ABSTRACT
Candida auris, an emerging multidrug-resistant fungal pathogen, predominately colonizes the human skin long term leading to subsequent life-threatening invasive infections. Fungal morphology is believed to play a critical role in modulating mucocutaneous antifungal immunity. In this study, we used an intradermal mouse model of C. auris infection to examine fungal colonization and the associated innate and adaptive immune response to yeast and filamentous C. auris strains. Our results indicate that mice infected with a filamentous C. auris had significantly decreased fungal load compared to mice infected with the yeast form. Mice infected with yeast and filamentous forms of C. auris stimulated distinct innate immune responses. Phagocytic cells (CD11b+Ly6G+ neutrophils, CD11b+Ly6Chi inflammatory monocytes, and CD11b+MHCII+CD64+ macrophages) were differentially recruited to mouse skin tissue infected with yeast and filamentous C. auris. The percentage and absolute number of interleukin 17 (IL-17) producing innate lymphoid cells, TCRγδ+, and CD4+ T cells in the skin tissue of mice infected with filamentous C. auris were significantly increased compared to the wild-type of yeast strain. Furthermore, complementation of filamentous mutant strain of C. auris (Δelm1 + ELM1) strain exhibited wild-type yeast morphology in vivo and induced comparable level of skin immune responses similar to mice infected with yeast strain. Collectively, our findings indicate that yeast and filamentous C. auris induce distinct local immune responses in the skin. The decreased fungal load observed in mouse skin infected with filamentous C. auris is associated with a potent IL-17 immune response induced by this morphotype.
IMPORTANCE
Candida auris is a globally emerging fungal pathogen that transmits among individuals in hospitals and nursing home residents. Unlike other Candida species, C. auris predominantly colonizes and persists in skin tissue resulting in outbreaks of systemic infections. Understanding the factors that regulate C. auris skin colonization and host immune response is critical to develop novel preventive and therapeutic approaches against this emerging pathogen. We identified that yeast and filamentous forms of C. auris induce distinct skin immune responses in the skin. These findings may help explain the differential colonization and persistence of C. auris morphotypes in skin tissue. Understanding the skin immune responses induced by yeast and filamentous C. auris is important to develop novel vaccine strategies to combat this emerging fungal pathogen.
KEYWORDS: Candida auris, skin immune response, yeast, filament
OBSERVATION
Candida auris is an emerging multidrug-resistant fungal pathogen that poses a serious threat to public health. The World Health Organization has recently placed C. auris in a critical priority group of fungal pathogens in addition to being classified as an urgent threat by the U.S. Centers for Disease Control and Prevention (CDC) Antibiotic Threats Report (1, 2). Unlike other Candida species such as Candida albicans that colonize the mammalian gastrointestinal tract, C. auris has a unique ability to colonize and persist in human skin long term resulting in nosocomial transmission (3–6). Individuals who have been colonized with C. auris can develop a high fungal burden on their skin, which has been positively correlated with environmental contamination, transmission, and outbreaks of systemic infections (7). Emerging evidence suggests that C. auris persists in murine skin for several months (3). Furthermore, recent findings from our laboratory suggest that skin immune responses to C. auris are different from C. albicans (4). However, the factors that regulate C. auris skin colonization and the local immune response are poorly understood.
Clinical isolates of C. auris can exhibit different morphotypes such as filamentous, pseudohyphal, and yeast forms (8–14). Previous findings in other Candida species suggest that fungal morphology plays a critical role in regulating the colonization, virulence, and immune response in different host niches including the skin (15). For example, C. albicans predominately exists as a yeast form on the stratum corneum of the skin, while pathogenic pseudohyphal and filamentous forms of C. albicans exist in the dermis. Furthermore, C. albicans yeast but not filament-locked mutant strain induces a potent Th17 immune response in the mouse skin (15). Given that C. auris preferentially colonizes the human and murine skin (3, 4, 6, 16), the role of yeast and filament forms in the colonization potential and skin immune responses is still not known. In this study, we examined the colonization and skin immune responses of yeast and filamentous forms of C. auris. We generated a filamentous mutant strain in C. auris by deleting ELM1 in wild-type (WT) South Asian AR0387 strain as described elsewhere (8). ELM1 is a regulator of C. auris morphology, where disruption of this gene results in filamentation (8). A markerless CRISPR-Cas9-mediated genome editing system was used to delete and complement the ELM1 gene in C. auris as previously described (17). We confirmed the gene deletion by PCR and sequencing (Fig. S1; Table S1). As expected, we identified that WT C. auris AR0387 strain exists as a typical round-shaped yeast form, whereas deletion of ELM1 gene (Δelm1) resulted in an elongated, filamentous morphology which is consistent with previous findings (Fig. 1A) (8). Furthermore, complement strain (Δelm1 + ELM1) retained the wild-type morphology as described elsewhere (8).
