Abstract
The lungs are a significant organ site of murine cytomegalovirus (mCMV) latency. We have shown that activity of the major immediate-early promoter (MIEP), which drives the transcription from the ie1-ie3 transcription unit, does not inevitably initiate the productive cycle (S. K. Kurz, M. Rapp, H.-P. Steffens, N. K. A. Grzimek, S. Schmalz, and M. J. Reddehase, J. Virol. 73:482–494, 1999). Thus, even though MIEP activity governed by the MIEP-enhancer is unquestionably the first condition for recurrence, regulation of the enhancer by transcription factors is not the only mechanism controlling latency. Specifically, during latency, focal and stochastic MIEP activity in lung tissue was found to selectively generate IE1 transcripts, while transactivator-specifying IE3 transcripts were missing. This suggested a control of mCMV latency that is effectual at IE1-IE3 precursor mRNA cotranscriptional processing. Here we have used this model for studying the kinetics of reactivation and recurrence in individual lung tissue pieces after hematoablative, genotoxic treatment. Notably, reactivation was triggered, but the number of transcriptionally active foci in the lungs did not increase over time. This result is not compatible with a model of spontaneous reactivations accumulating after withdrawal of immune control. Instead, the data support the idea that reactivation is an induced event. In some pieces, focal reactivation generated IE3 transcripts but not gB transcripts, while other pieces contained foci that had proceeded to gB transcription, and only a few foci actually reached the state of virus recurrence. This finding indicates the existence of several sequentially ordered control points in the transition from mCMV latency to recurrence.
The lungs are a relevant organ site of primary and recurrent human cytomegalovirus (hCMV) disease (for overviews, see references 21, 22, 31, 34, 39, and 44). Murine CMV (mCMV) can serve us as a model for studying CMV pneumonia in acute infection (6, 27, 33, 37) as well as for studying viral latency, reactivation, and recurrence in the lungs (2, 17, 18, 42, 43). We have shown recently that transcription from the major immediate-early (MIE) transcription unit ie1-ie3 (hereafter referred to as ie1/3), which is driven by a strong MIE promoter-enhancer (MIEPE) (3), occurs during pulmonary latency of mCMV but fails to initiate the productive cycle (17). Notably, the paralogous MIEPE of hCMV can functionally replace the MIEPE of mCMV for productive infection in vitro (1) and in vivo (4), suggesting that regulation of MIE gene expression via the enhancer element has some degrees of freedom and that a more stringent control is operative at subsequent checkpoints.
During productive infection, a 2.75-kb ie1 gene mRNA (2,305 nucleotides of exons 1, 2, 3, and 4 of ie1/3) and a 2.75-kb ie3 gene mRNA (2,303 nucleotides of exons 1, 2, 3, and 5 of ie1/3) specifying the mCMV IE1 (12) and IE3 (26) proteins, respectively, are thought to be generated from a precursor transcript by differential splicing (10–12, 26). While viral genomes were found to be evenly distributed in the latently infected lungs (17), a mosaic of transcriptionally active and transcriptionally silent pieces of lung tissue indicated that MIE transcriptional activity at any given time during latency was focal, randomly distributed, and, most probably, temporary. Notably, in the transcriptionally active lung tissue pieces, IE1 mRNA was generated selectively, while IE3 mRNA was missing (17). For the sake of clarity it should be emphasized that we refer to transcriptional activity with specific respect to transcription of the ie1/3 transcription unit, which is controlled by the MIEPE and is implicated in the initiation of the productive cycle. One must certainly consider the possibility of transcription occurring elsewhere in the latent viral genome. Representational difference analysis comparing transcription in latently infected lung tissue with that in normal lung tissue has indeed already indicated expression of mCMV genes outside the ie1/3 transcription unit, and the identities of these transcripts are currently under investigation (43).
The absence of IE3 mRNA explains the absence of gB early-late gene transcripts as well as of infectious virus, because the 88-kDa IE3 protein of mCMV (26), the functional analog of the hCMV 86-kDa IE2 protein (5, 14; reviewed in reference 41), is the main transactivator of early gene expression initiating the productive cycle. Specifically, previous work by Messerle et al. has demonstrated efficient activation of the e1 promoter by IE3 alone but not by IE1 alone, whereas IE1 did enhance the activity when coexpressed with IE3 (26). Accordingly, recurrent infection measured 14 days after immunoablative, genotoxic treatment was in fact associated with the generation of IE3 mRNA in addition to IE1 mRNA (17). We therefore concluded that transcription of the ie1/3 transcription unit, and thus an “on position” of the MIEPE, is a primary condition for virus recurrence and that there exists an additional, subsequent control point at the level of cotranscriptional processing defining the levels of IE1 and IE3 mRNAs. Actually, since the MIEPE was on or off during latency, cotranscriptional processing appears to provide a more stringent control of the latent state. In essence, these previous data have implied that IE3 rather than IE1 mRNA is indicative of productive reactivation. If this is true, the generation of IE3 mRNA should correlate with recurrence of infectious virus.
The original aim of the present work was to investigate the temporal association between the generation of IE3 mRNA and virus recurrence in the lungs and to estimate the incidences of transcriptional reactivation and virus recurrence after immunoablative, genotoxic treatment. One could envisage two alternative mechanisms of CMV reactivation and recurrence. We refer to these mechanisms as the model of spontaneous reactivation and the model of induced reactivation. The model of spontaneous reactivation assumes that MIEPE activity, which drives IE1/3 precursor transcription, occurs randomly, either spontaneously or as a result of endogenous and random signalling, and that recurrence of infectious virus is precluded in immunocompetent mice by antiviral effector cells eliminating cells in which reactivation leads to the presentation of antigenic viral peptides. This mechanism was previously our own favored idea (17, 42) and was proposed recently based on the kinetics of mCMV recurrence after combined NK-cell and T-cell depletion in latently infected B-cell-deficient mice (28). If this model applies, we should find an accumulation of reactivations over time after withdrawal of immune control. By contrast, the model of induced reactivation assumes that reactivation involves an external signalling that has to switch on the productive viral cycle. In that case, reactivation should be a quantal event that follows the rule of all or nothing.
Here we present data supporting the model of induced reactivation. Moreover, the pattern of transcriptional activity in the lungs after induction of reactivations reveals a hitherto unknown complexity of regulation involving multiple, sequential checkpoints on the way from mCMV latency to recurrence.
MATERIALS AND METHODS
Generation of latently infected mice with an intermediate viral DNA load.
Sex-matched, syngeneic bone marrow transplantation (BMT) was performed with female mice of the inbred strain BALB/c (major histocompatibility complex haplotype H-2d) used at the age of 8 weeks as bone marrow cell (BMC) donors and recipients. Hematoablative conditioning of the recipients was performed by total-body gamma irradiation with a single, sublethal dose of 6 Gy from a 137Cs gamma radiation source (OB58; Buchler, Braunschweig, Germany). Cell suspensions of donor femoral and tibial BMCs were obtained by flushing medium through the bone shafts, and contaminating vascular and sinusoidal CD8 T cells were depleted as described previously (42). BMT was performed by infusion of 2 × 107 donor BMCs into the tail veins of recipients at ca. 6 h after the irradiation. Compared to previous work, in which the BMC dose was only 5 × 106 (17), the increased BMC dose was chosen with the intention to improve the efficacy of reconstitution, resulting in an improved control of acute, primary infection and in a reduced load of latent viral genome in host tissues (42). Infection with 105 PFU of purified mCMV (strain Smith ATCC VR-194/1981) was performed subcutaneously at the left hind footpad at ca. 2 h after BMT. At 2-month intervals, tail vein blood was monitored for the presence of viral DNA by using a PCR specific for a 363-bp sequence within exon 4 of gene ie1 of mCMV, followed by dot blot hybridization with an internal, γ-32P-end-labeled oligonucleotide probe and by phosphorimaging (17). By 6 months after BMT and infection, under the conditions used here, the viral genome was cleared from blood cells.
