Abstract
The enhancer within the long terminal repeats (LTRs) of acquired somatic mouse mammary tumor viruses (MMTV) can activate juxtaposed genes and induce mammary tumors. In contrast, germ line proviral MMTV genomes are integrated in the host genome and considered to be genetically confined transcription units. Here we demonstrate that transcription initiated in an MMTV provirus proceeds into flanking host sequences. We discovered multiple polyadenylated transcripts which are induced in Stat5a null mice. These range from 1.5 kb to more than 8 kb and are specifically expressed in mammary tissue from pregnant and lactating mice from the 129 but not C57BL/6 strain. The RNAs emanate from both LTRs of the endogenous MTV-3 provirus on chromosome 11 and proceed at least 10 kb into the juxtaposed genomic territory. Transcripts originating in the 5′ LTR splice from the native splice site within the MMTV envelope gene into at least six exons, three of which contain functional internal splice sites. The combination of alternative splicing and the use of several polyadenylation sites ensure the generation of multiple transcripts. To date no significant open reading frame has been discovered. Furthermore, we demonstrate that transcription from the MMTV 5′ LTR is highly active in the absence of Stat5a, a transcription factor that had been shown previously to be required for transcription from the MMTV LTR.
Mouse mammary tumor virus (MMTV) can be transmitted through milk or through the germ line (1). Although most mouse strains carry several MMTV proviruses, only some transmit an exogenous virus. The nonpathogenic MMTV proviruses are transcribed, but little or no envelope protein is produced. However, the same subtype synthesizes superantigen (SAg) from the viral transcript, which in turn renders the host resistant to infections. Each provirus is characterized by a distinct SAg that interacts with the T-cell receptor Vβ element through its COOH-terminal portion (19). Different mouse strains are characterized by a distinct pattern of proviruses. For example, MTV-3 is found in 129 and GR mice (8, 19) but not in C57BL/6 and BALB/C mice (18).
Transcription of the proviral genome originates within the 5′ long terminal repeat (LTR) and proceeds to a transcriptional stop and polyadenylation site within the 3′ LTR. Both prolactin and glucocorticoids control transcription from the MMTV LTR through distinct regulatory sequences (6, 10, 15, 23, 26). As a result of hormonal stimulation during pregnancy, virus production dramatically increases during lactation (2). Three types of transcripts can be detected: a full-length RNA of approximately 8 kb encoding the gag and pol products, and two shorter transcripts encoding the envelope protein (env) and superantigen (sag), respectively (Fig. 4A). The latter two transcripts are splice products and share the 5′ splice donor site. MMTV lacks an oncogene and probably induces tumors by acting as an insertional mutagen that activates the expression of cellular oncogenes juxtaposed to the insertion site in the host chromosome (16, 17). A mammary-specific enhancer within both LTRs is responsible for the activation of promoters located outside the integration site. In some cases, transcription originating from the 3′ LTR has been linked to the activation of juxtaposed oncogenes.
FIG. 4.
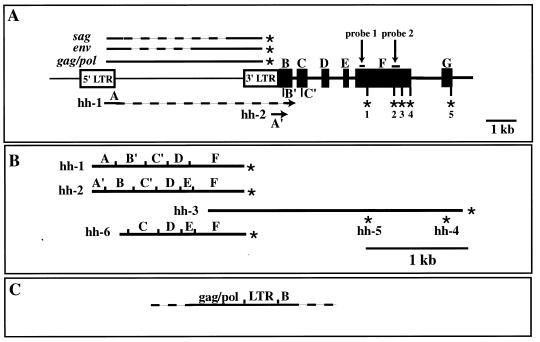
Structure of the hitchhiker locus and its transcripts. (A) Open rectangles represent the 5′ and 3′ LTRs of MTV-3. Exons located 3′ of MTV-3 are shown as solid boxes. B′ and C′ represent internal splice sites used in the transcript hh-1 (B). C′ is also used by hh-2, and C is used by hh-6. EST probes 1 and 2 are indicated (the coordinates of the probes are shown in Materials and Methods). The asterisks in exon F point to the polyadenylation sites used in different transcripts. MMTV transcripts are presented above the genomic structure, and hitchhiker transcripts are shown below. Dashed lines represent sequences removed from the primary transcripts by splicing. (B) Structures of the cDNAs isolated. hh-6 corresponds to an EST from the soars mammary gland NbMMG library. (C) Transcripts that contain sequences from the env gene and the hitchhiker locus. RT-PCR analyses were performed with a primer in exon B followed by nested PCRs (see Materials and Methods). This confirmed the existence of RNAs, which probably start in the 5′ LTR and continue through the MTV-3 genome and the hitchhiker locus, followed by various splicing patterns. These transcripts would account for the Northern signals that are larger than 4 kb and are specifically detected with hitchhiker probes.
The MMTV LTR has been a rich source for discoveries of transcription elements and mechanisms of transcriptional regulation. Transcription originating in the LTR is induced by steroid hormones and is controlled by numerous transcription elements, including binding sites for the glucocorticoid receptor, NF1, Oct proteins, and many others. Transcriptional activation in mammary tissue increases during pregnancy and peaks during lactation, similar to that seen for milk proteins, such as the whey acidic protein (WAP) (3). Transcriptional activation of WAP during pregnancy is controlled by prolactin through the Jak2/Stat5 pathway (13). Mutation of the Stat5a binding site (TTCNNNGAA) within the promoter fully abrogates transcription in transgenic mice (11), and deletion of the Stat5a gene from mice leads to downregulation of WAP (14). It has recently been shown that both Stat5a and Stat5b bind to the MMTV LTR and are required for its activation in mammary tissue in vivo (20).
We have generated mice with an inactive Stat5a gene and established its critical role in mammary gland development and function (12). In a quest to identify genes and signaling pathways that are controlled by the prolactin-Jak2/Stat5 axis (either directly or indirectly), we prepared subtractive cDNA libraries. This approach led us to the discovery of transcripts that originate in both LTRs of the MTV-3 locus, proceed into the juxtaposed host genome, and result in a family of splice and polyadenylation variants. Furthermore, our study provided insight into the capacity of Stat5a to negatively regulate transcription from the MMTV LTRs.
MATERIALS AND METHODS
Mice.
