Abstract
Transcription activator-like effectors (TALEs) in plant-pathogenic Xanthomonas bacteria activate expression of plant genes and support infection or cause a resistance response. PthA4AT is a TALE with a particularly short DNA-binding domain harboring only 7.5 repeats which triggers cell death in Nicotiana benthamiana; however, the genetic basis for this remains unknown. To identify possible target genes of PthA4AT that mediate cell death in N. benthamiana, we exploited the modularity of TALEs to stepwise enhance their specificity and reduce potential target sites. Substitutions of individual repeats suggested that PthA4AT-dependent cell death is sequence specific. Stepwise addition of repeats to the C-terminal or N-terminal end of the repeat region narrowed the sequence requirements in promoters of target genes. Transcriptome profiling and in silico target prediction allowed the isolation of two cell death inducer genes, which encode a patatin-like protein and a bifunctional monodehydroascorbate reductase/carbonic anhydrase protein. These two proteins are not linked to known TALE-dependent resistance genes. Our results show that the aberrant expression of different endogenous plant genes can cause a cell death reaction, which supports the hypothesis that TALE-dependent executor resistance genes can originate from various plant processes. Our strategy further demonstrates the use of TALEs to scan genomes for genes triggering cell death and other relevant phenotypes.
The modularity of transcription activator-like effectors enables regulating their specificity and potential target sites, revealing two cell death inducer genes in Nicotiana benthamiana.
Introduction
The hypersensitive response (HR) is a form of programmed cell death in plants that effectively prevents pathogen infection by sacrificing cells in the infected area. It is generally triggered upon recognition of virulence proteins called effectors from the pathogen (Toruño et al. 2016; Thordal-Christensen 2020). Often, resistance (R) proteins recognize these effectors once delivered into the cytoplasm of the plant cell by the pathogen, via the type III secretion system (T3SS) (Büttner and He 2009). Typically, these R proteins are cytoplasmic proteins possessing both a nucleotide-binding site (NBS) and leucine-rich (LRR) repeat domains known as NBS–LRR proteins or NLRs (Kourelis and Van Der Hoorn 2018; Tamborski and Krasileva 2020).
Transcription activator-like effectors (TALEs) are proteins from Xanthomonas pathogens that function as plant transcription factors. After being delivered into plant cells by the T3SS, TALEs are transported into the nucleus, where they specifically match with effector-binding elements (EBEs) present in host promoters, leading to the activation of target genes (Boch and Bonas 2010; Timilsina et al. 2020). In this way, native TALEs evolved different target specificities to induce the expression of different genes (termed susceptibility genes) supporting bacterial infection (Cernadas et al. 2014; Hu et al. 2014; Erkes et al. 2017; Schwartz et al. 2017; Mücke et al. 2019; Teper and Wang 2021; Zhang et al. 2022). Functional TALE domains comprise an N-terminal domain for T3SS secretion and translocation, a central DNA-binding domain, and a C-terminal domain with nuclear localization signals (NLSs) and a eukaryotic acidic transcriptional activation domain (Boch and Bonas 2010). The modular DNA-binding domain consists of tandem 34 amino acid repeats, each one binding to a single DNA nucleotide. Specificity is determined by two variable amino acids at positions 12 and 13 termed repeat-variable diresidue (RVD) (Boch et al. 2009; Moscou and Bogdanove 2009). In addition, the N-terminal region next to the first repeat recognizes an additional nucleotide, mainly thymine, though other nucleotide types at this “zeroth” position have been demonstrated to allow binding in certain contexts (Yu et al. 2011; Doyle et al. 2013; Schreiber and Bonas 2014). The last repeat is only conserved for 20 amino acids and therefore termed a half repeat. Thus, a simple “TALE code” enables to predict the optimal DNA target sequence from the sequence of RVDs within the TALE array (Boch et al. 2009; Moscou and Bogdanove 2009). In addition, the modular DNA-binding domain allows a simple rearranging of repeats to build designer TALEs with any desired DNA-binding specificity (Cermak et al. 2011; Geißler et al. 2011; Weber et al. 2011). Accordingly, TALEs have been employed as gene activators in synthetic biology (Li et al. 2015; Schreiber and Tissier 2017; Schreiber et al. 2019) and started the genome editing revolution as TALE-nuclease fusions (Christian et al. 2010; Miller et al. 2010; Li et al. 2011; Kim et al. 2015; Becker et al. 2022).
Plants can recognize TALEs and deploy a battery of defenses ultimately culminating in an HR to thwart pathogen colonization. Two different mechanisms for TALE recognition have been described. One occurs in the cytoplasm and involves NLR proteins which perceive TALE topology, irrespective of their DNA-binding specificity. For example, the tomato (Solanum lycopersicum) NLR protein Bs4 mediates recognition of both AvrBs3 and AvrBs4, two TALEs sharing 96% sequence identity but having distinct DNA-binding specificities (Boch et al. 2009). The other mechanism involves executor resistance genes (ex-R), which act as traps for TALEs and trigger cell death upon expression, thereby effectively blocking bacterial infection (Zhang et al. 2015; Nowack et al. 2022). Up to now, only six ex-R genes associated with the corresponding TALEs have been cloned: Bs3 and Bs4C-R from pepper and Xa7, Xa10, Xa23, and Xa27 from rice (Gu et al. 2005; Römer et al. 2009; Strauß et al. 2012; Tian et al. 2014; Wang et al. 2015; Chen et al. 2021; Luo et al. 2021). Besides their functional similarity to induce cell death, only Xa10 and Xa23 are similar on the protein level, suggesting that many different ways exist among ex-R genes to initiate cell death (Zhang et al. 2015; Nowack et al. 2022). In general, the molecular details of how these ex-R genes signal the cell to die are still unknown. The lack of homology between ex-R and NLR genes demonstrates that their initial mechanism is different, although they might eventually trigger related effects, like ion fluxes across membranes, in an example of convergent evolution (Balint-Kurti 2019).
Recently, we identified an unusually short TALE, named PthA4AT, in a variant of Xanthomonas citri subsp. citri (X. citri) named X. citri AT. This TALE harbors only 7.5 repeats and is both necessary and sufficient to trigger an HR in Citrus limon and to confer resistance to citrus canker (Roeschlin et al. 2017; Roeschlin et al. 2019). PthA4AT also triggers cell death when infiltrated via Agrobacterium in Nicotiana benthamiana (Roeschlin et al. 2019). Disrupting the three NLSs of PthA4AT by point mutations impairs the development of this response, indicating that nuclear localization is required. Moreover, PthA4AT is able to mediate transcriptional activation, and the HR is greatly attenuated when the C-terminal domain containing the activation domain is removed, pointing to a mechanism for PthA4AT perception operating in both C. limon and N. benthamiana based on host gene expression (Roeschlin et al. 2019).
A TALE with such a short DNA target sequence is expected to find multiple EBEs within the promoters of all Nicotiana genes. Different bioinformatic approaches have been developed to predict potential target sites of TALEs in genomic sequences according to RVD specificities (Doyle et al. 2012; Grau et al. 2013; Pérez-Quintero et al. 2013; Erkes et al. 2019, 2021). These prediction tools often yield numerous putative targets, which requires further experimental evidence of gene activation to identify genes triggering a specific phenotype (Boch et al. 2014).
Here, we employ designer TALEs in an innovative strategy to reduce the number of potential target sites and identify plant genes responsible for the PthA4AT-induced cell death in N. benthamiana. Our results reveal that some R genes are in principle not linked to immunity-related functions, revealing pathways leading to cell death activation in plants.
