Abstract
Background:
The therapeutic potential of Quercus infectoria (QI) gall, including its anti-inflammatory, antioxidant, and anticancer properties, is well-known. However, its impact on lung, gastric, and esophageal cancer cells remain unclear. This study aims to explore the effects of QI gall aqueous extract on cell viability, apoptosis, and gene expression in A549, BGC823, and KYSE-30 cell lines.
Methods:
A549, BGC823, and KYSE-30 cells were seeded in complete medium and incubated with different concentrations of QI gall extract for 24 hours. Cell viability was measured by an MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide] assay. The induction of apoptosis was assessed through flow cytometric analysis after the adding FITC-conjugated Annexin V (Annexin V-FITC) and propidium iodide (PI). The mRNA expression levels of CCND1, TP53, BCL2 and BAX genes were determined using Real-time Quantitative Polymerase Chain Reaction analysis.
Results:
The MTT assay demonstrated that treatment with QI gall extract significantly reduced the number of viable cells in the A549, BGC823, and KYSE-30 cell lines at IC50 concentrations of 440.1, 437.1, and 465.2 mg/ml, respectively. Additionally, compared to untreated cell population, the percentages of early apoptosis, late apoptosis, and necrosis in the A549, BGC823, and KYSE-30 cells significantly increased following treatment with QI gall extract (P< 0.05). Also, the treatment with QI gall extract influenced the expression of CCND1, TP53, BCL2 and BAX genes.
Conclusions
The present findings indicated that the gall extract of QI can inhibit the growth of A549, BGC823, and KYSE-30 cells by inducing apoptosis, which may be mediated via mitochondria-dependent pathway.
Key Words: Apoptosis, Cell viability, Esophageal cancer, Gastric cancer, Lung cancer, Quercus infectoria
Introduction
Quercus infectoria (QI) is a member of the Fagaceae family, recognized for the development of spherical galls resulting from infections caused by Cynipidae wasp (1). These galls are found in Asia Minor, Iran, Syria, and Greece and possess various therapeutic qualities, including astringency, anti-diabetic, antipyretic, antibacterial, antiviral, and anti-inflammatory properties (2). They contain components such as tannins, gallic acid, and ellagic acid, known for their anti-cancer and antioxidant attributes (3).
Cancer is a significant global cause of death especially in developing countries (4). The challenges in its diagnosis and treatment arise from the intricate diversity observed at the tissue level (5, 6). With modernization and an aging population, cancer-related morbidity and mortality, particularly from lung, gastric, and esophageal cancers, are on the rise (7).
Lung cancer is a prominent global health concern, leading to a significant number of fatalities. There are over 220,000 new cases and 157,000 annual deaths in the United States (8, 9). In 2012, it accounted for 19% of global cancer-related deaths, exceeding the combined mortality of prostate, breast, and colon cancers. Lung cancer is broadly categorized into non-small-cell lung cancer (NSCLC, 85%) and small-cell lung cancer (SCLC, 15%), each demanding distinct treatment strategies. NSCLC encompasses adenocarcinoma (40%–70%), squamous-cell carcinoma (20%–30%), and large-cell carcinoma (10%–15%), each with specific characteristics and locations in the lung. Lung cancer often carries a grim prognosis due to late diagnosis, partly attributed to the absence of early symptoms. Moreover, various genetic alterations, including amplifications, mutations, fusions, and deletions, contribute to the disruption of physiological processes and signaling pathways in NSCLC (10, 11).
Gastric cancer (GC) is a widespread global cancer with the second-highest mortality rate, particularly evident in East Asia, where it consistently has an incidence of 60% (12). It is categorized into cardia gastric cancer (CGC) and non-cardia gastric cancer (NCGC), each having distinct causes (13). Adenocarcinomas constitute the majority (around 90%) of GC cases. The incidence of GC varies globally, influenced by behaviors such as alcohol and tobacco use, which lead to genetic alterations (14). Despite numerous treatment options, such as surgery, chemotherapy, and radiotherapy, GC typically exhibits a poor prognosis and low survival rates (15). Therefore, it is strongly recommended to develop novel and highly efficient therapeutic strategies for GC (16).
Esophageal cancer (EC) is prevalent, especially in Asia, ranking sixth in global cancer-related mortality (17). It consists of two main types: esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), exhibiting differences in causative factors and distribution (18, 19). ESCC is more prevalent in low- to middle-income regions like Golestan, Iran, and central China, often associated with alcohol and tobacco use. EAC, prevalent in high-income areas, is linked to gastroesophageal reflux and obesity (20, 21). Several molecular factors influence the progression of ESCC, including genetic and epigenetic changes, although their precise impact on prognosis and initiation remains uncertain (22).
