Abstract
The receptor molecules for human and animal hepatitis B viruses have not been defined. Previous studies have described a 170 to 180 kDa molecule (p170 or gp180) that binds in vitro to the pre-S domain of the large envelope protein of duck hepatitis B virus (DHBV); cDNA cloning revealed the binding protein to be duck carboxypeptidase D (DCPD). In the present study, the DCPD cDNA was transfected into several nonpermissive human-, monkey-, and avian species-derived cell lines. Cells transfected with a plasmid encoding the full-length DCPD protein bound DHBV particles, whereas cells expressing truncated versions of DCPD protein that fail to bind the pre-S protein did not. The DHBV binding to DCPD-reconstituted cells was blocked by a monoclonal antibody that neutralizes DHBV infection of primary duck hepatocytes (PDH) and also by a pre-S peptide previously shown to inhibit DHBV infection of PDH. In addition to promoting virus binding, DCPD expression was associated with internalization of viral particles. The entry process was prevented by incubation of reconstituted cells with DHBV at 4°C and by the addition of energy-depleting agents known to block DHBV entry into PDH. These results demonstrated that DCPD is a DHBV receptor. However, the lack of complete viral replication in DCPD-reconstituted cells suggested that additional factors are required for postentry events in immortalized cell lines.
Hepatitis B virus (HBV) infects 400 million individuals worldwide and causes acute and chronic hepatitis as well as cirrhosis of the liver. Moreover, persistently infected individuals have an approximate 100-fold increase in the risk for development of primary hepatocellular carcinoma (1, 19). Studies on this important human pathogen are severely hampered by its narrow host range since only humans and chimpanzees are susceptible to HBV infection. In addition, none of the established hepatocyte-derived cell lines are permissive to productive HBV infection. A major barrier for HBV replication in laboratory animals resides at the step of virus attachment and entry since HBV transgenic mice support efficient viral replication (9). Thus, cloning of the HBV receptor would permit animal and cell reconstitution experiments to study the early events of the viral life cycle. In this regard, duck hepatitis B virus (DHBV), a related avian hepatotropic DNA virus (hepadnavirus), may provide a suitable system for receptor characterization since ducks and primary duck hepatocytes (PDH) are easily available for infection experiments and support viral replication. Cloning of the DHBV receptor may aid in the identification of the HBV human counterpart.
The pre-S domain of DHBV large envelope protein is believed to be the ligand for the viral receptor (11, 12, 22). Previous immunoprecipitation experiments have identified a cellular binding partner (gp180) for this portion of the large envelope protein (10, 15). Subsequent cDNA cloning revealed the binding protein to be a carboxypeptidase H-like molecule now classified as duck carboxypeptidase D (DCPD) (16). Using a pre-S protein tagged to glutathione S-transferase (GST), we have independently identified a similar-size interacting protein called p170, obtained partial peptide sequences and subsequently cloned its cDNA (25, 25a). Sequence analysis revealed that p170 and gp180 are the same protein. In the present study, we carried out reconstitution experiments in a variety of cell lines. The ability of DCPD-transfected cell lines to bind and internalize DHBV virion particles establishes DCPD as a DHBV receptor.
MATERIALS AND METHODS
The DCPD cDNA clones.
To prepare the N-25 deletion mutant of DCPD, a 0.86-kb PCR fragment of DCPD coding sequence (nucleotide 77 through the HindIII site) was assembled with a 0.95-kb HindIII-BamHI fragment and a 2.4-kb BamHI-NcoI fragment derived from duck liver cDNA libraries, and the resultant 4.2-kb DCPD cDNA was inserted into the NotI-AflII sites of pcDNA 3.1/Zeo(−) vector (Invitrogen). To allow the expression of the mutant protein, an artificial translational initiation codon was placed at the beginning of the cDNA (5′-GCGGCCGCCATGGATATTAAG-3′; NotI site italicized; initiation codon underlined). The full-length DCPD cDNA clone was derived from the N-25 mutant by replacing the 0.86-kb NotI-HindIII fragment with a 0.95-kb fragment containing the intact 5′ coding sequence.