The ES cell line used for gene targeting was derived from the 129SvEv strain. Stat5a-hemizygous (+/−) mice were subsequently bred with C57BL/6 mice, and the Stat5a-null allele was maintained in a mixed background. This is commonly done since 129 mice are poor breeders and have small litter sizes. The MTV-3 locus characterized in this report is present in the 129 but not C57BL/6 strain. It is located on chromosome 11 (70.5 centimorgams [cM]) approximately 10 cM distal to the Stat5a locus. We have observed a recombination frequency of less than 10% between the MTV-3 and Stat5a loci, which explains the cosegregation of the Stat5a-null and MTV-3 alleles.
cDNA subtraction library.
To identify genes overexpressed in Stat5a-null mice at parturition compared to control littermates, we generated a subtractive library. PCR-based subtractive hybridization was performed with the Clontech PCR-Select DNA subtraction kit (Catalog no. K1804-1). Briefly, in this procedure, tester cDNA (containing the cDNAs to be cloned) was synthesized from RNA obtained from mammary tissue of mice homozygous null for Stat5a. Driver cDNA (used in excess as a reference cDNA) was synthesized from RNA from littermates containing two intact copies of the Stat5a gene. After restriction of tester and driver cDNAs with RsaI to obtain shorter, blunt-end molecules, two tester populations with different adapters were created. The driver cDNA had no adapters. Subsequent hybridization steps led to the enrichment of differentially expressed sequences from which templates for PCR amplification were generated. By using suppression PCR, only differentially expressed sequences were amplified exponentially. The background was reduced, and differentially expressed sequences were further enriched in the second amplification step.
Initially, 96 clones were subjected to sequence analysis. Seventy-four clones were discarded because they contained only vector sequence, only linkers used in the cloning process, or only rRNA sequences. The 22 survivors were placed in three groups. Twelve inserts corresponded to known genes, two recognized matches to known expressed sequence tags (ESTs) in GenBank, and eight clones represented new sequences. Additional information on these clones can be obtained online (14a).
RNA isolation and Northern blotting.
To verify the differential expression between Stat5a-null and control mice, RNA blot analyses were performed (experimental details and extensive Northern analyses on 22 genes can be viewed online [14a]). RNA was prepared from mammary tissue of Stat5a-null mice, wild-type littermates, and C57BL/6 mice within 12 h after parturition and analyzed by Northern blotting. Liver RNA was used as a nonmammary control. Total RNA was extracted using the acid guanidinium thiocyanate-phenol-chloroform method (16). Poly(A)+ RNA was isolated by using a Poly(A) Quick mRNA isolation kit (Stratagene). The poly(A)+ RNA was used for both production of the cDNA library and Northern blot analyses. Twenty micrograms of total RNA of each sample was fractionated in a 1.3% formaldehyde gel. The RNA was transferred to GeneScreen Plus nylon membranes and UV cross-linked in a UV-Stratalinker 1800. PCR fragments amplified with specific internal primers for each clone were labeled with [α32P]dCTP, using a Prime-It II kit (Stratagene) according to the manufacturer’s protocol, and used as probes for RNA expression analysis.
Positions of the probes within the hitchhiker locus are shown in Fig. 1E and 4A. We precisely define the probes used in this study by referring to the sequence of hitchhiker locus (accession no. AF120673). Probe 1 was amplified by using an upstream primer, 5′-GGAAATGCCATGTCAACCTCG-3′, corresponding to bp 2606 to 2626 and a downstream primer, 5′-GCTGGGATTTGAACTCAGGG-3′, corresponding to bp 3724 to 3705; probe 2 was amplified by using an upstream primer, 5′-GCTAAGACTCAAAGGTGGGG-3′, corresponding to bp 4947 to 4965 and a downstream primer, 5′-GTACTTCCAGCTCATGTTAGG-3′, corresponding to bp 5487-5467. The probe for analyzing the expression of the sag gene was based on the complete MMTV proviral genomic sequence (accession no. AF033807) and corresponds to bp 7909 to 8209 bp of the reported sequence. Three plasmids isolated as a result of the subtraction procedure and containing three different inserts: probe 1, probe 2, and a sag probe were used as templates for PCRs. For amplification of the sag probe, we used universal M13 5′-GTTTTCCCAGTCACGAC-3′ and 5′-AGCGGATAACAATTTCACACAGGA-3′ primers. The PCR products were gel purified and extracted from the gel by using a Qiagen gel extraction kit according to the manufacturer’s protocol before radiolabeling the probes. WAP and β-casein mRNA levels were analyzed by using specific oligonucleotides labeled with [γ-32P]ATP, using a KinAce-It kit (Stratagene) as previously described (17). Expression of glyceraldehyde-3-phosphate dehydrogenase was analyzed with a 1.3-kb BamHI-HindIII fragment labeled with [α-32P]dCTP. Purification of the probes was performed with STE SELECT-D G-50 columns for the PCR and DNA fragments and G-25 for the oligonucleotide probes. Hybridizations with PCR and DNA fragments were performed at 65°C according to the manufacturer’s protocol for QuickHyb hybridization solution (Stratagene). Oligonucleotides were hybridized at 55°C.
FIG. 1.
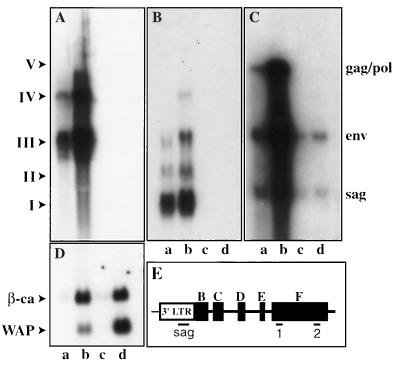
Transcripts from the hitchhiker locus and MMTV proviruses. Total RNA was isolated from mammary tissue of Stat5a-null mice (lanes a and b) and wild-type littermates (lanes c and d) at day 13 of pregnancy (lanes a and c) and at parturition (lanes b and d). The membrane was hybridized with probe 2 located at the 3′ end in exon F (A) and then hybridized with probe 1 located at the 5′ end in exon F (B), with the sag probe located in the 3′ LTR (C), and finally with oligonucleotides specific for WAP and β-casein (β-ca) RNA (D). (E) Positions of probes in the hitchhiker locus. Positions and coordinates of the probes are described in Materials and Methods. I, II, III, IV, and V are the major transcripts derived from the hitchhiker locus. Note that at parturition, WAP RNA is reduced in Stat5a-null mice as described elsewhere (14).
Isolation of cDNA clones containing sequences from probes 1 and 2.