The CCND1 (cyclin D1) gene, positioned on chromosome 11q13, plays roles in cell cycle regulation, DNA repair, and cell migration (23, 24). The TP53 (tumor protein p53) gene, located on chromosome 17, encodes protein P53, which is crucial for maintaining DNA integrity, controlling cell growth, and promoting apoptosis to eliminate damaged cells (25). BCL2 (B-cell leukemia/lymphoma 2) proteins influence apoptosis, with BAX (BCL2 associated X) initiating cell death, while Bcl-2 has the opposite function of inhibiting apoptosis and sustaining cells. The ratio of the two protein expressions can determine the cell’s fate (26, 27).
In this study, we aim to investigate the potential of Quercus infectoria gall aqueous extract in inhibiting cell proliferation and inducing apoptosis in common cancers of the armed forces. Moreover, we will examine the expression levels of key genes, including CCND1, TP53, BCL2 and BAX, to gain insights into the mechanism of action of the extract. The findings of this study could contribute to the development of novel therapeutic strategies for cancer treatment.
Materials and Methods
Plant material
The QI galls used in this study were obtained from the local herbal medicine shop and were identified based on their morphological appearance. The confirmed plant galls were subsequently subjected to extraction.
Preparation of aqueous extract
The QI galls were first washed with distilled water and allowed to dry at room temperature. They were then powdered using an electric grinder. Subsequently, 100 grams of the powdered material were combined with 500 ml of sterile distilled water and soaked for 1 hour using a shaker at 120 rpm. The mixture was placed in a 50 °C water bath for 72 hours and then centrifuged at 1600 G at 4 °C. The supernatant was filtered using suction pressure through Whatman No.1 filter paper fitted to a Buchner funnel. The filtrates were then evaporated in a rotary evaporator at 80 °C under reduced pressure. The resulting concentrated filtrates were freeze-dried at -50 °C under vacuum until they formed a fine crystal-like crude extract. These crude extracts were stored in airtight jars at 4 °C until they were used.
Phytochemical screenings of QI gall extract by GC-MS
GC-MS was employed to analyze the QI gall extract sample. The analysis initiates with the gas chromatograph, where the sample is efficiently vaporized into the gas phase and separated into its various components using a capillary column coated with a stationary phase (either liquid or solid). The phytochemical analysis of the aqueous extract was conducted using the Varian 4000 Ion Trap GC-MS/MS System.
Cell lines and culture conditions
The A549 cell line (lung adenocarcinoma cancer cells), BGC823 cell line (gastric cancer cells), and KYSE-30 cell line (esophageal cancer squamous cells) were obtained from the Pasteur Institute of Iran in Tehran, Iran. These cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Bio-IDEA, Bl-1003 (500ml)), supplemented with 10% fetal bovine serum (FBS, Bio-IDEA, Bl-1003 (500ml)), and 1% penicillin-streptomycin (Bio-IDEA, Bl-1203 (100ml)), at 37 °C and 5% CO2 in a humidified atmosphere.
Cell viability assay
Cell viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) tetrazolium reduction assay. Cells were cultured at a density of 10,000 cells per well in a 96-well plate and allowed to culture for 24 hours. Subsequently, they were treated with QI gall extract at various concentrations (1000, 500, 250, 125, 62.5, 31.25, and 15.625 mg/mL). After 24 hours of treatment, 50 μL of MTT solution (Sigma Aldrich, Germany) was added to each well, and the cells were further incubated for 4 hours at 37 °C. The medium was then removed, and 100 μL of dimethyl sulfoxide (DMSO) was added for 20 minutes to dissolve the purple formazan crystals formed at the bottom of the wells. The plates were gently shaken, and the optical density was measured at 570 nm using a microplate reader. The microplate reader recorded the absorbance at 570 nm for each well, and the IC50 of the test compound was calculated. The percent cell survival was determined as the relative absorbance of treated cells compared to untreated cells.
Cellular apoptosis assay
The Annexin V-FITC Apoptosis Detection Kit (BioLegend, USA) was used to evaluate cell apoptosis. This kit comprises Annexin V conjugated to the fluorochrome FITC, propidium iodide (PI), and binding buffer. A total of 1×106 A549, BGC823, and KYSE-30 cells were seeded into each well of a six-well plate and treated with the respective IC50 value of QI extract for each cell line for 24 hours. The control samples of cells received no treatment and were provided with an equal volume of culture medium. Following treatment, the cells were trypsinized and subjected to three washes with culture medium and cold PBS before being transferred to tubes. Subsequently, 100 μL of binding buffer, 3 μL of FITC-conjugated Annexin V (Annexin V-FITC), and 3 μL of PI were added to the cells, and they were incubated at room temperature in the dark for 15 minutes. The stained cells were then brought up to a final volume of 300 μL using binding buffer and were immediately analyzed with a flow cytometer.