High-sensitivity verification of latency and detection of virus recurrence.
The absence of infectious virus in the lungs was verified by testing tissue homogenates, usually in 2-ml aliquots representing 1/18 fractions of the lungs, for infectivity in the reverse transcriptase (RT-) PCR-based focus expansion assay (17, 18). This assay involves centrifugally enforced virus penetration, three rounds of virus replication in cell cultures of permissive mouse embryofetal fibroblasts (MEF), and detection of viral IE1 mRNA by RT-PCR. As documented in greater detail previously (18), 0.01 PFU, corresponding to five viral DNA molecules, can thus be detected, which defines the detection limit as <0.2 PFU for the whole lungs. Recurrence of infectious virus in nine individual 1/18 pieces of the lungs (seven of which were derived from the left lung and two of which were derived from the postcaval lobe) was monitored with the same assay at 4, 8, and 12 days after genotoxic, hematoablative treatment accomplished by total-body gamma irradiation with a single dose of 6.5 Gy.
Simultaneous isolation of DNA and poly(A)+ RNA from the lungs.
Pieces of lung tissue (three pieces per lobe derived from the superior, middle, and inferior lobes of the lungs) were immediately shock frozen in liquid nitrogen to minimize the risk of postmortem transcriptional reactivation, as specified in more detail previously (17). Frozen pieces were transferred to Eppendorf tubes containing standard extraction buffer with guanidinium thiocyanate. Extracted DNA was sedimented, and poly(A)+ RNA was isolated from the extraction supernatant based on affinity to an oligo(dT)-cellulose matrix by procedures specified previously (18). A 1/18 piece of the lungs represents ca. 3 × 106 to 4 × 106 lung cells. The average yields of DNA and poly(A)+ RNA per piece were found to be ca. 20 and ca. 2 μg, respectively.
Determination of the latent viral DNA load in lung tissue.
Viral DNA contained in lung tissue was quantitated by serially diluting the extracted tissue-derived total DNA, followed by performing a 96-well microplate format PCR specific for a 363-bp sequence within exon 4 of gene ie1 of mCMV, precisely as described previously (17). Plasmid pIE111, which encompasses gene ie1, served as the standard for the quantification. Specifically, 105 molecules of pIE111 were mixed with 6 μg of carrier DNA derived from the lungs of uninfected mice, and the mixture was titrated in parallel to the DNA derived from latently infected lung tissue specimens. Throughout, titrations were started with 600 ng of tissue DNA, which represents the DNA content of 105 diploid tissue cells. Amplification products (20 μl) were vacuum dot blotted, hybridized with an internal γ-32P-end-labeled oligonucleotide probe, and analyzed quantitatively by digital phosphorimaging (42). The number of viral copies in the lung cell DNA, and thus the latent viral DNA load in the lungs, was determined in a log-log plot from the linear portions of the experimental and standard titrations.
Analysis of viral transcription in lung tissue during latency and reactivation.
Transcripts of the viral genes ie1, ie3, and gB within lung tissue-derived poly(A)+ RNA (see above) were detected by RT-PCRs as described in detail recently (17). RNAs synthesized in vitro from respective recombinant plasmids (17) were used to control the sensitivity of detection. A map illustrating the locations of all primers and probes, as well as the positions and lengths of the in vitro-synthesized RNAs, is also given in reference 17. In essence, the IE1-specific RT-PCR amplifies a 188-bp DNA fragment detected with probe IE1-P directed against the exon 3-exon 4 splicing junction. The IE3-specific RT-PCR gives a 229-bp DNA fragment detected with probe IE3-P directed against the exon 3-exon 5 splicing junction. Finally, the gB-specific RT-PCR amplifies a 405-bp DNA fragment detected with probe gB-P directed against an internal sequence.
Because the gB gene does not possess an exon-intron structure, a possible problem caused by viral DNA contaminating the poly(A)+ RNA preparation must be considered. In our assays of transcription during pulmonary latency, we never detected mCMV gB-specific signals when RT was omitted from the RT-PCR mix (17), and the same was true here for the study of transcription during reactivation (not shown). Accordingly, in cases positive for gB-specific signals, detection was not abolished by pretreatment of the template poly(A)+ RNA preparation with RNase-free DNase (catalog no. M6101; Promega, Madison, Wis.) (not shown). It may be informative to note that contaminating viral DNA was in fact a problem in the analysis of poly(A)+ RNA isolated from MEF infected in vitro at a high multiplicity. The difference is certainly the ratio between viral DNA and cellular DNA within the contaminating DNA. Apparently, during latency and reactivation in tissues, only a minority of the cells contain viral DNA, and the viral DNA contamination in the poly(A)+ RNA preparations is thus below the detection limit.
Amplification products were analyzed by agarose (2%, wt/vol) gel electrophoresis, Southern blotting, and hybridization with the corresponding γ-32P-end-labeled probes, followed by autoradiography.
Estimation of frequencies of transcriptionally active and recurrently infected foci in the lungs.
The frequency of foci of mCMV reactivation or recurrence was estimated from the observed fraction of lung pieces negative for the parameter in question (i.e., IE1, IE3, or gB transcripts or infectious virus) [f(0)] by using the Poisson distribution equation λ = −ln f(0). The fraction of pieces containing n (n = 0, 1, 2, 3, and so on) foci, F(n), was calculated by using the formula F(n) = λn × e−λ/n! or the mathematically equivalent formula F(n) = λ/n × F(n − 1). An overview of the rationale and applications of the Poisson probability distribution is provided by Lefkovits and Waldmann (20). The frequency derived from the number of analyzed tissue pieces was then extrapolated to 18 pieces of a complete “statistically generated lung.” For simplicity, it was assumed that none of the foci was located at the experimental cut border between neighboring tissue pieces. This constraint may lead to a negligible overestimation of frequencies (17).
RESULTS
Approach to the analysis of pulmonary latency and reactivation of mCMV.
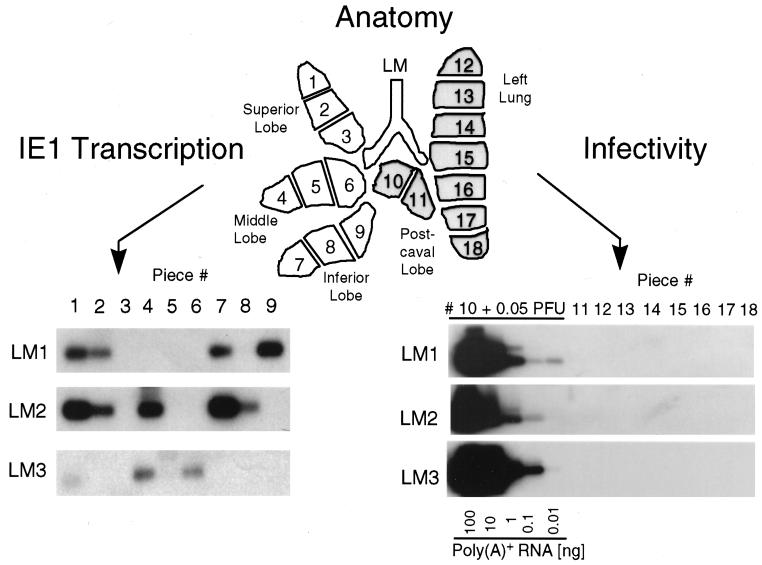
For a cohort of infected BMT recipients, clearance of mCMV DNA from the blood was monitored by ie1 gene-specific PCR at bimonthly intervals, essentially as documented in previous work (17), except that tail vein blood was used for the longitudinal analysis of individual mice. BMT recipients were regarded as being latently infected when the load of viral DNA in the blood had declined to below the detection limit of the assay, i.e., to <100 copies per 106 blood cells (not shown). After 8 months, three mice which were PCR negative for mCMV DNA in the blood and which were designated latent mouse 1 (LM1) to LM3 were used to verify the absence of infectious virus in their lungs. According to the “mosaic approach” published previously (17), the lobes of the lungs were cut into 18 pieces of nominally equal size. Pieces designated #1 to #9, derived from the superior, middle, and inferior lobes, were used for the simultaneous analysis of latent viral DNA load and viral transcription, whereas pieces designated #10 to #18, derived from the postcaval lobe and left lung, served to verify the latent state by demonstrating the absence of infectivity (Fig. 1).