Results
PthA4AT blocks disease progression of Xanthomonas campestris pv. campestris (Xcc8004) and tobacco necrosis virus in N. benthamiana
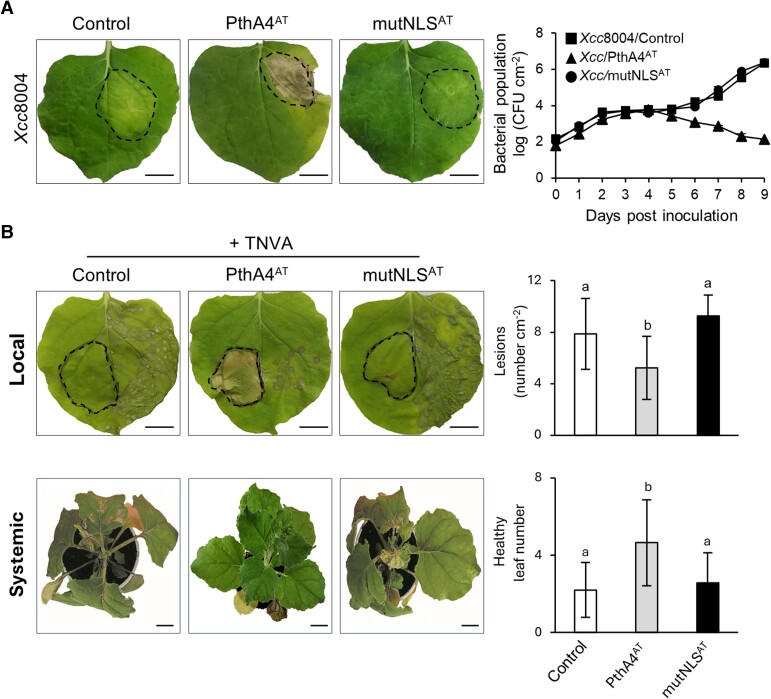
Previously, agroinfiltration assays demonstrated that nuclear localization of X. citri TALE PthA4AT triggered cell death in N. benthamiana (Roeschlin et al. 2019). We investigated whether this response may hinder the progress of pathogenic bacteria and viruses. Infiltration with Xanthomonas campestris pv. campestris strain 8004 (Xcc8004) causes macroscopic chlorosis coupled with bacterial growth in N. benthamiana (Yun et al. 2006; Fig. 1A). In contrast, Xcc8004 harboring PthA4AT (Xcc/PthA4AT) triggered a cell death response and halted disease progression, leading to a substantial reduction of Xcc8004 bacterial population in the infiltrated area (Fig. 1A). Similar to Citrus (Roeschlin et al. 2019), PthA4AT is translocated into the N. benthamiana nucleus (Supplementary Fig. S1), and the arrest of bacterial progression was not observed in leaves inoculated with an Xcc8004 expressing the PthA4AT derivative mutated in the NLSs (mutNLSAT) (Fig. 1A).
Figure 1.
PthA4AT confers protection against X. campestris pv. campestris (Xcc8004) and TNVA in N. benthamiana leaves. A) Macroscopic symptoms observed on leaves inoculated by infiltration with Xcc8004 harboring PthA4AT or its derivative mutated in the three NLSs (mutNLSAT) at 5 dpi. Bacterial growth on N. benthamiana leaves (CFU per cm2) after inoculation along 9 dpi. Values are the mean ±SD of three independent biological replicates. Control: Xcc8004 transformed with empty vector. B) Necrotic lesions observed at 3 dpi on leaves rub-inoculated with TNVA. N. benthamiana leaves were infiltrated via Agrobacterium transformed with PthA4AT or mutNLSAT on one half of the leaf, and the other half was inoculated with TNVA 4 days later. Upper panel: Local protection of PthA4AT was measured as the number of lesions per cm2 at 3 dpi using ImageJ software v1.4. Lower panel: Systemic protection was measured as the number of emerging leaves without necrosis (healthy leaf number) at 12 dpi. Values are expressed as means ± SD of three independent biological replicates. Letters indicate significant differences at P-value < 0.01 (Tukey's test). Control: Agrobacterium transformed with empty vector. Images were digitally extracted for comparison. Dashed lines indicate infiltrated area. Scale bar: 20 mm.
Similar results were obtained using a different plant–pathogen interaction. Agroinfiltration of PthA4AT into one half of the N. benthamiana leaf and subsequent rub inoculation with tobacco necrosis virus A (TNVA) into the neighboring half significantly reduced the number of virus-induced necrotic lesions, whereas using the mutNLSAT variant did not (Fig. 1B). Furthermore, the number of distal leaves with necrosis symptoms also decreased in PthA4AT-infiltrated plants relative to the controls (Fig. 1B). Taken together, these findings indicate that PthA4AT induces a cell death response that effectively impedes the disease progression of pathogenic bacteria Xcc8004 and TNVA virus in N. benthamiana.
PthA4AT-mediated cell death is dependent on specific RVDs
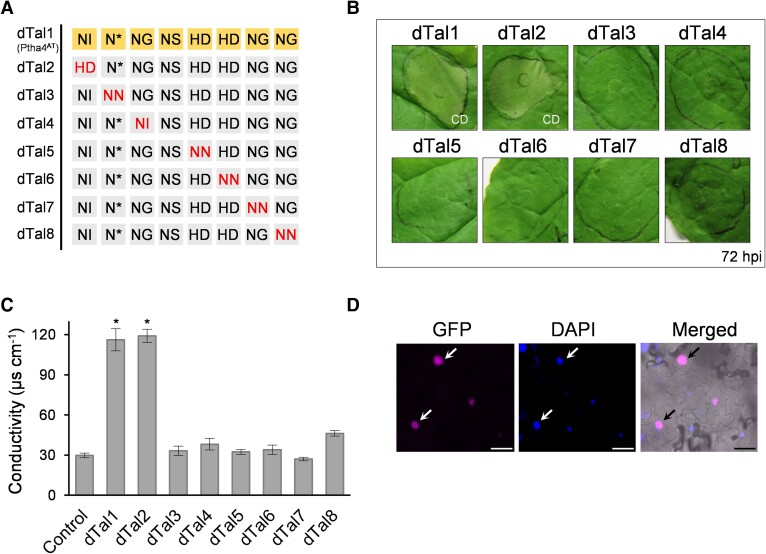
To test whether PthA4AT triggers cell death through the expression of one or several specific host genes, we exploited the modularity of the TALE DNA-binding specificity and engineered designer dTALEs (named dTals) with a controlled RVD composition (Geißler et al. 2011). The four most common RVDs in nature (NI, HD, NN, and NG) specify bases A, C, G/A, and T, respectively. Less common RVDs such as N* can target C/T, and NH can achieve guanine-specific recognition. NS, however, can target any of the four bases (Cong et al. 2012; Streubel et al. 2012). Initially, a designer TALE (dTal1) containing the same RVDs as PthA4AT was engineered. Similar to PthA4AT, dTal1 triggers cell death in the infiltrated area at 72 hours post-inoculation (hpi) and confers resistance to TNVA (Supplementary Fig. S2). Next, we generated seven dTALEs (dTal2 to dTal8) to substitute individual RVDs throughout dTal1 with RVDs targeting a different base, thereby altering their target specificity (Fig. 2A). Only dTal2, where NI was substituted by HD in the first repeat, was able to induce cell death at 72 hpi (Fig. 2B). To quantify this response, we measured ion leakage resulting from membrane damage. Conductivity was increased only in the tissue expressing dTal1 and dTal2, confirming that these dTALEs triggered a cell death response (Fig. 2C). GFP signals were detected within the nucleus for all dTALEs, indicating their proper expression and translocation (Fig. 2D).
Figure 2.
Substitution of individual RVDs of dTal1/PthA4AT. A) RVDs of dTALEs where PthA4AT-RVDs have been modified to target a different base. RVDs of dTal1 (PthA4AT) are shown in the first row. Red letters (HD in first column in dTal2; NN in second column in dTal3; NI in third column in dTal4; NN in fifth column in dTal5; NN in sixth column in dTal6; NN in seventh column in dTal7; NN in last column in dTal8) indicate RVD substitutions. B) Cell death development on N. benthamiana leaves infiltrated with the different constructs shown in A and photographed 72 hpi.C) Cell damage evaluated by conductivity measurement at 72 hpi. Values are the mean ± SD of three independent biological replicates. * significant compared to control (one-way ANOVA, Tukey's test, P-value < 0.01). Control: infiltration medium. D) Representative image of GFP (magenta) and DAPI (blue) signal in the nucleus (arrows) of N. benthamiana leaves expressing a GFP-dTALE fusion at 72 hpi. Left: fluorescence image; right: merged image (bright field). Scale bar: 40 µm. CD, cell death.
Additional experiments using the same strategy for target refinement of RVDs in positions two (N*) and four (NS) further suggest a sequence dependence for PthA4AT-mediated HR development, although two-base specificities are possible for position four (Supplementary Fig. S3). Taken together, our results demonstrate that re-targeting PthA4AT to alternative recognition sites by substituting specific RVDs disrupts cell death activation, suggesting that PthA4AT triggers cell death by one or several specific host genes. Our data also suggest that the designer TALE technology is a promising tool to identify cell death inducer genes in N. benthamiana using PthA4AT as a starting point.