Total RNA extraction and cDNA synthesis
Total RNA extraction was carried out from A549, BGC823, and KYSE-30 cell lines following treatment using the TRIzol™ (Genexco, Iran)/chloroform method, according to the standard protocol. Briefly, 1×106 cells per well were cultured in a 6-well plate. Subsequently, the cells were treated with QI gall extract for 24 hours in a 5% CO2 incubator at 37 °C. After the incubation, total RNA was extracted from the cells, and DNase I (Thermo Fisher Scientific) was employed to remove genomic DNA during the RNA extraction process. The concentration and purity of the extracted RNAs were determined by assessing the A260/A280 nm wavelength ratios using a Nanodrop (Thermo Fisher Scientific, USA) and agarose gel electrophoresis. For cDNA synthesis (RT-PCR), 1 μg of extracted RNA was used, and it was prepared with a cDNA synthesis kit (Solis BioDyne, Estonia), following the manufacturer's instructions.
Quantitative real-time PCR
Expression of CCND1, TP53, BCL2 and BAX genes was analyzed by Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) technique using the LightCycler®96 Instrument (Roch, Germany). The synthesized cDNA served as the template for RT-qPCR analysis. Each reaction mixture was composed of 7.5 μL of SYBR green master mix (RealQ Plus 2x Master Mix Green High ROX, Ampliqon, Denmark), 5 pmol/μL of both forward and reverse primers, and 1 μg of cDNA, adjusted with sterile distilled water to reach a final volume of 15 μL. All primers were sourced from Metabion (Metabion International AG, Germany), and their details are provided in Table 1. The cycling conditions were as follows: an initial denaturation step at 95 °C for 10 minutes, followed by 40 cycles of denaturation at 95 °C for 15 seconds and annealing at 62 °C for 1 minute. The analysis of relative gene expression was carried out using the 2-ΔΔCt method, with the housekeeping gene B2M serving as an internal control. All qPCR assays for the evaluation of CCND1 (Acc. No. AY893329.1), TP53 (Acc. No. MH011443.1), BCL2 (Acc. No. AC021803.8), and BAX (Acc. No. KC346257.1) genes expression levels were conducted in triplicate in this study.
Table 1.
The sequence of the primers used in the real-time PCR reaction.
| Gene name | Sequence 5'-3' | Tm | GC% | Product size (bp) |
|---|---|---|---|---|
| BAX | F: GAGCAGATTATGAAGACAGGGG | 58.26 | 50.00 | 200 |
| R: ACGGCGGCAATCATCCTC | 60.20 | 61.11 | ||
| BCL2 | F: TCGCCCTGTGGATGACTGA | 60.61 | 57.89 | 134 |
| R: CAGAGACAGCCAGGAGAAATCA | 59.76 | 50.00 | ||
| B2M | F: AGATGAGTATGCCTGCCGTGT | 61.58 | 52.38 | 106 |
| R: TGCGACATCTTCAAACCTCCAT | 60.29 | 45.45 | ||
| TP53 | F: TACTCCCCTGCCCTCAACA | 59.84 | 57.89 | 193 |
| R: GGACCAGACCATCGCTATCT | 58.67 | 55.00 | ||
| CCND1 | F: GGCGGAGGAGAACAAACAGA | 59.97 | 55.00 | 181 |
| R: TGTGAGGCGGTAGTAGGACA | 59.96 | 55.00 |
Statistical analysis
All the collected data were validated through a minimum of three independent experiments, and the results are presented as the mean ± standard deviation. Statistical analysis of MTT, Real-Time PCR, and flow cytometer data was performed using One-way ANOVA with GraphPad Prism 9 software Version 9.0.0. Statistical significance was considered for P< 0.05.
Results
GC-MS analysis for QI gall extract
After successfully extracting the plant material, the initial phytochemical analysis, as depicted in Figure 1 from the GC-MS device, revealed that the aqueous plant extract contains the compounds listed in Table 2.
Fig. 1.
GC-MS chromatogram of the aqueous extract from Quercus infectoria gall.
Table 2.