The C-16 mutant construct was generated by filling in the EcoRI digest of full-length DCPD cDNA clone with deoxyribonucleotides and Klenow fragment, followed by religation with T4 ligase, thus creating a frameshift mutation followed by premature termination of protein translation. The C-81 construct was obtained by subcloning the 3.5-kb NotI (flushed)-XhoI fragment of DCPD cDNA into the EcoRV-XhoI sites of pcDNA3 (Invitrogen). For the generation of mutants C-36 and C-54, two C-terminal fragments of DCPD cDNA were amplified by PCR using a common sense primer and specific antisense primers with engineered stop codons. The sequences are 5′-TGATCTAGACTAATCGTCGTGGTGCTGCCG-3′ for the C-36 mutant (XbaI site underlined) and 5′-TGATCTAGATTGAGCAGACACACCAGAT-3′ for the C-54 construct. The PCR products were double digested with XhoI and XbaI to exchange with the cognate fragment of C-81-pcDNA3. The mutant constructs were verified by sequencing analysis. Two independent clones from each mutant construct were tested, and identical results were obtained.
The DCPD and pre-S antibodies.
The native DCPD protein was purified from duck liver as previously described (25). After electrophoresis through a polyacrylamide gel, the DCPD band was removed and 200 μg of protein was used in complete Freund’s adjuvant to immunize rabbits. Rabbits were bled after three booster injections. To produce pre-S antibodies, the GST-pre-S (amino acids 1 to 161) fusion protein expressed in Escherichia coli (25) was cleaved with thrombin, and the pre-S portion was used to immunize rabbits and mice. Two mouse monoclonal antibodies (MAbs), designated MAb 15 and MAb 76, were used in the form of culture supernatant. Their binding sites on the pre-S domain were mapped by using a panel of GST-tagged pre-S deletion constructs (18a, 25).
Immunoprecipitation of DCPD and complex formation with GST–pre-S protein.
Bosc cells grown in 60-mm-diameter dishes were transfected with DCPD cDNA clones by the calcium phosphate precipitation method. Two days later, cells were metabolically labeled with [35S]methionine for 4 h and subsequently lysed in 1 ml of lysis buffer. The supernatant was precleared by incubation at 4°C for several hours with Staphylococcus aureus followed by centrifugation to remove the pellet. The precleared lysate (100 μl) was incubated with a 1:100 dilution of the DCPD antibodies and 4 μl of protein A beads. Another 100-μl aliquot of lysate was incubated with 3 μl of glutathione-Sepharose beads conjugated with GST–pre-S fusion protein (5 μg). Beadbound DCPD proteins were separated on 8% polyacrylamide gels and revealed by fluorography (25).
Immunofluorescent (IF) staining of DCPD expression and binding of DHBV particles.
To detect expression of DCPD, cells were grown on coverslips and fixed with ethanol-acetic acid (95:5). The fixed cells were incubated at 4°C for 1 h with phosphate-buffered saline (PBS)–3% bovine serum albumin (BSA), 1 h with a 1:1,000 dilution of the rabbit polyclonal DCPD antibody, and then 1 h with goat anti-rabbit antibodies conjugated to fluorescein isothiocyanate (FITC; Sigma). To detect DHBV bound to transfected cells, the slides were blocked with PBS–3% BSA and incubated with a 1:1,000 dilution of the rabbit anti-pre-S antibody followed by the addition of a FITC-conjugated secondary antibody.
Binding of DHBV to DCPD-transfected cells and PDH.
Virus binding experiments were carried out for 36 to 48 h after transfection or soon after plating of PDH, usually using 60 μl of viremic duck serum diluted 1:10 in culture medium per well of the six-well plate. The viremic duck sera were obtained by intravenous injection of 2 to 3-day-old ducklings with 200 μl of a high-titer viremic duck serum followed by a bleed 4 to 6 days later. After washing, the cells were scraped off the plates. For experiments investigating virus internalization, half of the cell pellet from each well was resuspended in 500 μl of trypsin-EDTA solution and incubated at 37°C for 5 min. The action of trypsin was stopped by addition of complete culture medium, and the cells were pelleted. The cell pellet was lysed in 300 μl of buffer containing 10 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1% NP-40.
Inhibition of DHBV infection of PDH by pre-S antibodies.