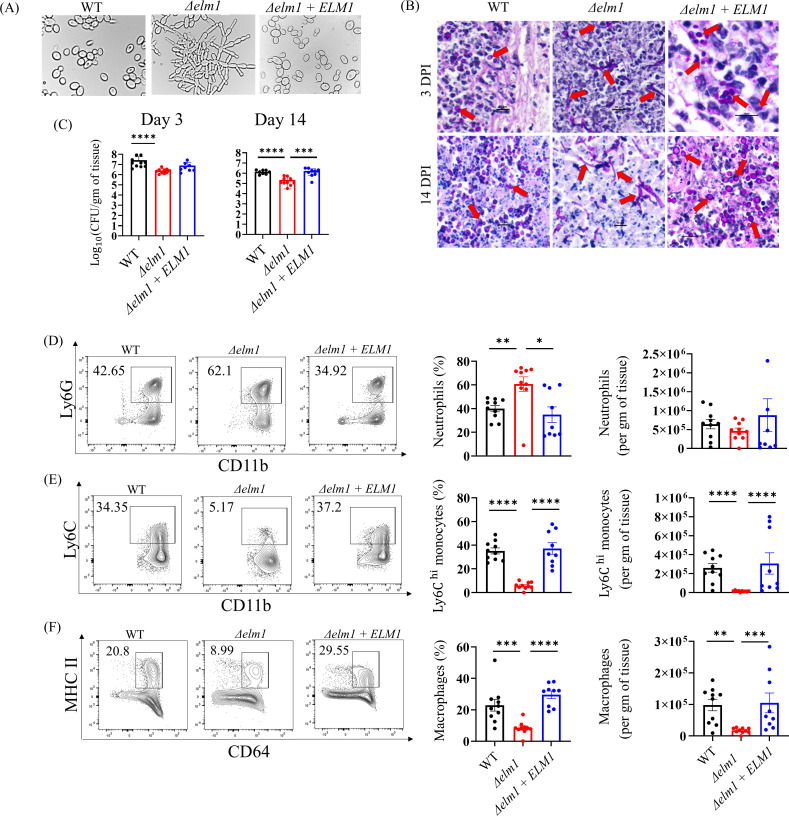
Fig 1.
Fungal colonization and innate immune responses in mouse skin tissue infected with yeast and filamentous C. auris. (A) Morphology of wild-type C. auris 0387, Δelm1 mutant, and Δelm1 + ELM1 complementstrain obtained via differential interference contrast microscopy after growth in vitro. (B) Representative periodic acid-Schiff-stained skin tissues from 3 days and 14 days post-infected mice with wild-type C. auris 0387, Δelm1 mutant, and Δelm1 + ELM1 complement strain (20× and 100× resolution). (C) Fungal load in the skin of mice intradermally infected with wild-type C. auris 0387, Δelm1 mutant, and Δelm1 + ELM1 complement strain following 3 days and 14 days post-infection. Representative flow cytometry plots, percentage, and absolute numbers of (D) CD11b+Ly6G+ neutrophils, (E) CD11b+Ly6Chi monocytes, and (F) CD11b+MHCII+CD64+ macrophage from mouse skin tissue infected with wild-type C. auris 0387, Δelm1 mutant, and Δelm1 + ELM1 complement strain. Mice (n = 8–10) were used for each group, and data were represented as the mean ± standard error of the mean for each group. Statistical significance was calculated using the Mann-Whitney U test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 were considered as significant.