A cDNA library was generated (Edge Bio Systems vector pEAK8) from mRNA isolated from mammary tissue of Stat5a-null mice within 12 h after parturition. Approximately 2 million nonamplified colonies were plated on 40 140-mm-diameter LB-ampicillin plates. The colonies were transferred to colony/plaque screen hybridization transfer membranes, air dried, fixed by UV irradiation, autoclaved for 5 min, and dried for 10 min in a steam autoclave. Hybridizations were performed for 12 h with [α-32P]dCTP-labeled probes 1 and 2 at 65°C overnight. Positive clones were isolated and colony purified in an additional hybridization screen. Individual positive colonies were transferred into LB broth, grown overnight, and used to inoculate 200 ml of terrific broth with 100 μg of ampicillin per ml. Plasmids were isolated by using a Maxi (Qiagen) plasmid purification kit.
Isolation and subcloning of genomic BAC clones.
Probe 2 was used to isolate corresponding bacterial artificial chromosome (BAC) clones from a 129SvJ library (Genome Systems). Hybridization of nylon filters containing individual BAC clones spotted at high density was performed at 65°C according to the protocol of the supplier (Genome Systems). Two BAC clones were isolated, and a restriction map was established by Southern blotting followed by hybridizations with probes 1 and 2. BamHI, HindIII, and EcoRI fragments of both isolated BAC clones were subcloned into the pBluescript II SK (Stratagene) and pZErO-1 (Invitrogen) cloning vectors.
Sequence analysis.
Sequencing of plasmid DNA and PCR products was performed by using a standard protocol for cycle sequencing with the ABI PRISM 310 Genetic Analyzer (Perkin-Elmer). Sequence analysis was performed with the EditView 1.0 and Sequencher 3.0 software packages.
Chromosomal localization.
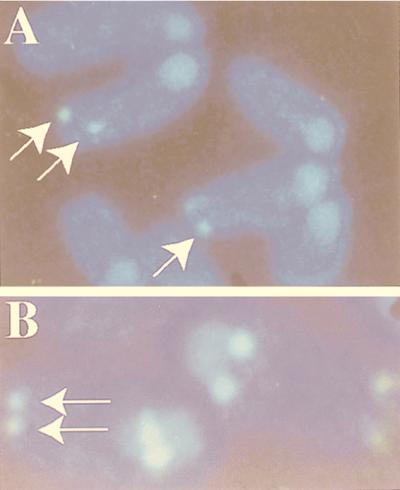
Genome Systems determined the chromosomal localization of the BAC clone. The BAC clone, which hybridized to the hh-1 and hh-2 transcripts of the hitchhiker locus, was used as a probe in a fluorescence in situ hybridization (FISH) analysis. BAC clone DNA was labeled with digoxigenin-dUTP by nick translation. Labeled probe was combined with sheared mouse DNA and hybridized to normal metaphase chromosomes derived from mouse embryo fibroblast cells in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Specific hybridization signals were detected by incubating the hybridized slides in fluorescein-conjugated antidigoxigenin antibodies followed by counterstaining with 4′,6-diamidino-2-phenylindole (DAPI). The initial experiment resulted in specific labeling of the terminus of a medium-sized chromosome, which was believed to be chromosome 11 on the basis of DAPI staining. We conducted a second experiment in which a probe specific for the centromeric region of chromosome 11 was cohybridized with the BAC clone. This experiment resulted in specific labeling of the centromere and the telomeric region of chromosome 11. This demonstrated that the DNA from the BAC clone is located in the telomeric of chromosome 11, in the area that corresponds to band 11E2. Of the total of 80 metaphase cells analyzed, 73 exhibited specific labeling.
RT-PCR.
Reverse transcription (RT) was performed with the Superscript preamplification system for first-strand cDNA synthesis (GIBCO BRL). Five micrograms of total RNA was transcribed with 1 μl of 2 μM primer at 50°C in total volume of 12 μl. Gene-specific primers were used for the hitchhiker locus (RT primer 5′-TGCTGTCTCCGCTTCCACTGG-3′) and for Stat5b (RT primer 5′-CTGGTCCATGTTGGCTGGC-3′). After cDNA synthesis, the RNA was removed with RNase T1. PCR amplifications of 1 μl of cDNA product from the above reactions were performed with gene-specific primers. The hitchhiker fragment was amplified by using 5′ primer 5′-GGCTGCTGCCTCATTAGG-3′ and 3′ primer 5′-GTGTTGTATCCCTGACTGC-3′ in 25 cycles at an annealing temperature of 53°C. The Stat5b fragment was amplified by using 5′ primer 5′-GACACTTGCTTCTGCTGG-3′ and 3′ primer 5′-CAGAGGCTGGTTCGCGAAGCC-3′ in 25 cycles at an annealing temperature of 55′C. PCR products were analyzed by electrophoresis in a 2.5% agarose gel.
The presence of transcripts that originate in the 5′ LTR and proceed through the provirus into the flanking genomic sequences was detected by RT-PCR with a primer located in exon B and reading 5′ (5′-TGCACTTCTGTATCAGG-3′) followed by nested PCR. Four micrograms of total RNA from Stat5a-null mouse mammary tissue at parturition was used to synthesize the first-strand cDNA with the primer located in exon B according to the manufacturer’s protocol for the 5′RACE System for Rapid Amplification of cDNA Ends, version 2.0 (GIBCO BRL). After first-strand cDNA synthesis, the original mRNA template was removed by treatment with RNase Mix (mixture of RNase H, which is specific for RNA-DNA heteroduplex molecules, and RNase T1; GIBCO BRL). Unincorporated deoxynucleoside triphosphates primers, and proteins were separated from the cDNA by using GLASSMAX spin cartridges (GIBCO BRL). Nested PCR amplification was accomplished by using Taq DNA polymerase in 30 cycles at an annealing temperature of 58°C with all possible combinations of the following primers: a primer (5′-GTAAGACAGCATCATGAGATGG-3′) that anneals to a site located in exon B within the cDNA molecule with two primers located 5′ of the 3′ LTR, corresponding to bp 7302 to 7320 (5′-CAGTGCCTTGCGAAGAGCC-3′) and bp 6720 to 6739 (5′-CGAGCTAAGCGATTCGTCGC-3′) of the complete MMTV proviral genomic sequence (accession no. AF033807); and a primer located 5′ of the 3′ LTR, corresponds to bp 7301 to 7319 (5′-GCTCTTCGCAAGGCACTGG-3′), with primer described above corresponding to bp 6720 to 6739 of the complete MMTV proviral genomic sequence (accession no. AF033807). PCR products of expected sizes were analyzed by electrophoresis in a 1.5% agarose gel. Control reactions in the absence of RT did not result in any DNA fragments, demonstrating the absence of contaminating genomic DNA.