FIG. 1.
Transcriptional status of mCMV during latency in the lungs. A scheme illustrating the lobular anatomy of the lungs in ventral view is shown at the top. The lobes were cut into equal pieces listed on the lung map as #1 through #18. Minor size (weight) differences were compensated for by adjustment of the aliquots included in the assays. (Lower right) Verification of mCMV latency. The absence of infectious virus in the left lungs and postcaval lobes of mice LM1 to LM3 was demonstrated at 8 months after BMT and infection by testing the infectivity of lung tissue homogenates with the RT-PCR-based focus expansion assay (18). As a positive control, 0.05 PFU of purified mCMV was added to the homogenate of piece #10 before centrifugal infection of an MEF culture. Poly(A)+ RNA derived from this culture after 72 h of viral replication was serially diluted as indicated, whereas in the case of indicator cultures infected with homogenates of pieces #11 through #18, a constant amount of 100 ng of poly(A)+ RNA was subjected to ie1 exon 3-exon 4 (hereafter referred to as exon 3/4)-specific RT-PCR, yielding an amplification product of 188 bp. Shown are the autoradiographs obtained after gel electrophoresis, Southern blotting, and hybridization with the γ-32P-end-labeled oligonucleotide probe IE1-P (17), which is directed against the exon 3/4 splice junction. (Lower left) Random pattern of IE1-specific transcripts. Poly(A)+ RNA isolated directly from lung tissue pieces #1 through #9 (a 1/10 aliquot thereof [ca. 200 ng]) of mice LM1 to LM3 was subjected to the ie1 exon 3/4-specific RT-PCR. The signal obtained for LM3 piece #1 was reproduced with a second aliquot, and, throughout, pieces negative for IE1 transcripts remained negative when a second aliquot was tested. Testing of third and fourth aliquots of all preparations for IE3 transcripts and gB transcripts, respectively, gave negative results throughout. The presence of mRNA in the preparations that corresponded to negative pieces was verified for a fifth aliquot by RT-PCR specific for hypoxanthine phoshoribosyltransferase transcripts (not shown), as documented in detail previously (17).
The analysis of IE1 transcripts by RT-PCR performed with tissue-derived poly(A)+ RNA reproduced the previously published random pattern of transcriptionally active and silent pieces for mice LM1 to LM3 (Fig. 1, lower left). RT-PCRs specific for IE3 or gB transcripts were negative throughout (not shown), thus confirming the previous conclusion that mCMV latency in the lungs is associated with a focal and selective generation of IE1 transcripts (17). For the same three individual mice, LM1 to LM3, absence of infectious virus is documented for pieces #11 to #18 by the RT-PCR-based focus expansion assay, an assay involving centrifugally enforced infection of MEF, three rounds of virus replication in MEF monolayer cultures, and sensitive detection of generated IE1 transcripts by RT-PCR (18). For controlling the sensitivity of the assay, the homogenate from piece #10 was supplemented with 0.05 PFU before infection of the MEF indicator culture. While IE1 transcripts were detected within 0.01 ng of poly(A)+ RNA derived from this virus-supplemented culture, IE1 transcripts were absent, without exception, in 100 ng of poly(A)+ RNA derived from cultures inoculated with the unsupplemented tissue homogenates. In conclusion, the latency phenotype defined in our recent report (17) was fully reproduced here for the cohort of mice that was then used for the analysis of mCMV reactivation from latency.
Determination of the load of latent viral DNA in the lungs.
The load of latent mCMV DNA in organs, specifically in the lungs, is known to be critical for the incidence of the recurrence of infectious virus (2, 32, 42). Members of our group have documented recurrence of virus in all lobes of latently infected lungs (all 25 lobes of five mice tested were recurrently infected) with a latent mCMV DNA load of ca. 5,000 copies per 106 lung cells, whereas reduction of the load to ca. 1,000 copies per 106 lung cells by experimental preemptive antiviral CD8 T-cell immunotherapy after BMT greatly reduced the risk of recurrence (2 of 25 lobes of five mice tested were recurrently infected) (42). Accordingly, the very high average load of 7,500 (range, 6,000 to 9,000) copies per 106 lung cells reached under the conditions used in our previous study on mCMV reactivation in the lungs (17) was indeed associated with recurrence of infectious virus in all tested pieces of the lungs. However, such conditions are supraoptimal for statistical analyses, because frequency estimates based on the Poisson distribution require the existence of a so-called zero fraction of negative samples, that is, in our case, of tissue pieces in which recurrence did not occur. We therefore modified the conditions of BMT by increasing the number of transplanted donor BMCs with the intention of reaching an intermediate load high enough to observe recurrence but low enough to also have negative pieces. Clearly, defining this condition was critical for all data described here, and it took us more than one trial to do so.
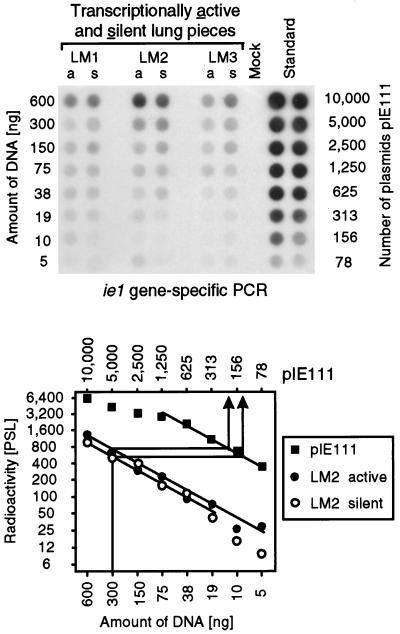
For the determination of the latent mCMV DNA loads in the lungs of mice LM1 to LM3, DNAs derived from transcriptionally silent and transcriptionally active tissue pieces (Fig. 1, lower left) were pooled separately (e.g., in the case of LM1, pieces #1, #2, #7, and #9 for the active pool and pieces #3, #4, #5, #6, and #8 for the silent pool), and the loads were then determined as described previously (17, 42) by DNA titration and ie1 gene exon 4-specific PCR in microplate format, followed by phosphorimaging (Fig. 2). The results show that (i) the latent mCMV DNA load varied only moderately between the three mice tested, (ii) this load did not significantly differ between transcriptionally active and silent lung tissue pieces of the same individual mouse, and (iii) the loads were on the order of 100 copies per 300 ng of lung cell DNA, which is 2,000 copies in 106 lung cells. Compared to the conditions discussed above, this was indeed the intended intermediate load predicted to give an intermediate incidence of recurrence.
FIG. 2.
