Abstract
The initial step of virus-cell interaction was studied by immunofluorescence microscopy. Single particles of murine leukemia virus (MLV) vectors and human immunodeficiency virus (HIV) were visualized by immunofluorescence. Fluorescent dots representing single virions could be localized by staining of capsid proteins (CA) or surface envelope proteins (SU) after fixation of virus supernatants. This technique can be used to determine particle concentration in viral supernatants and also to study virus-cell interaction. We investigated the role of the Env-receptor interaction for the initial binding event between the cell and the viral particles. Ecotropic MLV vector particles were shown to bind to human cells which do not express the specific viral receptor. In addition, MLV particles defective for Env were shown to bind the cells similarly to infectious MLV. Time course experiments of virus-cell binding and dissociation showed identical profiles for infectious and Env-defective MLV particles and suggested that MLV Env is not involved in the early phases of attachment of virus to cells. The possible implication of cellular factors in enhancing viral binding and infectivity is discussed.
Understanding of the mechanism by which retroviral particles infect target cells is useful both for the development of antiretroviral therapies and for the improvement of retroviral vectors. Virus-cell binding, virus entry, and subsequent trafficking of human immunodeficiency virus (HIV) and murine leukemia virus (MLV) particles have been actively studied, mainly by genetic approaches, by biochemical methods, or by fluorescence-activated cell sorter (FACS) analysis of individual target cells. For example, cell surface receptors for retroviruses have been identified and characterized by genetic approaches of DNA transfection and pseudotype infection and Env-mediated fusion assays (42). Specific binding of virus Env and its receptor can be studied by binding assays with Env labeled with radioisotopes (12, 18, 21) or antibody staining followed by FACS analyses (19). Trafficking of retroviruses in target cells has been studied mainly by localization of virus components by fractionation of subcellular compartments or microscopic observation of fluorescence-labeled proteins rather than virus particles (17, 23). While these approaches provide important information for the understanding of virus-cell interactions, microscopic visualization of single retrovirus particles in the process of infection would represent a powerful approach to directly observe initial events in virus infection.
Observation of single retrovirus particles has thus far been limited to electron microscopy (EM) techniques. EM has been used to count virus particles (45), to study virus morphology and the process of virus budding from producer cells, and occasionally to study virus-cell binding (33). However, relatively little information on retrovirus particle-cell interaction in the infection process has been obtained by EM techniques. Recent developments in fluorescence microscopy have raised interest in the visualization of single virus particles. Although many researchers considered that most viruses would be too small to detect by optical microscopy because of its limited resolution (250 nm), visualization of single particles of adenovirus, adeno-associated virus, and vaccinia virus by immunofluorescence microscopy has recently been reported. Nonenveloped adenovirus and adeno-associated virus have been chemically labeled with cyanine-based fluorophores, and their binding and entry into cells have been examined (4, 22). By directly conjugating fluorophores to the virus, there is no complication of antibody accessibility and infection can be analyzed in living cells and tissues. However, this method is unlikely to be feasible for many enveloped viruses, including retroviruses, which are more fragile and difficult to purify than adenovirus and adeno-associated virus. Binding of the enveloped vaccinia virus (approximately 250 to 350 nm in diameter) to cells was studied by confocal microscopy after indirect immunostaining of virus antigen in fixed samples (40). Individual vaccinia virus virions appeared as clear fluorescent spots, and it was suggested that the method may be applicable to viruses as small as 50 nm.
In this study, we have established a method to visualize single retrovirus particles by indirect immunostaining. By using this method, virus particles were enumerated and unexpected binding of noninfectious MLV vector particles to cells was observed.
MATERIALS AND METHODS
Cells and viruses.
TE671 (ATCC CRL8805), A431 (ATCC CRL-1555), NIH 3T3, and all packaging cell lines were grown in Dulbecco’s modified Eagle’s medium. Human umbilical vein endothelial cell (HUVEC) (HUV-EC-C CRL-1730) cultures were grown in endothelial growth medium (EGM-MV; TCS Clonetics Corp.). Igrov-1 (provided by M. Ford), CEM (ATCC CCL 119), Raji (ATCC CCL-86), U937 (ATCC CRL-1593.2), and H9 (ATCC HTB-176) cells were cultured in RPMI 1640 medium. Growth media were supplemented with 10% (vol/vol) heat-inactivated fetal calf serum, penicillin G (100 U/ml), and streptomycin (100 mg/ml), and cell cultures were maintained at 37°C under 5% CO2. TELCeB6, TELCeB6/AF7, and TELCeB6/MOF are helper-free packaging cells producing MLV-based retrovirus vector particles which carry the MFGnlslacZ genome and bear no envelope, 4070A amphotropic MLV envelope, and ecotropic Moloney MLV envelope glycoproteins, respectively (10, 24). The HIV-1(GUN-1) isolate (37) was produced by chronically infected H9 cells. Virus particles were harvested after cocultivation with fresh H9 cells for 3 days. Virus stocks were snap frozen and stored in liquid nitrogen. Viral supernatants were filtered through 450-nm-pore-size filters (Sartorius) before use.
Antibodies.
Goat polyclonal antibodies (Quality Biotech Inc.) were used to detect MLV capsid proteins (anti-RLV p30) and MLV surface envelope proteins (SU) (anti-RLV gp69/71). A rat monoclonal immunoglobulin G (IgG) antibody, 83A25, which recognizes MLV SU was kindly provided by L. H. Evans (13). A mouse monoclonal anti-HIV-1 p24 IgG antibody EVA365 (AIDS reagents, MRC program) and a rat monoclonal IgG antibody against the HIV-1(GUN-1) gp120 V3-loop, Aw (26), were used to stain HIV-1 virions. Fluorescein isothiocyanate (FITC)-conjugated donkey IgG against goat IgG (Jackson), FITC-conjugated rabbit IgG against mouse IgG (Dako), and Texas red-conjugated donkey IgG against rat IgG (Jackson) were used as secondary antibodies. For immunoelectron microscopy, 10-nm-gold particle-conjugated goat IgG against fluorescein (British Biocell International) was used as the tertiary antibody.
Immunofluorescent staining of viral particles.
Two methods of fixing virus particles onto glass slides or coverslips (BDH) were examined. Virus suspensions (5-μl volumes) were spread and air dried on a 1-cm2 area of glass slide and fixed with 4% paraformaldehyde for 15 min at room temperature. Alternatively, 50 μl of viral supernatants was incubated on glass slides for 1 h at 37°C in the presence of 8μg of Polybrene (Sigma) per ml. This method follows our observation that Polybrene promotes the adhesion of virus particles to plastic and glass surfaces. Fixed virus particles were permeabilized with 0.2% Triton X-100 for 15 min at room temperature and then washed three times with phosphate-buffered saline (PBS). Virus samples were incubated with anti-Rauscher leukemia virus p30 and 83A25 (MLV samples) or anti-HIV-1 p24 and anti-HIV-1 gp120 (HIV samples) primary antibodies for 45 min at room temperature, washed three times with PBS, and incubated with the appropriate secondary antibodies for 45 min at room temperature. After being washed three times with PBS, the slides were mounted with immunofluorescence mounting medium (Dako) and observed by confocal microscopy (MRC 1024 [Bio-Rad] equipped with a krypton-argon laser). All pictures were acquired by using Kalman filtration and analyzed with Lasersharp software (Bio-Rad).
Estimation of particle size, infectious titer, and physical number of MLV vectors.