Compounds identified in the aqueous extract of a Quercus infectoria gall via GC-MS. RT=Retention time, Total Fatty Acid: 16.046 + 12.527 (28.527%).
| RT | Compound name | Molecular formula | Molecular Weight (g/mol) | Compound nature |
|---|---|---|---|---|
| 12.472 | Formic acid | CH₂O₂ | 46.03 | carboxylic acid |
| 18.831 | 2,4-Dimethylimidazole | C5H10N2 | 96.13 | chemical compound |
| 23.724 | Methyl salicylate | C8H8O3 | 152.149 | organic compound |
| 30.684 | Phenol | C 6H5OH | 94.113 | aromatic organic compound |
| 33.488 | Maltol | C6H6O3 | 126.11 | Organic compound |
| 34.969 | Maltol | C6H6O3 | 126.11 | Organic compound |
| 36.127 | Palmitic acid | C16H32O2 | 256.4 | fatty acid |
| 37.174 | a-furfuryl alcohol | C5H6O2 | 98.1 | hydroxymethyl |
| 41.957 | Oleic acid | C18H34O2 | 282.47 | fatty acid |
The effect of QI gall extract on cell viability
The MTT assay was employed to assess the impact of QI gall extract on the viability of A549, BGC823, and KYSE-30 cells. Cells from these lines were exposed to varying concentrations (1000, 500, 250, 125, 62.5, 31.25, 15.625 mg/mL) of QI gall extract for 24 hours. Their viability rates were then compared with a control group that had not been exposed to QI gall extract. The MTT results were used to determine the concentration of the extract required to inhibit 50% of the cell population (IC50 value), which was found to be 440.1 mg/ml for A549 cells, 437.1 mg/ml for BGC823 cells, and 465.2 mg/ml for KYSE-30 cells (Fig. 2).
Fig. 2.

The graph depicting cell viability shows a correlation with the logarithm of concentration. As the concentration increases, the percentage of cell viability (%) decreases.
Cell viability decreased as the concentration of the extract increased in comparison to the control group, and this decrease in cell viability was found to be statistically significant across different cell groups compared to the control (Fig. 3).
Fig. 3.

Comparative chart illustrating the impact of various concentrations of extract on cell viability. With the increase in the concentration of the extract, the cell viability has decreased significantly compared to the control (*P< 0.01, **P< 0.001, ***P< 0.0001).
Apoptosis induction in A549, BGC823, and KYSE-30 cells by QI gall extract
In the current study, after confirming that treatment of human lung, gastric and esophageal cancer cells with QI extract leads to a decrease in cell viability, we investigated the relationship between QI gall extract and apoptosis.
The percentage of viable cells, early apoptosis, late apoptosis, and necrosis for untreated cells in the A549 cell line were 90.9%, 1.19%, 0.515%, and 7.37%, respectively (Fig. 4). In the BGC823 cells, they were 92.5%, 0.830%, 0.159%, and 6.5%, respectively. In the KYSE-30 cell line, they were 90.3%, 0.763%, 2%, and 6.97%, respectively. However, the percentage of viable population, early apoptosis, late apoptosis, and necrosis for treated cells and in the A549 cell line were 54.9%, 19.2%, 17.9%, and 8.06%, respectively. In the BGC823 cell line, they were 44.6%, 18.1%, 16.1%, and 21.1%, respectively. In the KYSE-30 cell line, they were 50%, 5.19%, 24%, and 20.8%, respectively.
Fig. 4.

Flow cytometry chart depicting results for all three cell lines, demonstrating induction of apoptosis in living cells by the aqueous extract. Significance is highlighted on the Graph, comparing control cells to treated cells (*P< 0.01).
The rates of early apoptosis, late apoptosis, and necrosis in treated cells showed a significant increase compared to untreated cells (P< 0.05). Therefore, the changes in cell populations demonstrated the ability of QI gall extract to induce apoptotic cell death in A549, BGC823, and KYSE-30 cells.
Changes in the expression levels of CCND1, TP53, BCL2, and BAX genes because of QI gall extract
Following the observation of the regulatory effect of QI gall extract on apoptosis in A549, BGC823, and KYSE-30 cells, we further investigated the impact of QI gall extract on the expression of the key apoptotic regulators, BAX and BCL2. Additionally, we assessed the mRNA expression levels of CCND1 and TP53 genes in A549, BGC823, and KYSE-30 cells treated with QI gall extract, given their crucial roles in cell cycle regulation and apoptosis induction.
To explore the connection between mRNA levels of CCND1, TP53, BCL2, and BAX genes and the occurrence of lung, gastric, and esophageal cancers, we conducted real-time quantitative PCR. The mRNA expression values of CCND1, TP53, BCL2, and BAX genes were normalized using the internal B2M gene (a housekeeping gene) as a control. As depicted in Figure 5, the expression of BAX gene (A) and TP53 gene (D) in the treated cell line samples exhibited a significant increase (P< 0.05) compared to the control sample. Moreover, the expression of BCL2 gene (B) and CCND1 (C) in the treated cell line samples showed a significant decrease (P< 0.05) compared to the control sample.