Quantitation of the latent viral DNA in transcriptionally active and silent tissue pieces. In parallel with the isolation of the poly(A)+ RNA used for the transcriptional analyses shown in Fig. 1, DNA was isolated from lung tissue pieces #1 through #9 derived from mice LM1 to LM3. For each mouse, DNA pools corresponding to transcriptionally active pieces (pool a) and transcriptionally silent pieces (pool s) were formed, according to the pattern of IE1-specific transcription shown in Fig. 1, lower left. The DNA pools were titrated as indicated and were subjected to an ie1 exon 4-specific PCR. A negative control was provided by DNA isolated from uninfected lungs (Mock). As a standard for the quantitation, this mock DNA was supplemented with plasmid pIE111, titrated in duplicate, and subjected to PCR accordingly. (Top) Autoradiograph of the dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging data for the same blot. For the sake of clarity, the computations are depicted as graphs only for mouse LM2. Log-log plots of radioactivity (mean of duplicates in the case of the standard) measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of sample DNA (abscissa) are shown. The upper line relates the amount of DNA to the number of plasmids in the pIE111 standard. Calculations were made from the linear portions of the graphs, as shown as an example for 300 ng of DNA containing 200 copies and 120 copies of viral DNA in the active and silent pools, respectively, of mouse LM2. The results, given as copies of the viral genome per 106 lung cells (6 μg of DNA) were as follows: for LM1, a = 1,320 and s = 1,480; for LM2, a = 4,000 and s = 2,400; and for LM3, a = 1,260 and s = 1,460.
Incidence of virus recurrence after immunoreductive treatment.
The prediction made from the load was tested by a method employed with success previously, i.e., facilitating virus recurrence by hematoablative, immunoreductive treatment accomplished by total-body gamma irradiation with a dose of 6.5 Gy (2, 17, 18, 32, 42). For estimating the frequency of focal recurrence events, it is crucial to confine recurrent infection to the site of its origination. It is therefore pertinent to our work that the infected BMT recipients mounted an effective antiviral antibody response that deterred the recurrent virus from spreading. Even though reconstitution of B cells is delayed by acute mCMV infection after BMT, high titers of neutralizing antibodies were eventually generated by the time when latency was established (not shown). That antibodies are indeed effectual in precluding virus dissemination after recurrence from latency has been documented in previous work (7, 32).
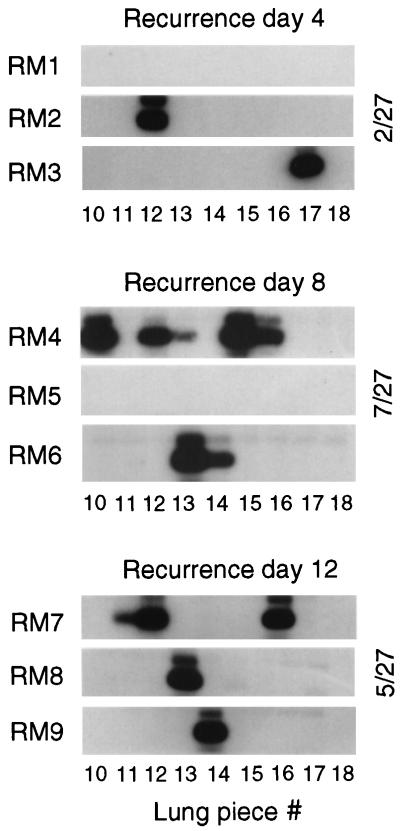
The recurrence assay was performed with nine additional mice of the same latently infected cohort, referred to as recurrent mouse 1 (RM1) to RM9. The results were determined on day 4 after the irradiation for RM1 to RM3, on day 8 for RM4 to RM6, and on day 12 for RM7 to RM9. Sensitive detection of infectivity in lung tissue pieces #10 to #18 derived from the postcaval lobes and left lungs was achieved by testing the tissue homogenates in the RT-PCR-based focus expansion assay (Fig. 3). Notably, as we had intended, there was a clear-cut distinction between positive pieces and negative pieces, demonstrating the focal and stochastic nature of recurrence events in the lungs after release from cellular immune control. Since completion of the productive viral cycle takes only 1 day in the case of mCMV, it was not unexpected to see recurrence already on day 4, but it was indeed a great surprise to notice that recurrences apparently did not accumulate over time. Specifically, mice with only a single recurrence were found on day 4 as well as on day 12, and RM5 was negative on day 8, even though, as has been proved by RM2 and RM3, 4 days is sufficient to generate a detectable amount of virus. We propose that the observed numerical variance in the incidences of recurrence per time point reflects individual differences in the latent viral DNA load (Fig. 2) rather than a time course of recurrence. The time-independent on-or-off pattern suggests that virus recurrence was induced once by the treatment and was thereafter maintained for the documented period of time, because withdrawal of cellular immune control prevented rapid remission to latency.
FIG. 3.
Kinetics of mCMV recurrence after hematoablative treatment. Mice of the same cohort for which the establishment of latency was verified in Fig. 1 were subjected to gamma irradiation with a dose of 6.5 Gy. The recurrence of infectivity was monitored by the RT-PCR-based focus expansion assay on days 4, 8, and 12 after the treatment for lung tissue pieces #10 to #18 derived from the postcaval lobes and left lungs of mice RM1 through RM9. Shown are the autoradiographs obtained after hybridization of the amplification products with probe IE1-P (see the legend to Fig. 1). The incidence of recurrence is indicated for each time point.
Patterns of transcriptional reactivation.
It is understood that pieces in which reactivation proceeded to recurrence of infectious virus must necessarily have also contained mRNAs representing all stages of the temporally regulated gene expression pertinent to the productive viral cycle. Specifically, one should find there IE1 mRNA, IE3 mRNA specifying the key transactivator of early genes, and the early-late gB mRNA. However, what about the pieces in which recurrence did not occur? One can envisage quite different scenarios. An extreme but attractive idea was that only cells in which the mCMV MIEPE is in the on position during latency, that is, foci with IE1 transcription, are susceptible to triggering of the viral gene expression cascade. Alternatively, the induction of reactivation could be independent of the transcriptional status during latency, and there may be other possibilities.
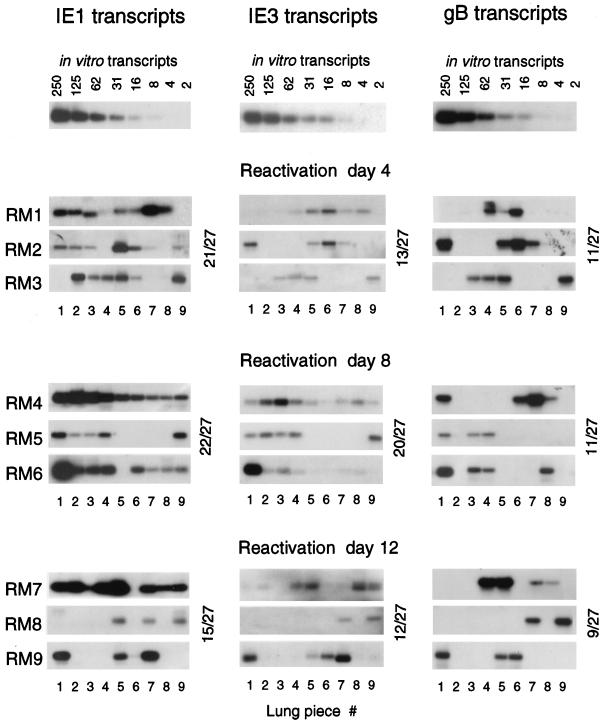
Because of the technical incompatibility of the respective assays, viral transcription and viral infectivity could not be analyzed for the same tissue pieces. Therefore, IE1, IE3, and gB transcription in tissue pieces derived from the remaining three lobes of the lungs was studied for mice RM1 to RM9 (Fig. 4). The sensitivity of detection was defined with the corresponding in vitro-synthesized transcripts. With all three RT-PCRs, 8 to 16 mRNA molecules could be clearly visualized (Fig. 4, top row). As we have elaborated in our recent report on IE1 expression during latency, transcriptionally active pieces are clearly positive if they contain between 100 and 12,000 transcripts per piece, and pieces scored negative by testing a 1/10 aliquot of the yield of poly(A)+ RNA, that is, ca. 200 ng, did not turn positive when larger aliquots were subjected to the RT-PCR (17). Admittedly, we cannot completely exclude the possibility of missing a very low expression, but this technical limitation and unavoidable cutoff does not seriously interfere with our conclusion that transcriptional activity differs significantly between individual tissue pieces, thus resulting in a mosaic pattern.