A LacZ(MLV-A) vector was harvested from TELCeB6/AF7 and sequentially passed through syringe filters with the following pore sizes: 450 nm (Sartorius), 200 nm (Sartorius), 100 nm (Millipore), and 20 nm (Millipore). After each filtration, aliquots of filtrates were examined by titer determination for LacZ infection on TE671 cells as described previously (36) and by virus particle immunostaining. To determine the physical virus particle number, 110-nm-diameter red fluorescent carboxylate-modified microspheres (FluoSpheres; Molecular Probes) were added to the filtrates at a final concentration of 2.7 × 108 microspheres/ml. Alternatively, the same concentration of microspheres was added to the viral supernatant before filtration, in order to confirm the retention capacity of the filters. Mixtures of virus particles and fluorescent microspheres were immobilized on glass slides and processed for immunostaining as described above. The virus particle number in the vector preparation was calculated as the product of the microsphere concentration and the ratio of counts of fluorescent dots for virus and microsphere particles on the assumption that viral particles and microspheres were immobilized and fixed with similar efficiency, as suggested by the following observations. Microsphere-virus particle ratios were similar when the two different methods described were used to immobilize virions and microspheres or when a 50:50 mixture of acetone and methanol was used instead of paraformaldehyde to fix the samples. In addition, a similar ratio was found when the same mixture was immobilized and fixed on nickel grids and analyzed by EM (see below).
EM.
To directly compare our immunofluorescence method with EM, a mixture of viral supernatant and microspheres was immobilized on coated nickel grids by the same procedure used for immobilization on glass slides. The mixture of virus and beads was incubated for 1 h at 37°C in the presence of 8 μg of Polybrene per ml. Alternatively 200 μl of virus-bead mixture was ultracentrifuged onto nickel grids at 100,000 × g for 10 min in an EM-90 rotor (Beckman Airfuge) by the method described by Zheng et al. (45). Specimens were then fixed on grids with 4% paraformaldehyde in PBS for 15 min at room temperature and permeabilized by treatment with 0.2% Triton X for 15 min at room temperature. After 10 min of blocking treatment with 0.1% bovine serum albumin (Sigma) and 0.1% fish gelatin in Tris-buffered saline (TBS; pH 8.2), capsid immunostaining was performed by incubation with anti-RLV p30 for 45 min at room temperature. After being washed three times with TBS, the grids were incubated with FITC-conjugated anti-goat antibody for 45 min at room temperature, washed three times in TBS, and incubated with gold-labeled anti-FITC antibody for 45 min at room temperature. After three washes in TBS and one wash in water, the grids were negatively stained for 2 min in 2% aqueous uranyl acetate or 3% phosphotungstic acid (PTA). The virus particles were immunolabeled with gold-conjugated antibodies to accurately differentiate virus particles from negatively stained cellular debris. The grids were observed with a IL20 Philips transmission electron microscope. The ratio between viral particles and beads was determined by the number of virions found during the counting of 500 beads on duplicate grids. Viral particle number was estimated by multiplying the bead/virion ratio by the concentration of beads in the mixture analyzed.
Virus-cell microscopy binding assay.
Cells were seeded onto 13-mm glass coverslips in a 24-well plate at 5 × 104 cells/well. After overnight incubation, 0.5 ml of viral suspensions, containing about 4 × 108 physical virus particles/ml, were added for 1 h at 37°C. The cells were then washed five times with PBS supplemented with 1 mM MgCl2 and 1 mM CaCl2 to prevent cell detachment from the glass surface. The cells were fixed with acetone-methanol (1:1) for 3 min at 4°C. The cell monolayer was air dried and rinsed with PBS. Alternatively, coverslips were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.2% Triton X for 15 min at room temperature. The samples were then blocked with a 1% solution of bovine serum albumin in PBS for 15 min, washed with PBS, and incubated with primary antibodies for 45 min at room temperature. The coverslips were washed three times with PBS, incubated with secondary antibodies for 45 min at room temperature, and extensively washed with PBS. After a final wash with distilled water, the coverslips were mounted with immunofluorescence mounting medium (Dako) and analyzed by confocal microscopy.
To study virus binding to cells in suspension, adherent cells were harvested as a suspension by using 10 mM EDTA solution in PBS. Then 106 cells were incubated with 1 ml of virus suspension for 1 h at 37°C. After the cells were washed three times in PBS, each sample was resuspended in 50 μl of PBS, fixed with 250 μl of acetone-methanol (1:1) for 3 min, and centrifuged onto 13-mm coverslips in 24-well plates. After a further wash with PBS, the coverslips were processed for immunostaining as described above.
Alternatively, cells in suspension were fixed with a 4% solution of paraformaldehyde for 15 min at room temperature, permeabilized with 0.2% Triton X for 15 min, and processed for immunostaining in suspension. The cells were then concentrated by centrifugation, resuspended in 10 μl of mounting medium, and mounted on glass microscope slides.
FACS analysis.
Cells were washed in PBS and detached with 10 mM EDTA in PBS. Then 106 cells were incubated with 1 ml of virus suspensions for 1 h at 37°C, washed three times with ice-cold PBS, and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. For p30 staining, cells and virions were also permeabilized with 0.2% Triton X in PBS for 15 min. The cells were washed extensively in PBS, incubated with primary antibody, washed three times with PBS, and incubated with the secondary antibody for 45 min at room temperature. After a final wash, samples were subjected to FACScan (Becton-Dickinson) analysis. The same samples were also processed for analysis by confocal microscopy as described above.
RESULTS
Visualization of retrovirus particles.
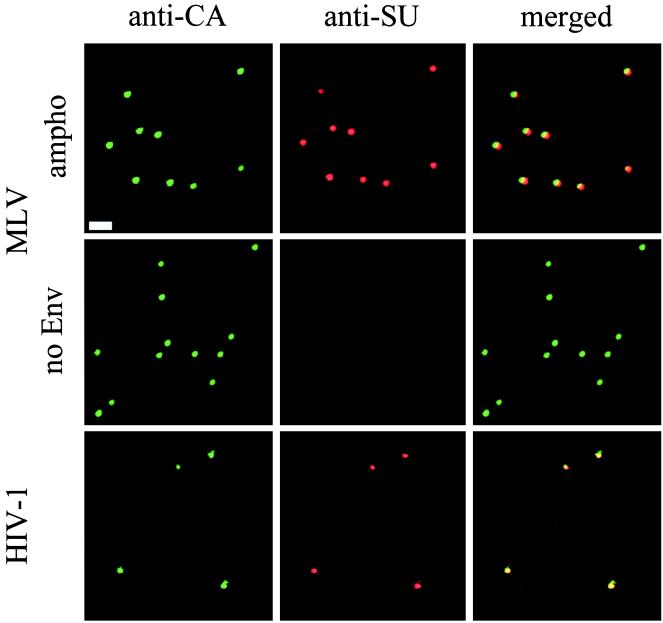
To test if retrovirus particles could be visualized by immunostaining, MLV vector particles were immobilized on glass slides by drying the viral supernatant onto the slide surface or by incubation at 37°C in the presence of 8 μg of Polybrene per ml. Immobilized virions were fixed with paraformaldehyde and permeabilized with Triton X. Double staining was then performed with anti-CA and anti-Env antibodies followed by FITC- and Texas red-labeled secondary antibodies, respectively. Doubly stained spots were observed for MLV vectors bearing MLV-A Env proteins (Fig. 1, MLV ampho), while noninfectious MLV vectors, devoid of Env proteins, produced spots stained with anti-CA but not anti-Env antibodies (Fig. 1, MLV no Env). As a negative control, no spots were observed for supernatants harvested from TEL cells which do not contain MLV structural genes (data not shown). These results demonstrated that complexes, which contain both MLV CA and Env, can be specifically detected in the supernatant from packaging cells producing infectious MLV vectors. Figure 1 also shows that similar complexes containing both CA and Env for HIV-1 can be visualized with anti-HIV antibodies. The specificity of staining was further demonstrated by staining different HIV-1 strains with strain-specific anti-Env antibodies and that anti-HIV antibodies do not stain MLV particles and vice versa (data not shown).