Stepwise extension of the RVD domain at the C- and N-terminal ends refines specificity for PthA4AT-mediated cell death activation
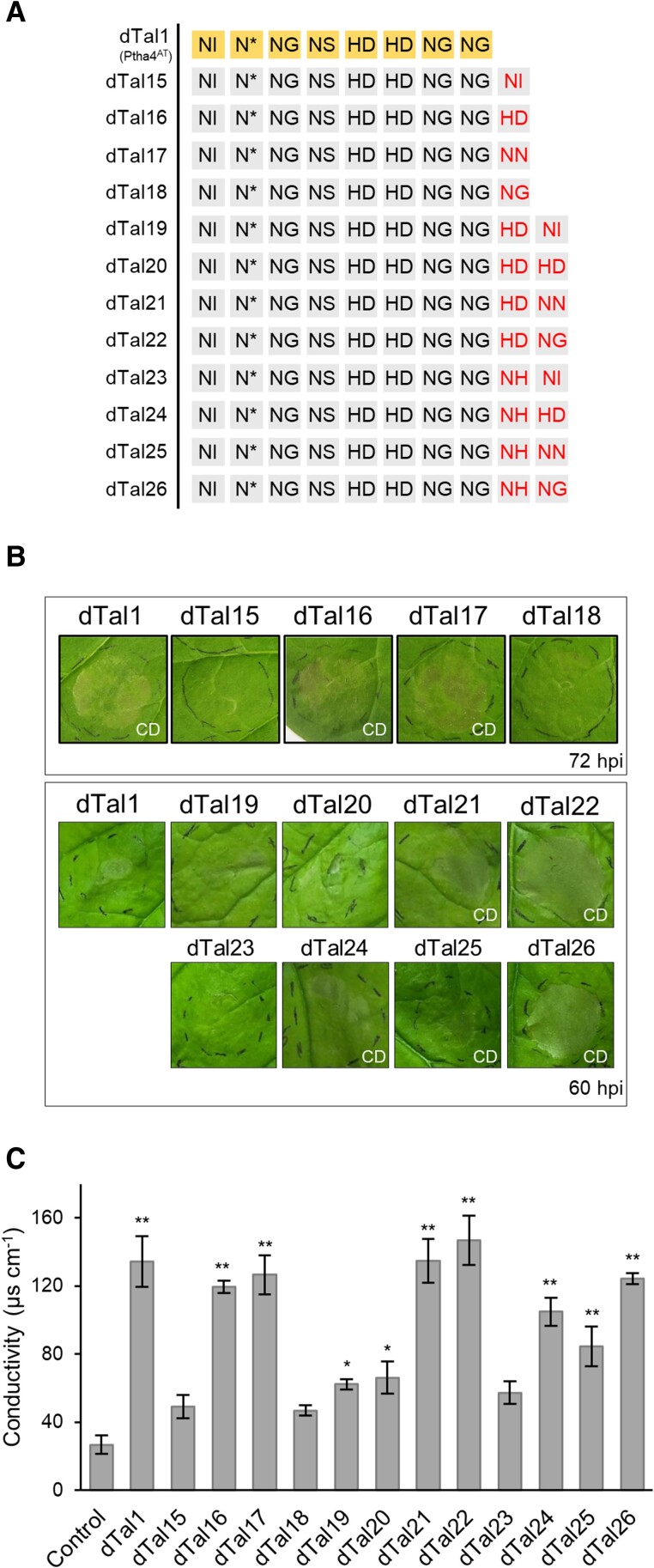
PthA4AT is predicted to bind to a target sequence of 9 bp in promoter regions (8 bp via the RVD region plus the preceding base, Fig. 2A), resulting in the activation of target genes. This short sequence will occur frequently by chance in the N. benthamiana genome. As the number of candidate genes is expected to decrease with an increasing number of RVDs, additional PthA4AT derivatives with additional repeats were built that specify longer target DNA sequences. This should eventually facilitate the identification of those genes responsible for the cell death phenotype. Individual repeats were added to the C-terminal end of the dTal1 repeat region to specify either A, C, G, or T. The ability of these longer dTALEs to still trigger cell death was then assayed in N. benthamiana. dTal15, dTal16, dTal17, and dTal18 added NI, HD, NN, or NG to dTal1 (Fig. 3A). Infiltration of dTal16 and dTal17, but not of dTal15 and dTal18, resulted in cell death at 72 hpi (Fig. 3B). These observations were confirmed by conductivity measurements (Fig. 3C). dTal16 and dTal17 were then used for another round of extension. dTal16 was extended by one repeat, resulting in dTal19, dTal20, dTal21, and dTal22. A modified version of dTal17 (with NN in its last position substituted by NH, to confer better specificity for G) was also extended, resulting in dTal23, dTal24, dTal25, and dTal26 (Fig. 3A). Cell death was observed for dTal21 and dTal22 and also for dTal24, dTal25, and dTal26 at 60 hpi, earlier than dTal1-induced cell death (Fig. 3B). Base preference for T in position two was confirmed in this longer RVD context (Supplementary Fig. S4). Quantitative differences were confirmed by electrolyte leakage measurements (Fig. 3C). We noticed that PthA4AT derivatives with different RVD combinations activate cell death. This indicates either that some RVD-base mismatches are tolerated or that these dTALEs target different cell death inducer genes.
Figure 3.
Extensions on the C-terminal end of the repeat domain of dTal1. A) RVDs of the different dTALEs. RVDs of dTal1 (PthA4AT) are shown on yellow background in the first row. Red letters in the last two columns indicate RVD extensions. B) Cell death response on leaves infiltrated via Agrobacterium photographed at 72 hpi. C) Cell death quantification by conductivity measurements at 72 hpi. Values are the mean ± SD of three independent biological replicates. **: significant compared to control (one-way ANOVA, Tukey's test, P-value < 0.01). Control: infiltration medium. CD, cell death.
We wondered if it is also possible to add repeats to the N-terminal end of the repeat region, because the N-terminal repeats have a higher impact on the overall DNA-binding capability of TALEs than C-terminal ones (Meckler et al. 2013). Nevertheless, extending at the N-terminal end is not straightforward, because the N-terminal domain of TALEs typically specifies a T preceding the target sequence of the repeat region (Boch et al. 2009). To allow extension of our dTALEs on the N-terminal end of the repeat region, we used an N-terminal domain which can recognize all four bases (Lamb et al. 2013). Using this, we extended dTal27 (10 RVDs) which causes a strong cell death, by two N-terminal repeats specifying AT, CT, GT, and TT, respectively. Following infiltration, the dTALE specifying TT (dTal60) caused a strong cell death, while AT- or CT-specifying dTALEs (dTal57 and dTal58, respectably) caused an intermediate and the dTALE specifying GT (dTal59) only a low reaction (Supplementary Fig. S5). A further round of N-terminal extension of one repeat specified for TTT and GTT causing the strongest cell death (dTals 63 and 64; Supplementary Fig. S5). Taken together, these results show that stepwise addition of repeats to the C- and N-terminal ends of the RVD region results in dTALEs with longer target sequences that show a differential cell death induction at individual target sites.
Transcriptomic profiling and TALE target prediction identify PAT1 as a cell death inducer gene in N. benthamiana
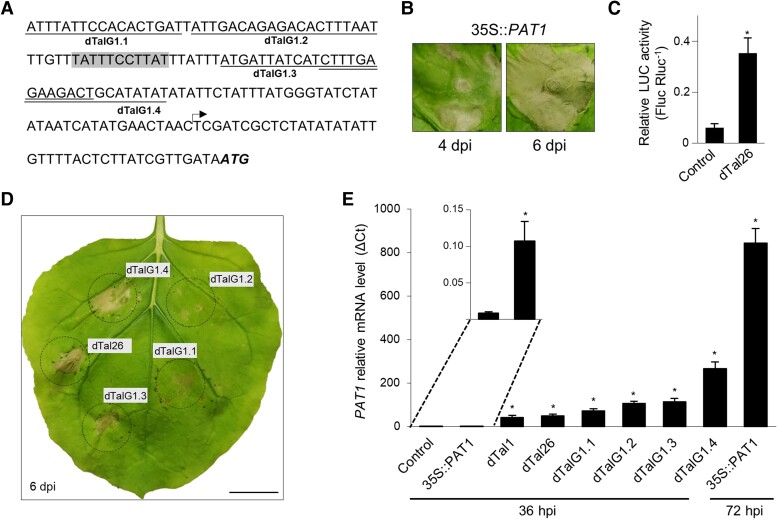
Target genes can be identified by analyzing plant transcripts that are activated in the presence of specific TALEs, in combination with in silico prediction of TALE target sequences in promoter regions (Hu et al. 2014; Schwartz et al. 2017). dTal26 was used to identify cell death inducer genes. Similar to dTal1 and PthA4AT, infiltration via Agrobacterium of dTal26 to Nicotiana leaves protects against subsequent infection by TNVA virus (Supplementary Fig. S6). dTal26 triggers cell death faster than dTal1, covering the whole infiltrated area at 60 hpi (Fig. 3B), suggesting that extending the binding site confers better ability to induce gene expression of cell death inducer genes. RNA of N. benthamiana leaves infiltrated with Agrobacterium delivering dTal26 was sampled at 36 hpi, when the effector was expressed but cell death was not yet visible (Supplementary Fig. S7). A dTALE that does not trigger cell death was used as a negative control. To predict dTal26-binding sites, promoter regions spanning 400 nt upstream to 200 nt downstream of the transcriptional start site from dTal26-induced genes were obtained from the N. benthamiana genome, and PrediTALE (Erkes et al. 2019) was used (with dTal26 RVDs as input and N* substituted by NG in the second position, to refine the actual requirement for T at this position, Supplementary Fig. S4). Although longer than PthA4AT, the relatively short length of dTal26 posed a challenge due to its low binding specificity (no target with P-value < 10−6, Supplementary Table S1), hindering the filtering of candidates based on robust DNA-binding predictions. Nonetheless, applying stringent expression criteria for target prioritization identified 34 genes (Supplementary Table S1). Out of these, five genes fulfilled the promoter sequence requirements at the 5′ end of dTal26-predicted binding site for a cell death inducer gene, as outlined in the previous section, and were selected for overexpression (Supplementary Fig. S8 and Fig. 4A). Of these, only the NBlab18G05420 gene, encoding the patatin-like protein PAT1, showed a cell death response at 4 days post-inoculation (dpi) when overexpressed in N. benthamiana (Fig. 4B), suggesting a role as a cell death inducer gene.