FIG. 4.
Patterns of transcriptional reactivation. Transcripts of viral genes ie1, ie3, and gB were detected by respective RT-PCRs (17) for poly(A)+ RNAs derived from lung tissue pieces #1 through #9 (superior, middle, and inferior lobes) of mice RM1 through RM9, analyzed in groups of three on days 4, 8, and 12 after induction by gamma irradiation with a dose of 6.5 Gy. For standards to determine the sensitivity of detection, carrier poly(A)+ RNA derived from uninfected lung tissue was supplemented with a defined number of the respective in vitro-synthesized RNA molecules and titrated as indicated (top row). Throughout, RT-PCRs were performed with ca. 200 ng of sample poly(A)+ RNA, which represents 1/10 of the yield from one tissue piece. Faint or otherwise questionable signals (such as, e.g., IE1 in RM1 #4, IE1 in RM9 #6, and IE3 in RM7 #1) were either confirmed or rejected after testing of a second and, if necessary, a third aliquot. Note that some decisions need to be made from the original autoradiographs. Throughout, negative samples remained negative when further aliquots were tested. The reason for the apparently smaller size of the IE1 amplification product in RM1 #3 is under investigation. Shown are the autoradiographs obtained after hybridization with γ-32P-end-labeled probes IE1-P, IE3-P, and gB-P (for a map, see reference 17). Incidences of positive pieces are indicated for each time point.
Notably, analysis of poly(A)+ RNA derived from tissue pieces #1 to #9 resulted in on-or-off patterns for all three types of transcripts, indicating the focal nature of transcriptional reactivation. As was the case for recurrence of infectious virus (Fig. 3), the patterns of transcription did not reveal any alteration with respect to time. It was actually not unexpected to find pieces containing only IE1 transcripts or pieces containing IE1 and IE3 transcripts, but not yet gB transcripts, at any particular point in time. However, we would have expected a shift from a high incidence of IE-specific transcription at early times to a high incidence of gB transcription at later times. Clearly, this was not the case. Specifically, the incidence of pieces expressing gB transcripts did not increase over time (Fig. 4, right column).
Patchwork pattern model of mCMV reactivation and recurrence.
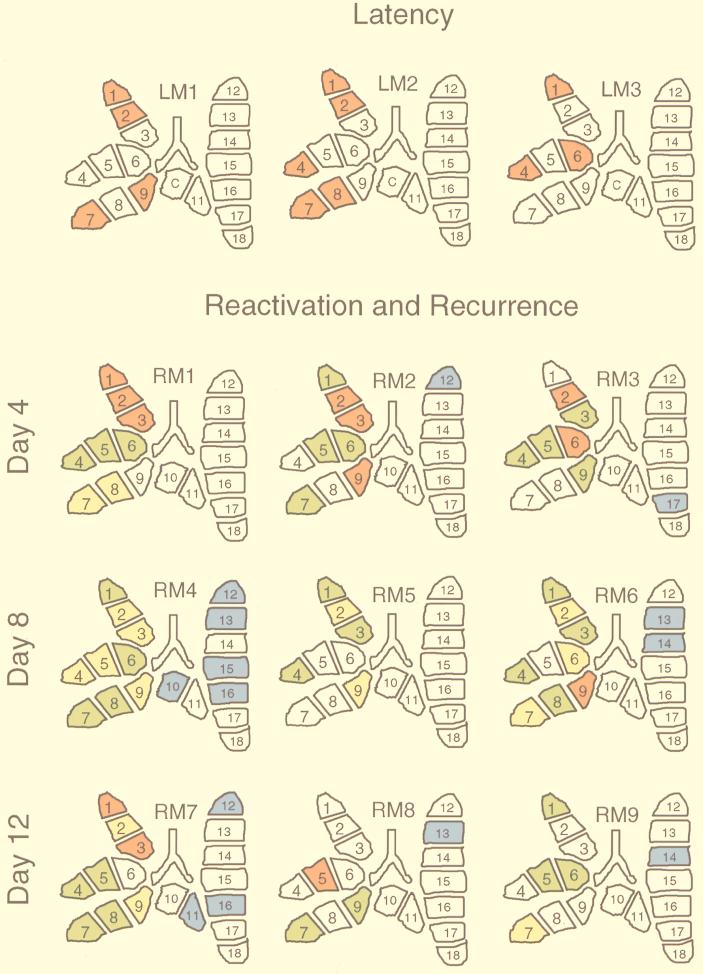
It is fairly laborious to recognize a rule in the complicated patterns revealed by the autoradiographs. We have therefore converted all data into color-coded topographical maps of the lungs (Fig. 5). Tissue pieces that were silent with respect to productive-cycle viral transcription (pieces #1 to #9) or infectivity (pieces #10 to #18) were left uncolored. In the superior, middle, and inferior lobes, red symbolizes selective IE1 transcription, yellow indicates the presence of IE1 and IE3 transcripts, and green stands for the presence of IE1, IE3, and gB transcripts. In the postcaval lobe and left lung, the presence of infectious virus is indicated by blue. At a glance, the distribution of uncolored and red pieces illustrates the selective and stochastic nature of IE1-specific transcription during latency (Fig. 5, top row). By contrast, all colors are represented in the lungs at all time points of reactivation and recurrence, which gave us the impression of a patchwork.
FIG. 5.
Patchwork pattern of mCMV transcriptional reactivation and recurrence in the lungs. (Top) Pattern typical of latency. (Bottom) Pattern observed during reactivation and recurrence. The data shown as autoradiographs in Fig. 1, 3, and 4 are here compiled and illustrated as topographical maps of the lungs. Numbers 1 to 18 are assigned to individual tissue pieces. C, piece #10 was used as a positive control for the verification of latency (see Fig. 1, lower right). Pieces derived from superior, middle, and inferior lobes (#1 to #9) were tested for transcripts, and pieces derived from postcaval lobes and left lungs (ventral view) (#10 to #18) were tested for infectivity. Color code: uncolored, negative in the respective assays; red, positive for IE1 transcripts only; yellow, simultaneously positive for IE1 and IE3 transcripts; green, simultaneously positive for IE1, IE3, and gB transcripts; blue, Pieces containing recurrent virus. LM, latent mouse. RM, recurrent/reactivating mouse.
The results for mouse RM1, showing a red superior lobe, a green middle lobe, and a yellow inferior lobe, might have suggested a coordinated transcriptional reactivation within the anatomical unit of a lung lobe, but this interpretation was quickly proven wrong by the random patterns observed for the lungs of the remaining eight mice. It is important to emphasize that random does not mean arbitrary. The detection of transcripts in fact followed a rule, namely, the sequential order in which transcripts appear during the cycle of viral gene expression. Specifically, with no exception, pieces containing gB transcripts (that is, the green-coded pieces) always also contained IE3 and IE1 transcripts. In a like manner, pieces containing IE3 transcripts (that is, the yellow-coded pieces) always also contained IE1 transcripts. This rule of sequential order gave us confidence that our data are valid.
In conclusion, during reactivation, some tissue pieces maintained or reacquired a latency phenotype, some proceeded to the generation of IE3 mRNA, others proceeded to gB transcription, and only few reached the state of virus recurrence.
Foci of mCMV transcriptional reactivation and recurrence in the lungs.