FIG. 1.
Visualization of MLV vector and HIV-1 particles. Virus supernatants of MLV vectors bearing MLV-A Env (ampho) and no Env and HIV-1 GUN-1 isolate were air dried and fixed with 4% paraformaldehyde on glass slides. After permeabilization, immunostaining was performed with anti-CA antibodies followed by FITC-labeled secondary antibodies (anti-CA) and with anti-SU antibodies followed by Texas red-labeled secondary antibodies (anti-SU). Images for green and red fluorescence were acquired separately and overlaid (merged). Bar, 2 μm.
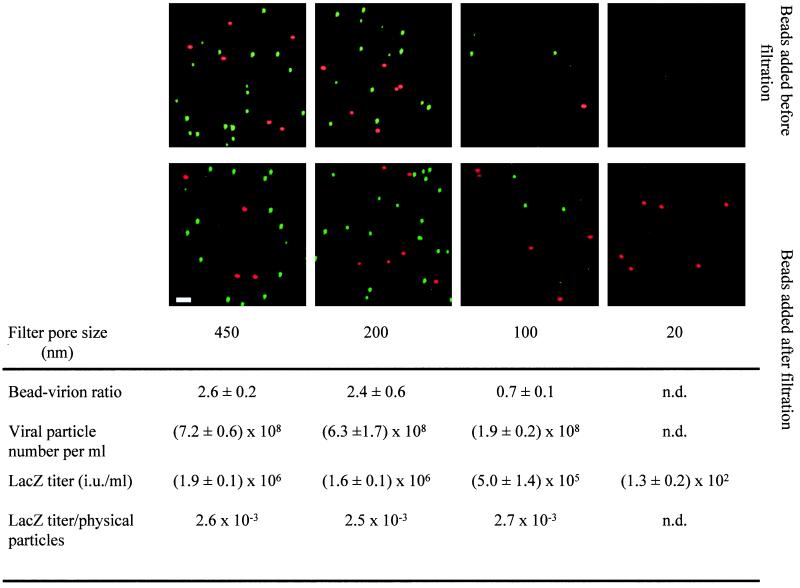
To examine if the above-stained spots represent single virus particles, their size was estimated by their removal following filtration through different pore sizes. Virus supernatant was mixed with a known concentration of 110-nm-diameter fluorescent microspheres, filtered, and then processed for capsid immunostaining (Fig. 2, top panels). No removal of immunostained spots or fluorescent microspheres was observed after filtration through the 200-nm-pore-size filter, while removal was significant and similar for both fluorescent microspheres and immunostained particles following filtration through the 100- and 20-nm-pore-size filters (Fig. 2, top panels). To accurately estimate the number of immunostained spots remaining after filtration, a constant concentration of fluorescent microspheres was added after filtration of the supernatant (Fig. 2, bottom panels). The ratio between immunostained spots and microspheres was determined. No removal of immunostained complexes was observed after filtration through the 200-nm-pore-size filter, while filtration through the 100- and 20-nm-pore-size filters resulted in about 70% and more than 99% removal, respectively (Fig. 2, bottom panels). LacZ infectious titer was also similarly removed by filtration (Fig. 2). These results indicate that the sizes of immunostained complexes and infectious LacZ vectors were between 20 and 200 nm, consistent with the reported size of retrovirus particles as estimated by EM (see Fig. 3) (15, 29). We therefore conclude that the immunofluorescent spots, which stained positively for both CA and Env proteins and were about 100 nm in diameter, represent single retrovirus particles.
FIG. 2.
Estimation of size and physical number of MLV particles. A LacZ(MLV) supernatant was filtered through 450-, 200-, 100-, and 20-nm-pore-size filters in succession. Red-fluorescent microspheres were added to the supernatant before filtration (top panels) or after filtration (bottom panels) at 2.7 × 108/ml and processed for immunostaining for MLV CA proteins as in Fig. 1. Stained viral particles and fluorescent microspheres from the sample filtered before microsphere addition (bottom panel) were counted in five random fields of 5,500 μm2, and the microsphere/viral particle ratio was measured. Results are expressed as mean values of the estimated ratios ± standard errors of the mean. LacZ titer was measured on TE671 cells. Bar, 2 μm. n.d., not determined.
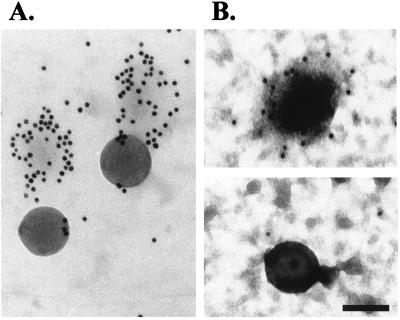
FIG. 3.
Negative staining and immuno-EM of viral particles. A mixture of virus particles and beads was adsorbed to an EM nickel grid by ultracentrifugation (A) or by a 1-h incubation at 37°C in the presence of 8 μg of Polybrene per ml (B). Samples were then processed for capsid immunostaining and negative staining with PTA (A) or uranyl acetate (B). The beads measure 110 nm in diameter. Bar, 100 nm.
Physical counting of virus particles.
We investigated the use of immunofluorescence microscopy as alternative method to estimate the physical number of retrovirus particles. Such a method would be much less time-consuming and less labor-intensive than EM techniques and could be used routinely. We therefore added a known concentration of fluorescent microspheres (final concentration, 2.7 × 108 microspheres/ml) to virus suspensions and processed the samples as for immunofluorescence observations as described above. The virus particle number was estimated from the ratio between immunostained virus particles and fluorescent microspheres. By this method, the physical particle concentration of the sample in the above experiment was estimated to be about 7 × 108/ml (Fig. 2). To test the reliability of this counting method, a virus-microsphere mixture was quantified by transmission electron microscopy and immunofluorescence in parallel. By using the same procedure as for immunofluorescence microscopy, microspheres and virus were immobilized onto EM grids and processed for capsid immunostaining. Specimens on EM grids were additionally treated with gold-labeled anti-FITC antibody and negatively stained with uranyl acetate or PTA. The viral particle number calculated from the microsphere/virus ratio was 0.34 × 109 particles/ml when viral particles were immobilized to the grid by incubation with Polybrene and 1.42 × 109 particles/ml when viral particles were immobilized by the ultracentrifugation method. The same viral suspension was estimated to contain 1.25 × 109 particles per ml by immunofluorescence. This result confirmed the reliability of immunofluorescence as method for the quantification of viral particles in an MLV suspension. Transmission electron microscopy observations also confirmed that viral particles and microspheres are of similar size (100 to 120 nm in diameter) and do not form aggregates during the fixation step (Fig. 3).