Next, using an intradermal murine model of skin infection, we examined the morphology of the WT, Δelm1 mutant, and complement Δelm1 + ELM1 strains in vivo. Our results indicate that mouse skin infected with the WT strain features the yeast form, whereas mice infected with Δelm1 exhibited pseudohyphae/filamentous fungal forms at 3 days and 14 days post-infection (Fig. 1B). Histologically, all skin samples contain a dermal abscess comprising a well-demarcated and partially circumscribed mass of necrotic cellular debris and inflammatory cells admixed with varying numbers of fungal yeast. The dermal inflammation does not extend past the abscess. The intralesional WT yeast is 7–10 µm in greatest dimension, ovoid, rarely cluster and occasionally can be seen budding. Δelm1 intralesional fungi have hyphal and pseudohyphal morphology. The hyphal forms are approximately 2 µm wide and approximately 50–100 µm long with mostly parallel walls. The pseudohyphal forms are similar in size to hyphae but appear as rounded walls reminiscent of chains of connected yeast. The mice infected with WT yeast are abundant in the dermal abscess at 3 days and 14 days post-infection. However, Δelm1-infected mouse skin tissue had filamentous/hyphae/pseudohyphae present in moderate numbers in the abscess at 3 days post-infection and had decreased numbers in the 14 days post-infection skin sections. Δelm1 + ELM1 yeast have similar morphology to WT yeast; they are 7–10 µm, are ovoid, and rarely cluster. Next, we examined the skin colonization of mice infected with the WT, Δelm1, and Δelm1 + ELM1 strains. Groups of mice were infected intradermally with 1–2 × 106 colony forming units (CFU) of either strain, and the fungal load was determined from skin tissue collected after 3 days and 14 days post-infection (Fig. 1C). Mice infected with the Δelm1 mutant strain had significantly lower fungal load in the skin tissue in comparison to mice infected with the WT strain (Fig. 1C). The average fungal load of WT C. auris-infected skin tissue after 3 days was 7.42 ± 0.69 log10 CFU/g of tissue, whereas that of Δelm1-infected skin was 6.41 ± 0.56 log10 CFU/g of tissue (Fig. 1C). The Δelm1 + ELM1 strain had an average of 6.89 ± 0.64 log10 CFU/g of tissue following 3 days post-infection (Fig. 1C). After 14 days post-infection, WT C. auris-infected skin tissue had an average of 6.14 ± 0.53 log10 CFU/g of tissue, while Δelm1-infected mice skin tissue had an average fungal load of 5.34 ± 0.48 log10 CFU/g of tissue (Fig. 1C). Mice infected with Δelm1 + ELM1 strain had an average of 6.21 ± 0.55 log10 CFU/g of tissue 14 days post-infection (Fig. 1C). We also utilized qPCR to determine fungal DNA copies from mice skin tissue infected with WT (or) Δelm1 strains to verify CFU data, as previously described (18). A strong correlation (r2 = 0.7793) between cycle threshold (Ct) and respective CFU values was observed, regardless of morphology (Fig. S2). This correlation between CFU and fungal DNA copies confirms the fungal burden in mice infected with yeast and filamentous morphotypes. Collectively, our findings show that mice infected with filamentous C. auris had significantly decreased fungal load compared to mice infected with yeast form.
Next, we compared the skin immune responses of mice infected with WT, Δelm1, and Δelm1 + ELM1 strains of C. auris. Innate immune cells such as neutrophils, monocytes, and macrophages are crucial in host defense against Candida infection (19–22). We examined three major phagocytic cells using flow cytometry as described previously (4), (Fig. S3A); CD11b+Ly6G+ neutrophils, CD11b+Ly6Chi inflammatory monocytes, and CD11b+MHCII+CD64+ macrophages in the skin of mice infected with WT, Δelm1, and Δelm1 + ELM1 C. auris strains. We found that the percentage but not the absolute number of CD11b+Ly6G+ neutrophils was significantly increased in the mouse skin tissue infected with the Δelm1 strain compared to WT C. auris-infected groups (Fig. 1D). On the other hand, the percentage and absolute number of CD11b+Ly6Chi monocytes and CD11b+MHCII+CD64+ macrophages were significantly decreased in the skin tissue of mice infected with the Δelm1 strain (Fig. 1E and F). The percentage and absolute number of neutrophils, monocytes, and macrophages in mice skin infected with Δelm1 + ELM1 were comparable to WT, as no significance was observed between these strains (Fig. 1D and E). These results suggest that the myeloid phagocyte response is different in mouse skin infected with yeast and filamentous C. auris.
