Abstract
Background.
Measles elimination (interruption of endemic measles virus transmission) in the United States was declared in 2000; however, the number of cases and outbreaks have increased in recent years. We characterized the epidemiology of measles outbreaks and measles transmission patterns after elimination to identify potential gaps in the US measles control program.
Methods.
We analyzed national measles notification data from 1 January 2001 to 31 December 2019. We defined measles infection clusters as single cases (isolated cases not linked to additional cases), 2-case clusters, or outbreaks with ≥3 linked cases. We calculated the effective reproduction number (R) to assess changes in transmissibility and reviewed molecular epidemiology data.
Results.
During 2001–2019, a total of 3873 measles cases, including 747 international importations, were reported in the United States; 29% of importations were associated with outbreaks. Among 871 clusters, 69% were single cases and 72% had no spread. Larger and longer clusters were reported since 2013, including 7 outbreaks with >50 cases lasting >2 months, 5 of which occurred in known underimmunized, close-knit communities. No measles lineage circulated in a single transmission chain for >12 months. Higher estimates of R were noted in recent years, although R remained below the epidemic threshold of 1.
Conclusions.
Current epidemiology continues to support the interruption of endemic measles virus transmission in the United States. However, larger and longer outbreaks in recent postelimination years and emerging trends of increased transmission in underimmunized communities emphasize the need for targeted approaches to close existing immunity gaps and maintain measles elimination.
Keywords: measles, measles outbreak, MMR vaccine
Notwithstanding the commitment by all 6 World Health Organization (WHO) regions toward measles elimination (ie, absence of endemic measles virus transmission in a defined geographic area in the presence of well-performing surveillance), the incidence of measles increased in all WHO regions during 2016–2019, with the highest number of global cases in 23 years reported in 2019 (>800 000 cases) [1]. In the United States measles was declared eliminated in 2000, a result of high coverage with 2 doses of measles, mumps, and rubella (MMR) vaccine, the first dose given routinely at 12–15 months of age and the second dose at 4–6 years of age. However, upsurges in measles activity abroad increase opportunities for importations and spread when the virus is introduced into communities with low vaccination coverage [2]. After elimination, the reported annual incidence of measles in the United States has remained low (<1 case per million population), yet a gradual increase in the number of transmissions per importation was noted between 2001 and 2015 [3]. More recently, 2 prolonged outbreaks in underimmunized communities in New York during 2018 and 2019 threatened the measles elimination status of the United States, and contributed to a record number of cases; >1200 cases were reported in 2019, the highest number of cases since 1992 [4].
Because of its high transmissibility, remarkably high measles immunity levels (>92%–94%) are needed to prevent sustained measles virus transmission [5]. As such, herd immunity for measles can be easily breached. In the Americas, the first region to have achieved measles elimination, endemic measles virus transmission was reestablished following outbreaks in Venezuela in 2018 and Brazil in 2019 [1]. Several countries in Europe also lost their measles elimination status in recent years, including the United Kingdom in 2019 [6]. Until measles eradication is reached, no country is invulnerable to losing its measles-free status, and recent outbreaks in the United States have raised concerns about the United States veering in this direction. In this article, we describe the epidemiology of measles outbreaks and measles virus transmission patterns after elimination in the United States to assess the risk of reestablishment of endemic measles virus circulation and inform prevention strategies.
METHODS
We analyzed data on confirmed measles cases reported in the United States from 1 January 2001 to 31 December 2019. Cases are considered imported if some of the exposure period (7–21 days before rash onset) occurs outside the United States and the rash occurs within 21 days of entering the United States, with no known measles exposure in the United States during that time [7]. For brevity, we use the term infection cluster to broadly describe single cases (isolated cases not epidemiologically linked to other cases), 2-case clusters (with known epidemiological links to each other), and outbreaks (≥3 epidemiologically linked cases). Generations of spread were assigned to each cluster based on the duration of transmission, calculated by subtracting the dates of rash onset of the first and last cases in the cluster, with single cases assigned 0-day duration. Outbreaks were classified as having a source if the importation that led to the outbreak was identified.
We report temporal trends in imported and total measles cases, measles outbreaks and outbreak source, outbreak size and duration, and genotyping data. We describe the geographic distribution of cases and summarize the largest outbreaks (>50 cases) reported after elimination.
To assess changes in the transmissibility of measles over time, we calculated the annual effective reproduction number (R), or the average number of secondary cases generated per infected individual. Expanding on a prior analysis that covered the years 2001–2014 [8], we calculated R using 2 methods. First, we fitted the expected distribution of chain sizes for a given value of R and dispersion parameter k from a negative binomial branching process model [9], to the yearly observed distribution of cluster sizes; k represents the degree of transmission heterogeneity, with values <1 indicating high transmission heterogeneity [9]. Second, we adapted [10] an existing algorithm (Wallinga and Teunis method) [11, 12] that infers R for each case in a cluster, based on the time in days between the rash onset of cases and the probability density of the serial interval (time between the onset of symptoms in primary cases and their secondary cases). We used a serial interval with a gamma probability distribution and a mean (standard deviation) of 11.1 (2.47) days [13]. Because containment measures can lessen transmission and the final sizes of outbreaks, we also used this algorithm to measure the yearly R of index cases (Ri), or cases with the earliest rash onset in each cluster; that is, we assessed transmissibility early in the outbreak, before implementation of control measures, to assess baseline population immunity. Details on these methods are found elsewhere [9, 10].
Because outbreaks in New York during 2018 and 2019 threatened US measles elimination, we further examined cases associated with these outbreaks. We focused on their importation status, the reporting jurisdiction (and connection to other jurisdictions), the duration of transmission, and whether cases had a direct known source (ie, whether a direct epidemiological link to another confirmed case could be ascertained). We evaluated the level of measles control during these outbreaks by estimating daily R using the Wallinga-Teunis method. Any measles genotype found repeatedly should be carefully investigated as a potential endemic genotype. To assess whether any endemic measles genotypes had been established in the United States during this period, we reviewed measles molecular epidemiology from 30 September 2018 (date of rash onset for the first case in these outbreaks) to 31 December 2019. Measles genotypes were determined based on analysis of the sequence of the 450 nucleotides coding for the COOH terminal 150 amino acids of the nucleoprotein gene (N-450), using standardized methods [14]. Measles sequencing was performed at the Centers for Disease Control and Prevention or Vaccine Preventable Disease Reference Centers of the Association of Public Health Laboratories. Sequences were submitted to the WHO global measles sequence database, MeaNS, and assigned to WHO-named strains in MeaNS [15]. Additional analyses were done to assess surveillance sensitivity and the potential for an unrecognized endemic transmission chain.
RESULTS
During 2001–2019, a total of 3873 measles cases, including 747 importations, were reported in the United States (Table 1). The number of importations increased from 123 in 2001–2004 to 194 in 2017–2019, while the total number of cases increased from 253 to 1775 during the corresponding periods. Twenty-nine percent of importations were associated with outbreaks. The number of outbreaks increased from 18 in 2001–2004 to 48 in 2017–2019; 72% of outbreaks had a known source. Among 872 unique infection clusters, 602 (69%) were single cases, 112 (13%) were 2-case clusters, and 158 (18%) were outbreaks with a total of 3047 cases; 723 (83%) had ≤1 generation of spread. The median size of outbreaks (5 cases; range, 4–6 cases) and their median duration (22.5 days; range, 20–28 days) did not change prominently over time, although the distribution generally tended toward larger and longer clusters over the years (Table 1). Genotype data were obtained from 43% of single cases, 70% of 2-case clusters, and 88% of outbreaks; measles genotype diversity decreased over time.