Figure 4.
Cell death activation by patatin-like protein PAT1. A)PAT1 promoter sequence showing the binding site of different dTALEs. Arrow, TSS. Start codon ATG is shown in bold and italics. The predicted dTal26-binding site is shown in gray. PAT1-specific designer dTALEs are underlined. B) Cell death response in N. benthamiana leaves infiltrated via Agrobacterium containing the PAT1 cDNA under the control of the CaMV35S promoter. Pictures were taken at 6 and 8 dpi. C) Relative luciferase activities of LUC (luciferase) reporter under the control of the PAT1 promoter (pPAT1::LUC) in N. benthamiana leaves co-infiltrated with dTal26 or with Agrobacterium transformed with empty vector (Control). Data are presented as Fluc Rluc-1 ± SD. * significant after Tukey's test (P-value < 0.05). Three biological replicates were performed. Each replicate consists of one 8-mm-diameter disc per infiltrated leaf collected at 36 hpi. D) Cell death response on N. benthamiana leaves infiltrated with PAT1-dTALEs constructs and photographed at 6 dpi. Images were digitally extracted for comparison. Black circles delimit infiltrated regions. Dashed lines: accurate infiltrated area. Scale bar: 20 mm. E)PAT1 mRNA levels analyzed by RT-qPCR at 36 and 72 hpi. Results are presented as mean ± SD. *, significant after t test (P-value < 0.005). Control: infiltration medium. Three replicates per sample were performed.
PAT1 expression is not sufficient to trigger the entire dTal26-mediated cell death response
The involvement and dynamics of PAT1 in cell death activation was studied through several experimental approaches. First, a dual-luciferase reporter assay was used to test activation of PAT1 by dTal26. A promoter region of PAT1, containing the predicted target sequence for dTal26, was cloned to direct expression of luciferase (pPAT1::LUC). Co-expression of dTal26 and pPAT1::LUC by agroinfiltration demonstrated that dTal26 is able to induce PAT1 expression 5-fold at 36 hpi (Fig. 4C). Next, four dTALEs targeting the PAT1 promoter were generated (Fig. 4A). Agroinfiltration, delivering each one of them individually, induced cell death visible at 6 dpi (Fig. 4D), confirming that TALE-mediated induction of PAT1 triggers cell death. Notably, PAT1-induced cell death is visible at later times than that triggered by dTal26. Consequently, we investigated the levels of PAT1 expression under the different activation conditions. Expression of PAT1 increases at 36 hpi after delivering dTal1 or dTal26 (Fig. 4E), confirming RNA-seq data. Similar levels were observed at 36 hpi after infiltration with PAT1-specific dTALEs, although cell death was observed much later. Levels of expression were low at 36 hpi after agroinfiltration with 35S::PAT1 (Fig. 4E inset), but high at 72 hpi, even though cell death will not be visible yet (Fig. 4B), suggesting that the strong dTal26-mediated cell death is likely caused by a combination of PAT1 and additional genes.
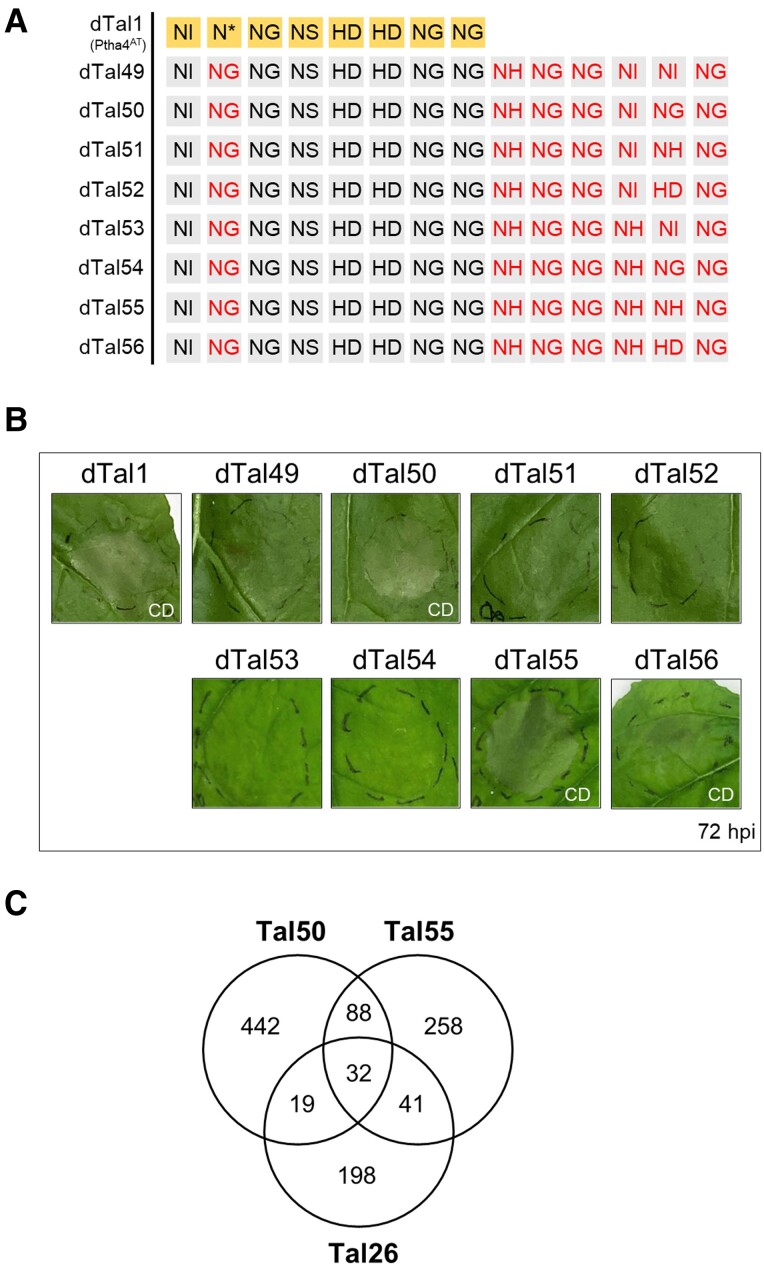
PthA4AT-extended dTALEs with 14 RVDs retain the cell death phenotype and allow the identification of an additional cell death inducer gene
We aimed to identify additional cell death inducer genes that are triggered by PthA4AT besides PAT1. To further reduce the number of predicted targets, we added repeats C-terminally to the dTal27 repeats (10 RVDs) to make the dTALE more specific. Following the same strategy as above, additional dTALEs were constructed and assayed for cell death response after infiltration. (Supplementary Fig. S9 and Fig. 5A). These experiments revealed four 14 RVD-long dTALEs still capable of developing cell death (Fig. 5B). To search for the corresponding cell death inducer genes, we performed a second transcriptomic assay to obtain genes induced by two of them (dTal50 and dTal55). The expression data were combined with PrediTALE results for binding prediction to get a new list of putative targets. To avoid candidates being discarded during the filtering process, more relaxed expression criteria than those used in the dTal26 analysis above were applied. In this new analysis, 290 genes were considered putative targets of dTal26, 580 of dTal50, and 419 of dTal55. Among them, 32 genes were predicted targets of all three dTALEs (Fig. 5C), whereas 60 genes were predicted targets of dTal26 and only one of the two other dTALE derivatives. Twenty-seven of these last 61 genes, however, contain a binding site for both dTALEs (dTal50 and dTal55) in its promoter region when considering a P-value 10−4 in PrediTALE, suggesting that they could also be targets for the three dTALEs. A list of 92 genes was generated of potential cell death inducer genes (Supplementary Table S2).
Figure 5.
Extensions of six repeats on the C-terminal end of the RVD domain of dTal1. A) RVDs of the different dTALEs derived from dTal46 and dTal47. RVDs of dTal1 (PthA4AT) are shown on yellow background in the first row. Red letters indicate RVD substitutions (NG in second column in all dTals) or extensions (six last columns). B) Cell death response on N. benthamiana leaves infiltrated via Agrobacterium Pictures were taken at 72 hpi. C) Venn diagram of induced genes in N. benthamiana leaves infiltrated with dTal26, dTal50, or dTal55 and sampled at 36 hpi (Log2FoldChange > 1; adjusted P-value < 0.05) combined with PrediTALE predictions (P < 10−5). CD, cell death.