As shown in Fig. 2, a LacZ(MLV-A) preparation of about 7 × 108 physical particles/ml contained about 2 × 106 LacZ infectious units/ml. Therefore, the infectious unit/physical particle number ratio was estimated to be 3 × 10−3. This is in agreement with previous estimations that the infectious titer is only a small fraction of the physical retrovirus particle number (20, 35, 44).
Virus particle binding to the cell surface is independent of specific Env-receptor interaction.
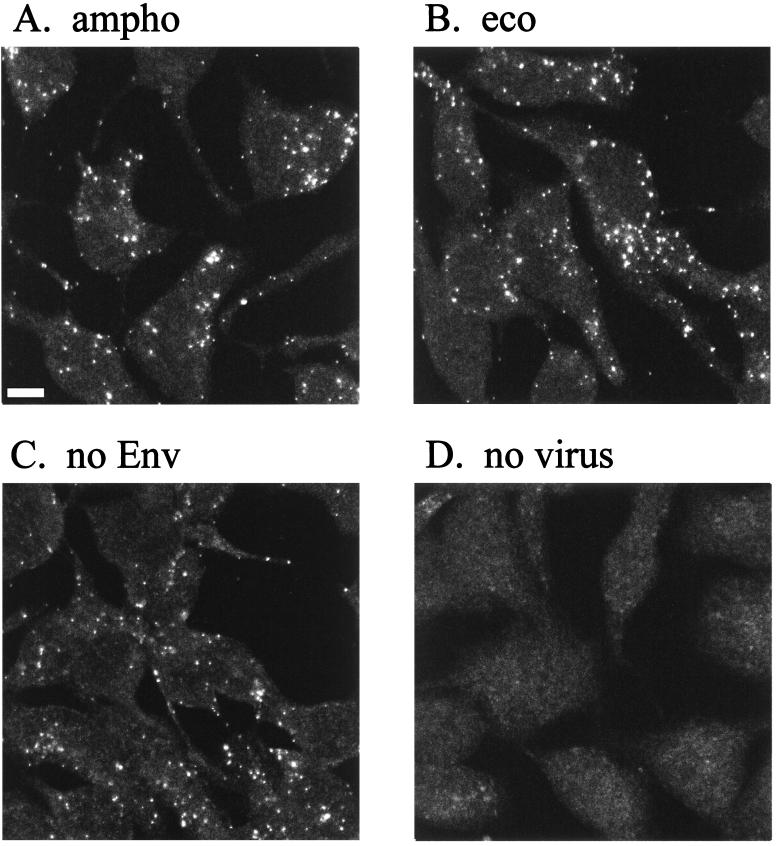
Virus particles bound to the cell surface were visualized by immunostaining to study virion-cell interaction. Human TE671 cells, which express the receptor for MLV-A but not for MLV-E, were grown on glass coverslips and incubated with viral supernatants. Virus particles attached to the cells were stained after fixation and permeabilization. Figure 4 shows immunofluorescence staining with anti-CA antibody. Fluorescent spots were detected on cells incubated with LacZ(MLV-A) (Fig. 4A) but not on cells incubated with a control cell supernatant containing no virus (Fig. 4D). Analysis of sequential optical sections of the specimen indicated that the great majority of viral particles were located on the cell surface (data not shown). Similar viral binding was observed under conditions preventing capping and endocytosis, such as at 4°C or in the presence of sodium azide (data not shown). This suggests that the observed dots represented single virions rather than aggregates formed by capping after membrane binding.
FIG. 4.
Binding of MLV vector particles to TE671 cells. Cell supernatant of TE671 cells producing LacZ(MLV-A) (A), LacZ(MLV-E) (B), LacZ pseudotype without Env proteins (C), and no virus particle (D) was added to TE671 cells. The cells were washed extensively after a 1-h incubation and then fixed with acetone-methanol (1:1). Samples were stained for MLV CA proteins with FITC-labeled secondary antibodies. Bar, 5 μm.
Surprisingly, LacZ pseudotypes bearing ecotropic MLV Env (Fig. 4B) or no Env (Fig. 4C) bound to TE671 cells at similar levels to LacZ(MLV-A) despite the fact that they cannot infect TE671 cells. LacZ(MLV-A) and LacZ(MLV-E) but not Env-defective pseudotypes could also be stained with anti-Env on TE671 cells in double-staining experiments (data not shown). These experiments show the ability of MLV particles to bind to the cell surface in the absence of the specific viral receptor, as shown by LacZ(MLV-E) binding to human cells, or in the absence of viral glycoprotein, as shown by Env-defective virus binding. As shown later (see Fig. 8), this phenomenon is not peculiar to TE671 cells. Furthermore, LacZ(MLV-E) particles produced from a murine packaging cell line also bound efficiently to TE671 cells, which are resistant to MLV-E infection (data not shown). These results indicate that the virion-cell binding is independent of specific receptor-Env interaction and is not peculiar to the initial pair of virus producer and target cell types tested.
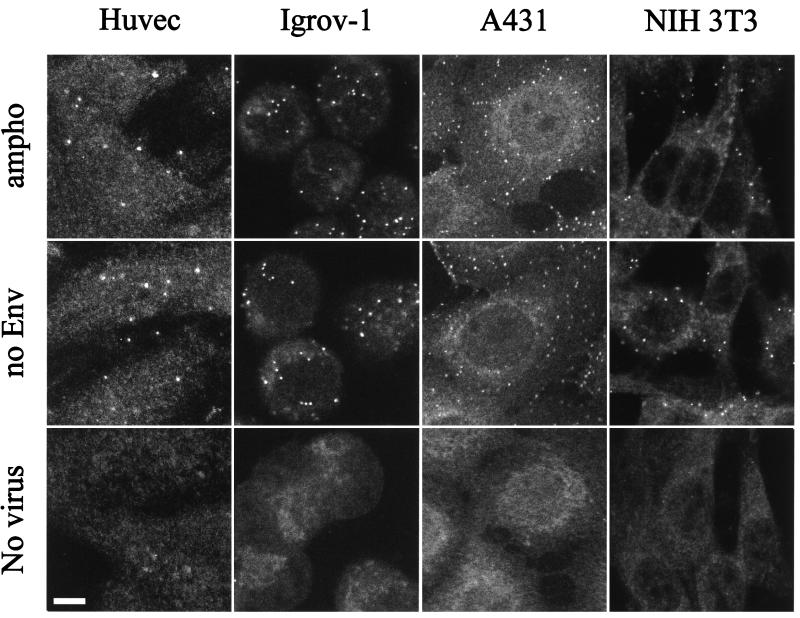
FIG. 8.
Binding of MLV vector particles to different adherent cells. Cell cultures were incubated with MLV-A (ampho), MLV Env defective (no Env), or mock viral supernatants (no virus). After a 1-h incubation, the cells were washed, fixed, permeabilized, and stained for capsid proteins. Bar, 5 μm.
Comparison of virus binding assays by FACS and microscopy analyses.