Table 1.
Reported Measles Importations, Cases, Clusters, Outbreaks, and Genotypes—United States, 2001–2019
| Measles Cases, Outbreaks, and Clusters | 2001–2004 | 2005–2008 | 2009–2012 | 2013–2016 | 2017–2019 | Total (2001–2019) |
|---|---|---|---|---|---|---|
| Measles cases | ||||||
| Total cases, no. | 253 | 304 | 410 | 1131 | 1775 | 3873 |
| International importations, no.a | 123 | 109 | 161 | 160 | 194 | 747 |
| Importations associated with outbreaks, no. (% of importations)b | 43 (35) | 23 (21) | 34 (21) | 46 (29) | 68 (35) | 214 (29) |
| Outbreak-related cases, no. (% of total cases) | 116 (46) | 189 (62) | 196 (48) | 947 (84) | 1599 (90) | 3047 (79) |
| Measles outbreaksc | ||||||
| Total outbreaks, no. | 18 | 18 | 32 | 42 | 48 | 158 |
| Outbreaks with known source, no. (% of total outbreaks) | 15 (83) | 14 (78) | 20 (63) | 28 (67) | 37 (77) | 114 (72) |
| Outbreak size, median (range), no. of cases | 4 (3–14) | 4.5 (3–34) | 4.5 (3–21) | 5.5 (3–383) | 6 (3–697) | 5 (3–697) |
| Outbreak duration, median (range), d | 20 (3–61) | 28 (4–79) | 19 (5–68) | 22.5 (4–121) | 23.5 (5–322) | 22.5 (3–322) |
| Measles clustersd | ||||||
| Cluster size, no. (%) | ||||||
| Isolated or single cases | 101 (74) | 91 (75) | 164 (74) | 120 (62) | 126 (63) | 602 (69) |
| 2-Case clusters | 18 (13) | 12 (10) | 25 (11) | 32 (16) | 25 (13) | 112 (13) |
| 3–5-Case outbreaks | 11 (8) | 10 (8) | 17 (8) | 21 (11) | 23 (12) | 82 (9) |
| 6–24-Case outbreaks | 7 (5) | 6 (5) | 15 (7) | 15 (8) | 20 (10) | 63 (7) |
| 25–49-Case outbreaks | 0 (0) | 2 (2) | 0 (0) | 3 (2) | 1 (1) | 6 (1) |
| ≥50-Case outbreaks | 0 (0) | 0 (0) | 0 (0) | 3 (2) | 4 (2) | 7 (1) |
| Cluster duration, no. (%) | ||||||
| 0 Generationse | 104 (76) | 95 (79) | 170 (77) | 126 (65) | 134 (67) | 629 (72) |
| 1 Generation | 20 (15) | 9 (7) | 22 (10) | 23 (12) | 20 (10) | 94 (11) |
| 2 Generations | 4 (3) | 7 (6) | 19 (9) | 24 (12) | 23 (12) | 77 (9) |
| 3–5 Generations | 8 (6) | 8 (7) | 8 (4) | 16 (8) | 14 (7) | 54 (6) |
| ≥6 Generations | 1 (1) | 2 (2) | 2 (1) | 5 (3) | 8 (4) | 18 (2) |
| Genotyped, no. (%) | ||||||
| Isolated or single cases | 16 (16) | 24 (26) | 66 (40) | 62 (52) | 91 (72) | 259 (43) |
| 2-Case clusters | 6 (33) | 7 (58) | 17 (68) | 26 (81) | 22 (88) | 78 (70) |
| 3–5-Case outbreaks | 5 (45) | 5 (50) | 15 (88) | 20 (95) | 22 (96) | 67 (82) |
| ≥6-Case outbreaks | 6 (86) | 8 (100) | 14 (93) | 19 (90) | 25 (100) | 72 (95) |
| All measles clusters | 33 (24) | 44 (36) | 112 (51) | 127 (66) | 160 (80) | 476 (55) |
| Genotypes identified | B3, D3, D4, D5, D6, D7, D8, H1, H2 | B3, D4, D5, D6, D8, D9, H1 | B3, D4, D8, D9, G3, H1 | B3, D4, D8, D9, H1 | B3, D4, D8, H1 | B3, D3, D4, D5, D6, D7, D8, D9, G3, H1, H2 |
International importations are those arising in persons who acquired measles outside the United States and brought their infection into the United States.
Includes outbreaks associated with >1 importation; 36 (23%) of 158 outbreaks were associated with >1 importation (median, 3 importations per outbreak; range 2–13).
An outbreak is defined as ≥3 epidemiologically linked cases
Measles infection clusters include isolated or single cases, 2-case clusters, and outbreak of ≥3 cases.
Single cases were assigned a transmission duration of 0 days. Transmissions lasting ≤6, 7–14, or 15–24 days were considered to have 0, 1, or 2 generations of spread, respectively; subsequent generations were added every 10 additional days [9].
Measles cases were reported in 48 states and Washington, DC (Figure 1); 5 states (New York, California, Ohio, Washington, and Minnesota) accounted for 2592 cases (67%), 2209 outbreak-related cases (72%), and 62 outbreaks (39%). Seven outbreaks with >50 cases (<5% of outbreaks but 48% of all cases) had a median size of 147 cases (range, 59–697 cases) and a median duration of 3 months (range, 2–11 months) (Table 2). Five of these outbreaks affected known undervaccinated, close-knit communities [16–21], and 79%–100% of these cases occurred in persons who were unvaccinated or had an unknown vaccination status.
Figure 1.
