 10.3892/ol.2015.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan TL, Goh YY. The role of group IIA secretory phospholipase A2 (sPLA2-IIA) as a biomarker for the diagnosis of sepsis and bacterial infection in adults—A systematic review. PLoS ONE. 2017;12:e0180554. doi: 10.1371/journal.pone.0180554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu C, Song H, Shen B, Wu L, Liu F, Liu X. Promoting effect of hepatitis B virus on the expressoin of phospholipase A2 group IIA. Lipids Health Dis. 2017;16:5. doi: 10.1186/s12944-016-0400-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snider JM, You JK, Wang X, et al. Group IIA secreted phospholipase A2 is associated with the pathobiology leading to COVID-19 mortality. J Clin Invest. 2021;131. doi: 10.1172/JCI149236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guidet B, Piot O, Masliah J, et al. Secretory non-pancreatic phopholipase A2 in severe sepsis: relation to endotoxin, cytokines and thromboxane B2. Infection. 1996;24:103-108. doi: 10.1007/BF01713312 [DOI] [PubMed] [Google Scholar]
- 20. Nyman KM, Uhl W, Forsström J, Büchler M, Beger HG, Nevalainen TJ. Serum phospholipase A2 in patients with multiple organ failure. J Surg Res. 1996;60:7-14. doi: 10.1006/jsre.1996.0003 [DOI] [PubMed] [Google Scholar]
- 21. Vadas P. Elevated plasma phospholipase A2 levels: correlation with the hemodynamic and pulmonary changes in gram-negative septic shock. J Lab Clin Med. 1984;104:873-881. [PubMed] [Google Scholar]
- 22. Holmes MV, Simon T, Exeter HJ, et al. Secretory phospholipase A(2)-IIA and cardiovascular disease: a Mendelian randomization study. J Am Coll Cardiol. 2013;62:1966-1976. doi: 10.1016/j.jacc.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580-585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar S, Ambrosini G, Bucher P. SNP2TFBS—a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017;45:D139-D144. doi: 10.1093/nar/gkw1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo Y, Hitz BC, Gabdank I, et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882-D889. doi: 10.1093/nar/gkz1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cano-Gamez E, Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soskic B, Cano-Gamez E, Smyth DJ, et al. Chromatin activity at GWAS loci identifies T cell states driving complex immune diseases. Nat Genet. 2019;51:1486-1493. doi: 10.1038/s41588-019-0493-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farh KK-H, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337-343. doi: 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishigaki K. Beyond GWAS: from simple associations to functional insights. Semin Immunopathol. 2022;44:3-14. doi: 10.1007/s00281-021-00894-5 [DOI] [PubMed] [Google Scholar]
- 33. Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714-719. doi: 10.1038/nature09266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nevalainen TJ, Eerola LI, Rintala E, Laine VJO, Lambeau G, Gelb MH. Time-resolved fluoroimmunoassays of the complete set of secreted phospholipases A2 in human serum. Biochim Biophys Acta. 2005;1733:210-223. doi: 10.1016/j.bbalip.2004.12.012 [DOI] [PubMed] [Google Scholar]
- 35. Akinkuolie AO, Lawler PR, Chu AY, et al. Group IIA secretory phospholipase A(2), vascular inflammation, and incident cardiovascular disease. Arterioscler Thromb Vasc Biol. 2019;39:1182-1190. doi: 10.1161/ATVBAHA.118.311894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benson MD, Yang Q, Ngo D, et al. Genetic architecture of the cardiovascular risk proteome. Circulation. 2018;137:1158-1172. doi: 10.1161/CIRCULATIONAHA.117.029536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wootton PTE, Drenos F, Cooper JA, et al. Tagging-SNP haplotype analysis of the secretory PLA2IIa gene PLA2G2A shows strong association with serum levels of sPLA2IIa: results from the UDACS study. Hum Mol Genet. 2006;15:355-361. doi: 10.1093/hmg/ddi453 [DOI] [PubMed] [Google Scholar]
- 38. Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carayol J, Chabert C, Di Cara A, et al. Protein quantitative trait locus study in obesity during weight-loss identifies a leptin regulator. Nat Commun. 2017;8:2084. doi: 10.1038/s41467-017-02182-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morales J, Pujar S, Loveland JE, et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022;604:310-315. doi: 10.1038/s41586-022-04558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hurt-Camejo E, Camejo G, Peilot H, Oörni K, Kovanen P. Phospholipase A(2) in vascular disease. Circ Res. 2001;89:298-304. doi: 10.1161/hh1601.095598 [DOI] [PubMed] [Google Scholar]
- 42. Smith JW, Barlas RS, Mamas MA, et al. Association between serum secretory phospholipase A2 and risk of ischaemic stroke. Eur J Neurol. 2021;28:3650-3655. doi: 10.1111/ene.15004 [DOI] [PubMed] [Google Scholar]
- 43. Talvinen KA, Kemppainen EA, Nevalainen TJ. Expression of group II phospholipase A2 in the liver in acute pancreatitis. Scand J Gastroenterol. 2001;36:1217-1221. doi: 10.1080/00365520152584879 [DOI] [PubMed] [Google Scholar]
- 44. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinform. 2009;7:147-154. doi: 10.1016/S1672-0229(08)60044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramírez-Bello J, Jiménez-Morales M. Functional implications of single nucleotide polymorphisms (SNPs) in protein-coding and non-coding RNA genes in multifactorial diseases. Gac Med Mex. 2017;153:238-250. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-2-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-3-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-4-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights
Supplemental material, sj-docx-5-bbi-10.1177_11779322241261427 for Identification of an Allele-Specific Transcription Factor Binding Interaction that May Regulate PLA2G2A Gene Expression by Aki Hara, Eric Lu, Laurel Johnstone, Michelle Wei, Shudong Sun, Brian Hallmark, Joseph C Watkins, Hao Helen Zhang, Guang Yao and Floyd H Chilton in Bioinformatics and Biology Insights