It is understood that the experimental number of tissue pieces in our mosaic approach is arbitrary and defines the resolution of the analysis. Reactivation and recurrence do not, of course, follow borderlines of experimentally defined tissue pieces. By definition, a green piece is one in which a reactivation event has led to a detectable amount of gB transcripts in one cell or, possibly, in a group of cells. Operationally, we call this a focus of reactivation that has proceeded to gB transcription. A piece is scored green if it contains at least one green focus, but it may also contain several green foci. Obviously, a green piece may contain in addition yellow and red foci, which will remain invisible for us because, as a rule, transcripts of the highest rank in the order of viral gene expression define the “color” of the whole tissue piece. Accordingly, we can conclude that a yellow piece does not contain a green focus, but it may contain invisible red foci. Likewise, a red piece contains only red foci, with one red focus as the minimum. Thus, by counting the number of yellow and red pieces, the number of yellow and red foci, respectively, will necessarily be underestimated. If we extrapolate this basic understanding to a model with a higher overall incidence of reactivation, all pieces were likely to contain a green focus, and consequently, we would under such a condition have failed to recognize that yellow and red foci existed at all. If a green piece were to be subdivided into smaller pieces, green, yellow, and red “subpieces” would be likely to appear. However, there are technical limitations to such an approach. First, increasing the resolution of the analysis by increasing the number of pieces is quite laborious. Second, and more fundamental, the yield of poly(A)+ RNA is the limiting factor. It was therefore very fortunate indeed that the low overall incidence of reactivation achieved under the particular conditions of our experimental setting resulted in green, yellow, and red pieces among only nine pieces derived from three lobes of each lung. Because there are zero fractions for all colors, we can use Poisson statistics for calculating the frequencies of all considered types of foci present during latency as well as during transcriptional reactivation and virus recurrence.
(i) Frequency of IE1-expressing foci during latency.
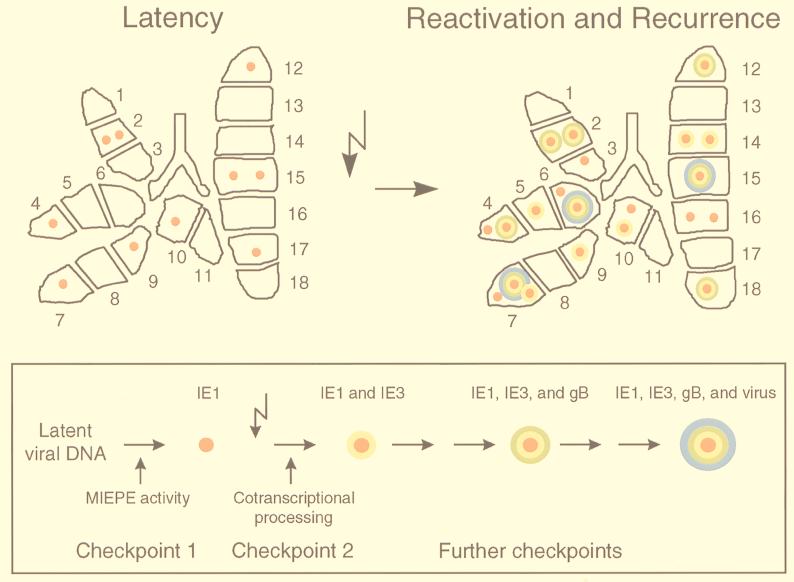
The estimate is based on the data shown in Fig. 1 (original autoradiographs) and Fig. 5 (color-coded topographical maps) for the superior, middle, and inferior lobes of three latently infected mice analyzed. The Poisson distribution parameter λ-red is calculated by the formula λ = −ln f(0), in which the zero fraction f(0) is the fraction of uncolored (transcriptionally silent) pieces among the number of pieces tested, that is 15/27. From the formula F(n) = λ/n × F(n − 1), and extrapolated to 18 pieces, a statistically generated prototypic latent lung contained 10 foci of IE1 transcription located in eight positive pieces; that is, six pieces contained a single focus and two pieces contained two foci (Fig. 6, upper left).
FIG. 6.
Images of focal activities during mCMV latency and reactivation: the orbital model. (Upper left) Prototypic latently infected lung. (Upper right) Prototypic reactivating/recurrently infected lung. Shown are topographical maps of the statistically generated prototypic lungs, with the number and type of foci indicated by symbols illustrating the sequential order of viral gene expression, which is ascending from IE1-specific transcription (red core) via IE3-specific cotranscriptional processing (yellow shell) and gB-specific transcription (green shell) to the production of infectious progeny virions (blue shell). (Box) Orbital model of mCMV reactivation, with sequentially ordered gene expression symbolized by colors that represent increasing “energy.” Checkpoints thought to be involved in the transition from latency to recurrence are indicated. The flash symbol represents the induction by gamma rays.
(ii) Frequency of productively infected foci during recurrence.
Because, as discussed above, the incidence of recurrence did not depend on the time of analysis, the estimate could be based on 81 pieces derived from the postcaval lobes and left lungs of nine mice tested at three time points after immunoreductive treatment (Fig. 3 and 5). For the calculation of λ-blue, the zero fraction f(0) was 67/81. Accordingly, a statistically generated prototypic recurrent lung contained three foci of recurrent infection located in three pieces (Fig. 6, upper right, blue shell).
(iii) Frequency of transcriptionally active foci during reactivation.
The estimates are based on 81 pieces derived from the superior, middle, and inferior lobes of the same nine mice (Fig. 4 and 5) for which the incidence of recurrence in the postcaval lobes and left lungs was tested (see above). The calculations are a bit more sophisticated, because for estimating a particular color-specific frequency, the corresponding zero fraction includes all pieces with colors of lower rank, whereas pieces with colors of higher rank do not enter the calculation. As outlined above, the sequential order of transcription is uncolored < red < yellow < green. Thus, for the calculation of λ-red, f(0) = 23/35, that is, the number of uncolored pieces divided by the sum of uncolored and red pieces. For the calculation of λ-yellow, f(0) = 35/50, that is, the sum of uncolored and red pieces divided by the sum of uncolored and red and yellow pieces. Likewise, for the calculation of λ-green, f(0) = 50/81, that is, the sum of uncolored and red and yellow pieces divided by the total number of pieces. For each λ color, the resulting number and distribution of foci was calculated. Of eight green foci calculated, three had to be considered as actually being blue and thus had to be subtracted. Figure 6 (upper right) illustrates the patchwork pattern on the level of foci for a statistically generated prototypic lung during transcriptional reactivation and mCMV recurrence.
Our present view of the transition from mCMV latency to recurrence, as it appears from the available data, is sketched for summary as an orbital model (Fig. 6, box).
DISCUSSION
Molecular mechanisms of CMV latency and recurrence are almost a “blank spot” in our knowledge of CMV biology. For hCMV latency in myelomonocytic progenitor cells, the group of Mocarski has identified latency-associated novel types of transcripts specified in the IE region (15). However, these transcripts are found only in a very low percentage of latently infected cells (38), and it is not known whether latently infected nonhematopoietic cell types express the same latency-associated transcripts. A possible role for these transcripts in the establishment or maintenance of latency, or in the induction of reactivation, has yet to be defined. Even less is known about the molecular regulation of reactivation and recurrence, but the focus of thinking has long been on regulation by cellular transcription factors addressing binding sites in the enhancer elements that govern the MIE transcription units ie1/2 and ie1/3, specifying the transactivator proteins IE2 and IE3 in hCMV and mCMV, respectively. However, the existence of latency-associated IE transcripts of hCMV (15) in concert with our recent finding of focal generation of IE1 transcripts during pulmonary latency of mCMV (17) implied that checkpoints for latency must exist beyond the regulation of the enhancer.