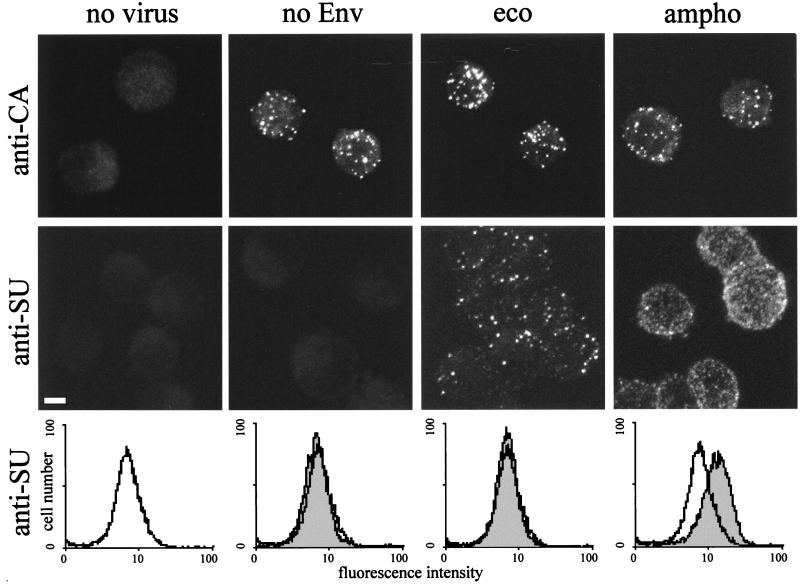
A standard method used to study virus-cell binding is FACS analysis of cells which are incubated with virus supernatants and then subjected to surface staining with anti-Env antibodies. In this assay, positive fluorescence shifts represent specific receptor-Env interactions and no shift has been observed in the absence of cognate receptors and Env, such as the case of MLV-E with receptor-negative human cells (19). The observations in our microscopy study, showing that retrovirus particles bind to cells in the absence of specific receptor-Env interaction, appear to contradict the results of the FACS analyses. Therefore, we directly compared FACS and microscopy analyses of virus-cell binding. Suspensions of TE671 cells were incubated with amphotropic, ecotropic, and Env-defective viruses and control supernatant. After fixation, cells were processed for CA staining or for Env staining. The same samples were then analyzed by both FACS and confocal microscopy. Figure 5 shows micrographs of anti-CA and anti-Env (SU) staining and FACS histograms of anti-Env staining. FACS analysis of anti-CA staining showed only minimal shifts for all three viruses (data not shown), consistent with the microscopy observation, which showed binding of similar numbers of virus particles for amphotropic, ecotropic, and Env-defective viruses (Fig. 5). Following SU staining, a significant shift by FACS was detected only with amphotropic virus, while confocal microscopy also detected the presence of virus particles on cells which had been incubated with MLV-E. However, cells incubated with amphotropic virus, but not ecotropic virus, demonstrated a diffuse surface staining following anti-SU staining, in addition to brighter spots of fluorescence, while CA staining produced only bright spots. Such diffuse fluorescence is likely to be caused by specific binding of soluble SU proteins present in virus supernatants to their receptors (8, 25, 43) and results in significant shifts during FACS analyses. These results suggest that, although FACS analysis can efficiently detect the specific Env-receptor interaction, it reflects soluble SU binding rather than virus particle binding.
FIG. 5.
Comparison of FACS and confocal microscopy analyses of MLV vector binding to TE671 cells. Suspended TE671 cells were incubated with supernatants of TE671 cells producing LacZ(MLV-A) (ampho), LacZ(MLV-E) (eco), LacZ pseudotype without Env proteins (no Env), and no virus particle (no virus) for 1 h, washed, and then fixed with 4% paraformaldehyde. The cells were stained with either anti-MLV CA (anti-CA) antibodies after permeabilization or anti-MLV SU antibodies without permeabilization. The micrographs are projected optical sections performed every 1 μm perpendicular to the z axis throughout the sample. Bar, 5 μm. Histograms of cells incubated with vector particles are shaded and shown together with histograms of cells incubated with virus-free cell supernatant (white).
Kinetics of virion binding to cells.
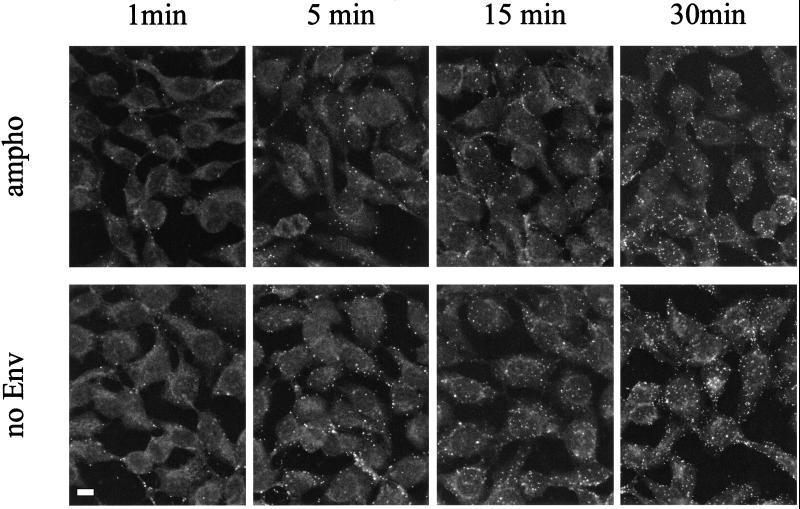
The kinetics of virus binding to TE671 cells in the presence and absence of specific receptor-Env interaction was compared. The virus-cell association rate was examined by incubating target cells with amphotropic or Env-defective virus suspensions for variable times at 37°C and then staining them for CA (Fig. 6). When both viruses were used, bound virions could be detected after only 1 min and the maximum number of virions was detected after a 30-min incubation. No increase in the number of bound viruses was detected beyond 30 min (60 and 120 min [data not shown]). It has been reported that the large majority of viral particles added to cell cultures may not meet the target cells because their movement is restricted by Brownian motion (7). There is no method yet to directly measure the rate of collision between the virus particle and the cell. However, we calculated a theoretical number of virions that would collide with a single cell during 60 min under our experimental conditions as 100, according to previously described formulae (27). It is likely that a large proportion of virions stably bind to the cell surface when they collide with the cell, because more than 30 fluorescent spots were visualized on a single cell after a 60-min incubation. No quantitative difference was detected at any time point between virions with and without Env (Fig. 6). Similar kinetics of virus attachment to TE671 cells was also observed at 4°C for both amphotropic and Env-defective viruses (data not shown). These results suggested that the rate of virion attachment to the cell surface is similar in the presence and absence of specific receptor-Env interaction and that such attachment is mediated by components on virus and cell surfaces other than viral Env and its cognate receptor.
FIG. 6.
Kinetics of binding of MLV vector particles to adherent cells. TE671 cells were incubated with MLV vector particles with MLV-A Env (ampho) and without any Env protein (no Env) for 1, 5, 15, and 30 min and then processed for immunostaining with anti-MLV CA antibodies followed by FITC-labeled secondary antibodies. Bar, 5 μm.