Nucleotide sequence accession numbers.
The following accession numbers have been assigned to the sequences submitted to GenBank: AF120673 (genomic sequence of the hitchhiker locus without the MTV-3 insertion), AF118272 (hh-1 cDNA), AF118273 (hh-2 cDNA), AF118558 (cDNAs for hh-3, hh-4, and hh-5), AF118847 (hh-6 cDNA, full-length sequence of EST 1333114), AF119341 (5′ LTR of MTV-3), and AF119342 (3′ LTR of MTV-3).
RESULTS
Isolation of transcripts from Stat5a-null mammary tissue.
In an attempt to isolate genes and identify prolactin controlled signaling pathways in mammary tissue, we prepared subtractive cDNA libraries from Stat5a-null and control mice (9a). A PCR-based subtraction approach was used to clone those mRNAs which are preferentially expressed in mammary tissue from Stat5a-null mice at parturition compared to control mice (see Material and Methods). For this purpose, RNA was prepared from littermates which were either homozygous null for Stat5a or contained two intact copies of the Stat5a gene. The ES cell line containing the targeted Stat5a allele had a 129 genotype, and Stat5a +/− mice were bred with C57BL/6 (see Materials and Methods for the rationale). Initially we identified 96 differentially expressed cDNA clones (see Material and Methods) and further analyzed clones (named probe 1 and probe 2 in subsequent experiments) whose expression appeared to be confined to mammary tissue of Stat5a-null mice (Fig. 1). Probe 1 (see Fig. 1E and 4A for locations of the probes) hybridized to three major transcripts (transcripts I, II, and III) with approximate sizes of 1.5, 2.5, and 4.5 kb (Fig. 1B). Transcripts IV and V were detectable but substantially weaker (Fig. 1B). Probe 2 preferentially hybridized to transcripts III and IV (Fig. 1A). Transcript V was weak but detectable. Neither probe 1 nor 2 hybridized to any transcripts from mammary tissue of C57BL/6 mice and wild-type littermates from the Stat5a-null mice (Fig. 1A and B). A sag probe was used to identify transcripts that contain endogenous MMTV sequences (Fig. 1C). sag, env, and gag/pol transcripts were abundant in mammary tissue from Stat5a-null mice and less abundant in wild-type littermates (Fig. 1C). Hybridization of the blot with probes specific for WAP and β-casein demonstrated that β-casein is expressed at similar levels in Stat5a-null mice and control littermates, whereas expression of WAP is reduced in the absence of Stat5a (14) (Fig. 1D). A tissue survey revealed that the presence of the transcripts was confined to mammary tissue (data not shown). No sequence similarity was detected between probes 1 and 2, and no matching sequence was found in GenBank.
Developmental regulation.
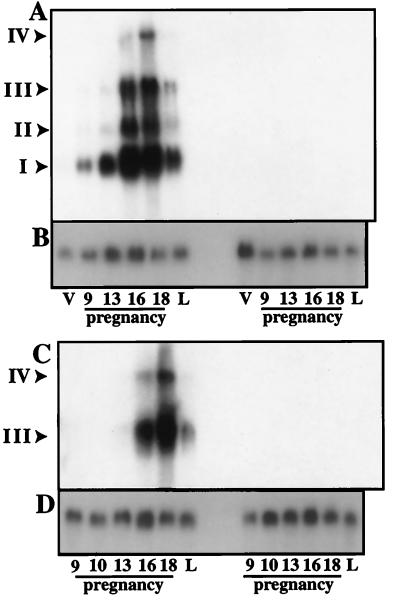
The activities of genes at different stages of mammary gland development provides insight into their transcriptional regulation and potentially into their function. Thus, we determined levels of the transcripts corresponding to probes 1 and 2 during pregnancy and lactation. The developmental patterns obtained with probes 1 and 2 were strikingly similar (Fig. 2). Expression in mammary tissue was low during the early stages of pregnancy but sharply increased around day 13 and further increased until day 18 of pregnancy (Fig. 2). Expression declined dramatically within 1 day after parturition.
FIG. 2.
Expression of RNAs from the hitchhiker locus at different stages of mammary development. Total RNA was isolated from mammary tissue of Stat5a-null (left) and wild-type littermates (right) from the virgin state (V), days 9, 13, 16 and 18 of pregnancy, and at parturition (L). (A) Hybridization with probe 1 located at the 5′ end of exon F (Fig. 1 and 4). (C) Hybridization with probe 2 located at the 3′ end in exon F (Fig. 1 and 4). (B and D) Hybridization with glyceraldehyde-3-phosphate dehydrogenase for normalization purposes. I, II, III, IV, and V are the major transcripts derived from the hitchhiker locus.
Chromosomal localization.
Deletion of genes by homologous recombination results in major territorial changes, including the addition of a transcriptionally active neomycin gene cassette in the targeted locus. To exclude that the novel transcripts were derived from the targeted Stat5a locus, we determined the chromosomal localization of the genes represented by probes 1 and 2. We screened a BAC library with probe 2 and identified two clones. Southern blot analyses showed that the two BAC clones also hybridized to probe 1, suggesting for the first time that probes 1 and 2 were part of one genetic locus. We used one of the BAC clones in a FISH analysis and determined that it hybridized to chromosome 11E (Fig. 3). This region contains a number of genes, including Grb2 and the MTV-3 provirus. The MTV-3 provirus is present in several mouse strains, such as GR and 129, but not in C57/BL6. Since the Stat5 locus resides on chromosome 11E60, approximately 10 cM from the FISH signal, it is unlikely that the new transcripts are the result of the Stat5a deletion.
FIG. 3.
Chromosomal localization of the hitchhiker locus at the telomere of chromosome 11. (A) Hybridization of BAC clone DNA containing the hitchhiker locus to metaphase chromosomes derived from mouse embryo fibroblast cells resulted in specific labeling (arrows) of the distal end of a medium-sized chromosome which was believed to be chromosome 11 on the basis of DAPI staining. (B) The second hybridization of metaphase chromosomes with both the BAC clone DNA containing the hitchhiker locus (specific signals shown by arrows) and a probe which is specific for the centromeric region of chromosome 11 resulted in the specific labeling of the centromere and the telomeric region of chromosome 11.
Transcripts.