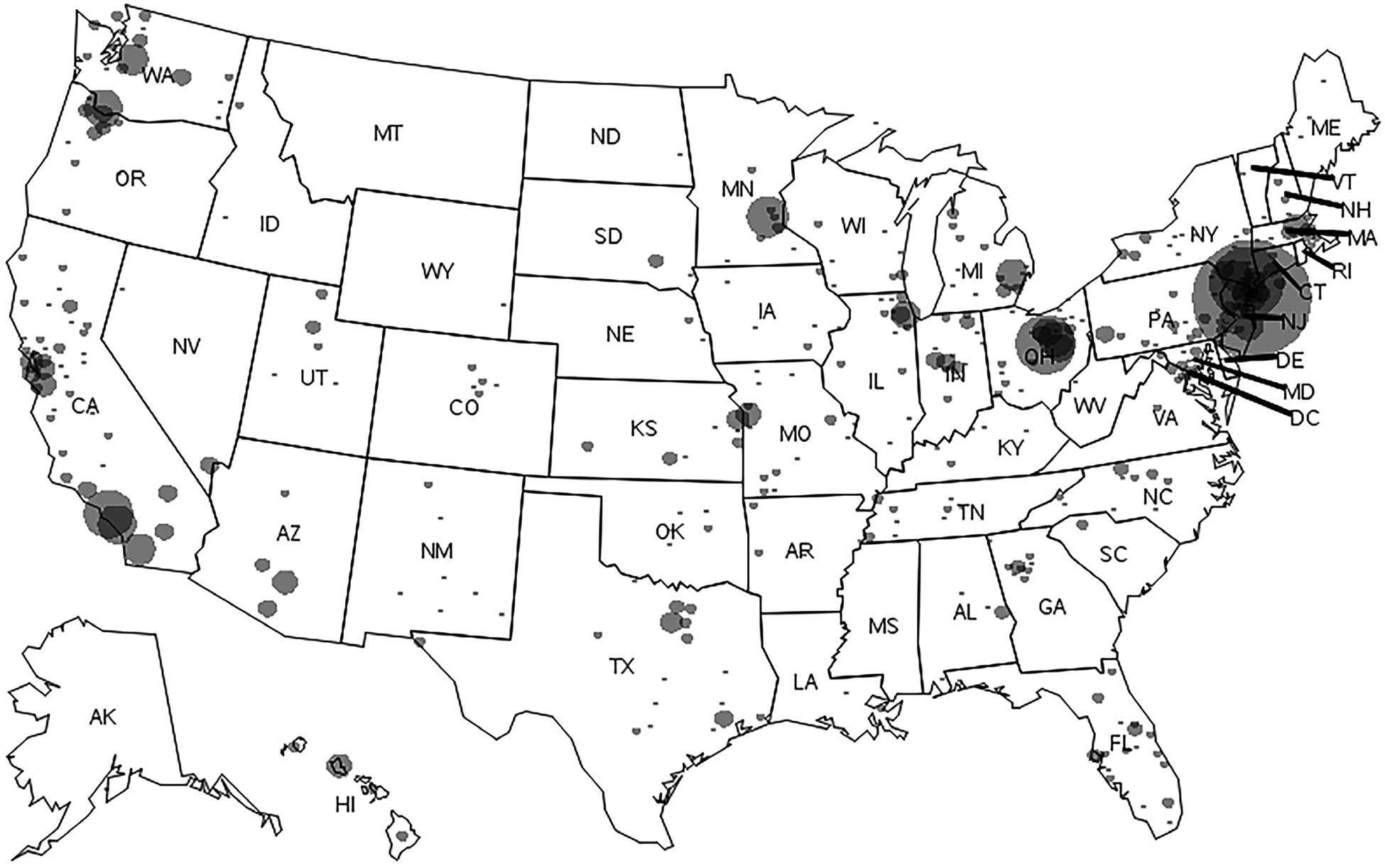
Measles cases by county of residence, United States, 2001–2019. Measles cases have been reported in 48 states, including the District of Columbia (DC), with a range of 1–1352 cases by state. Circle size is proportional to the number of cases. Abbreviations: AK, Alaska; AL, Alabama; AR, Arkansas; AZ, Arizona; CA, California; CO, Colorado; CT, Connecticut; DE, Delaware; FL, Florida; GA, Georgia; HI, Hawaii; IA, Iowa; ID, Idaho; IL, Illinois; IN, Indiana; KS, Kansas; KY, Kentucky; LA, Louisiana; MA, Massachusetts; MD, Maryland; ME, Maine; MI, Michigan; MN, Minnesota; MO, Missouri; MS, Mississippi; MT, Montana; NC, North Carolina; ND, North Dakota; NE, Nebraska; NH, New Hampshire; NJ, New Jersey; NM, New Mexico; NV, Nevada; NY, New York; OH, Ohio; OK, Oklahoma; OR, Oregon; PA, Pennsylvania; RI, Rhode Island; SC, South Carolina; SD, South Dakota; TN, Tennessee; TX, Texas, UT, Utah; VA, Virginia; VT, Vermont; WA, Washington; WI, Wisconsin; WV, West Virginia; WY, Wyoming.
Table 2.
Measles Outbreaks With ≥50 Cases—United States, 2001–2019
| Year | Primary Reporting State/Jurisdiction | Source Country (Genotype) | Outbreak Size, No. of Cases | Outbreak Duration, mo | Unvaccinated or Unknown Vaccination Status, No. (%) | Undervaccinated, Close-Knit Community | Cases From Affected Community, No. (%) |
|---|---|---|---|---|---|---|---|
| 2018–2019 | NYC + 3 states | Israel, UK, Belgium, Ukraine (D8) | 697 | 9.5 | 610 (88) | Yesa | 606 (93)b |
| 2018–2019 | NYS + 1 state | Israel, Ukraine (D8) | 408 | 10.6 | 371 (91) | Yesc | - |
| 2014 | OH | Philippines (D9) | 383 | 4.0 | 378 (99) | Yesd | 380 (99) |
| 2014–2015 | CA + 6 states, Canada, Mexico | Unknown (B3) | 147 | 2.1 | 116 (79) | No | - |
| 2019 | WA + 2 states | Ukraine (D8) | 79 | 2.4 | 74 (94) | Unknowne | - |
| 2017 | MN | Unknown (B3) | 75 | 3.4 | 69 (92) | Yesf | 61 (81) |
| 2013 | NYC + 1 state | UK (D8) | 59 | 2.9 | 59 (100) | Yesa | 58 (100)g |
Abbreviations: CA, California; MN, Minnesota; NYC, New York City; NYS (New York State, outside NYC); OH, Ohio; WA, Washington; UK, United Kingdom.
Orthodox Jewish community; ≥1-dose measles, mumps, and rubella (MMR) vaccine coverage among children aged 12–59 months was 79.5% in the Williamsburg area of Brooklyn in 2018 [20].
Based on the 649 cases of measles reported by NYC [20].
Orthodox Jewish community; measles vaccination coverage was 77% in schools in the outbreak area in 2018 [19].
Amish community; measles vaccination coverage with ≥1 dose was 14% in affected Amish households in 2014, and 59.3% of parents reported that they did not give their child any vaccines, based on a survey in Holmes county in 2007 [18, 21].
MMR vaccination coverage was 10%–14% lower than the statewide average of 88% in Clark county since 2013 [17].
Somali-Minnesotan community; MMR vaccination coverage among 24-month-old Somali Minnesotan children statewide was 42% in 2016 [16].
Based on the 58 cases of measles reported by NYC [28].
Estimates of R for measles across postelimination years were 0.78 (95% confidence interval, .72–.83; k = 0.28), and 0.74 (.71–.77), based on the distribution of cluster sizes and the Wallinga-Teunis method, respectively; Ri was 0.60 (95% confidence interval, .55–.66). R and Ri varied from year to year, with higher estimates noted in more recent years (Figure 2). R point estimates remained <1 throughout the years, while Ri point estimates were >1 in 2013 and 2018.
Figure 2.