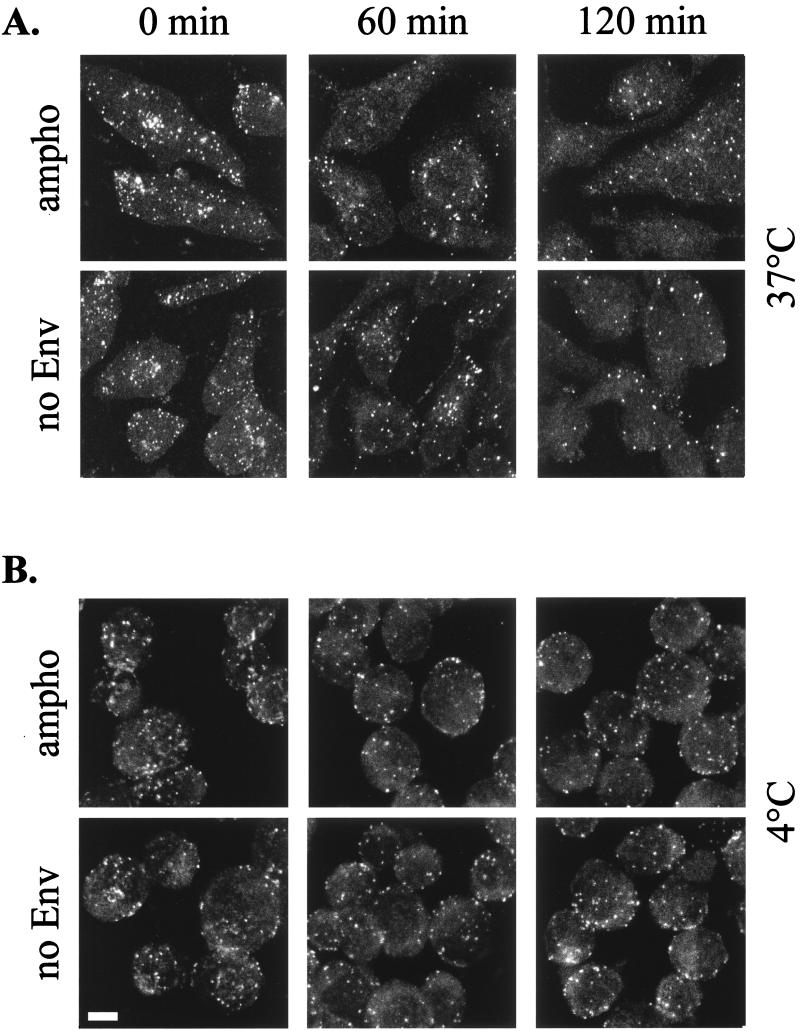
The virus-cell dissociation rate was then analyzed in two ways. First, adherent TE671 cells incubated with vector particles were washed and kept at 37°C in the normal medium supplemented with 0.1% sodium azide in order to prevent particle internalization. Second, suspensions of TE671 cells were incubated with amphotropic and Env-defective virus at 37°C. After 60 min, the cells were washed and kept in PBS at 4°C. The number of viral particles bound to the cell surface after different times (from 1 to 120 min) was detected. Similar numbers of virions were detected on cells incubated with amphotropic or Env-defective viruses at each time point analyzed (Fig. 7). The number of virus particles decreased by about 10 and 70% at 4 and 37°C, respectively.
FIG. 7.
Kinetics of MLV vector particles dissociation from cells. TE671 cells were incubated with MLV vector particles with MLV-A Env (ampho) and without any Env protein (no Env). After 1 h, the cells were washed and kept in fresh medium at 37°C and 0.1% sodium azide (A) or in PBS at 4°C (B) for the indicated time. Samples were then processed for capsid immunostaining. The micrographs are projected optical sections performed every 1 μm. Bar, 5 μm.
Inefficient Env-independent binding on suspension cell lines.
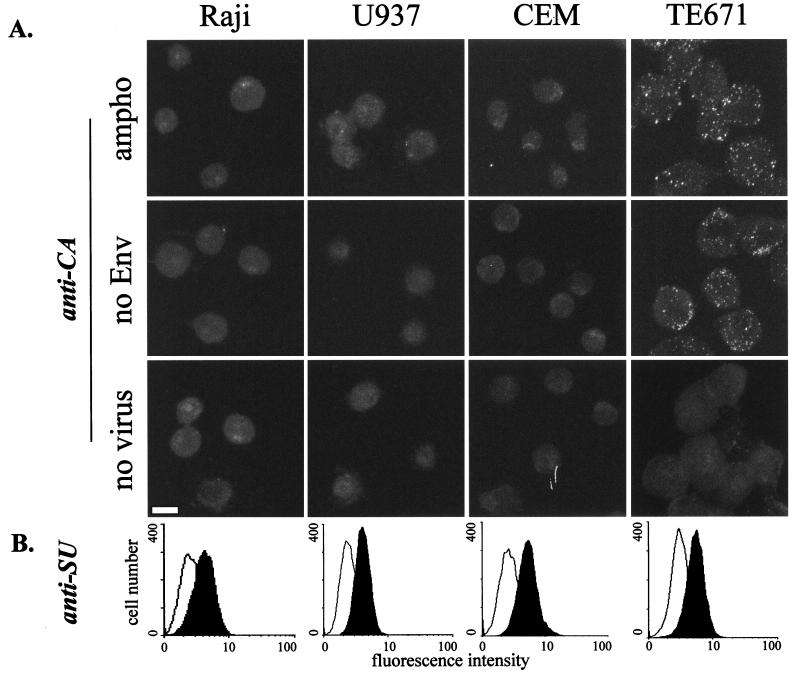
Viral binding of virus particles with or without Env proteins was studied on several cell lines. Adherent cell types, including murine NIH 3T3 fibroblasts, human epithelial A431 cells, human ovarian Igrov-1 cells, human endothelial HUVEC (Fig. 8) and human microvasculature endothelial cells (HMEC) cells, and human glioblastoma NP2 cells (data not shown) were analyzed as described above. The levels of binding of amphotropic and Env-defective MLV were similar on each cell line tested, suggesting that many adherent cell lines have the ability to adsorb virus particles in an Env-receptor-independent manner, albeit at somewhat different levels (Fig. 8). In contrast, poor virus binding was detected on several cell lines growing in suspension. A viral binding assay for cells in suspension was performed with the human T-cell lines CEM and Jurkat; B-cell lines Namalwa, DG75, and Raji; monocytic cell line U937; and adherent cells suspended by EDTA treatment. Cells in suspension were incubated with MLV vectors with or without MLV-A Env and then stained with anti-CA antibodies. Efficient virus particle binding, for both enveloped and nonenveloped vectors, was shown for TE671 cells (Fig. 9) and the other suspensions of adherent cells analyzed (data not shown). In contrast, only small numbers of fluorescent spots were detected on the suspension cell lines (CEM, U937, and Raji [Fig. 9], and DG75, Jurkat, and Namalwa [data not shown]). These results indicate that some cell types lack the ability to efficiently adsorb MLV particles in an Env-receptor-independent way.
FIG. 9.
Binding of MLV vector particles and MLV-A Env to suspension cells. Adherent TE671 cells harvested with EDTA and three suspension cultures were incubated with MLV-A particles (ampho), MLV Env-defective particles (no Env), or mock supernatant (no virus). (A) Microscopy analyses. Cells were permeabilized and stained for MLV-CA. Bar, 10 μm. (B) FACS analysis. Cells were stained for SU and analyzed by FACS. White and black histograms show samples incubated with Env-defective and MLV-A supernatants, respectively. Micrographs are projected optical sections performed every 0.5 μm.
We also noticed that LacZ(MLV-A) infected all suspension cells tested much less efficiently than it infected adherent cells, such as TE671 and NIH 3T3 cells; LacZ titers on suspension cells were typically between 100- and 1,000-fold lower than those on TE671 cells. This observation may suggest that Env-receptor-independent binding is significant in the process of MLV infection.