Previous experimental models of mCMV reactivation were based on the detection of the end point of reactivation, namely, the recurrence of infectious virus in tissue explant cultures (8, 24, 25, 29) or, in vivo, after immunoablative treatment of latently infected mice (2, 9, 17, 23, 28, 32, 42) or after implantation of latently infected tissue into naive, immunodeficient scid recipients (36). Thus, in these approaches, events of molecular reactivation of viral gene expression that did not proceed to the completion of the productive cycle remained invisible. The mosaic approach of analyzing pieces of lung tissue made it possible to study in vivo transcriptional reactivation and virus recurrence simultaneously. This advantage gave us novel insights into the transition from mCMV latency to recurrence. The following information was obtained: (i) mCMV reactivations do not accumulate over time after withdrawal of cellular immune control; (ii) control of latency involves regulation of cotranscriptional IE precursor transcript processing; (iii) there exist multiple, sequential checkpoints in the viral transcriptional program that have to be passed on the way from latency to recurrence; (iv) at any given time point after induction of reactivation, foci exist in different stages of transcriptional progression; and (v) only a few instances of molecular reactivation proceed to recurrence of infectious virions.
Spontaneous versus induced reactivation.
It has long been a question whether molecular reactivation of CMV occurs spontaneously or whether it is triggered by exogenous signalling. Evidence to support the model of spontaneous reactivation was provided for mCMV by the observation of almost instant virus recurrence occurring after withdrawal of immune control by different modes of immunoablation, such as by treatment with an immunosuppressive drug (23) or with gamma rays (2, 17, 18, 32, 42), and by immune cell depletion affecting lymphocytes in general (9) or distinct subsets or combinations of subsets thereof (28). Furthermore, it is long established that mCMV recurrence occurs spontaneously from latently infected tissue explants in culture (8). It thus appeared to be straightforward to propose that withdrawal of immune control was the common trait in all of these examples. Recent work by Polic et al. has dissected the contributions of NK cells and T-lymphocyte subsets to the prevention of mCMV recurrence in B-cell-deficient μMT/μMT mutant mice (28). Similar to the control of primary mCMV infection (reviewed in reference 16), their data indicated a control of recurrent mCMV infection by redundant and hierarchical control mechanisms, with a predominant contribution made by CD8 T cells. Notably, cases of recurrently infected mice increased over time after withdrawal of immune control, thus at first glance supporting the hypothesis that reactivations do occur spontaneously and randomly during latency and that progression to recurrence is actively prevented by immune effector functions, probably involving gamma interferon (30). Since exclusive control of latency by the immune system is not in agreement with widely accepted concepts of herpesvirus latency that have been established for alpha- and gammaherpesviruses (13, 35), it was concluded that CMVs, members of the betaherpesvirus subfamily, were special in this respect (28).
Our data presented here are in obvious conflict with previous views on mCMV latency and recurrence. However, as discussed above, the parameter assessed in previous studies was not transcriptional reactivation but virus recurrence, and this difference needs to be considered. We saw a fundamental, qualitative difference in the transcriptional patterns between latency and reactivation (Fig. 6), but we did not see any influence of the time after induction on the pattern of reactivated gene expression. Specifically, we did not observe an accumulation of recurrences over time after withdrawal of cellular immune control. This finding suggests an inductive, quantal event responsible for shifting the pattern of random and selective IE1-specific transcription typical of latency to a pattern typical of reactivation. Only some foci of reactivation made their way to the generation of viral progeny, that is, to recurrence.
The most notable difference between our approach and the studies with B-cell-deficient mice (28) is the presence or absence, respectively, of antiviral antibodies. It is established knowledge from primary mCMV infection in immunocompromised hosts that virus spreads rapidly from a local site of experimental inoculation to all susceptible target organs (33). Likewise, Schmader et al. (36) have demonstrated that transplantation of latently infected tissue into immunodeficient scid mice leads to rapid reactivation of mCMV in the transplanted tissue and to subsequent dissemination of recurrent mCMV to susceptible target organs in the transplantation recipient. It was thus obvious that recurrent virus replicating in the salivary glands, lungs, livers, or spleens of the recipients had originated from the transplanted donor tissue and not from the sites at which it was eventually detected (36). It is also established knowledge that antiviral antibodies are effectual in preventing hematogenic spreading of the infection (32). Accordingly, from a comparison of mCMV recurrence in latently infected μMT/μMT mutant mice and the respective heterozygotes, Jonjic et al. (7) concluded that antiviral antibodies did not prevent recurrence but limited the dissemination of recurrent virus. Our data presented here clearly documented the absence of virus dissemination. There are several examples illustrated in Fig. 5 in which recurrent virus detected in a particular piece of lung tissue did not spread to directly neighboring pieces (e.g. in RM3 piece #17, RM8 piece #13, and RM9 piece #14). We are therefore sure that we detected recurrence at the site of its origination, and this is the essential basis for estimating the true frequency of recurrences. We therefore propose that an accumulation of positive cases over time in the absence of antiviral antibodies describes the kinetics of virus dissemination, whereas the original events of recurrence are focal and occur with an incidence that does not increase over time after the inductive treatment.
The hypothesis of an induced reactivation of mCMV is in better agreement with the strong evidence for inducible reactivation of herpes simplex viruses, such as by physical trauma, transient hyperthermia, emotional stress, hormonal imbalance, epinephrine, cadmium, and so forth (reviewed in reference 35). Likewise, the lytic cycle of Epstein-Barr virus is induced in latently infected B cells by phorbol esters, which are thought to operate via transcription factor AP-1 binding sites in the IE enhancer (19; reviewed in reference 13). Inductive signalling pathways may well be linked to the immune system. Thus, lymphokines associated with allogeneic immune stimulation appear to be involved in the reactivation of latent hCMV from a cell type of the myelomonocytic lineage that is related to the dendritic cell (40). The inductor in our model of reactivation of mCMV was gamma rays. We do not yet know the molecular mechanism of the proposed induction. The treatment disturbs the cytokine network by radiation-mediated apoptosis of hematopoietic and immune cells, but it may also directly induce a genotoxic stress response in latently infected cells, which is then likely to have an influence on transcription factors and splicing regulators also involved in the molecular regulation of mCMV gene expression. This issue will become a focus of future research on CMV latency and reactivation.
Sequential order of viral gene expression in foci of reactivation.
The order in which viral genes are expressed during reactivation is an open question in herpesvirus research, not only in the case of betaherpesviruses, but also for alphaherpesviruses (35). Our observation of foci of reactivation representing sequential stages of productive-cycle gene expression indicates that reactivated gene expression follows the order known for productive primary infection of permissive cell types. Under the conditions used and for the particular group of mice analyzed here, the statistical average number of IE1-expressing foci in the lungs was 10 during latency. During reactivation in the same cohort of latently infected mice, the total statistical average number of foci increased to 21, of which 7 still displayed the selective IE1 expression typical of latency. One interpretation could be that reactivation had started from IE1-expressing foci that all proceeded to more advanced stages in the viral gene expression cascade and that the pool of IE1-expressing foci was refilled by the same unknown random mechanism that had generated these foci during latency. The data are compatible with this idea, because such a mechanism would indeed predict a doubling of foci, precisely as actually observed. However, we cannot formally exclude the alternative that IE1-expressing foci were not involved in reactivation at all (note that the difference between 10 foci in latency and 7 foci during reactivation is not significant) and that reactivation started from transcriptionally silent cells. We favor the first alternative, but we do admit that this is presently a mere feeling. Let us nonetheless speculate a bit along this line of thinking. Stochastic activity of the MIEPE during latency could be the first step of reactivation by committing the dormant viral genome for the trigger that is effectual at the second checkpoint, the cotranscriptional processing of IE1/3 transcripts generating the IE3 transactivator. In the absence of the inductive signal, IE1-expressing foci are thought to fall back to quiescence, whereas the induction enables them to move on in the gene expression cascade to the next checkpoint.