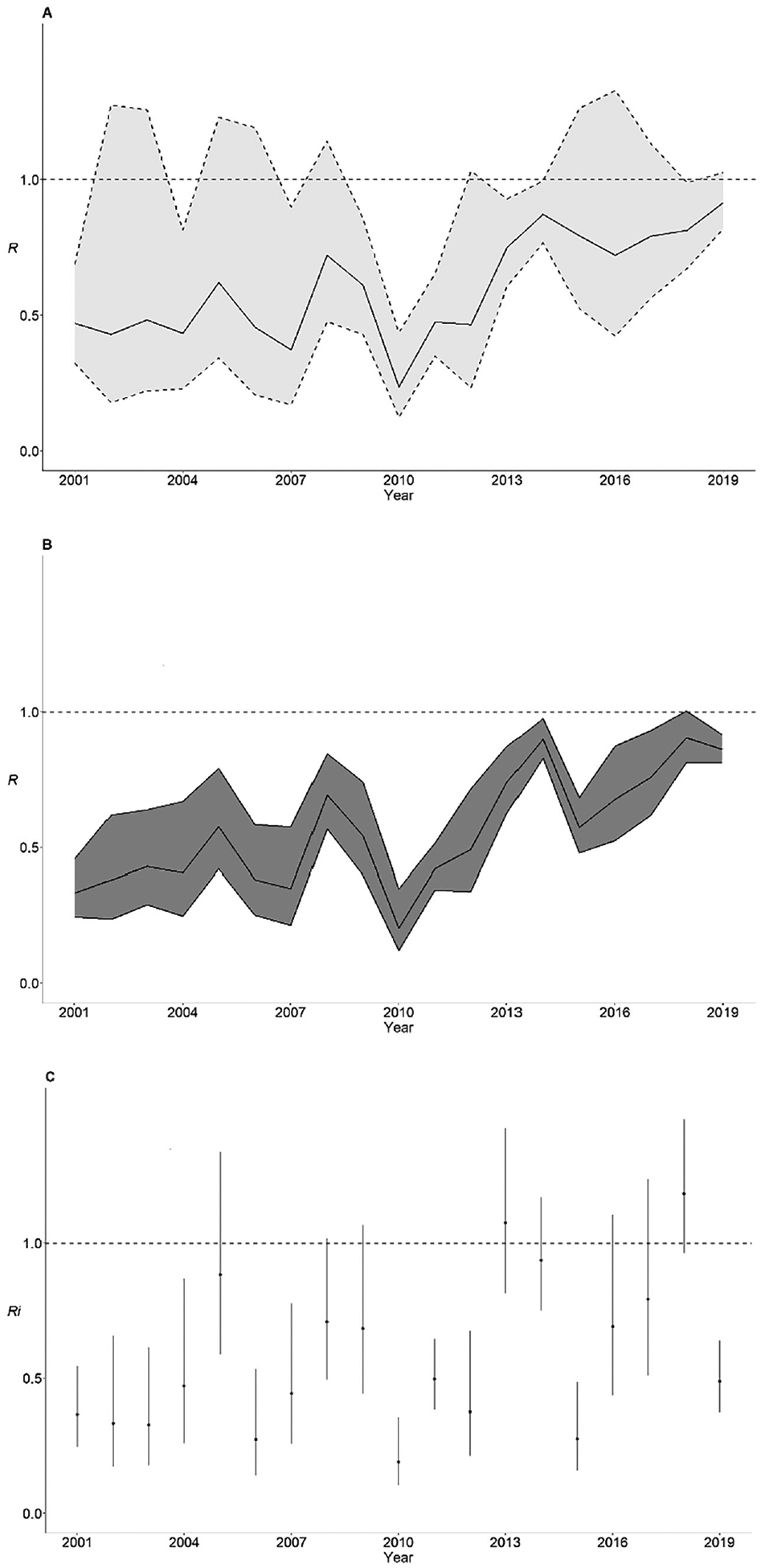
Estimates of the effective reproduction number (R) for measles by year in the United States, 2001–2019. A, Estimates of R according to the distribution of cluster sizes. B, C, Estimates of R for all cases (B) and index cases (Ri) (C), according to the Wallinga-Teunis method. Shaded light gray areas represent 95% confidence intervals obtained by likelihood profiling; shaded dark gray areas and vertical bars, 95% confidence intervals given by the quantiles of the distribution of reproduction numbers; and horizontal dashed lines, threshold value R = 1 (elimination is demonstrated by maintenance of R < 1 and is accomplished by keeping high levels of population immunity). For estimation of R based on the distribution of cluster sizes, cases in clusters spanning 2 years were divided based on their year of rash onset. For estimation of R and Ri based on the Wallinga-Teunis method, the algorithm assigns singleton cases an R value of 0.
Two outbreaks among related Orthodox Jewish communities in New York City and New York State, beginning 30 September 2018, involved 18 separate importations (14 from Israel and 1 each from Ukraine, the United Kingdom, United Kingdom/Belgium, and Ukraine/Israel, with rash onsets between September 2018 and April 2019) and 1105 cases (29% of cases reported during 2001–2019) (Figure 3). The outbreaks spread to Orthodox Jewish communities in 4 other jurisdictions. Because direct epidemiological linkages between the majority of these cases could not be ascertained (51% of cases lacked a direct known source of infection), the duration of individual chains of transmission were unknown; however, transmission ended 10.6 months after rash onset of the first case. The value of R among cases associated with the 2018–2019 New York outbreaks showed a quick decline, from a maximum value of 6.2 during the first week of transmission to an average value of 2.0 in the following 2 weeks (Figure 4). Subsequently, R fluctuated around 1 for the next 10 months, until transmission ended in mid-August 2019.
Figure 3.