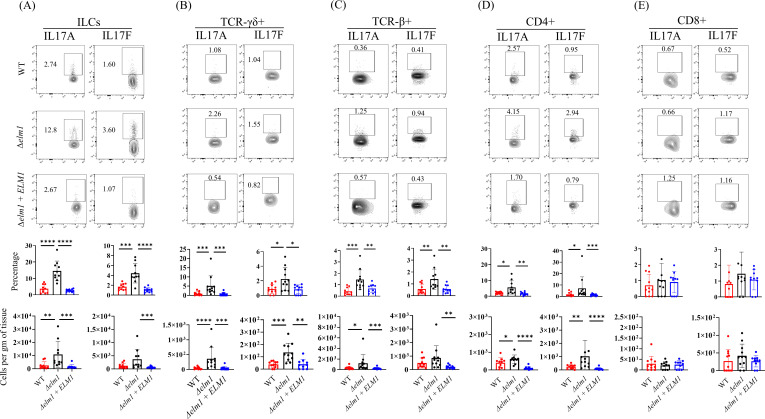
IL-17 producing innate lymphoid cells (ILCs) and T cells plays a crucial role in barrier defense against fungal infection (3, 21). Thus, we examined IL-17A and IL-17F producing ILCs, TCRγδ+, TCRβ+, CD4+, and CD8+ T cells as described previously (4) (Fig. S3B and C). We found that mice infected with the Δelm1 strain induced more potent IL-17-mediated host responses compared to mice infected with the WT C. auris strain (Fig. 2). The percentage of IL-17A and IL-17F producing ILCs, TCRγδ+, TCRβ+, and CD4+ T cells were significantly increased in the skin tissue of mice infected with Δelm1 compared to WT C. auris (Fig. 2A through D). Moreover, the absolute numbers of IL-17A+ILCs, IL-17A+TCRγδ+ IL-17A+TCRβ+, IL-17A+CD4+, IL-17F+TCRγδ+, and IL-17F+CD4+ T cells were significantly increased in the skin tissue of Δelm1-infected mice compared to WT groups (Fig. 2A through D). No significant differences in the percentage and absolute number of IL-17A and IL-17F producing CD8+ T cells were observed between Δelm1 and WT C. auris-infected mouse skin tissue (Fig. 2E). IL-17 responses in mice skin infected with Δelm1 + ELM1 were comparable to WT, as no significance was observed between these strains (Fig. 2D and E). Taken together, these findings suggest that mice infected with Δelm1 strain induce potent IL-17 responses compared to mice infected with WT C. auris strain.
Fig 2.
Filamentous C. auris induces potent IL-17 responses in the mouse skin. Representative flow cytometry plots, percentage, and absolute numbers of IL17A+ and IL17F+ producing (A) ILCs, (B) TCRγδ+ cells, (C) TCRβ+ cells, (D) CD4+ T cells, and (E) CD8+ T cells collected from mouse skin tissue infected intradermally with wild-type C. auris 0387, Δelm1 mutant, and Δelm1 + ELM1 complement strain. Mice (n = 8–10) were used for each group, and data were represented as the mean ± standard error of the mean for each group. Statistical significance was calculated using the Mann-Whitney U test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 were considered as significant.
Collectively, our findings reveal that yeast and filamentous C. auris strains induce distinct immune responses in the skin. Previous findings used topical mouse model of skin infection and C. auris 0387 yeast strain and identified that IL-17-mediated immune response is critical for protection (3). However, in this study, our goal was to compare the innate and adaptive immune responses between yeast and filamentous forms of C. auris 0387 strains using intradermal mouse model of infection. We used intradermal mouse model as C. auris can penetrate deep into skin tissue and persist for several months (3). Furthermore, skin immune responses are different between topical and intradermal mouse models of bacterial infection (23). Collectively, our findings suggest that immune responses induced by yeast and filamentous forms of C. auris are different. Our findings along with others (15) suggest that yeast and filamentous forms of C. auris and C. albicans induce different immune responses in the skin. C. albicans yeast but not filament-locked mutant strain induces a potent Th17 immune response in the mouse skin (15). Recent findings from our lab along with others indicate that fungal colonization was significantly higher in mice skin infected with yeast form of C. auris 0387 strain compared to mice infected with wild-type C. albicans SC5314 which can exist as both yeast and hyphal forms (3, 4). These findings suggest that yeast and filamentous morphologies of C. albicans and C. auris may differentially regulate the fungal colonization, persistence, and host immune response in the skin. Furthermore, we predict that distinct skin immune responses observed between yeast and filamentous C. auris and C. albicans may in part due to the differences in the cell wall composition (24, 25). Future studies using different clades of C. auris clinical strains are required to understand how yeast and filament forms of C. auris induce distinct skin immune responses which is critical to develop novel therapeutic and vaccine strategies to control and treat this emerging fungal pathogen.