Fig. 5.
Gene expression charts for BAX (A), BCL2 (B), CCND1 (C), and TP53 (D) in both control and treated cell lines. The relative changes in gene expression in treated cells were statistically compared to those in control cells (*P< 0.01, **P< 0.001, ***P< 0.0001, *****P< 0.00001).
Discussion
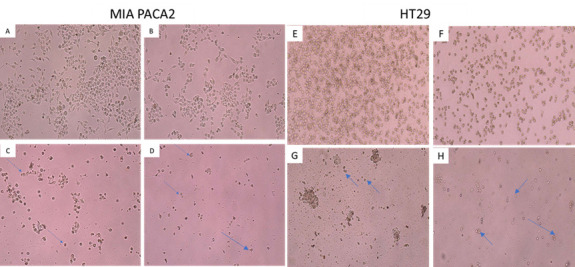
Our investigation has uncovered significant findings in the realm of cancer treatment. In this study, our primary objective was to explore the potential of QI gall aqueous extract for cancer treatment, with a specific focus on its direct cytotoxic effects on A549, BGC823, and KYSE-30 cells. The characteristics associated with apoptotic cells encompass chromatin aggregation, nuclear and cytoplasmic condensation, and the partition of cytoplasm and nucleus into membrane-bound vesicles. Notably, the nuclear morphology of apoptotic cells is characterized by chromatin condensation and nuclear fragmentation, which can be observed using various techniques, including the use of DNA-binding stains like 4',6-diaminido-2-phenylindole, Hoechst, and others (28).
Morphological and biochemical changes in cells play a crucial role in the apoptotic process, preventing tumor development, and treatment resistance (29). According to Kerr et al., (1972) morphological alterations in apoptotic cells include chromatin aggregation, nuclear and cytoplasmic condensation, and the partitioning of cytoplasm and nucleus into membrane-bound vesicles (30). The cells treated with QI aqueous extract displayed characteristic apoptotic changes, such as early apoptotic cells with condensed nuclear structures, cell shrinkage, and the formation of apoptotic bodies, as previously described. Late apoptotic cells exhibited condensed chromatin areas in the nucleus, as mentioned earlier. Additionally, the treated cells showed viable cells with intact nucleus structures (31). In our current study, we utilized MTT assay, real-time PCR and flow cytometry techniques to evaluate cell death.
The results of cytotoxicity assay revealed a dose-dependent reduction in the viability of three human cancer cell lines, A549, BGC823, and KYSE-30, following treatment with QI gall aqueous extract. According to the MTT assay results, the antiproliferative effects of QI gall aqueous extract on these cell lines displayed significant differences (P< 0.05). Additionally, the IC50 values for A549 (440.1 mg/mL) and BGC823 (437.1 mg/mL) cell lines were lower than that of KYSE-30 (465.2 mg/mL) cells.
To determine whether the cytotoxicity of QI gall aqueous extract involved the induction of apoptosis, A549, BGC823, and KYSE-30 cells were treated with the corresponding IC50 concentrations of the preparations. These cells were stained with propidium iodide (PI) and Annexin V-fluorescein isothiocyanate (FITC), and subsequently analyzed using flow cytometry. The results indicated that QI gall aqueous extract induced cell death through total apoptosis. Specifically, the A549, BGC823, and KYSE-30 cell lines exhibited higher Annexin V-FITC staining compared to the control cell line.
In the A549 cell line, the percentages of viable cells, early apoptosis, late apoptosis, and necrosis were 54.9%, 19.2%, 17.9%, and 8.06%, respectively. In the BGC823 cell line, the percentages were 44.6% viable cells, 18.1% early apoptosis, 16.1% late apoptosis, and 21.1% necrosis. In the KYSE-30 cell line, the percentages were 50% viable cells, 5.19% early apoptosis, 24% late apoptosis, and 20.8% necrosis. The rates of early apoptosis, late apoptosis, and necrosis in treated cells increased significantly compared to untreated cells (P< 0.05). Notably, the percentage of necrotic cells remained low in the A549 cell line. These results indicated that QI gall aqueous extract treatment led to an increased number of apoptotic cells without significantly elevating the percentage of necrotic cells.
According to Nasiri et al., (2018) the examination of BAX, BCL-2, Fas, and P53 gene expressions for determining cell viability under cancer treatment, using the real-time method, yields reliable findings (32). Mohammedi et al., (2021) conducted a study to assess oxidative stress, apoptosis, and the expression of microRNA in cardiovascular patients. The study involved measuring the expression of miR-208 and miR-1, as well as the mRNA levels of inflammatory mediators (TNFα, iNOS/eNOS) and apoptotic factors (Bax and Bcl-2) via quantitative real-time PCR (33).