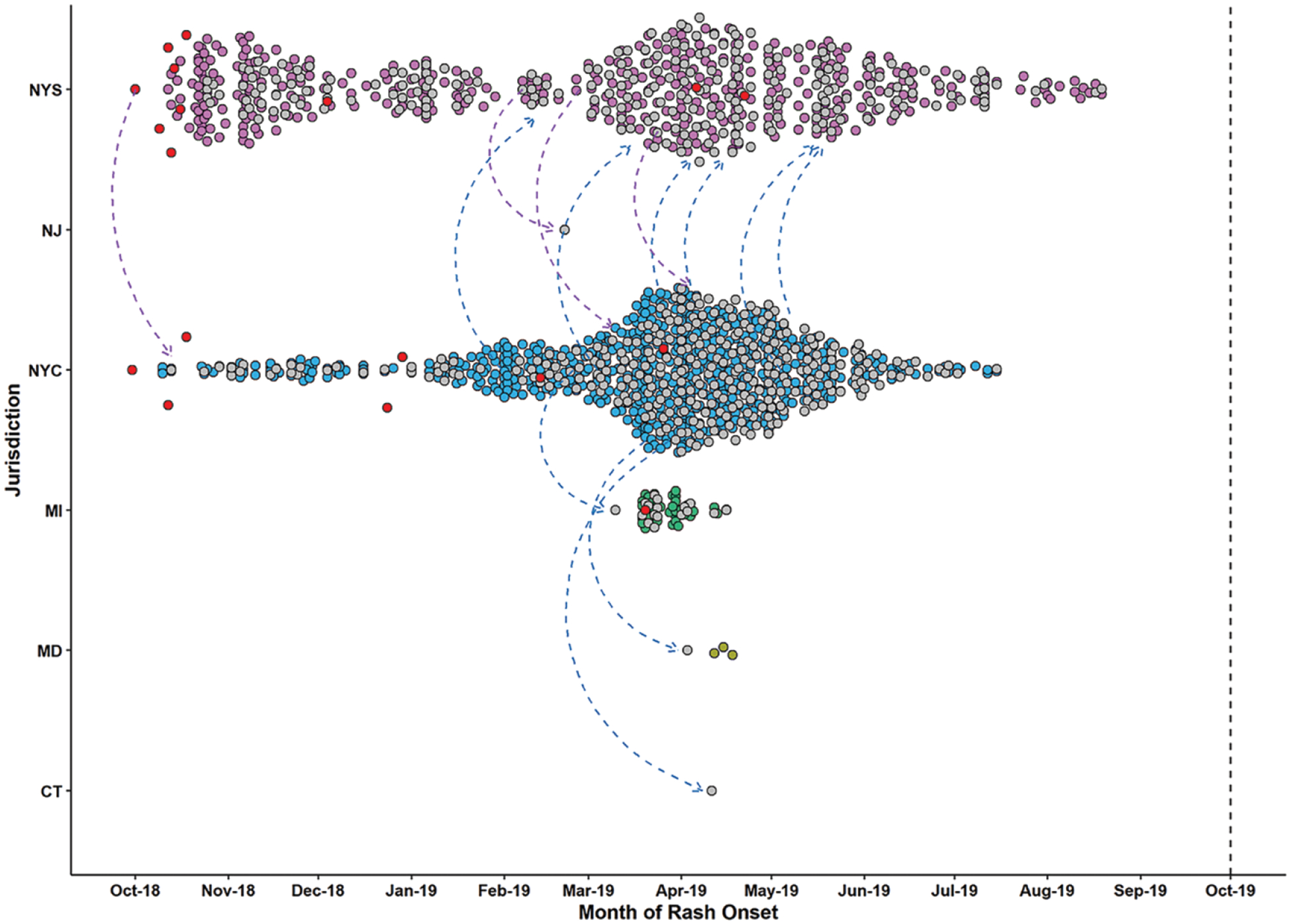
Measles transmission in the United States during the 2018–2019 New York State (NYS) and New York City (NYC) outbreaks. Included are all cases known to be related to these outbreaks (1105 cases from 6 jurisdictions: New York City [649 cases], New York State [407 cases], Michigan [MI; 42 cases], Maryland [MD; 5 cases], Connecticut [CT; 1 case], and New Jersey [NJ; 1 case]; 2 cases [1 from New York State and 1 from Maryland] are not pictured because they did not have information on the date of rash onset). Each case is represented by a circle and plotted based on the reporting jurisdiction (y-axis) and the rash onset date (x-axis). Cases reported by distinct jurisdictions are shown in separate colors. Internationally imported cases are shown in red (14 from Israel and 1 each from Ukraine, the United Kingdom, United Kingdom/Belgium, and Ukraine/Israel). Gray circles represent cases with an unknown source (ie, cases without a direct epidemiological link to another case); arrows, known or potential exportation of measles from one jurisdiction to another; vertical dashed line, date for 12 months of continuous transmission.
Figure 4.
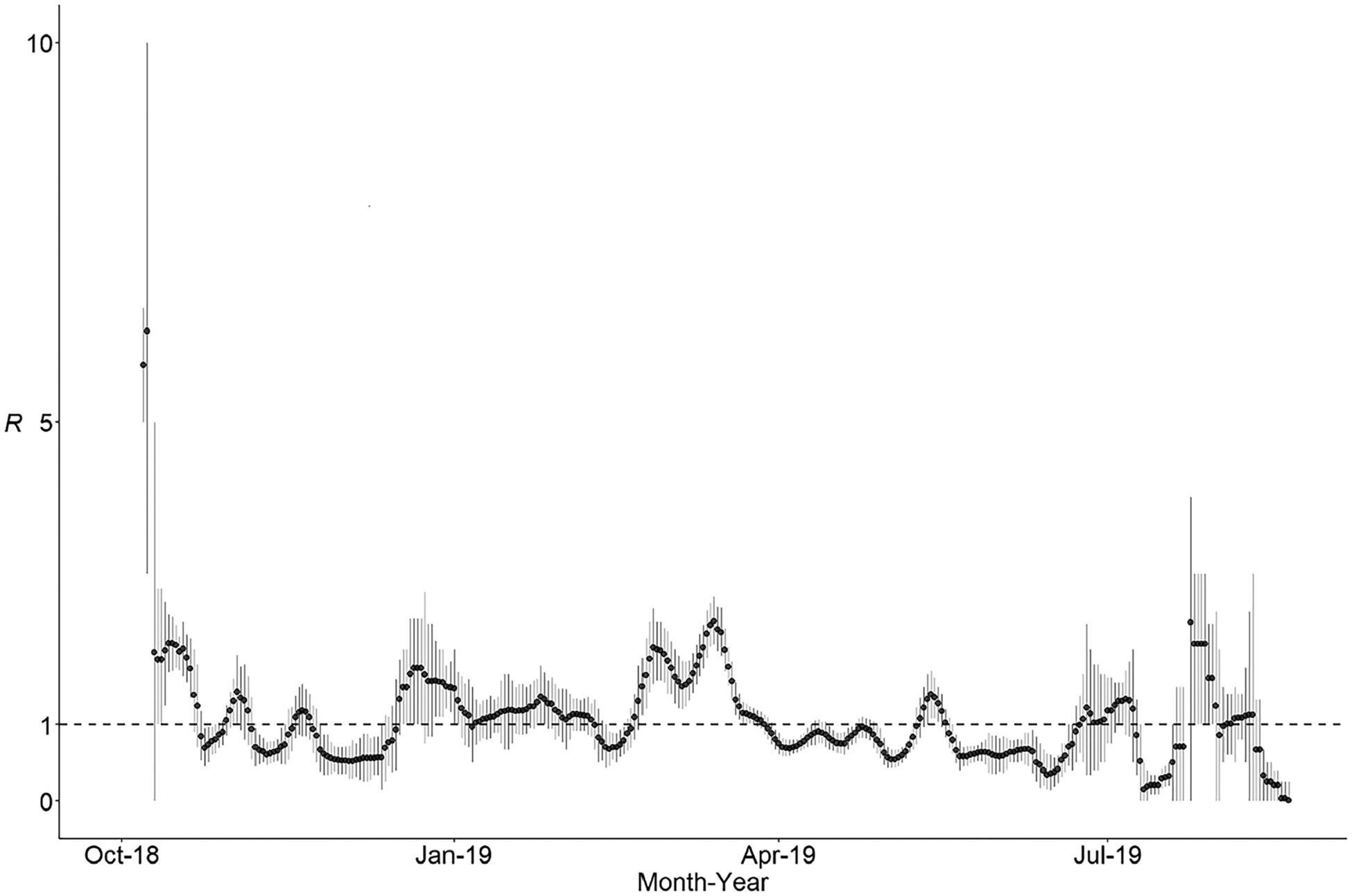
Daily estimates of the effective reproduction number (R) during the 2018 and 2019 New York outbreaks. Calculations were based on all cases known to be related to these outbreaks; a total of 1105 cases from 6 jurisdictions: New York City (649 cases), New York State (407 cases), Michigan (42 cases), Maryland (5 cases), Connecticut (1 case), and New Jersey (1 case). Two cases (1 from New York State and 1 from Maryland) were not included in this analysis because they did not have information on the date of rash onset. Black circles show the mean estimate; vertical bars, 0.25 and 0.75 quantiles; and horizontal dashed line, the threshold value R = 1 (control of an outbreak is demonstrated by maintenance of R < 1). Estimates are presented over sliding 7-day windows, with the estimates plotted on the last day of the window. Here we accounted for the 18 separate importations associated with these outbreaks in the calculation of R; importations could not be attributed to any of the prior cases but could be the source of any subsequent case.