The PDH was cultured in six-well plates and incubated at 37°C overnight with 5 μl of viremic duck serum diluted in 800 μl of culture medium, together with different dilutions of the rabbit antiserum against the pre-S domain or culture supernatants containing the pre-S-specific MAbs. After a thorough wash, cells were cultured for an additional 7 days and then Southern blot analysis of DHBV DNA was performed.
Western blot analysis.
Cell lysates were electrophoresed through 8% (for DCPD protein) or 12% (for pre-S protein) sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The blots were blocked at room temperature for 3 h with 3% BSA in PBS containing Tween 20 (0.05%) and then incubated overnight with a 1:1,000 dilution of the rabbit polyclonal anti-DCPD antibody or a 1:1,000 dilution of the rabbit pre-S antibody. After thorough washing, the blots were incubated for an additional 3 h with a 1:1,000 dilution of 125I-labeled protein A (New England Nuclear). The blot were washed again and exposed to X-ray films.
Southern blot analysis.
The cell lysates were treated at 37°C for several hours with proteinase K (0.5 mg/ml) in the presence of sodium dodecyl sulfate (0.5%). The DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in Tris-EDTA (pH 8.0). After electrophoresis through a 1.5% agarose gel and staining with ethidium bromide, the DNA was transferred to nylon membranes and hybridized with a randomly primed DHBV DNA probe. The 2.8-kb DHBV DNA (positions 1996 to 1745) used for probe preparation was obtained by three rounds of successive PCR amplification of a DHBV genome using three pairs of nested primers.
Nucleotide sequence accession number.
The GenBank accession number of the sequence reported is AF039749.
RESULTS
Design of reconstitution experiments.
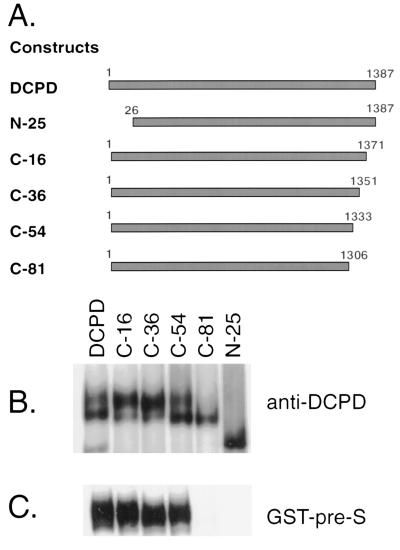
We anticipated that the ability of DCPD to mediate DHBV binding in transfected cells would require a physical interaction with the pre-S domain of the viral large envelope protein. Therefore, cells transfected with mutant constructs of DCPD that have lost ability to bind the pre-S protein should not have an affinity for DHBV particles. Such mutant constructs serve as appropriate negative controls in the reconstitution experiments. As illustrated in Fig. 1A, we constructed five deletion mutants of DCPD which lack the C-terminal 16, 36, 54, or 81 amino acid residues or the N-terminal 25 residues containing the signal peptide. The cDNA clones encoding for full-length DCPD and the truncation mutants were transfected into Bosc cells, a retroviral packaging cell line derived from the 293 human embryonic kidney cell line (20). This cell line supports a high transfection efficiency (Fig. 2a) and allows enhanced expression of foreign proteins as a result of the expression of simian virus 40 large T antigen. All constructs expressed similar amounts of protein, as revealed by immunoprecipitation with anti-DCPD antibodies (Fig. 1B). However, the C-81 and N-25 deletion mutants have lost the ability to bind GST-tagged pre-S protein (Fig. 1C).
FIG. 1.
Illustration of DCPD constructs, protein expression, and affinity for DHBV pre-S envelope protein. (A) Cartoon illustrating the structure of various DCPD constructs used in this study. (B) Expression of DCPD protein and truncation mutants. (C) Affinity with GST–pre-S fusion protein. Transfected Bosc cells were labeled with [35S] methionine, and DCPD proteins present in cell lysates were either immunoprecipitated by a polyclonal antibody (B) or pulled down by a GST–pre-S construct (C). Note that the N-25 and C-81 deletion mutants of DCPD were unable to bind the pre-S protein (C). The failure of the N-25 mutant protein to interact with GST–pre-S constructs may be due to protein degradation, since the N-25 construct migrated faster than the expected size (B).