Combining natural substances with medical interventions can enhance the effectiveness of cancer treatments. This strategy can reduce the need for multiple medications, lower costs, and is well-tolerated. Consequently, the incorporation of natural products into combination therapy presents a promising approach for the treatment of cancer (34). Our study underscores the anti-apoptotic potential of QI gall extract, with increased levels of BAX and TP53 observed following treatment. TP53 gene expression in A549, BGC823, and KYSE-30 cell lines increased by 4.5, 3.3, and 3.7-fold, respectively, with significant upregulation in each cell line. Similarly, BAX gene expression in these cell lines increased by 3.8, 2, and 2.3-fold, respectively, with significant enhancement in each cell line. In contrast, the levels of CCND1 and BCL2 decreased following treatment, with CCND1 gene expression in A549, BGC823, and KYSE-30 cell lines decreasing by 0.22, 0.58, and 0.48-fold, respectively, and BCL2 gene expression in these cell lines decreasing by 0.22, 0.37, and 0.27-fold, respectively, all exhibiting significant decreases in each cell line.
The early phase of apoptosis is frequently characterized by phosphatidylserine externalization (35). During apoptosis, phosphatidylserine is translocated and presented on the outer surface of the cell membrane, where it associates with annexin V. Cells undergoing late-apoptosis or necrosis, characterized by compromised cell membrane integrity, can be identified through staining with propidium iodide. Abdullah's investigation confirmed phosphatidylserine externalization in HeLa cells treated with QI gall aqueous extract through AO/PI staining, indicating apoptosis-induced cell death (36).
As outlined in earlier research, apoptosis, is a genetically controlled mechanism of cellular demise that occurs in living cells (37). It is triggered by two distinct death signaling pathways: the intrinsic mitochondria-dependent and the extrinsic death receptor-dependent pathways (38). The production of apoptosomes and the activation of caspase 3 depend on the release of cytochrome C from mitochondria into the cytosol in the intrinsic mitochondria-dependent pathway (39). In the presence of apoptotic stimuli, the Bcl-2 family members play a crucial role as mediators of cytochrome C release. Bax is a pro-apoptotic protein, and Bcl-2 is an anti-apoptotic protein, both belonging to the Bcl-2 family. Bax or Bcl-2 can regulate mitochondrial permeability and help the flow of cytochrome C (40). The fate of many cells is consequently determined by the Bax/Bcl-2 ratio (41). Loss of mitochondrial membrane potential (MMP) and the release of cytochrome C result in an imbalance between Bax and Bcl-2 proteins, triggering the activation of caspase 3 and inducing apoptosis (42). Cyclin-dependent kinase inhibitors (CKIs) play a crucial role in the physiological regulation of cell cycle control. Members of Cip (CDK interacting protein/Kinase inhibitory protein) family (P21 Cip1) attach to cyclin-CDK complexes and inhibit their function. P21 Cip1, a transcriptional target of P53, is vital for initiating G1 arrest. Furthermore, the CCND1/CDK4 complex phosphorylates serine residues on Rb, consequently nullifying its growth-inhibitory impacts. These effects are caused by Rb's negative regulation of the E2F family of heterodimeric transcriptional regulators (43, 44). Unphosphorylated Rb inhibits cell division through interacting with and inactivating E2Fs. Upon phosphorylation of Rb by CDK/cyclin complexes, it results in the liberation of active E2F entities. These entities, in turn, facilitate the transcription of genes essential for the G1-S transition and the progression into the S-phase (45).
In conclusion, the current study highlights the potential of Quercus infectoria gall aqueous extract as an alternative treatment option for common cancers in the armed forces. The extract showed promising results in inhibiting cell proliferation and inducing apoptosis, which are essential processes in the development and progression of cancer. Furthermore, the extract was found to modulate the expression levels of key genes involved in cell cycle regulation and apoptosis. These findings contribute to the growing body of evidence supporting the use of natural products in cancer treatment. Further research is necessary to elucidate the exact mechanism of action of Quercus infectoria gall aqueous extract and to determine its potential as a therapeutic agent in clinical settings.
Statement of Ethics
This research, along with all the protocols used, has been approved by the Ethics committee of AJA University of Medical Science (IR.AJAUMS.REC.1400.197).
Financial support
The present study was supported by AJA University of Medical Sciences.
Conflict of Interest
The authors declare that there is no conflict of interest that could inappropriately influence this research.
Acknowledgments
I would like to thank my supesrvisor, Dr. Mohsen Ebrahimi, a respected and resourceful scholar, who provided valuable guidance during every stage of the writing of this article.