From 30 September 2018 to 31 December 2019, genotyping data were obtained from 821 of 1512 total cases (54%) and 42 of 43 2-case clusters and outbreaks (98%). Two measles virus genotypes were identified: D8 in 763 genotyped cases (93%) and B3 in 58 (7%); 34 unique N-450 sequences for genotype D8 and 13 unique N-450 sequences for genotype B3 were reported (Supplementary Figure 1). The most frequent genotype D8 sequence (71% of D8 specimens), identical to the named strain MVs/Gir Somnath.IND/42.16, was identified in 50 separate clusters throughout the United States, including the New York outbreaks (Figure 5), and in 14 of the 15 genotyped importations related to these outbreaks. During the same period, viruses with N-450 sequences identical to MVs/Gir Somnath.IND/42.16 were detected in Israel, Ukraine, Belgium, and the United Kingdom (MeaNS; www.who-measles.org). From 19 August 2019 (the rash onset of the last case in New York State) to 31 December 2019, identical MVs/Gir Somnath. IND/42.16 sequences were detected in 8 unique clusters, 5 in which the importation was identified and 3 with an unknown source. The unknown source clusters began >42 days after the last case in the New York outbreaks, and the index and any subsequent cases were not associated with travel to or connection with affected communities in New York.
Figure 5.
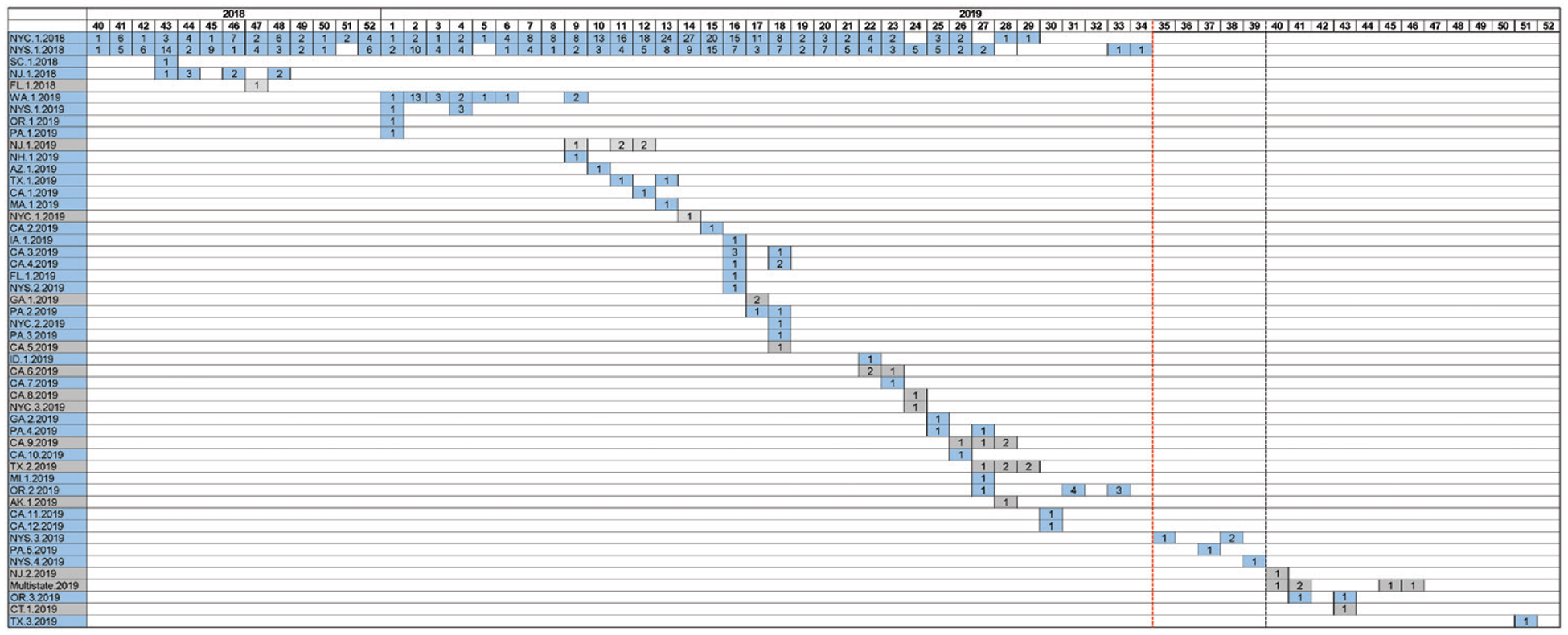
MVs/Gir Somnath.IND/42.16 D8 measles virus sequences identified in the United States, from 30 September 2018 to 31 December 2019 (n = 545). The numbers shown within individual squares indicate the total number of cases in which the named strain, MVs/Gir Somnath.IND/42.16, was identified, plotted based on the reported epidemiological week and year (columns) and the individual infection cluster (rows). Individual clusters are labeled using the reporting jurisdiction letter abbreviations, a chronological number per reporting jurisdiction, and the year in which the cluster started. Clusters with known sources (ie, in which an importation was identified) are shown in blue, and clusters with no known source are shown in gray. The red vertical dashed line represents the date of rash onset for the last case in New York State; the black vertical dashed line, the date that outbreak was declared to be over (ie, 42 days or 2 maximum incubation periods after the date of rash onset for the last case). Eight infection clusters were identified after the last case in the New York State outbreak; 3 did not have a known source (2 single cases and a 12-case multistate outbreak). Abbreviations: AK, Alaska; AZ, Arizona; CA, California; CT, Connecticut; FL, Florida; GA, Georgia; IA, Iowa; ID, Idaho; MA, Massachusetts; MI, Michigan; NH, New Hampshire; NJ, New Jersey; NYC, New York City; NYS, New York State (outside New York City); OR, Oregon; PA, Pennsylvania; SC, South Carolina; TX, Texas; WA, Washington.
Instances in which cases were missed, indicated by a gap in case detection within infection clusters of >21 days (1 maximum incubation period), were observed in <3% of clusters (Supplementary Data). Evaluation of unknown source cases based on time and space did not reveal the possibility of an unrecognized endemic transmission chain (Supplementary Table 1).