In an attempt to define the nucleotide sequences of the novel transcripts and to deduce the amino acid sequence, cDNA libraries were generated from mammary tissue of Stat5a-null mice. Five cDNA clones were isolated with probe 2, and two cDNA clones were isolated with probe 1. Sequence analyses revealed that probes 1 and 2 were derived from transcripts with alternative splice sites, alternative exons, and different polyadenylation sites (Fig. 4A). Based on the sequence of the seven cDNA clones, we predicted the existence of at least seven different transcripts (Table 1).
TABLE 1.
Structures and molecular components of transcripts from the hitchhiker locusa
| Name submitted to GenBank | Size (kb), blot | Exons | Poly(A) site | GenBank accession no. |
|---|---|---|---|---|
| hh-1 | 1.5, I | A, B′, C′, D, F | 1 | AF118272 |
| hh-2 | 1.5, I | A′, B, C′, D, E, F | 1 | AF118273 |
| hh-3 | 4, III | ?, F | 4 | AF118558 |
| hh-4 | 3.8, III | ?, F | 3 | AF118558 |
| hh-5 | 3.4, II | ?, F | 2 | AF118558 |
| hh-6 | 1.5, I | A′, B, C, D, E, F | 1 | AF118847 |
| MTV-3 5′ LTR | AF119341 | |||
| MTV-3 3′ LTR | AF119342 |
Sizes of the transcripts are based on Northern blots (designated I to III) and sequence analyses of the cloned cDNAs. The exon structure and poly(A) sites are shown in Fig. 4.
Clones hh-1 and hh-2 had been isolated with probe 1 and represent the 1.5-kb transcripts. While the 1.5-kb transcripts are the predominant ones, this probe also detected the larger transcripts II, III, IV, and V (Fig. 1B). Sequence analysis revealed that clones hh-1 and hh-2 share several features and exons (Fig. 4A). No open reading frame was detected. The five clones identified with probe 2 span a region of 2.5 kb, and their 5′ part overlaps with the 3′ part of hh-1 and hh-2 (Fig. 4B). Based on the position of their polyadenylation sites, these five clones can be placed in three groups (hh-3, hh-4, and hh-5 in Fig. 4B). None of these cDNAs contained an open reading frame. Since probe 2 detected transcripts of 4 kb and larger, we predict that these clones represent the 3′ part of the message. Sequence analyses revealed that all clones represented novel transcripts, which exhibit distinct identities with other transcripts over specific sequences. The first 250 bp of clone hh-1 match all MMTV LTR-derived transcripts up to the natural MMTV splice site. The first 113 bp of clone hh-2 match the 5′ part of transcripts initiating within the 5′ MMTV LTR, which is identical to the 3′ LTR. Furthermore, five ESTs from a library prepared from virgin C57BL/6 mammary tissue showed matches to some parts of hh-1 and hh-2 (Fig. 4B).
Probes 1 and 2 detected transcripts larger than 4 kb (Fig. 1A and B), suggesting the presence of RNA species containing additional MMTV sequences. We used RT-PCR to identify such transcripts. A cDNA was generated from total RNA with a primer located in exon B. Various sets of primers (see Materials and Methods) were used to identify transcripts that contain sequences from the MMTV gag/pol region and exon B (data not shown). Based on these experiments, we predict the presence of transcripts that originate in the 5′ LTR and proceed through the provirus into the flanking genomic sequences (Fig. 4C). Based on all available data, we predict the presence of transcripts that contain any of the MMTV RNAs followed by any combination of flanking exonic sequences.
Genomic structure.
Two BAC clones from a 129SvJ library were isolated and sequenced, and the genetic map of the locus was determined (Fig. 4A; accession no. AF120673). Based on the sequence of 13,000 nucleotides (nt) from the genomic region and sequences of the seven cDNA clones and seven additional ESTs retrieved from GenBank, we propose the structure depicted in Fig. 4. The locus consists of the MTV-3 provirus and at least six additional exons, two of which (exons B and C) harbor additional functional internal splice sites, as well as four distinct polyadenylation sites within exon F and one in exon G (Fig. 4A). Exon A coincides with the first 250 nt of the MMTV transcript which originates within the 5′ MMTV-LTR and proceeds into the MMTV splice site located 130 nt downstream of the 5′ LTR. Transcript hh-2 contains exon A′ which contains the transcribed sequences from the 5′ or 3′ LTR. Since exon A′ is followed by exon B which is linked directly to the 3′ LTR, it is likely that this transcript originates in the 3′ LTR. Using 5′RACE, we determined that the start sites of hh-1 and hh-2 are located in the 5′ LTR and 3′ LTR, respectively (not shown). Exon B is used in its entirety in clone hh-2 (Fig. 4). An alternative splice site inside exon B is used in clone hh-1. Both clone hh-1 and clone hh-2 contain the 3′ part of exon C and the entire exon D. While clone hh-1 splices directly into exon F, clone hh-2 picks up exon E before it splices into exon F (Fig. 4). The 3′ ends of clones hh-1 and hh-2 coincide at the same polyadenylation site within exon F. However, based on the isolated cDNA clones, there are at least three additional polyadenylation sites within exon F, and use of the 3′-most one results in a transcript of approximately 4 kb (Fig. 4). Exon G has been identified as a match of the genomic sequence with an EST from mammary tissue (see below). All splice sites used adhered to the consensus sequence (Table 2).
TABLE 2.
Intron/exon junctions in the hitchhiker locus
| Transcript | Junctionsa |
|---|---|
| hh-1 | |
| A to B′ | GAGGAGAG/gtaggtta, aatcttag/TCTGCACC |
| B′ to C′ | CACTGGAG/gtgagtct, cctcacag/GCCTGGGA |
| C′ to D | GGCAGCAG/gtaagca, ccccttag/CCCATGGT |
| D to F | GAAAAAAG/gtactgtt, ttccccag/ATGATGTT |
| hh-2 | |
| A′ to B | TGCGGCAG/ctggcgcc, attaagag/TCTGCACC |
| B to C′ | CACTGGAG/gtgagtct, cctcacag/GCCTGGGA |
| C′ to D | GGCAGCAG/gtaagca, ccccttag/CCCATGGT |
| D to E | GAAAAAAG/gtactgtt, gtcttaag/AGATGCTT |
| E to F | TGGCTAAG/gtagggtt, ttccccag/ATGATGTT |
| hh-6 | |
| B to C | CACTGGAG/gtgagtct, tctttttc/AGAGAAAC |
| C to D | GGCAGCAG/gtaagca, ccccttag/CCCATGGT |
| D to E | GAAAAAAG/gtactgtt, gtcttaag/AGATGCTT |
| E to F | TGGCTAAG/gtagggtt, ttccccag/ATGATGTT |
Uppercase and lowercase letters denote exons and introns, respectively.