References
- 1.Elham A, Arken M, Kalimanjan G, Arkin A, Iminjan M. A review of the phytochemical, pharmacological, pharmacokinetic, and toxicological evaluation of Quercus Infectoria galls. J Ethnopharmacol. 2021;273:113592. doi: 10.1016/j.jep.2020.113592. [DOI] [PubMed] [Google Scholar]
- 2.Kaur G, Athar M, Alam MS. Quercus infectoria galls possess antioxidant activity and abrogates oxidative stress-induced functional alterations in murine macrophages. Chem Biol Interact. 2008;171(3):272–82. doi: 10.1016/j.cbi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Vermani A, Navneet, Prabhat Screening of Quercus infectoria gall extracts as anti-bacterial agents against dental pathogens. Indian J Dent Res. 2009;20(3):337–9. doi: 10.4103/0970-9290.57380. [DOI] [PubMed] [Google Scholar]
- 4.Nair M, Sandhu SS, Sharma AK. Cancer molecular markers: A guide to cancer detection and management. Semin Cancer Biol. 2018;52(Pt 1):39–55. doi: 10.1016/j.semcancer.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–85. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn BK, Wagner PD, Anderson D, Greenwald P. Molecular markers for early detection. Semin Oncol. 2010;37(3):224–42. doi: 10.1053/j.seminoncol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol. 2012;7(5):924–33. doi: 10.1097/JTO.0b013e31824cc334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shivapurkar N, Reddy J, Chaudhary PM, Gazdar AF. Apoptosis and lung cancer: a review. J Cell Biochem. 2003;88(5):885–98. doi: 10.1002/jcb.10440. [DOI] [PubMed] [Google Scholar]
- 10.Liu G, Pei F, Yang F, Li L, Amin AD, Liu S, et al. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int J Mol Sci. 2017;18(2):367. doi: 10.3390/ijms18020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 12.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 13.Lv L, Liang X, Wu D, Wang F, Zhang Y, Cang H, et al. Is cardia cancer a special type of gastric cancer? A differential analysis of early cardia cancer and non-cardia cancer. J Cancer. 2021;12(8):2385–2394. doi: 10.7150/jca.51433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilic M, Ilic I. Epidemiology of stomach cancer. World J Gastroenterol. 2022;28(12):1187–1203. doi: 10.3748/wjg.v28.i12.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haghighat P, Bekaii-Saab T. An update on biochemotherapy of advanced gastric and gastroesophageal adenocarcinoma. J Natl Compr Canc Netw. 2008;6(9):895–900. doi: 10.6004/jnccn.2008.0068. [DOI] [PubMed] [Google Scholar]
- 16.Dang Y, Liu T, Yan J, Reinhardt JD, Yin C, Ye F, Zhang G. Gastric cancer proliferation and invasion is reduced by macrocalyxin C via activation of the miR-212-3p/Sox6 Pathway. Cell Signal. 2020;66:109430. doi: 10.1016/j.cellsig.2019.109430. [DOI] [PubMed] [Google Scholar]
- 17.Yang CS, Chen X, Tu S. Etiology and Prevention of Esophageal Cancer. Gastrointest Tumors. 2016;3(1):3–16. doi: 10.1159/000443155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuwano H, Kato H, Miyazaki T, Fukuchi M, Masuda N, Nakajima M, et al. Genetic alterations in esophageal cancer. Surg Today. 2005;35(1):7–18. doi: 10.1007/s00595-004-2885-3. [DOI] [PubMed] [Google Scholar]
- 19.Lao-Sirieix P, Caldas C, Fitzgerald RC. Genetic predisposition to gastro-oesophageal cancer. Curr Opin Genet Dev. 2010;20(3):210–7. doi: 10.1016/j.gde.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Melhado RE, Alderson D, Tucker O. The changing face of esophageal cancer. Cancers (Basel). 2010;2(3):1379–404. doi: 10.3390/cancers2031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islami F, Kamangar F, Aghcheli K, Fahimi S, Semnani S, et al. Epidemiologic features of upper gastrointestinal tract cancers in Northeastern Iran. Br J Cancer. 2004;90(7):1402–6. doi: 10.1038/sj.bjc.6601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210, 215. doi: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Holm K, Staaf J, Jönsson G, Vallon-Christersson J, Gunnarsson H, Arason A, et al. Characterisation of amplification patterns and target genes at chromosome 11q13 in CCND1-amplified sporadic and familial breast tumours. Breast Cancer Res Treat. 2012;133(2):583, 94. doi: 10.1007/s10549-011-1817-3. [DOI] [PubMed] [Google Scholar]