Strains and reagents
The Candida auris strain used in this study is the South Asian AR0387 isolate sourced from the CDC AR Isolate Bank, USA. Reagents used to generate C. auris ELM1 deletion strain include the following: pCE27 and pCE35 plasmids purchased from Addgene, DreamTaq Green DNA Polymerase (5 U/µL) (Thermo Scientific, Cat. no. FEREP0712), Phusion High Fidelity Polymerase (Thermo Scientific), 10 mM dNTPs (Thermo Scientific), and restriction enzyme FastDigest MssI (Thermo Scientific). Antibodies and staining reagents used in this study for immune cell isolation and analysis were used as previously described (4).
Generation of ELM1 mutant strain
C. auris ELM1 deletion mutant and complement strain was generated by CRISPR-mediated genome editing via the LEUpOUT system as previously described (8, 17). Briefly, transformants were selected using nourseothricin (NAT) resistance marker. All oligonucleotides used in the study are provided in Table S1. The biotech software Benchling was used to design the 20 bp gRNA for ELM1 (B9J08_002849) with the reference genome C. auris B8441. The plasmid pCE27 was used to generate the gRNA cassette via amplification and PCR stitching of fragments A and B. A 60-bp forward primer featuring 20 bp overhangs on either side of the gRNA, having homology with the extant pCE27 sequence for stitching, is used to include the gRNA during the amplification of fragment A. The transformant fragment C is produced following fragment B amplification from pCE27 plasmid and subsequent stitching with fragment A. The pCE35 plasmids were digested with restriction enzyme MssI in order to produce the CRISPR-CAS9 cassette under the following conditions: 10 minute digestion period at 37°C, then 60°C with inactivated enzyme 10 minutes prior to transformation. The ELM1 exon was replaced by donor DNA (dDNA) designed using upstream and downstream portions of the ELM1 gene at an intersection point possessing approximately 23 bp sequence homology. The genomic DNA of C. auris AR0387 was used to amplify and join the dDNA fragments. The complement strain was generated using the hygromycin B (HYG) marker approach of the LEUpOUT system, as described in reference 17. A distinct gRNA cassette was produced to target the donor DNA inserted in place of the deletion. The ELM1 gene to be re-introduced in the mutant genome was amplified from WT C. auris genomic DNA. Primers that flank this region were used to make approximately 80 bp upstream and downstream flanking homology to the ELM1 region of interest.
Intradermal infection in mice
All animal studies were approved by the Institutional Animal Care and Use Committee at Purdue University. Female and male C57BL/6J mice of 6–8 weeks of age were used. Mice were anesthetized with isoflurane and shaved with an electronic trimmer. Hair follicles in the trimmed area were removed using a topical chemical depilatory cream and subsequently wiped and cleaned using sterile water. A dosage of 0.1 mL of 1–2 × 107 CFU of live C. auris AR0387 or Δelm1 was used to infect each mouse intradermally using 27-G 1″ hypodermic needle as described before (4).
Fungal burden determination in skin tissue
Mice were euthanized humanely, and infected skin tissues were excised following 3 days and 14 days post-infection. Tissue weights were recorded and mechanically homogenized in sterile 1× phosphate-buffered saline (PBS). The homogenate was then serially diluted and plated onto antibiotic containing (50 µg/mL kanamycin, 100 µg/mL streptomycin, and 100 µg/mL ampicillin) YPD-agar plates and incubated at 37°C for 24 hours to determine the fungal burden in the skin (CFU/g of tissue). Additionally, homogenates were plated onto CHROMagar to confirm C. auris colonies (4).
Histopathology and periodic acid-Schiff staining
Mouse skin was fixed in 4% paraformaldehyde and embedded in paraffin. Sections 4 µm thick were stained with hematoxylin and eosin and periodic acid-Schiff (PAS) according to standard methods. Microscopic examination was performed by a board-certified veterinary pathologist, and the interpretation was based on standard histopathological morphology.