ESTs from GenBank.
Searching GenBank with the seven cDNAs isolated in our screen resulted in two kinds of matches. Several hundred ESTs matched a 150-bp sequence within exon F. This sequence represents a middle repetitive element. More informative was the match to six ESTs from a normalized cDNA library obtained from virgin mammary tissue of the C57BL/6 strain. In addition to confirming the genomic structure, it extended the number of splice sites and exons used. Of particular interest is clone hh-6 (originally accession no. AI159140; updated to AF118847), which has its 5′ end within exon B. At the end of exon B it splices into the 5′ end of exon C. In contrast, both hh-1 and hh-2 use a splice site within exon C (Fig. 4). Thus, hh-6 represents a splice variant not detected in our clones. Another EST with genomic sequences 3′ of the known exon F was detected and is now defined as exon G (Fig. 4). Given the multiplicity of exons as well as intra or exon splice sites, we predict further transcripts.
Mouse strains 129SvJ and C57BL/6.
The 129 but not the C57BL/6 strain contains the MTV-3 provirus (7). Given that the EST clones that match the hitchhiker transcripts were derived from a mammary cDNA library from C57BL/6 mice, it is possible that transcription at this locus is independent of the MMTV provirus. Alternatively, the source of the library may not have been C57BL/6 but a strain that contains MTV-3. We used RT-PCR assays to establish whether C57BL/6 mice express transcripts from the hitchhiker locus. The RT primers matched sequences in exon C′ and the second primer was located in the transcribed part of the MMTV LTR. Using this primer set, we expected two fragments, one corresponding to the hh-1 transcript and the second to corresponding hh-2. RNA from mammary tissue of postpartum Stat5a-null, 129SvJ, and C57BL/6 mice was analyzed. The expected fragments were detected in tissue from the Stat5a-null and 129SvJ mice but not the C57BL/6 mice (Fig. 5). As a control we amplified Stat5b, which can be detected in mammary tissue from all three mice (Fig. 5). This finding strongly suggests that the mammary gland cDNA library (library 403; European IMAGE Consortium) was not derived from a C57BL/6 mouse. In the process of sequencing the hitchhiker locus from the two BAC clones from the 129SvJ genomic library, it became clear that they represented two different alleles, one containing and one lacking the MTV-3 provirus. We sequenced 20,000 bp from both alleles and were able to determine the point of proviral insertion (GenBank accession no. AF120673). Importantly, our results from the BAC clone demonstrate that the genomic library in question is not pure 129.
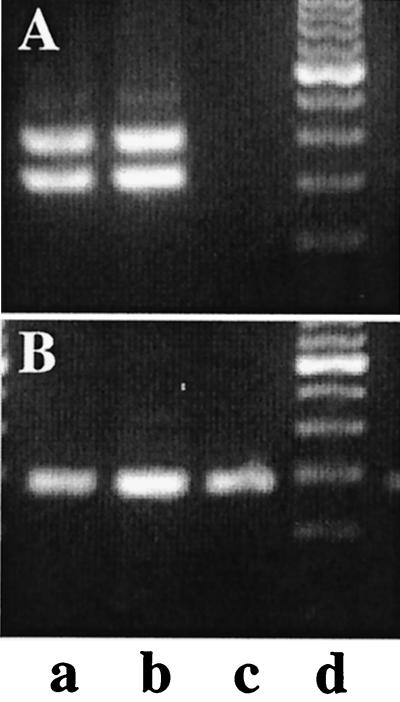
FIG. 5.
Absence of MTV-3–hitchhiker fusion transcripts in C57BL/6 mice. RT-PCR assays were performed as described in Materials and Methods. Mammary tissue from postpartum Stat5a-null (a), Stat5a +/− (b), and C57BL/6 (c) mice was analyzed for the presence of hitchhiker (A) and Stat5b (B) transcripts. The two bands in panel A represent the hh-1 and hh-2 transcripts.
Although we used mRNA from a (129 × C57BL/6)F1 cross in the differential cloning approach, we isolated transcripts that were specific for the 129 strain. Why did this happen? Originally, we detected the transcripts only in Stat5a-null mice and not in control littermates because we worked in a mixed background. We have now determined that the hitchhiker locus is on chromosome 11E70 and is closely linked to the Stat5 locus at 11E60.5. ES cells are derived from the 129 strain, which has suboptimal reproductive features. Thus it is customary to breed hemizygous mice derived from the targeted 129 ES cells into strains that have large litter sizes and lactate well. We bred the hemizygous 129 mice with Black Swiss mice. The PCR-based subtraction was performed with cDNA from Stat5a-null mice and their littermates that had two Stat5a wild-type alleles. Since the Stat5a gene and the hitchhiker locus are as close as 10 cM, only little recombination was observed, and our subtraction strategy resulted in the isolation of cDNAs specific for genes close to the Stat5a locus. Thus, our original molecular screening resulted in the isolation of transcripts specific to the 129 strain that carries the MTV-3 locus on chromosome 11.
Influence of Stat5a on transcription from MMTV LTRs.
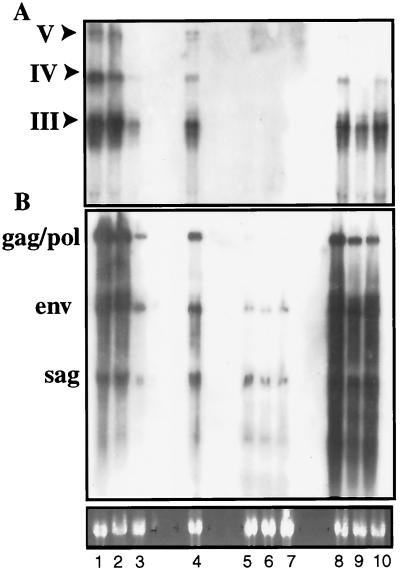
Recently Qin and coworkers reported that transcription from the MMTV LTR in mammary tissue is dependent on a Stat5a binding site within the LTR and the presence of Stat5a (20). Since the hitchhiker cDNAs were isolated from a library enriched for transcripts preferentially expressed in the absence of Stat5a, we evaluated this aspect. Specifically, we determined the transcriptional activity of the MTV-3 hitchhiker locus and that of other endogenous MMTVs in mammary tissue from Stat5a-null mice, control 129 mice, and C57BL/6 mice. Analysis with probe 2 demonstrated the expected RNA fragments in the Stat5a-null mice (Fig. 6A). Fragments of the same size and similar intensity were detected in mammary RNA from lactating 129 mice. As expected, no signal was detected in C57BL/6 mice. Since the transcripts were detected in Stat5a-null mice, activation of the novel transcripts within the MTV-3 locus does not depend on the presence of Stat5a. We further analyzed expression of all MMTV proviruses by hybridizing the RNA with a sag probe, which detects all proviral transcripts. Levels of expression of sag, env, and gag/pol were similar in Stat5a-null and control 129 mice (Fig. 6B) but much lower in C57BL/6 mice.