- 24.Pestell RG. New roles of cyclin D1. Am J Pathol. 2013;183(1):3, 9. doi: 10.1016/j.ajpath.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ara N, Atique M, Ahmed S, Ali Bukhari SG. Frequency of p53 gene mutation and protein expression in oral squamous cell carcinoma. J Coll Physicians Surg Pak. 2014;24(10):749, 53. doi: 10.2014/JCPSP.749753. [DOI] [PubMed] [Google Scholar]
- 26.Fiebig AA, Zhu W, Hollerbach C, Leber B, Andrews DW. Bcl-XL is qualitatively different from and ten times more effective than Bcl-2 when expressed in a breast cancer cell line. BMC Cancer. 2006;6:213. doi: 10.1186/1471-2407-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74(4):609–19. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 28.Ziegler U, Groscurth P. Morphological features of cell death. News Physiol Sci. 2004;19:124–8. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]
- 29.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY). 2016;8(4):603–19. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26(4):239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngabire D, Seong YA, Patil MP, Niyonizigiye I, Seo YB, Kim GD. Induction of apoptosis and G1 phase cell cycle arrest by Aster incisus in AGS gastric adenocarcinoma cells. Int J Oncol. 2018;53(5):2300–2308. doi: 10.3892/ijo.2018.4547. [DOI] [PubMed] [Google Scholar]
- 32.Sayed Mahdi N, Azarbani F, Pirnia A, Abbaszadeh A, Gholami M. The Effect of Caffeic Acid on Spermatogonial Stem Cell-type A Cryopreservation. Rep Biochem Mol Biol. 2018;7(1):85–93. [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammadi A, Balizadeh Karami AR, Dehghan Mashtani V, Sahraei T, Bandani Tarashoki Z, Khattavian E, et al. Evaluation of Oxidative Stress, Apoptosis, and Expression of MicroRNA-208a and MicroRNA-1 in Cardiovascular Patients. Rep Biochem Mol Biol. 2021;10(2):183–196. doi: 10.52547/rbmb.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yusof WNSW, Abdullah H. Phytochemicals and Cytotoxicity of Quercus infectoria Ethyl Acetate Extracts on Human Cancer Cells. Trop Life Sci Res. 2020;31(1):69–84. doi: 10.21315/tlsr2020.31.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig LM, Maxcy KL, LaBelle JL. Flow Cytometry-Based Detection and Analysis of BCL-2 Family Proteins and Mitochondrial Outer Membrane Permeabilization (MOMP). Methods Mol Biol. 2019;1877:77–91. doi: 10.1007/978-1-4939-8861-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdullah H, Ismail I, Suppian R. Induction of apoptosis in HeLa cervical cancer cells treated with aqueous and supercritical fluid extracts of Quercus infectoria. Res J Pharmacogn. 2021;8(4):63–77. [Google Scholar]
- 37.Wyllie AH. Apoptosis: an overview. Br Med Bull. 1997;53(3):451–65. doi: 10.1093/oxfordjournals.bmb.a011623. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Luo Q, Xu H, Gong F, Zhou X, Sun Y, et al. Mitochondria-dependent apoptosis of activated T lymphocytes induced by astin C, a plant cyclopeptide, for preventing murine experimental colitis. Biochem Pharmacol. 2011;82(3):260–8. doi: 10.1016/j.bcp.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Mohan S, Abdelwahab SI, Kamalidehghan B, Syam S, May KS, Harmal NS, et al. Involvement of NF-κB and Bcl2/Bax signaling pathways in the apoptosis of MCF7 cells induced by a xanthone compound Pyranocycloartobiloxanthone A. Phytomedicine. 2012;19(11):1007–15. doi: 10.1016/j.phymed.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Lindsten T, Zong WX, Thompson CB. Defining the role of the Bcl-2 family of proteins in the nervous system. Neuroscientist. 2005;11(1):10–5. doi: 10.1177/1073858404269267. [DOI] [PubMed] [Google Scholar]
- 41.Bivik CA, Larsson PK, Kågedal KM, Rosdahl IK, Ollinger KM. UVA/B-induced apoptosis in human melanocytes involves translocation of cathepsins and Bcl-2 family members. J Invest Dermatol. 2006;126(5):1119–27. doi: 10.1038/sj.jid.5700124. [DOI] [PubMed] [Google Scholar]
- 42.Green DR. At the gates of death. Cancer cell. 2006;9(5):328–30. doi: 10.1016/j.ccr.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–42. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 45.Paggi MG, Baldi A, Bonetto F, Giordano A. Retinoblastoma protein family in cell cycle and cancer: a review. J Cell Biochem. 1996;62(3):418–30. doi: 10.1002/(SICI)1097-4644(199609)62:3%3C418::AID-JCB12%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]