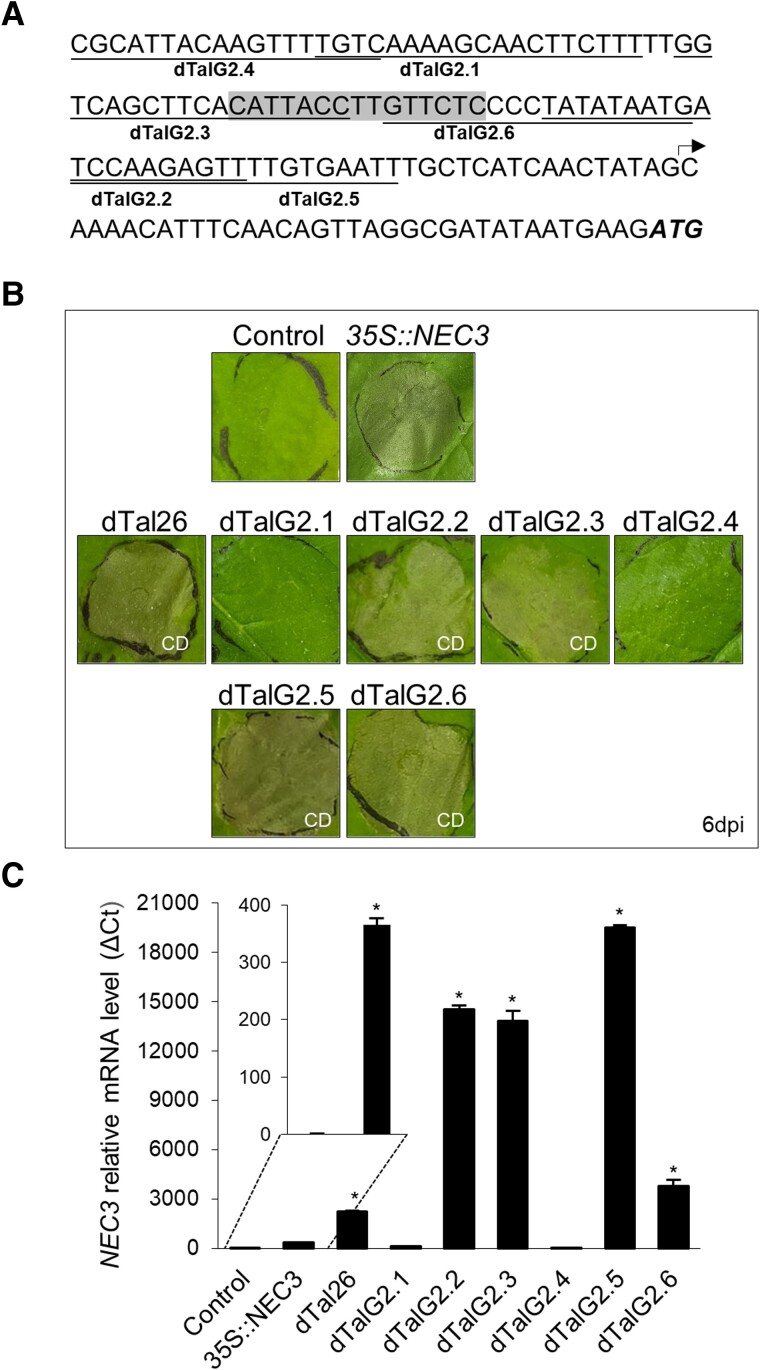
We selected genes from this list (based on gene function, expression data, and sequence motifs) and engineered dTALEs targeting their promoters to test them individually for cell death induction in N. benthamiana. Four dTALEs designed to activate the NBlab07G09180 gene, encoding a possible bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3 (NEC3), were able to activate the gene and trigger cell death (Fig. 6, A, B and C). In addition, overexpression of the NBlab07G09180 gene using a 35S promoter fusion also triggered cell death (Fig. 6B), indicating that the NEC3 gene is another cell death inducer gene in N. benthamiana.
Figure 6.
NBlab07G09180, encoding a bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3 (NEC3), mediates cell death induction in N. benthamiana. A) Graphical representation of the NEC3 promoter region. Gene-specific dTALEs (dTalG2.1 to dTalG2.6) are underlined. dTal26/dTal50/dTal55 effector-binding element is highlighted in gray. Arrow indicates TSS. The start codon (ATG) is shown in bold italics. B) Cell death response on leaves infiltrated via Agrobacterium with 35S::NEC3 and gene-specific dTALEs. Pictures were taken at 6 dpi.C)NEC3 mRNA levels analyzed by RT-qPCR at 48 hpi for dTALEs and 72 hpi for 35S::NEC3. Results are presented as mean ± SD. * significant after t test (P-value < 0.005). Control: infiltration medium. Three replicates per sample were performed. CD, cell death.
Discussion
The modularity of TALEs opens the possibility to use them as probes for screening strategies, where the endogenous activation of specific genes by a given TALE translates into a visible phenotype. In a strategy starting from less stringent short TALEs, longer dTALEs with increased target specificity are engineered by stepwise extension of the RVD domain and those still retaining the phenotype of interest are selected (Supplementary Fig. S10). A classical strategy used to identify TALE target genes, based on the combination of global gene expression and binding-site prediction, is then used to identify and validate genes. This strategy can now be used for other screening strategies where a visible phenotype can be tracked.
In this study, we exploited this strategy to identify target genes involved in cell death activation in N. benthamiana by the short TALE PthA4AT (Roeschlin et al. 2019). PthA4AT, a natural TALE triggering HR in C. limon (Roeschlin et al. 2017), also triggers cell death via ex-R genes when ectopically expressed in N. benthamiana (Roeschlin et al. 2019). Moreover, the possibility of PthA4AT-mediated cell death being triggered after recognition of the effector in the cytoplasm in a Bs4-like matter is discarded, as disrupting the three NLSs or removing the activation domain impairs the development of this response. This experimental system was exploited here as a proof of concept to identify additional genes regulating this biological process. We could clarify that PthA4AT causes cell death via expression of specific genes. Furthermore, this work supports the concept that genes involved in normal physiological processes can cause cell death when their expression is misregulated by a TALE, which is significant for understanding ex-R gene evolution and developing novel plant resistances.
Up to date, six cell death inducer genes, induced by natural TALEs in specific plant–pathogen interactions, have been identified which function as R genes (Gu et al. 2005; Römer et al. 2009; Strauß et al. 2012; Tian et al. 2014; Wang et al. 2015; Chen et al. 2021; Luo et al. 2021). PthA4AT is an unusual TALE with only 7.5 repeats. Short TALEs with few repeats will statistically bind to more target sites in a genome than longer ones with more repeats, which in principle increases their potential as discovery tools. However, this also increases the possibility of these short TALEs binding to multiple genes and generating an unspecific cytotoxic effect. This does not seem to be the case for PthA4AT. Despite harboring only 7.5 repeats, the strong cell death visible at 72 hpi is disrupted or delayed when the RVD composition is modified, indicating that it is the induced expression of specific target genes which causes cell death. The short length of PthA4AT imposed, however, a difficulty to the identification of these host genes, beyond the degeneracy of the code for some RVDs and the mismatch tolerance common to all TALEs (Meckler et al. 2013; Rogers et al. 2015; Rinaldi et al. 2017). The high number of putative target genes in the Nicotiana genome did not allow a computational prediction of the one/s responsible for the cell death response. We approached this problem by stepwise extensions of the repeat domain of PthA4AT either on the C-terminal or N-terminal side. This strategy allowed to generate longer PthA4AT derivatives extended up to six repeats C-terminally or three repeats N-terminally that still were able to trigger cell death, while significantly reducing the number of possible targets.
The hypothesis of PthA4AT activating more than one executor gene became apparent as our study advanced. This possibility was initially revealed when the substitution of the ambiguous NS by other RVDs with affinities for individual bases identified two possibilities for this position to trigger cell death. Eventually, we analyzed the transcriptional response to three different designer TALEs (dTal26, dTal50, dTal55) and identified a cell death inducer genes (NBlab07G09180, encoding a possible bifunctional monodehydroascorbate reductase and NEC3) that was induced by all three TALEs and an additional one (NBlab15G13810, encoding a patatin-like proteins) that was a predicted target of dTal26, but only of one of its two longer derivatives (dTal55). This confirmed that PthA4AT and the shorter dTALEs targeted more than one cell death inducer gene. Accordingly, the cell death response observed at 72 hpi for dTal26 was stronger than that observed at the same timepoint for the longer dTal50 and dTal55 or any of the identified cell death inducer genes themselves. Taken together, our results indicate that there is likely an additive effect with the contribution of several high-effect and low-effect executor genes activated following PthA4AT inoculation.