The precise molecular regulation at the second checkpoint is not yet elucidated, except that it occurs after transcription initiation. One can think of differential splicing of a common IE1/3 precursor mRNA, a hypothesis suggested by the detection of the predicted 5.1-kb IE1/3 precursor mRNA species in productively infected cells (10, 26). Alternatively, regulation could occur by differential usage of a polyadenylation signal located in ie1 exon 4, even though a predicted 3.35-kb IE1 precursor mRNA species has not yet been found (10). Specifically, exclusive usage of this polyadenylation site in exon 4 would explain selective generation of IE1 transcripts during latency.
An unexpected new finding is the apparent existence of at least two further checkpoints in the viral gene expression cascade, one located before and one located after gB gene expression. As a consequence, only a few molecular reactivation events proceeded to the generation of progeny virions (in our example, a statistical average of 3 foci of recurrence [Fig. 6, right, blue shells] of 14 foci of reactivation [Fig. 6, right, all except red foci]). It should be noted that our selection of marker genes in the mCMV gene expression cascade was arbitrary. Inclusion of further genes in the analysis of gene expression might reveal the existence of even more checkpoints. Considering the complexity of CMV genomes, molecular identification of all checkpoints will be a laborious task. What we hope to have shown here is not more but also not less than a principle.
Conclusion.
Our data have provided evidence supporting the model of induced reactivation of CMVs for the example of mCMV. In this respect, CMVs are now back home in the herpesvirus family.
ACKNOWLEDGMENTS
We thank Susanne Schmalz for expert technical assistance, Rafaela Holtappels for performing the BMT, and Hans-Peter Steffens for help with computer-generated images.
Support was provided by a grant to M.J.R. from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMBF), Collaborative Research Project on CMV, individual project 01KI 9607/5, and by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 311.
REFERENCES
- 1.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balthesen M, Messerle M, Reddehase M J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J Virol. 1993;67:5360–5366. doi: 10.1128/jvi.67.9.5360-5366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsch-Häsler K, Keil G M, Weber F, Jasin M, Schaffner W, Koszinowski U H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:8325–8329. doi: 10.1073/pnas.82.24.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grzimek N K A, Podlech J, Steffens H-P, Holtappels R, Schmalz S, Reddehase M J. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J Virol. 1999;73:5043–5055. doi: 10.1128/jvi.73.6.5043-5055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtappels R, Podlech J, Geginat G, Steffens H-P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonjic S, Pavic I, Polic B, Crnkovic I, Lucin P, Koszinowski U H. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J Exp Med. 1994;179:1713–1717. doi: 10.1084/jem.179.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan M C, Mar V L. Spontaneous activation of latent cytomegalovirus from murine spleen explants: role of lymphocytes and macrophages in release and replication of virus. J Clin Invest. 1982;70:762–768. doi: 10.1172/JCI110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan M C, Shanley J D, Stevens J G. Immunosuppression reactivates and disseminates latent murine cytomegalovirus. J Gen Virol. 1977;37:419–423. doi: 10.1099/0022-1317-37-2-419. [DOI] [PubMed] [Google Scholar]
- 10.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate-early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keil G M, Ebeling-Keil A, Koszinowski U H. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 14.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koszinowski U H, Reddehase M J, Jonjic S. The role of T-lymphocyte subsets in the control of cytomegalovirus infection. In: Thomas D B, editor. Viruses and the cellular immune response. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 429–445. [Google Scholar]
- 17.Kurz S K, Rapp M, Steffens H-P, Grzimek N K A, Schmalz S, Reddehase M J. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J Virol. 1999;73:482–494. doi: 10.1128/jvi.73.1.482-494.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurz S K, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 20.Lefkovits I, Waldmann H. Limiting dilution analysis of cells in the immune system. Cambridge, England: Cambridge University Press; 1979. pp. 38–59. [Google Scholar]
- 21.Ljungman P. Cytomegalovirus pneumonia: presentation, diagnosis, and treatment. Semin Respir Infect. 1995;10:209–215. [PubMed] [Google Scholar]
- 22.Ljungman P, Einsele H. Cytomegalovirus infection. Curr Opin Hematol. 1994;1:418–422. [PubMed] [Google Scholar]
- 23.Mayo D R, Armstrong J A, Ho M. Reactivation of murine cytomegalovirus by cyclophosphamide. Nature (London) 1977;267:721–723. doi: 10.1038/267721a0. [DOI] [PubMed] [Google Scholar]
- 24.Mayo D R, Armstrong J A, Ho M. Activation of latent murine cytomegalovirus infection: cocultivation, cell transfer, and the effect of immunosuppression. J Infect Dis. 1978;6:890–896. doi: 10.1093/infdis/138.6.890. [DOI] [PubMed] [Google Scholar]
- 25.Mercer J A, Wiley C A, Spector D H. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J Virol. 1988;62:987–997. doi: 10.1128/jvi.62.3.987-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle M, Bühler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podlech J, Holtappels R, Wirtz N, Steffens H-P, Reddehase M J. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J Gen Virol. 1998;79:2099–2104. doi: 10.1099/0022-1317-79-9-2099. [DOI] [PubMed] [Google Scholar]
- 28.Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Lucin P, Jonjic S, Koszinowski U H. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med. 1998;188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter K R, Starnes D M, Hamilton J D. Reactivation of latent murine cytomegalovirus from kidney. Kidney Int. 1985;28:922–925. doi: 10.1038/ki.1985.218. [DOI] [PubMed] [Google Scholar]
- 30.Presti R M, Pollock J L, Dal Canto A J, O’Guin A K, Virgin H W., IV Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J Exp Med. 1998;188:577–588. doi: 10.1084/jem.188.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quabeck K. The lung as a critical organ in marrow transplantation. Bone Marrow Transplant. 1994;14(Suppl. 4):S19–S28. [PubMed] [Google Scholar]
- 32.Reddehase M J, Balthesen M, Rapp M, Jonjic S, Pavic I, Koszinowski U H. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J Exp Med. 1994;179:185–193. doi: 10.1084/jem.179.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddehase M J, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski U H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riddell S R. Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts. Semin Respir Infect. 1995;10:199–208. [PubMed] [Google Scholar]
- 35.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2296. [Google Scholar]
- 36.Schmader K, Henry S C, Rahija R J, Yu Y, Daley G G, Hamilton J D. Mouse cytomegalovirus reactivation in severe combined immune deficient mice after implantation of latently infected salivary gland. J Infect Dis. 1995;172:531–534. doi: 10.1093/infdis/172.2.531. [DOI] [PubMed] [Google Scholar]
- 37.Shanley J D, Pesanti E L. The relation of viral replication to interstitial pneumonitis in murine cytomegalovirus lung infection. J Infect Dis. 1985;151:454–458. doi: 10.1093/infdis/151.3.454. [DOI] [PubMed] [Google Scholar]
- 38.Slobedman B, Mocarski E. Quantitative analysis of latent human cytomegalovirus. J Virol. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth R L, Sinclair J, Scott J P, Gray J J, Higenbottam T W, Wreghitt T G, Wallwork J, Borysiewicz L K. Infection and reactivation with cytomegalovirus strains in lung transplant recipients. Transplantation. 1991;52:480–482. doi: 10.1097/00007890-199109000-00017. [DOI] [PubMed] [Google Scholar]
- 40.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 41.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 42.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin H W., IV . Plenary lecture at the 7th International Cytomegalovirus Workshop, Brighton, England, 28 April 1999. 1999. [Google Scholar]
- 44.Zaia J A. Prevention and treatment of cytomegalovirus pneumonia in transplant recipients. Clin Infect Dis. 1993;17(Suppl. 2):S392–S399. doi: 10.1093/clinids/17.supplement_2.s392. [DOI] [PubMed] [Google Scholar]