FIG. 6.
Transcription from the MMTV 5′ LTR in the absence of Stat5a. RNA from mammary tissue from postpartum Stat5a-null (lanes 1 to 3), Stat5a +/− (lane 4), C57BL/6 (lanes 5 to 7), and 129SvJ (lanes 8 to 10) mice was analyzed for hitchhiker transcripts by using probe 2 (A) and for MMTV transcripts by using a sag probe (B). Locations of the probes are shown in Fig. 1E. Probe 2, containing no MMTV sequences, detects the major hitchhiker transcripts III, IV, and V.
DISCUSSION
The MTV-3 provirus is located on chromosome 11 and contains a complete yet defective genome (9). Viral transcripts are detected, but the Env proteins are present at low amounts, suggesting a defect in translation or posttranslational processing (9). We have discovered that a large portion of transcripts originating in the 5′ LTR of MTV-3 do not encode viral transcripts but splice into the juxtaposed mouse genome. In addition, transcripts originate in the 3′ LTR of MTV-3 and proceed into 10 kb of flanking genomic sequence and pick up hitchhiking exons. These findings imply that the insertion of proviruses can have consequences in the evolution of the genome.
Hitchhiked exons in the MTV-3 locus.
We detected two classes of transcripts, those that originate within the 5′ LTR of MTV-3 and those that originate in the 3′ LTR. Both classes proceed for at least 10 kb into the genomic flanking sequences. Transcripts originating in the 5′ LTR used the native MMTV splice site, picked up at least six different exons, and utilized at least five different polyadenylation sites. Transcripts originating in the 3′ LTR proceeded directly into exon B and essentially followed the same splice pattern as transcripts originating in the 5′ LTR. All splice donor and acceptor sites shared appropriate consensus sequences. None of the transcripts exhibited any significant open reading frame, suggesting that they do not encode mature peptides. From Northern blot analyses, it appears that a large portion of the transcripts from the 5′ LTR proceed into the hitchhiker locus and do not form mature MMTV transcripts. Since it is not possible to generate unique hybridization probes for MTV-3, we cannot determine accurately the percentage of transcripts that enter the hitchhiker locus. Similarly, a large number of transcripts originate from the 3′ LTR. The majority of hitchhiker transcripts have sizes between approximately 1.5 and 4 kb, which can be accounted for by the MMTV and genomic exons identified over a stretch of 20 kb. However, additional RNA species were seen on Northern blots, and both large-scale genomic sequencing and EST data mining should provide more details on the full structure of this locus.
Evolutionary considerations.
Acquired retroviruses can induce tumors through the deregulation of juxtaposed genes (17). In contrast to these selection events, no apparent selection occurs for proviral genomes that are passed on through the germ line. Transcription in the hitchhiker locus originates in both the 5′ and 3′ LTRs and proceeds past the 3′ LTR for at least another 10 kb. The primary transcript contains at least six exons, and the combination of different splice donor and acceptor sites and several polyadenylation sites leads to a plethora of polyadenylated transcripts. This raises several questions about the influence of proviral genomes in the evolution of the genome. Several scenarios can be envisioned. First, transcription from the LTRs in conjunction with the generation of multiple polyadenylated RNAs is of no consequence, even over large time spans. Second, the observed transcription interferes with native genes in this region. Third, the transcripts serve as a pool for future genes. At this point there is no experimental evidence for the latter two hypotheses. We have to assume that the presence of hitchhiker transcripts is the result of (i) weak transcriptional stop sites in the 3′ LTR and (ii) the presence of splice donor and acceptor sites and polyadenylation sites in the juxtaposed genomic territory.
MTV-3 is carried by mice of the 129 strain but not by C57BL/6 mice. Since the MTV-3 SAG is responsible for the deletion of Vβ3+, Vβ5.1+, and Vβ5.2+ T cells, strains carrying this provirus are protected from MTV-3-induced mammary tumors (9). Our studies have demonstrated that a large portion of transcripts originating in the MTV-3 LTR contains flanking host genomic sequences as the consequence of alternative splicing. Theoretically, this presents the opportunity for the virus to rapidly shift its transcripts and alter the ratio of viral versus nonviral transcripts. However, there is no experimental evidence for such a scenario. A selective advantage of having transcripts originating in the 3′ LTR is not apparent. We have not detected any obvious open reading frame in any of the transcripts. However, such transcripts could serve as a source for future genes. Proviral genomes are scattered throughout the genome and may serve as an evolutionary driving force. Once thought to be rather stable, the genome is now known to exhibit great plasticity. Specifically, transposable elements can affect genome evolution at several levels. Insertion mutations by transposable elements account for up to 80% of mutations in Drosophila (4). As shown in this study, the possibility to capture genomic transcripts by means of transcription from endogenous retroviral LTRs may open new avenues for genome evolution. With the availability of the human and mouse genomic sequences within the next decade and large-scale EST sequencing efforts, we will obtain a deeper insight into the transcriptional activity of proviral insertion sites.
Transcriptional regulation by Stat5.