Our data support the recently postulated hypothesis that some TALE-induced ex-R genes may not originally function in plant defense (Nowack et al. 2022). Instead, the presence of TALE target sites in their promoter might have possibly occurred by chance, and such plants were further selected by humans because of their pathogen resistance. This is nicely exemplified by the Xa23 rice resistance gene, functional against the TALE AvrXa23 from Xanthomonas oryzae pv. oryzae. Only in the resistant rice cultivar, a transposon and the AvrXa23 target sequence are inserted into the promoter of a small open reading frame, encoding a possible transmembrane protein (Wang et al. 2015). XA23 is similar to XA10, which also functions as an executor, involved in calcium transport across the endoplasmic reticulum membrane (Tian et al. 2014; Wang et al. 2015). The physiological functions of Xa10 and Xa23 are unknown, but it is plausible that they still contain, or originally contained, one that is distinct from their role as resistance gene. The function of our identified targets as genes participating in normal cellular processes, unrelated to classical immune responses, reinforces the view that executor genes are primarily involved in cellular processes including developmental programmed cell death and inadvertently trigger uncontrolled cell death when activated by an external TALE (Nowack et al. 2022). Patatin-like proteins are phospholipases involved in lipid peroxidation and represent the major storage protein in potato tubers. Intriguingly, patatin-like proteins are also induced in plant–Xanthomonas interactions (Cacas et al. 2009; Kim et al. 2014). Nectarins are proteins from the floral nectar. NEC3 is a major constituent of ornamental Nicotiana nectar and was found to contain carbonic anhydrase and monodehydroascorbate reductase activities (Carter and Thornburg 2004). A biological interpretation for any of the two candidates is speculative at this point. In summary, the targets identified in this study shed light on distinct cellular processes that can cause cell death.
To our knowledge, PthA4AT is the only 7.5 RVDs TALE with a biological function. In TALEs of this length, transcriptional activation is still occurring (Boch et al. 2009; Roeschlin et al. 2019). The strain containing PthA4AT is still infective in the host from which it was isolated (Citrus clementina), and cell death was observed only in other Citrus hosts (C. limon and Citrus sinensis) (Chiesa et al. 2013; Roeschlin et al 2019). The transcriptional program triggered by PthA4AT in C. limon resembles an immune response and protects the plant from future bacterial infections (Roeschlin et al. 2017), and we showed here that the cell death response in N. benthamiana also results in plant protection to subsequent attacks of bacteria and viruses. This suggests that physiological cell death reactions could reconstitute a functional pathogen resistance and unveil avenues for leveraging cell death inducer genes to engineer resistances in economically pivotal crops. Noteworthy, the overexpression of patatin-like genes in Arabidopsis shows cell death responses (La Camera et al. 2009) and conferred resistance to Pseudomonas syringae pv. tomato (Pst) and Hyaloperonospora arabidopsidis infection (Kim et al. 2014). Moreover, addition of TALE target sequences to the promoter of Xa27 or Xa10 resulted in resistance against additional Xanthomonas strains (Hummel et al. 2012; Zeng et al. 2015).
Screening strategies performed in Nicotiana, searching for cell death inducer genes by overexpression, were carried out in the past, but the designs were biased to paraquat or defense-related genes (Nasir et al. 2005; Coemans et al. 2008). Our approach using unbiased gene activation by PthA4AT and dTALEs allowed the identification of genes involved in cell death that remained undiscovered in the former approaches. The success of our strategy validates the use of TALE proteins to scan genomes in search of additional cell death inducers and multiple other phenotypes.
Materials and methods
Bacterial growth conditions, plant material, and inoculation assays
Strains and plasmids used in this study are listed in Supplementary Tables S3 and S4, respectively. Escherichia coli and Agrobacterium tumefaciens (strain GV3101) were grown in LB medium at 37°C and 28°C, respectively. Xanthomonas were grown in peptone–yeast–malt (PYM) medium at 28°C. TNVA was propagated in N. benthamiana by mechanical inoculation as the source of virus. N. benthamiana plants were grown at 23°C and a photoperiod of 16-h light/8-h dark. Leaves of 6-week-old plants were inoculated by pressure infiltration with Xcc8004 or Agrobacterium. For Xcc8004 inoculation, bacterial suspensions were prepared in 10 mM MgCl2 [106 colony-forming unit (CFU) ml−1], according to Chiesa et al. (2013). For Agrobacterium inoculation, overnight cultures were adjusted to OD600 = 0.4 in infiltration buffer (10 mM MES, 10 mM MgCl2, and 200 μM acetosyringone in DMSO). Before infiltration, cultures were shaken for 2 h at room temperature. Overexpression constructs were co-infiltrated with a strain containing the P19 gene, a suppressor of gene silencing. For TNVA inoculation 0.5 to 1 infective virions/μl, crude leaf extract were used to rub-inoculate N. benthamiana plants at the two- to four-leaf stage (Garcia et al. 2023). Plants were monitored for up to 8 days at 22°C and 50% to 60% relative humidity.
Microscopy
Xcc8004 expressing PthA4AT: an cherry was monitored for 72 hpi using a confocal laser scanning microscope (Zeiss LSM 880; Carl Zeiss Microscopy GmBH Jena, Germany). The excitation of an cherry was accomplished with the 543 nm laser, and the emission was detected between 579 and 624 nm. Gain detection was set at 880. Chlorophyll was excited with the 405 nm laser line, and fluorescence was detected in the 649–704 nm range. Gain detection was set at 658. Simulated three-dimensional images and sections were generated by the ZEN 3.1 blue edition Free Viewer software. For nuclear co-localization, six N. benthamiana leaves were infiltrated with 10 mM Tris–KCl pH 7.4 buffer containing 10 µg ml−1 DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) and incubated for 10 min. DAPI was excited with the 405-nm laser line, and the fluorescence was detected in the 438- to 479 nm range. Gain detections were set at 590. GFP was excited with the 488-nm laser line, and the fluorescence was detected in the 499 to 552 nm range. Chlorophyll was excited with the 405 nm laser line (15%) and fluorescence was detected in the 630 to 735 nm range. Simulated three-dimensional images and sections were generated by the ZEN 3.1 blue edition Free Viewer software. Nuclear localization for designer TALEs was monitored using a fluorescence lupe (MacroFluo MZ16F Leica Microsystems, USA). Two independent experiments were performed. In each experiment, three different leaves from three different plants per sample were used.
Cloning strategies
dTals were constructed using the Golden TAL technology kit (Geißler et al. 2011). The selected RVDs were assembled to the destination vector pSKA2 adding the N- and C-terminal modules of the Hax3 gene. To add repeats to the N-terminal end of the repeat region, the dTALEs require an N-terminal domain recognizing all four bases. Three amino acid changes (Q231R, W232G, S233A; Lamb et al. 2013) were incorporated into the Hax3 N-terminal domain by overlap extension PCR to build a generally applicable module for dTal assembly. Subsequently, pSKA2 constructions were transformed into A. tumefaciens. The pSKA2 vector fuses an N-terminal GFP epitope to the dTal. Coding sequences for dTal26-induced candidate targets and for the PAT1 promoter were assembled using GoldenBraid (Vazquez-Vilar et al. 2017). Plasmids, constructs, and primers used are listed in Supplementary Tables S4 and S5.
Conductivity assays
For ion leakage experiments, four 12-mm-diameter leaf discs were collected at 72 hpi in 5 ml of sterilized ultrapure water (Milli-Q) and incubated for 1 h in an orbital incubator (Stuart SI50, LabMakelaar Benelux B.V., The Netherlands) at 130 rpm and 25°C. Conductivity was measured using an ion conductivity meter (Basic C30, Crison, Spain). Three biological replicates per sample were performed. Each replicate contains four discs from four different leaves coming from two different plants.
Luciferase activity measurements
The Dual-Glo Luciferase Assay System kit (Promega, USA) was used in this analysis. Firefly luciferase (FLuc) activity was measured using a GloMax Multi Detection System (Promega, USA), setting 10 s for measurement. After measuring, 40 µl of Stop&Glo reagent was added per sample to quantify Renilla luciferase (RLuc) activity. FLuc/RLuc ratios were calculated as the mean value of three biological replicates. Each replicate consists of one 8-mm-diameter disc per infiltrated leaf collected at 36 hpi. Relative transcriptional activity was expressed as the FLuc/RLuc ratios normalized to the FLuc/RLuc ratios of the reference reporter Pnos:Luc (Supplementary Table S4).
RNA purification
Total RNA was extracted using E.Z.N.A Plant RNA kit following RNase-free DNase (Omega Bio-Tek, USA) treatment. Three replicates per biological condition were performed. Each replicate contains leaf samples (100 mg) from three independent inoculated N. benthamiana plants randomly harvested at 36 hpi and pooled.