DISCUSSION
The current epidemiological features of measles in the United States support the conclusion that measles elimination has been sustained in the country for 20 years. First, <30% of importations were associated with outbreaks. Second, most measles clusters were small and of short duration; approximately 70% were single cases or had no spread. Third, larger outbreaks of longer duration were primarily circumscribed to a few underimmunized communities [16–20]. Fourth, epidemiological and molecular data indicated no lineage circulated in a transmission chain for >12 months. Finally, R remained <1 throughout postelimination years. These results show that measles virus transmission is not self-sustaining in settings where 2-dose vaccination coverage is high and mitigation is promptly implemented [8]. Of note, the paucity of measles since 2020, following coronavirus disease 2019 (COVID-19)–related travel restrictions, provides additional evidence of the lack of endemic measles virus transmission in the United States (only 15 cases were reported from January 2020 to August 2021).
There are, nevertheless, indications of increased measles virus transmission in the United States through 2019. The proportion of importations that have led to outbreaks has been higher, the distribution of cluster sizes and durations have tended toward larger and longer chains, and the largest outbreaks (>50 cases) all occurred in the last 7 years of the study period. These changes are reflected in increases in R over time. While concurrent increases in the number of importations could explain some increased transmission, the number of United States–acquired cases (ie, cases not associated with travel with exposure occurring in the United States) has increased at a relatively faster rate, suggesting more transmissions per measles introduction.
Vaccine coverage for measles has remained high in the United States—1- and 2-dose immunization rates have been ≥90% since 1996 and 2010, respectively [22, 24]—yet the US measles immunity profile is not homogenous, and national coverage does not capture local areas of underimmunization which allow for measles virus transmission. Heterogeneity in transmission is evident by the small number of outbreaks (n = 7) that accounted for half of all reported cases, and supported by the low value of k. Recent increased transmissibility might imply that the size of underimmunized communities could be growing from an accumulation of susceptible persons (eg, in Minnesota, MMR vaccine coverage among 24-month-old Somali children declined from >90% in 2004 to 42% in 2016) [16], although it is also possible that recent increases in importations are only revealing these already vulnerable communities more often (ie, R is biased upward by the chance occurrence of importations landing in larger susceptible pockets in recent years).
The occurrence of larger outbreaks could also be related to multiple importations into a locality, increasing the potential for spread, or to delays in the recognition of measles and implementation of control efforts. These possibilities underscore the need for a decisive approach to offset misinformation and increase confidence in vaccines, as well as to counteract missed opportunities and poor access to vaccination to close immunity gaps [4, 25]. Importantly, past measles outbreaks provide a road map to underimmunized subpopulations for targeted interventions [26]. In fact, prior (albeit smaller) measles outbreaks had occurred in a few of the same communities affected by recent large outbreaks [27, 28]. The utility of serosurveys in elimination settings to complement data from outbreaks and better characterize immunity gaps needs to be further studied [23, 29].
A series of challenges to sustaining measles elimination were encountered in 2019. In New York, despite ongoing transmission and awareness of measles, reticence toward vaccination in a subset of the affected population persisted and broadened into poor acceptance of other mitigation recommendations; importations continued 7 months into the outbreak, and nonadherence to exclusions, “measles parties,” and avoidance of medical care were reported [20]. As a result, and owing to the magnitude of the outbreak, direct epidemiological links between cases were incompletely ascertained, impeding control efforts. Prolonged transmission permitted repeated spread to related communities in neighboring jurisdictions.
The US measles experience also uncovered other key aspects related to sustaining elimination. WHO defines reestablishment of endemic transmission as the “presence of a chain of transmission that continues uninterrupted for ≥12 months in a defined geographical area where measles had previously been eliminated” [30]. Interestingly, elimination might lapse not because a community size that perpetuates transmission is reached, but rather when mitigation is insufficient to halt transmission, keeping R near unity, and leading to a steady occurrence of a low number of cases [31]. Likewise, endemic transmission could occur in a confined geographic area, as opposed to transmission across a region or country due to more homogenous underimmunization; in the United States, sustained transmission was mostly circumscribed to specific areas in 1 county (Rockland) and 2 neighborhoods (Williamsburg and Borough Park, Brooklyn) [19, 20]. Furthermore, documenting the duration of transmission when multiple importations occur into a community is extremely challenging. This requires countries to discern each transmission chain by identifying individual links between locally acquired cases; otherwise, a worst case-scenario is assumed (ie, that all local cases arose from the earliest importation).
In New York, incomplete data on individual links blurred the actual number and duration of the transmission chains that composed the outbreak (the longest chain might have been considerably <11 months). Finally, sequencing N-450 has proved insufficient to map transmission pathways, and this has been exacerbated by a decline in the number of circulating measles genotypes globally [15]. Specifically, we show how the same N-450 sequence was introduced multiple times from multiple sources, so identical N-450 sequences do not necessarily indicate a single transmission chain. It is often possible to distinguish between viruses that share the same N-450 sequence by comparing the results of whole genome sequencing; therefore, efforts to sequence larger genome segments might prove helpful [32–35], yet we show that multiple importations from the same endemic setting can occur over a short period of time, and the variation in imported sequences could be limited.
Several limitations should be considered. Some underreporting is expected, yet US surveillance consistently detects a range of cluster sizes, including isolated cases, so sustained transmission is unlikely to go undetected. Furthermore, linkage of unknown source clusters based on time and space did not reveal the possibility of endemic chains. A sensitive surveillance system is particularly important at the end of outbreaks to rule out continued transmission. In the United States, an outbreak is considered to have ended if there are no additional outbreak-related cases for ≥2 maximum incubation periods (ie, 42 days) after rash onset in the last case, under enhanced surveillance (including monitoring of susceptible contacts and increased awareness of measles in the community and among healthcare providers). As an example, several cases with sequences identical to MVs/Gir Somnath.IND/42.16 were identified following the New York outbreaks, both within and outside affected jurisdictions. These cases were explained by separate importations or had no connection to the affected community and presented after these outbreaks were declared over. Gaps in measles surveillance are rarely noted in the United States; 2% of clusters had >1 and none had >2 maximum incubation periods elapse between cases. In calculating annual R, we assumed that clusters were single transmission chains, which might overestimate R [36, 37]. However, the majority of US clusters arise from single importations, and overall, the number of cases generated per importation has increased over time.
In summary, epidemiological and laboratory data support the maintenance of measles elimination in the United States Recent increases in the transmission of measles underscore the importance of equitable high immunization coverage, the need to strengthen confidence in vaccination and close existing immunity gaps, and the importance of preparedness for effective mitigation, in preserving elimination. These are critical issues of particular relevance in the midst of the COVID-19 pandemic, which is disrupting immunization systems here [38, 39] and elsewhere [40].
Supplementary Material
Acknowledgments.
The authors thank the epidemiology, clinical, and laboratory staff at local and state health departments throughout the United States, who collect these data, and staff at the Vaccine Preventable Disease Reference Centers of the Association of Public Health Laboratories, who conducted measles genotyping and sequencing.