Stat5 was originally discovered by Wakao and coworkers and described as a mammary gland-specific factor that could activate transcription from the β-casein gene promoter in HC-11 cells (25). It had been speculated that Stat5 could be the global activator of genes in mammary tissue. The two isoforms of Stat5, Stat5a and Stat5b, exhibit conservation of 96% and are activated by cytokines such as prolactin. Both Stat5a (14) and Stat5b (24) genes have been deleted from the mouse genome, and the observed physiological consequences are different. This is strong support that these molecules, despite their conservation, elicit different responses in vivo. In particular, Stat5a but not Stat5b is required for mammary development and function (14, 22). Although many genes that contain GAS sites can be activated by Stat5a or Stat5b in transfected tissue culture cells, no in vivo target gene other than the WAP gene (14) has been identified. Recently, Qin and coworkers reported the presence of a Stat5 binding site within the MMTV LTR and demonstrated that transcription from the LTRs of MTV-17 and MTV-3 is dependent on both Stat5a and Stat5b (20). They showed that transcription from the LTRs in mammary tissue, but not lymphoid tissue, of either Stat5a-null or Stat5b-null mice was reduced to less than 10% compared to control mice. These findings were important since they demonstrated for the first time that heterodimers formed between Stat5a and Stat5b were required for transcription of a specific promoter. However, we could not repeat these results in over 30 Stat5a-null mice. The amounts of transcripts derived from the MTV-3 locus and from all MMTV proviruses combined are the same in Stat5a-null and control 129 mice. Transcription from the MTV-3 locus may even be higher in the absence of Stat5a. We cloned and sequenced the MTV-3 locus from the 129 mice and could confirm the presence of GAS (Stat5 binding) sites in both LTRs (AF119341 and AF119342). At this point, we cannot explain these differences. Our finding that transcription from the MMTV LTRs is not affected, or is only marginally altered, by the presence of Stat5a fully agrees with our earlier studies of Stat5a-null mice (14). The presence of GAS site in a promoter, and its functionality in tissue culture cells, does not necessarily reflect its relevance in vivo.
REFERENCES
- 1.Bentvelzen P. Host-virus interactions in murine mammary carcinogenesis. Biochim Biophys Acta. 1974;355:236–259. doi: 10.1016/0304-419x(74)90012-2. [DOI] [PubMed] [Google Scholar]
- 2.Bittner J J. Genetic concepts in mammary cancer in mice. Ann N Y Acad Sci. 1958;71:943–975. doi: 10.1111/j.1749-6632.1958.tb46816.x. [DOI] [PubMed] [Google Scholar]
- 3.Burdon T, Sankaran L, Wall R J, Spencer M, Hennighausen L. Expression of a whey acidic transgene during mammary development. Evidence for different mechanisms of regulation during pregnancy and lactation. J Biol Chem. 1991;266:6909–6914. [PubMed] [Google Scholar]
- 4.Capy P. Evolutionary biology. A plastic genome. Nature. 1998;396:522–523. doi: 10.1038/25007. . (News; comment.) [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Cordingley M G, Riegel A T, Hager G L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- 7.Fairchild S, Knight A M, Dyson P J, Tomonari K. Co-segregation of a gene encoding a deletion ligand for Tcrb-V3+ T cells with Mtv-3. Immunogenetics. 1991;34:227–230. doi: 10.1007/BF00215257. [DOI] [PubMed] [Google Scholar]
- 8.Golovkina T V, Prakash O, Ross S R. Endogenous mouse mammary tumor virus Mtv-17 is involved in Mtv-2-induced tumorigenesis in GR mice. Virology. 1996;218:14–22. doi: 10.1006/viro.1996.0161. [DOI] [PubMed] [Google Scholar]
- 9.Hainaut P, Castellazzi M, Gonzales D, Clausse N, Hilgers J, Crepin M. A congenic line of the BALB/c mouse strain with the endogenous mouse mammary tumor virus proviral gene Mtv-3: tissue-specific expression and correlation with resistance to mouse mammary tumor virus infection and tumorigenesis. Cancer Res. 1990;50:3754–3760. [PubMed] [Google Scholar]
- 9a.Hennighausen, L., et al. Unpublished data.
- 10.Kusk P, John S, Fragoso G, Michelotti J, Hager G L. Characterization of an NF-1/CTF family member as a functional activator of the mouse mammary tumor virus long terminal repeat 5′ enhancer. J Biol Chem. 1996;271:31269–31276. doi: 10.1074/jbc.271.49.31269. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Rosen J M. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–2070. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Gallego M I, Smith G H, Robinson G W, Hennighausen L. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ. 1998;9:795–803. [PubMed] [Google Scholar]
- 13.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Robinson G W, Wagner K-U, Garrett L, Wynshaw-Boris A, Henninghausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 14a.Mammary Genome Anatomy Project. 8 January 1999, revision date. cDNA libraries prepared from mammary tissue. [Online.] http://mammary.nih.gov/mgap/slides/libraries.html. Laboratory of Genetics and Physiology, National Institute of Diabetes and Digestive and Kidney Diseases National Institutes of Health, Bethesda, Md. [30 July 1999, last date accessed.]
- 15.Mink S, Hartig E, Jennewein P, Doppler W, Cato A C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters G. Oncogenes at viral integration sites. Cell Growth Differ. 1990;1:503–510. [PubMed] [Google Scholar]
- 18.Peterson D O, Kriz K G, Marich J E, Toohey M G. Sequence organization and molecular cloning of mouse mammary tumor virus DNA endogenous to C57BL/6 mice. J Virol. 1985;54:525–531. doi: 10.1128/jvi.54.2.525-531.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pullen A M, Choi Y, Kushnir E, Kappler J, Marrack P. The open reading frames in the 3′ long terminal repeats of several mouse mammary tumor virus integrants encode V beta 3-specific superantigens. J Exp Med. 1992;175:41–47. doi: 10.1084/jem.175.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin W, Golovkina T V, Peng T, Nepomnaschy I, Buggiano V, Piazzon I, Ross S R. Mammary gland expression of mouse mammary tumor virus is regulated by a novel element in the long terminal repeat. J Virol. 1999;73:368–376. doi: 10.1128/jvi.73.1.368-376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson G W, McKnight R A, Smith G H, Hennighausen L. Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development. 1995;121:2079–2090. doi: 10.1242/dev.121.7.2079. [DOI] [PubMed] [Google Scholar]
- 22.Teglund S, McKay C, Schuetz E, van Deursen J M, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle J N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 23.Toohey M G, Lee J W, Huang M, Peterson D O. Functional elements of the steroid hormone-responsive promoter of mouse mammary tumor virus. J Virol. 1990;64:4477–4488. doi: 10.1128/jvi.64.9.4477-4488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S-H, Ram P A, Waxman D J, Davey H W. Requirement of Stat5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakao H, Schmitt-Ney M, Groner B. Mammary gland-specific nuclear factor is present in lactating rodent and bovine mammary tissue and composed of a single polypeptide of 89 kDa. J Biol Chem. 1992;267:16365–16370. [PubMed] [Google Scholar]
- 26.Yamamoto K R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]