RT-qPCR
cDNA was synthesized from 1 μg of DNase-treated total RNA using Maxima First Strand cDNA Synthesis (Thermo Fisher Scientific, USA). RT-qPCRs were carried out using 1 µg cDNA, 0.4 µM primers, and 1× Pyrotaq EvaGreen qPCR Master Mix (Cultek Molecular Bioline, Spain). Three replicates per sample were performed. Relative transcript abundance was normalized against the reference F-Box gene using the ΔΔCt method (Livak and Schmittgen 2001). Primers are shown in Supplementary Table S5.
RNA-Seq analysis and prediction of effector targets
Forty million 100 (for dTal26) or 150 (for dTal50 and dTal55) nucleotide paired-end reads per sample were obtained. Raw data were filtered using FastQC and Cutadapt v.2.10. Sequences were then mapped to the N. benthamiana genome (https://www.nbenth.com/) using HISAT2. Differential expression was estimated using DESEQ2 using RPKM (reads per kilobase per million reads) for normalization. PrediTALE (Erkes et al. 2019) was used for prediction of effector targets, considering regions spanning from 400 nt upstream to 200 nt downstream of the transcription start site (TSS). Promoter sequences were retrieved from the N. benthamiana (LAB330, version 3.01, https://www.nbenth.com/) reference genome.
Accession numbers
Sequence data from this article can be found in GEO Omnibus database under the following accession numbers: GSE221293 and GSE221441. NEC3 accession number is NBlab07G09180; PAT1 accession number is NBlab18G05420. Sequences are available for downloading at www.nbenth.com.
Supplementary Material
Acknowledgments
The authors thank Dr. Javier Forment and the Bioinformatic Unit at IBMCP for assistance in RNA-seq analysis and data mining.
Contributor Information
Roxana A Roeschlin, Instituto de Biología Molecular y Celular de Rosario (IBR)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Ocampo y Esmeralda S/n, S2002LRK, Rosario, Argentina.
Sepideh M Azad, Instituto de Biología Molecular y celular de Plantas (IBMCP), Universidad Politécnica de Valencia-CSIC, Ingeniero Fausto Elio S/N., 46022, Valencia, España.
René P Grove, Institute of Plant Genetics, Leibniz Universität Hannover, 30419 Hannover, Germany.
Ana Chuan, Instituto de Biología Molecular y celular de Plantas (IBMCP), Universidad Politécnica de Valencia-CSIC, Ingeniero Fausto Elio S/N., 46022, Valencia, España.
Lucila García, Instituto de Biología Molecular y Celular de Rosario (IBR)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Ocampo y Esmeralda S/n, S2002LRK, Rosario, Argentina; Área Virología, Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario (UNR), Suipacha 590, S2002LRK, Rosario, Argentina.
Regina Niñoles, Instituto de Biología Molecular y celular de Plantas (IBMCP), Universidad Politécnica de Valencia-CSIC, Ingeniero Fausto Elio S/N., 46022, Valencia, España.
Facundo Uviedo, Instituto de Biología Molecular y Celular de Rosario (IBR)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Ocampo y Esmeralda S/n, S2002LRK, Rosario, Argentina.
Liara Villalobos, Instituto de Biología Molecular y Celular de Rosario (IBR)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Ocampo y Esmeralda S/n, S2002LRK, Rosario, Argentina.
Maria E Massimino, Instituto de Biología Molecular y celular de Plantas (IBMCP), Universidad Politécnica de Valencia-CSIC, Ingeniero Fausto Elio S/N., 46022, Valencia, España.
María R Marano, Instituto de Biología Molecular y Celular de Rosario (IBR)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Ocampo y Esmeralda S/n, S2002LRK, Rosario, Argentina; Área Virología, Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario (UNR), Suipacha 590, S2002LRK, Rosario, Argentina.
Jens Boch, Institute of Plant Genetics, Leibniz Universität Hannover, 30419 Hannover, Germany.
José Gadea, Instituto de Biología Molecular y celular de Plantas (IBMCP), Universidad Politécnica de Valencia-CSIC, Ingeniero Fausto Elio S/N., 46022, Valencia, España.
Author contributions
R.A.R., A.C., R.P.G., S.M.A., L.G., L.V.-P., F.U., R.N., and M.E.M. performed the experiments. M.R.M., J.B., and J.G. designed and supervised the work and wrote the manuscript.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Subcellular localization of PthA4AT in Nicotiana benthamiana.
Supplementary Figure S2. Cell death response and tobacco necrosis virus A (TNVA) protection by dTal1 in Nicotiana benthamiana.
Supplementary Figure S3. Ambiguous repeat-variable diresidues (RVD) replacement to refine dTal1 specificity.
Supplementary Figure S4. Repeat-variable diresidues (RVDs) replacement over dTal26.
Supplementary Figure S5. Two rounds of repeat-variable diresidue (RVDs) extension at the N-terminal end of dTal27.
Supplementary Figure S6. Protection induced by dTal26 on Nicotiana benthamiana leaves rub-inoculated with tobacco necrosis virus A (TNVA).
Supplementary Figure S7. GFP signals in the nucleus of Nicotiana benthamiana leaves expressing dTal26 and photographed at 36 h post-inoculation (hpi).
Supplementary Figure S8. Promoter sequences of genes selected for overexpression in Nicotiana benthamiana.
Supplementary Figure S9. Repeat-variable diresidue (RVDs) extension on dTal27 C-terminal end.
Supplementary Figure S10. Strategy for refinement of dTALE target specificity used in this study.
Supplementary Table S1. dTal26-activated genes.
Supplementary Table S2. dTal-activated genes for dTal26, dTal50, and dTal55.
Supplementary Table S3. Bacterial strains and virus used in this study.
Supplementary Table S4. Plasmids used in this study.
Supplementary Table S5. List of primers used in this study.
Funding
M.R.M. thanks the Agencia Nacional de Promoción Científica y Tecnológica (PICT-2020-01624) and J.G. and J.B. thank the CA16107-EuroXanth COST Action (European Cooperation in Science and Technology) for funding.
Data availability
RNA-Seq data have been deposited in GEO Omnibus database under the following accession numbers: GSE221293 and GSE221441.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol Plant Pathol. 2019:20(8):1163–1178. 10.1111/mpp.12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Mücke S, Grau J, Boch J. Flexible TALEs for an expanded use in gene activation, virulence and scaffold engineering. Nucleic Acids Res. 2022:50(4):2387–2400. 10.1093/nar/gkac098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010:48(1):419–436. 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U, Lahaye T. TAL effectors—pathogen strategies and plant resistance engineering. New Phytol. 2014:204(4):823–832. 10.1111/nph.13015 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009:326(5959):1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Büttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009:150(4):1656–1664. 10.1104/pp.109.139089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas JL, Marmey P, Montillet JL, Sayegh-Alhamdia M, Jalloul A, Rojas-Mendoza A, Clérivet A, Nicole M. A novel patatin-like protein from cotton plant, GhPat1, is co-expressed with GhLox1 during Xanthomonas campestris-mediated hypersensitive cell death. Plant Cell Rep. 2009:28(1):155–164. 10.1007/s00299-008-0622-x [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Mol Biol. 2004:54(3):415–425. 10.1023/B:PLAN.0000036373.84579.13 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia N V, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011:39(17):1–11. 10.1093/nar/gkr739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas RA, Doyle EL, Niño-Liu DO, Wilkins KE, Bancroft T, Wang L, Schmidt CL, Caldo R, Yang B, White FF, et al. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 2014:10(2):e1003972. 10.1371/journal.ppat.1003972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu P, Mei L, He X, Chen L, Liu H, Shen S, Ji Z, Zheng X, Zhang Y, et al. Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice. Plant Commun. 2021:2(3):100143. 10.1016/j.xplc.2021.100143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa MA, Siciliano MF, Ornella L, Roeschlin RA, Favaro MA, Delgado NP, Sendín LN, Orce IG, Ploper LD, Vojnov AA, et al. Characterization of a variant of Xanthomonas citri subsp. citri that triggers a host-specific defense response. Phytopathology. 2013:103(6):555–564. 10.1094/PHYTO-11-12-0287-R [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010:186(2):756–761. 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coemans B, Takahashi Y, Berberich T, Ito A, Kanzaki H, Matsumura H, Saitoh H, Tsuda S, Kamoun S, Sági L, et al. High-throughput in planta expression screening identifies an ADP-ribosylation factor (ARF1) involved in non-host resistance and R gene-mediated resistance. Mol Plant Pathol. 2008:9(1):25–36. 10.1111/j.1364-3703.2007.00440.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012:3(1):1–6. 10.1038/ncomms1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012:40(W1):W117–W122. 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Hummel AW, Demorest ZL, Starker CG, Voytas DF, Bradley P, Bogdanove AJ. TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1. PLoS One. 2013:8:e82120. 10.1371/journal.pone.0082120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes A, Mücke S, Reschke M, Boch J, Grau J. PrediTALE: a novel model learned from quantitative data allows for new perspectives on TALE targeting. PLoS Comput Biol. 2019:15(7):e1007206. 10.1371/journal.pcbi.1007206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes A, Mücke S, Reschke M, Boch J, Grau J. Epigenetic features improve TALE target prediction. BMC Genomics. 2021:22(1):1–18. 10.1186/s12864-021-08210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkes A, Reschke M, Boch J, Grau J. Evolution of transcription activator-like effectors in Xanthomonas oryzae. Genome Biol Evol. 2017:9(6):1599–1699. 10.1093/gbe/evx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia L, Gerhardt N, Martin AP, Martínez MF, Alemano S, Marano MR. Tobacco necrosis virus A overcomes local cell death response in Nicotiana tabacum. Plant Pathol. 2023:72(1):154–169. 10.1111/ppa.13629 [DOI] [Google Scholar]