Footnotes
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Dixon MG, Ferrari M, Antoni S, et al. Progress toward regional measles elimination - worldwide, 2000–2020. MMWR Morb Mortal Wkly Rep 2021; 70:1563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee AD, Clemmons NS, Patel M, Gastañaduy PA. International importations of measles virus into the United States during the postelimination era, 2001–2016. J Infect Dis 2019; 219:1616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemmons NS, Wallace GS, Patel M, Gastanaduy PA. Incidence of measles in the United States, 2001–2015. JAMA 2017; 318:1279–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel M, Lee AD, Clemmons NS, et al. National update on measles cases and outbreaks—United States, January 1-October 1, 2019. MMWR Morb Mortal Wkly Rep 2019; 68:893–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truelove SA, Graham M, Moss WJ, Metcalf CJE, Ferrari MJ, Lessler J. Characterizing the impact of spatial clustering of susceptibility for measles elimination. Vaccine 2019; 37:732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wise J MMR vaccine: Johnson urges new impetus to increase uptake as UK loses measles-free status. BMJ 2019; 366:l5219. [DOI] [PubMed] [Google Scholar]
- 7.Gastanaduy P, Redd S, Clemmons N, et al. Manual for the surveillance of vaccine-preventable diseases. Chapter 7: Measles. Available at: https://www.cdc.gov/vaccines/pubs/surv-manual/chpt07-measles.html. Accessed 6 May 2021. [Google Scholar]
- 8.Gastanaduy PA, Paul P, Fiebelkorn AP, et al. Assessment of the status of measles elimination in the United States, 2001–2014. Am J Epidemiol 2017; 185:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumberg S, Funk S, Pulliam JR. Detecting differential transmissibilities that affect the size of self-limited outbreaks. PLoS Pathog 2014; 10:e1004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gastanaduy PA, Funk S, Lopman BA, et al. Factors associated with measles transmission in the United States during the postelimination era. JAMA Pediatr 2020; 174:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol 2013; 178:1505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol 2004; 160:509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klinkenberg D, Nishiura H. The correlation between infectivity and incubation period of measles, estimated from households with two cases. J Theor Biol 2011; 284:52–60. [DOI] [PubMed] [Google Scholar]
- 14.Bankamp B, Byrd-Leotis LA, Lopareva EN, et al. Improving molecular tools for global surveillance of measles virus. J Clin Virol 2013; 58:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown KE, Rota PA, Goodson JL, et al. Genetic characterization of measles and rubella viruses detected through global measles and rubella elimination surveillance, 2016–2018. MMWR Morb Mortal Wkly Rep 2019; 68:587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee E, Griffith J, Kenyon C, et al. Containing a measles outbreak in Minnesota, 2017: methods and challenges. Perspect Public Health 2020; 140:162–71. [DOI] [PubMed] [Google Scholar]
- 17.Carlson A, Riethman M, Gastanaduy P, et al. Notes from the field: community outbreak of measles—Clark County, Washington, 2018–2019. MMWR Morb Mortal Wkly Rep 2019; 68:446–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gastanaduy PA, Budd J, Fisher N, et al. A measles outbreak in an underimmunized Amish community in Ohio. N Engl J Med 2016; 375:1343–54. [DOI] [PubMed] [Google Scholar]
- 19.McDonald R, Ruppert PS, Souto M, et al. Notes from the field: measles outbreaks from imported cases in Orthodox Jewish Communities—New York and New Jersey, 2018–2019. MMWR Morb Mortal Wkly Rep 2019; 68:444–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zucker JR, Rosen JB, Iwamoto M, et al. Consequences of undervaccination—measles outbreak, New York City, 2018–2019. N Engl J Med 2020; 382:1009–17. [DOI] [PubMed] [Google Scholar]
- 21.Scott EM, Stein R, Brown MF, Hershberger J, Scott EM, Wenger OK. Vaccination patterns of the northeast Ohio Amish revisited. Vaccine 2021; 39:1058–63. [DOI] [PubMed] [Google Scholar]
- 22.Hill HA, Elam-Evans LD, Yankey D, Singleton JA, Kang Y. Vaccination coverage among children aged 19–35 months—United States, 2017. MMWR Morb Mortal Wkly Rep 2018; 67:1123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariyarajah A, Crowcroft NS. Measles serosurveys: a solution in search of the right problem. Front Public Health 2021; 9:539325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney CG, Zhou F, Singleton J, Schuchat A; Centers for Disease Control and Prevention. Benefits from immunization during the vaccines for children program era—United States, 1994–2013. MMWR Morb Mortal Wkly Rep 2014; 63:352–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Mbaeyi S, Cohn A, Messonnier N. A call to action: strengthening vaccine confidence in the United States. Pediatrics 2020; 145:e20200390. [DOI] [PubMed] [Google Scholar]
- 26.Durrheim DN. Measles elimination—using outbreaks to identify and close immunity gaps. N Engl J Med 2016; 375:1392–3. [DOI] [PubMed] [Google Scholar]
- 27.Gahr P, DeVries AS, Wallace G, et al. An outbreak of measles in an undervaccinated community. Pediatrics 2014; 134:e220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen JB, Arciuolo RJ, Khawja AM, Fu J, Giancotti FR, Zucker JR. Public health consequences of a 2013 measles outbreak in New York City. JAMA Pediatr 2018; 172:811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durrheim DN. Measles virus is unforgiving where immunity gaps exist. J Infect Dis 2017; 216:1183–4. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. Monitoring progress towards measles elimination. Wkly Epidemiol Rec 2010; 85:490–4. [PubMed] [Google Scholar]
- 31.Gastanaduy PA, Funk S, Paul P, et al. Impact of public health responses during a measles outbreak in an Amish community in Ohio: modeling the dynamics of transmission. Am J Epidemiol 2018; 187:2002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardy JL, Naus M, Amlani A, et al. Whole-genome sequencing of measles virus genotypes H1 and D8 during outbreaks of infection following the 2010 Olympic winter games reveals viral transmission routes. J Infect Dis 2015; 212:1574–8. [DOI] [PubMed] [Google Scholar]
- 33.Harvala H, Wiman A, Wallensten A, Zakikhany K, Englund H, Brytting M. Role of sequencing the measles virus hemagglutinin gene and hypervariable region in the measles outbreak investigations in Sweden during 2013–2014. J Infect Dis 2016; 213:592–9. [DOI] [PubMed] [Google Scholar]
- 34.Penedos AR, Myers R, Hadef B, Aladin F, Brown KE. Assessment of the utility of whole genome sequencing of measles virus in the characterisation of outbreaks. PLoS One 2015; 10:e0143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas S, Hiebert J, Gubbay JB, et al. Measles outbreak with unique virus genotyping, Ontario, Canada, 2015. Emerg Infect Dis 2017; 23:1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blumberg S, Lloyd-Smith JO. Inference of R(0) and transmission heterogeneity from the size distribution of stuttering chains. PLoS Comput Biol 2013; 9:e1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson RN, Stockwin JE, van Gaalen RD, et al. Improved inference of time-varying reproduction numbers during infectious disease outbreaks. Epidemics 2019; 29:100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santoli JM, Lindley MC, DeSilva MB, et al. Effects of the COVID-19 pandemic on routine pediatric vaccine ordering and administration—United States, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:591–3. [DOI] [PubMed] [Google Scholar]
- 39.Patel Murthy B, Zell E, Kirtland K, et al. Impact of the COVID-19 pandemic on administration of selected routine childhood and adolescent vaccinations—10 U.S. jurisdictions, March-September 2020. MMWR Morb Mortal Wkly Rep 2021; 70:840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson R COVID-19 disrupts vaccine delivery. Lancet Infect Dis 2020; 20:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