Immune cell isolation and staining of murine skin tissue
Single-cell suspension was prepared from murine skin tissue, and immune cells were stained as described previously (4). Briefly, whole dorsal skin tissue samples were excised and placed in 1.5 mL microcentrifuge tubes containing 1 mL cold sterile digestion media (RPMI-1640 with 1 µg/mL DNase and 0.25 mg/mL Liberase TL). Each tissue sample was then transferred into a 12-well plate and subsequently finely minced with ophthalmic scissors. Samples were digested with trypsin and 1 mM EDTA, and filtered using 40 µm cell strainers. Single-cell suspensions were washed with cell staining buffer (CSB) prior to surface antibody staining. Live/Dead Fixable Yellow Dead stain was used to determine live cell populations, while anti-mouse CD16/32 was used to block Fc receptors. Except MHCII (1:2,000), indicated surface antibodies were used to stain markers at a ratio of 1:200. After fixing cells with fixation buffer, samples were stained with CD207 (1:200) in 1× intracellular staining permeable wash buffer (ISPWB) for 1 hour. For intracellular staining, cell suspensions were stimulated with cell activation cocktail without brefeldin (1:500) and monensin (1:000) for 4.5 hours at 37°C (5% CO2). The cell activation cocktail used is composed of phorbol 12-myristate 13-acetate (PMA) and ionomycin lacking brefeldin A, with monensin-protein transport inhibitor to enhance intracellular cytokine staining signals by blocking transport processes during cell activation. Surface markers were stained with respective surface antibodies and treated with fixation buffer overnight. ISPWB (1×) was used to wash and subsequently incubate cell suspensions in indicated intracellular antibodies (1:50) the following day. After washing and resuspension in CSB, cells were acquired using Attune NxT Flow Cytometer (Invitrogen, Carlsbad, CA, USA). FlowJo software (Eugene, OR, USA) was used for data analysis.
Statistics analysis
All data included in this study are shown as mean ± standard error of the mean. Statistical analysis of the data was done by the Mann-Whitney U test using GraphPad Software. P values of ≤0.05 were considered significant.
ACKNOWLEDGMENTS
This study was supported by NIAID (1R01AI177604 to S.T.) and Division of Intramural Research of the NIAID, NIH (ZIA AI001175 to M.S.L.).
S.T. designed the experiments and supervised the project. G.B. performed the experiments and analyzed data. A.C. assisted in histopathology and PAS staining. G.B., M.S.L., and S.T. reviewed data analysis and wrote the manuscript. All authors contributed and approved the final version of the manuscript.
Contributor Information
Shankar Thangamani, Email: sthangam@purdue.edu.
Aaron P. Mitchell, University of Georgia, Athens, Georgia, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00055-24.
Supplemental tables and figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Ahmad S, Alfouzan W. 2021. Epidemiology, diagnosis, pathogenesis, antifungal susceptibility, and infection control measures to combat the spread of infections in healthcare facilities. Microorganisms 9:807. doi: 10.3390/microorganisms9040807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO . 2022. WHO fungal priority pathogens list to guide research, development and public health action. Geneva: World Health Organization. https://iris.who.int/bitstream/handle/10665/363682/9789240060241-eng.pdf?sequence=1. [Google Scholar]
- 3. Huang X, Hurabielle C, Drummond RA, Bouladoux N, Desai JV, Sim CK, Belkaid Y, Lionakis MS, Segre JA. 2021. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe 29:210–221. doi: 10.1016/j.chom.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Datta A, Das D, Nett JE, Vyas JM, Lionakis MS, Thangamani S. 2023. Differential skin immune responses in mice intradermally infected with Candida auris and Candida albicans. Microbiol Spectr 11:e0221523. doi: 10.1128/spectrum.02215-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Proctor DM, Drummond RA, Lionakis MS, Segre JA. 2023. One population, multiple lifestyles: commensalism and pathogenesis in the human mycobiome. Cell Host Microbe 31:539–553. doi: 10.1016/j.chom.2023.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK, NISC Comparative Sequencing Program . 2021. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27:1401–1409. doi: 10.1038/s41591-021-01383-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, Noble-Wang J, Pacilli M, McPherson TD, Black S, Kemble SK, Herzegh O, Ahmad A, Forsberg K, Jackson B, Litvintseva AP. 2021. Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin Infect Dis 73:1142–1148. doi: 10.1093/cid/ciab327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santana DJ, O’Meara TR. 2021. Forward and reverse genetic dissection of morphogenesis identifies filament-competent Candida auris strains. Nat Commun 12:7197. doi: 10.1038/s41467-021-27545-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan S, Yue H, Zheng Q, Bing J, Tian S, Chen J, Ennis CL, Nobile CJ, Huang G, Du H. 2021. Filamentous growth is a general feature of Candida auris clinical isolates. Med Mycol 59:734–740. doi: 10.1093/mmy/myaa116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown JL, Delaney C, Short B, Butcher MC, McKloud E, Williams C, Kean R, Ramage G. 2020. Candida auris phenotypic heterogeneity determines pathogenicity in vitro. mSphere 5:e00371-20. doi: 10.1128/mSphere.00371-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du H, Bing J, Hu T, Ennis CL, Nobile CJ, Huang G. 2020. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog 16:e1008921. doi: 10.1371/journal.ppat.1008921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seiser S, Arzani H, Ayub T, Phan-Canh T, Staud C, Worda C, Kuchler K, Elbe-Bürger A. 2024. Native human and mouse skin infection models to study Candida auris-host interactions. Microbes Infect 26:105234. doi: 10.1016/j.micinf.2023.105234 [DOI] [PubMed] [Google Scholar]
- 13. Yue H, Bing J, Zheng Q, Zhang Y, Hu T, Du H, Wang H, Huang G. 2018. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect 7:188. doi: 10.1038/s41426-018-0187-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bravo Ruiz G, Ross ZK, Gow NAR, Lorenz A. 2020. Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint. mSphere 5:e00151-20. doi: 10.1128/mSphere.00151-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kashem SW, Igyarto BZ, Gerami-Nejad M, Kumamoto Y, Mohammed JA, Jarrett E, Drummond RA, Zurawski SM, Zurawski G, Berman J, Iwasaki A, Brown GD, Kaplan DH. 2015. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity 42:356–366. doi: 10.1016/j.immuni.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santana DJ, Anku JAE, Zhao G, Zarnowski R, Johnson CJ, Hautau H, Visser ND, Ibrahim AS, Andes D, Nett JE, Singh S, O’Meara TR. 2023. A Candida auris-specific adhesin, Scf1, governs surface association, colonization, and virulence. Science 381:1461–1467. doi: 10.1126/science.adf8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ennis CL, Hernday AD, Nobile CJ. 2021. A markerless CRISPR-mediated system for genome editing in Candida auris reveals a conserved role for Cas5 in the caspofungin response. Microbiol Spectr 9:e0182021. doi: 10.1128/Spectrum.01820-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunker C, Polke M, Schulze-Richter B, Schubert K, Rudolphi S, Gressler AE, Pawlik T, Prada Salcedo JP, Niemiec MJ, Slesiona-Künzel S, Swidergall M, Martin R, Dandekar T, Jacobsen ID. 2021. Rapid proliferation due to better metabolic adaptation results in full virulence of a filament-deficient Candida albicans strain. Nat Commun 12:3899. doi: 10.1038/s41467-021-24095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID. 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359:232–236. doi: 10.1126/science.aao1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB. 2008. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog 4:e35. doi: 10.1371/journal.ppat.0040035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kashem SW, Kaplan DH. 2016. Skin immunity to Candida albicans. Trends Immunol 37:440–450. doi: 10.1016/j.it.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lionakis MS, Drummond RA, Hohl TM. 2023. Immune responses to human fungal pathogens and therapeutic prospects. Nat Rev Immunol 23:433–452. doi: 10.1038/s41577-022-00826-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino AV, Sen SK, Shaik J, Smelkinson M, Tamoutounour S, Collins N, Bouladoux N, Dzutsev A, Rosshart SP, Arbuckle JH, Wang C-R, Kristie TM, Rehermann B, Trinchieri G, Brenchley JM, O’Shea JJ, Belkaid Y. 2018. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172:784–796. doi: 10.1016/j.cell.2017.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horton MV, Johnson CJ, Zarnowski R, Andes BD, Schoen TJ, Kernien JF, Lowman D, Kruppa MD, Ma Z, Williams DL, Huttenlocher A, Nett JE. 2021. Candida auris cell wall mannosylation contributes to neutrophil evasion through pathways divergent from Candida albicans and Candida glabrata. mSphere 6:e0040621. doi: 10.1128/mSphere.00406-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Zou Y, Chen X, Li H, Yin Z, Zhang B, Xu Y, Zhang Y, Zhang R, Huang X, et al. 2022. Innate immune responses against the fungal pathogen Candida auris. Nat Commun 13:3553. doi: 10.1038/s41467-022-31201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables and figures.