Like our observation on TE671 cells described above (Fig. 5), we noticed a discrepancy between FACS and microscopy analyses of MLV-A particle binding to suspension cells. Suspension cultures, including detached TE671, were incubated with amphotropic and Env-defective viral supernatants, processed for Env staining, and subjected to FACS analysis (Fig. 9B). Significant and similar fluorescent shifts were obtained when cells were incubated with MLV-A supernatants, indicating that a similar amount of Env SU binds the cell surface of all cell types. Therefore, this result suggests that the viral receptor is similarly expressed and that a difference in viral infectivity and virus particle binding for adherent and suspension cells cannot be explained by different levels of the virus receptor expression.
DISCUSSION
Image analysis of single retrovirus particles has so far been limited to TEM studies. In this study, we show that retrovirus particles can be visualized as fluorescent spots following indirect immunofluorescent staining. Viral particles could be stained for both a virus core protein, CA, and a virus envelope protein, SU. By using filters with different pore sizes to retain the particles, we estimated that the diameters of the visualized spots were less than 200 nm but that the particles were largely retained by 100-nm-pore-size filters. TEM observation of the same virus preparations confirmed that the visualized particles were single virions of about 100 nm in diameter.
A method to count physical number of virus particles was developed by adding a known concentration of fluorescent microspheres to virus samples to act as internal controls. The virus particle concentration could be calculated from the ratio of virus particles and microspheres measured by fluorescence microscopy. The reliability of this method was confirmed by direct comparison with a TEM-based counting method (45). The advantage of immunofluorescence over TEM methods is that sample processing and analysis is much easier and less time-consuming. Therefore, we think that this technique will be useful in any experiment which requires virus quantification, such as screening for packaging cell lines that efficiently produce retrovirus vectors, monitoring the level of virus concentration during virus purification and concentration, and quantifying virus particle contamination of biological products.
The ratio of infectious units to particle number estimated by our immunostaining method was about 3 × 10−3. This result is in agreement with previous reports that the infectious titer is only a small fraction of the physical retrovirus particle number (20, 35, 44). A major reason for such a low infectious titer compared with the physical number of particles may be the rate of particle collision with the cell surface. It has been reported that the large majority of viral particles plated on cells may not meet the target cells because their movement is restricted by Brownian motion (1–3, 6, 7). Indeed, little or no reduction in infectivity was recorded when a virus inoculum recovered from the first round infection was plated on fresh cells in the second round (38, 41). One additional possibility is that not all the virus particles are fully functional, but to date it is impossible to examine the function and the stoichiometric composition of individual virus particles. It is noteworthy that more than 90% of the spots for both MLV vectors and HIV were costained for CA and SU and significant numbers of virus particles devoid of Env, which were previously hypothesized to result from inefficient Env incorporation and Env shedding (25, 43), were not detected (Fig. 1). Another possibility is that fully functional virus particles are guided to noninfectious pathways in some circumstances (8, 23) or are lost at various steps in the infection process.
Unexpectedly, virus-cell binding was detected to a similar extent in both the presence and the absence of specific receptor-Env interaction. Virus binding in the absence of cognate Env contradicted previous reports (5, 8, 19, 24) analyzing viral binding by using FACS as well as the concept that Env-receptor interaction promotes virus binding to cells. Comparison of FACS and microscopy observations of the same samples (Fig. 5) demonstrated that FACS analysis detects the binding of soluble SU proteins, shed from packaging cells or viral particles (8), to specific receptors and therefore indicates the levels of receptor on the cell surface. In contrast to microscopy observations, however, FACS did not detect the binding of virions bearing irrelevant Env (MLV-E on human cells). These results indicate that virus binding on certain cell types does not require specific Env-receptor interaction. Furthermore, comparison of MLV-A binding on suspension cell cultures by FACS and microscopy observations (Fig. 9) demonstrated that certain suspension cell lines can bind virus particles only poorly, despite the presence of similar levels of the MLV-A receptor Pit2 to those found on adherent cell lines. This result suggests that the Env-receptor interaction is not sufficient for efficient virion-cell binding and that such adsorption of the virus to the cell is mediated by cell surface components other than the specific virus receptor. The kinetics of virus binding on TE671 cells was similarly rapid under both conditions, i.e., where cognate receptors and Env are present and where virus bears no Env (Fig. 6). A time course of virus-cell dissociation showed that virion-cell binding was equally stable in the presence and absence of Env (Fig. 7). Therefore, specific Env-receptor interaction did not affect the kinetics of such rapid, stable binding between virus particles and cells. Overall, these results indicate that Env-receptor-independent binding could precede the recognition of the cognate receptors by virus particles.
Nonspecific, viral receptor-independent binding of virus particles to cell membranes has been observed for vesicular stomatitis virus (34), and Rous sarcoma virus (30, 32). However, the significance of such virus adsorption has been unclear. It is possible that viral particles first bind to the cell surface without the involvement of specific viral receptors and subsequently interact with their cognate receptors or functional virus entry sites in many cases of MLV infection. Particles bound to the cell surface could screen for the receptor molecules in two dimensions, while unbound particles must seek their receptors in the three-dimensional space moving with Brownian motion. We therefore suggest that receptor-Env-independent binding of the virus to the cell plays an important role by helping MLV particles to reach the specific receptor and subsequently enter the cell. It is intriguing that the suspension cell lines tested in this study have poor ability to adsorb virus particles and are less infectible than adherent cell lines, which can bind virus particles efficiently. This observation supports our hypothesis that virus binding independent of specific Env-receptor interaction enhances infection, although many other cellular components may affect efficient infection at various stages in the process, such as provirus integration (11, 16).
The ability of MLV and other retroviruses to assemble components of cellular origin in their envelope lipid bilayer is well documented (42). It is therefore likely that one or more of these components are responsible for the Env-independent binding and subsequent enhancement of infection. This hypothesis is supported by recent reports that HIV particles, which bear adhesion molecules and therefore can bind to cells more efficiently in a CD4-independent manner, infect cells more efficiently than does HIV without such adhesion molecules (14, 31). In addition, binding and internalization of HIV to cells lacking the binding receptor CD4 has been described (23, 28, 39). Finally, some cellular components, such as integrin molecules and proteoglycans, were suggested to be involved in CD4-independent HIV binding. It is presently unknown if any of these molecules can mediate the binding of MLV particles; some CD4-independent binding of HIV also seems to require HIV SU proteins under some circumstances (28, 39).
There is considerable interest in the development of targeted retrovirus vectors for in vivo gene therapy. In several studies, specific ligands have been incorporated into vector envelopes to retarget vectors to specific cell types or to enhance vector infection of target cells, but without much success (9). Our finding that retroviruses can bind to the cell in a manner independent of Env-receptor interaction may be relevant to targeting. First, vector particles may be wasted by binding to cells which lack targeted receptors in in vivo settings. Second, incorporation of specific ligands to the virus envelope may not necessarily result in significant enhancement of binding of the vector particle to the target cell, although such modifications can redirect Env proteins to cells bearing their specific receptors, as shown by FACS assays for Env binding (8, 24). Our results suggest the need for cautious interpretations of FACS analyses and indicate that targeting of retrovirus vectors at the later stages of vector infection, such as virus-cell fusion rather than virus-cell binding, may be more effective.
In conclusion, we have established that single retrovirus particles can be visualized by optical microscopy with immunofluorescent staining. This method has further demonstrated that MLV particles can bind to cells with rapid kinetics in a mechanism independent of specific receptor-Env interaction.