- Geißler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011:6(5):e19509. 10.1371/journal.pone.0019509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau J, Wolf A, Reschke M, Bonas U, Posch S, Boch J. Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol. 2013:9(3):e1002962. 10.1371/journal.pcbi.1002962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005:435(7045):1122–1125. 10.1038/nature03630 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer WB, Yang B, White FF, Wang N, Jones JB. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci U S A. 2014:111(4):E529. 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel AW, Doyle EL, Bogdanove AJ. Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 2012:195(4):883–893. 10.1111/j.1469-8137.2012.04216.x [DOI] [PubMed] [Google Scholar]
- Kim DS, Jeun Y, Hwang BK. The pepper patatin-like phospholipase CaPLP1 functions in plant cell death and defense signaling. Plant Mol Biol. 2014:84(3):329–344. 10.1007/s11103-013-0137-x [DOI] [PubMed] [Google Scholar]
- Kim YH, Kim HO, Baek EJ, Kurita R, Cha HJ, Nakamura Y, Kim H. Rh D blood group conversion using transcription activator-like effector nucleases. Nat Commun. 2015:6(1):1–13. 10.1038/ncomms9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, Van Der Hoorn RAL. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018:30(2):285–299. 10.1105/tpc.17.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, Roby D, Heitz T. The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant-Microbe Interact. 2009:22(4):469–481. 10.1094/MPMI-22-4-0469 [DOI] [PubMed] [Google Scholar]
- Lamb BM, Mercer AC, Barbas CF. Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013:41(21):9785. 10.1093/nar/gkt754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011:39(1):359–372. 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jiang Y, Chen H, Liao W, Li Z, Weiss R, Xie Z. Modular construction of mammalian gene circuits using TALE transcriptional repressors. Nat Chem Biol. 2015:11(3):207–213. 10.1038/nchembio.1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo D, Huguet-Tapia JC, Raborn RT, White FF, Brendel VP, Yang B. The Xa7 resistance gene guards the rice susceptibility gene SWEET14 against exploitation by the bacterial blight pathogen. Plant Commun. 2021:2(3):100164. 10.1016/j.xplc.2021.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsäesser J, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 2013:41(7):4118–4128. 10.1093/nar/gkt085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2010:29(2):143–148. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009:326(5959):1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Mücke S, Reschke M, Erkes A, Schwietzer CA, Becker S, Streubel J, Morgan RD, Wilson GG, Grau J, Boch J. Transcriptional reprogramming of rice cells by Xanthomonas oryzae tales. Front Plant Sci. 2019:10:162. 10.3389/fpls.2019.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir KH, Takahashi Y, Ito A, Saitoh H, Matsumura H, Kanzaki H, Shimizu T, Ito M, Fujisawa S, Sharma PC, et al. High-throughput in planta expression screening identifies a class II ethylene-responsive element binding factor-like protein that regulates plant cell death and non-host resistance. Plant J. 2005:43(4):491–505. [DOI] [PubMed] [Google Scholar]
- Nowack MK, Holmes DR, Lahaye T. TALE-induced cell death executors: an origin outside immunity? Trends Plant Sci. 2022:27(6):536–548. 10.1016/j.tplants.2021.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Quintero AL, Rodriguez-R LM, Dereeper A, López C, Koebnik R, Szurek B, Cunnac S. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS One. 2013:8(7):e68464. 10.1371/journal.pone.0068464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi FC, Doyle LA, Stoddard BL, Bogdanove AJ. The effect of increasing numbers of repeats on TAL effector DNA binding specificity. Nucleic Acids Res. 2017:45(11):6960–6970. 10.1093/nar/gkx342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeschlin RA, Favaro MA, Chiesa MA, Alemano S, Vojnov AA, Castagnaro AP, Filippone MP, Gmitter FG, Gadea J, Marano MR. Resistance to citrus canker induced by a variant of Xanthomonas citri ssp. citri is associated with a hypersensitive cell death response involving autophagy-associated vacuolar processes. Mol Plant Pathol. 2017:18(9):1267–1281. 10.1111/mpp.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeschlin RA, Uviedo F, García L, Molina MC, Favaro MA, Chiesa MA, Tasselli S, Franco-Zorrilla JM, Forment J, Gadea J, et al. PthA4AT, a 7.5-repeats transcription activator-like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance. Mol Plant Pathol. 2019:20(10):1394–1407. 10.1111/mpp.12844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JM, Barrera LA, Reyon D, Sander JD, Kellis M, Keith Joung J, Bulyk ML. Context influences on TALE-DNA binding revealed by quantitative profiling. Nat Commun. 2015:6(1):7440. 10.1038/ncomms8440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci U S A. 2009:106(48):20526–20531. 10.1073/pnas.0908812106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T, Bonas U. Repeat 1 of TAL effectors affects target specificity for the base at position zero. Nucleic Acids Res. 2014:42(11):7160–7169. 10.1093/nar/gku341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T, Prange A, Hoppe T, Tissier A. Split-TALE: a TALE-based two-component system for synthetic biology applications in planta. Plant Physiol. 2019:179(3):1001–1012. 10.1104/pp.18.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T, Tissier A. Generation of dTALEs and libraries of synthetic TALE-activated promoters for engineering of gene regulatory networks in plants. Methods Mol Biol. 2017:1629:185–204. 10.1007/978-1-4939-7125-1_13 [DOI] [PubMed] [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci U S A. 2017:114(5):E897-E903. 10.1073/pnas.1620407114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauß T, Van Poecke RMP, Strauß A, Römer P, Minsavage G V, Singh S, Wolf C, Strauß A, Kim S, Lee HA, et al. RNA-Seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc Natl Acad Sci U S A. 2012:109(47):19480–19485. 10.1073/pnas.1212415109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blücher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012:30(7):593–595. 10.1038/nbt.2304 [DOI] [PubMed] [Google Scholar]
- Tamborski J, Krasileva K V. Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol. 2020:71(1):355–378. 10.1146/annurev-arplant-081519-035901 [DOI] [PubMed] [Google Scholar]
- Teper D, Wang N. Consequences of adaptation of TAL effectors on host susceptibility to Xanthomonas. PLoS Genet. 2021:17(1):e1009310. 10.1371/journal.pgen.1009310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H. A holistic view on plant effector-triggered immunity presented as an iceberg model. Cell Mol Life Sci. 2020:77(20):3963–3976. 10.1007/s00018-020-03515-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, et al. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 2014:26(1):497–515. 10.1105/tpc.113.119255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat Rev Microbiol. 2020:18(8):415–427. 10.1038/s41579-020-0361-8 [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016:54(1):419–441. 10.1146/annurev-phyto-080615-100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Quijano-Rubio A, Fernandez-Del-Carmen A, Sarrion-Perdigones A, Ochoa-Fernandez R, Ziarsolo P, Blanca J, Granell A, Orzaez D. GB3.0: a platform for plant bio-design that connects functional DNA elements with associated biological data. Nucleic Acids Res. 2017:45(4):2196–2209. 10.1093/nar/gkw1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M, et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant. 2015:8(2):290–302. 10.1016/j.molp.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011:6(5):e19722. 10.1371/journal.pone.0019722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol Plant-Microbe Interact. 2011:24(9):1102–1113. 10.1094/MPMI-11-10-0254 [DOI] [PubMed] [Google Scholar]
- Yun MH, Torres PS, El OM, Rigano LA, Gonzalez-Lamothe R, Marano R, Castagnaro AP, Dankert MA, Bouarab K, Vojnov AA. Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 2006:141(1):178–187. 10.1104/pp.105.074542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Tian D, Gu K, Zhou Z, Yang X, Luo Y, White FF, Yin Z. Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol J. 2015:13(7):993–1001. 10.1111/pbi.12342 [DOI] [PubMed] [Google Scholar]
- Zhang B, Han X, Yuan W, Zhang H. TALEs as double-edged swords in plant–pathogen interactions: progress, challenges, and perspectives. Plant Commun. 2022:3(3):100318. 10.1016/j.xplc.2022.100318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yin Z, White F. TAL effectors and the executor R genes. Front Plant Sci. 2015:6:641. 10.3389/fpls.2015.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited in GEO Omnibus database under the following accession numbers: GSE221293 and GSE221441.