ACKNOWLEDGMENTS
We are grateful to Robin Weiss and Áine McKnight for critical reading of the manuscript and for helpful discussions. We thank Hugh Paterson for helpful advice on confocal microscopy.
This work is supported by Glaxo-Wellcome and the Medical Research Council. M.P. is funded by an EU TMR grant (no. FMBICT961804).
REFERENCES
- 1.Allison A C, Valentine R C. Virus particle adsorption. I. Theory of adsorption and experiments on the attachment of particles to non-biological surfaces. Biochim Biophys Acta. 1959;34:10–23. doi: 10.1016/0006-3002(59)90228-8. [DOI] [PubMed] [Google Scholar]
- 2.Allison A C, Valentine R C. Virus particle adsorption. II. Adsorption of vaccinia and fowl plaque viruses to cells in suspension. Biochim Biophys Acta. 1959;40:393–399. doi: 10.1016/0006-3002(60)91379-2. [DOI] [PubMed] [Google Scholar]
- 3.Allison A C, Valentine R C. Virus particle adsorption. III. Adsorption of viruses by cell monolayers and effect of some variables on adsorption. Biochim Biophys Acta. 1959;40:400–410. doi: 10.1016/0006-3002(60)91380-9. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett J S, Samulski R J. Fluorescent viral vectors: a new technique for the pharmacological analysis of gene therapy. Nat Med. 1998;4:635–637. doi: 10.1038/nm0598-635. [DOI] [PubMed] [Google Scholar]
- 5.Battini J L, Rodrigues P, Muller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chillakuru R A, Ryu D D, Yilma T. Propagation of recombinant vaccinia virus in HeLa cells: adsorption kinetics and replication in batch cultures. Biotechnol Prog. 1991;7:85–92. doi: 10.1021/bp00008a002. [DOI] [PubMed] [Google Scholar]
- 7.Chuck A S, Clarke M F, Palsson B O. Retroviral infection is limited by Brownian motion. Hum Gene Ther. 1996;7:1527–1534. doi: 10.1089/hum.1996.7.13-1527. [DOI] [PubMed] [Google Scholar]
- 8.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosset F L, Russell S J. Targeting retrovirus entry. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]
- 10.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel R, Katz R A, Skalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 12.DeLarco J, Todaro G J. Membrane receptors for murine leukemia viruses: characterization using the purified viral envelope glycoprotein, gp71. Cell. 1976;8:365–371. doi: 10.1016/0092-8674(76)90148-3. [DOI] [PubMed] [Google Scholar]
- 13.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortin J F, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol. 1997;71:3588–3596. doi: 10.1128/jvi.71.5.3588-3596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank H, Schwarz H, Graf T, Schafer W. Properties of mouse leukemia viruses. XV. Electron microscopic studies on the organization of Friend leukemia virus and other mammalian C-type viruses. Z Naturforsch Ser C. 1978;33:124–138. doi: 10.1515/znc-1978-1-224. [DOI] [PubMed] [Google Scholar]
- 16.Gaken J A, Tavassoli M, Gan S U, Vallian S, Giddings I, Darling D C, Galea-Lauri J, Thomas M G, Abedi H, Schreiber V, Menissier-de Murcia J, Collins M K, Shall S, Farzaneh F. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J Virol. 1996;70:3992–4000. doi: 10.1128/jvi.70.6.3992-4000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly K, Kalyanaraman V S, Sarngadharan M G. Analysis of the interaction between Rauscher murine leukemia virus and murine cell membrane receptor by in vitro binding assay. Cancer Lett. 1983;18:79–86. doi: 10.1016/0304-3835(83)90120-9. [DOI] [PubMed] [Google Scholar]
- 19.Kadan M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leopold P L, Ferris B, Grinberg I, Worgall S, Hackett N R, Crystal R G. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 23.Marechal V, Clavel F, Heard J M, Schwartz O. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J Virol. 1998;72:2208–2212. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin F, Kupsch J, Takeuchi Y, Russell S, Cosset F L, Collins M. Retroviral vector targeting to melanoma cells by single-chain antibody incorporation in envelope. Hum Gene Ther. 1998;9:737–746. doi: 10.1089/hum.1998.9.5-737. [DOI] [PubMed] [Google Scholar]
- 25.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKnight A, Weiss R A, Shotton C, Takeuchi Y, Hoshino H, Clapham P R. Change in tropism upon immune escape by human immunodeficiency virus. J Virol. 1995;69:3167–3170. doi: 10.1128/jvi.69.5.3167-3170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nermut M V, Frank H, Schafer W. Properties of mouse leukemia viruses. 3. Electron microscopic appearance as revealed after conventional preparation techniques as well as freeze-drying and freeze-etching. Virology. 1972;49:345–358. doi: 10.1016/0042-6822(72)90487-4. [DOI] [PubMed] [Google Scholar]
- 30.Notter M F, Leary J F, Balduzzi P C. Adsorption of Rous sarcoma virus to genetically susceptible and resistant chicken cells studied by laser flow cytometry. J Virol. 1982;41:958–964. doi: 10.1128/jvi.41.3.958-964.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paquette J S, Fortin J F, Blanchard L, Tremblay M J. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J Virol. 1998;72:9329–9336. doi: 10.1128/jvi.72.11.9329-9336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piraino F. The mechanism of genetic resistance of chick embryo cells to infection by Rous sarcoma virus-BRYAN strain (BS-RSV) Virology. 1967;32:700. doi: 10.1016/0042-6822(67)90046-3. [DOI] [PubMed] [Google Scholar]
- 33.Pudney J, Song M J. Electron microscopic analysis of HIV-host cell interactions. Tissue Cell. 1994;26:539–550. doi: 10.1016/0040-8166(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 34.Schlegel R, Willingham M C, Pastan I H. Saturable binding sites for vesicular stomatitis virus on the surface of Vero cells. J Virol. 1982;43:871–875. doi: 10.1128/jvi.43.3.871-875.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith R E. High specific infectivity avian RNA tumor viruses. Virology. 1974;60:543–547. doi: 10.1016/0042-6822(74)90348-1. [DOI] [PubMed] [Google Scholar]
- 36.Tailor C S, Takeuchi Y, O’Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavoloni N. A simple procedure to determine the biological titer of recombinant retroviral vectors. Gene Ther. 1997;4:150–155. doi: 10.1038/sj.gt.3300370. [DOI] [PubMed] [Google Scholar]
- 39.Valenzuela A, Blanco J, Krust B, Franco R, Hovanessian A G. Neutralizing antibodies against the V3 loop of human immunodeficiency virus type 1 gp120 block the CD4-dependent and -independent binding of virus to cells. J Virol. 1997;71:8289–8298. doi: 10.1128/jvi.71.11.8289-8298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Paul R, Burgeson R E, Keene D R, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991;65:6468–6477. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss R A. Cellular receptor and viral glycoproteins involved in retrovirus entry. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 1–108. [Google Scholar]
- 43.Yu H, Empig C, Xia J, Anderson W F. Quantitation of MoMuLV envelope protein on the cell surface. Virology. 1998;243:415–422. doi: 10.1006/viro.1998.9073. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Soong N, Anderson W F. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J Virol. 1995;69:6557–6562. doi: 10.1128/jvi.69.10.6557-6562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Y Z, Webb R, Greenfield P F, Reid S. Improved method for counting virus and virus like particles. J Virol Methods. 1996;62:153–159. doi: 10.1016/s0166-0934(96)02100-3. [DOI] [PubMed] [Google Scholar]