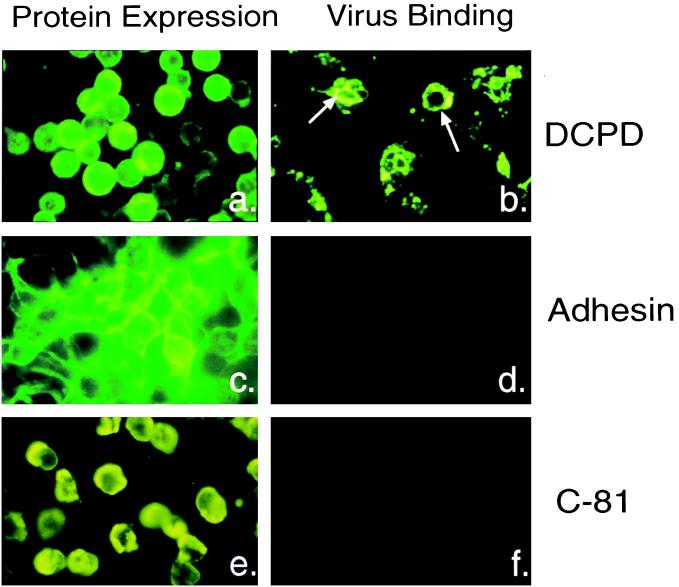
FIG. 2.
Intracellular localization of transfected proteins and subsequent virus binding properties of the cells. (a, c, and e) Cellular localization and distribution of full-length DCPD (a), sialoadhesin (c), and DCPD truncation mutant C-81 (e) in transfected Bosc cells; (b, d, and f) binding of DHBV particles to Bosc cells transfected with full-length DCPD (b), sialoadhesin (d), and the C-81 mutant of DCPD (f). Sialoadhesin expression was detected by polyclonal rat antiserum provided by D. Sgroi. Bound viral particles were revealed by an antibody raised against the pre-S domain of viral large envelope protein. The different patterns of IF signal between panels b and a was caused by capping and internalization events following virus binding.
DCPD-reconstituted cells bind DHBV particles.
As demonstrated by indirect IF staining, the DCPD protein appears on the cell surface (Fig. 2a). Cell surface expression was also confirmed by confocal microscopy (data not shown). Quantitative analysis suggest that about 50% of the DCPD protein is accessible since Western blot analysis revealed a 50% reduction of the DCPD protein if cells were trypsin treated prior to cell lysis (data not shown). Incubation of DCPD transfected Bosc cells with viremic duck serum produced binding of DHBV particles as revealed by IF staining with antiserum directed against the pre-S domain of viral large envelope protein (Fig. 2b). Such IF signals were undetectable in Bosc cells transfected with vector backbone (data not shown) or with nonrelevant cell surface proteins such as sialoadhesin (6) (Fig. 2c and d). More importantly, transfection with the C-81 deletion mutant of DCPD also failed to confer virus binding (Fig. 2e and f).
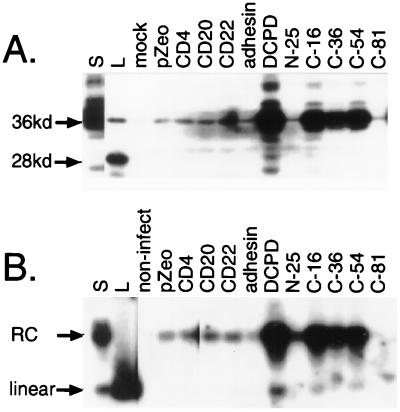
The IF staining results were confirmed by Western blot analysis. The DHBV large envelope protein was found to be efficiently retained only in Bosc cells transfected with cDNAs encoding the full-length DCPD and the three C-terminal deletion mutants that maintained affinity for the GST–pre-S fusion construct (C-16, C-36, and C-54) (Fig. 1C and 3A). Cells transfected with the empty vector or other irrelevant cell surface proteins, such as CD4, CD20 (8), CD22 (21), sialoadhesin (6), and the N-25 or C-81 mutant of DCPD, displayed nonspecific background levels of binding similar to that of mock-transfected cells. Moreover, Southern blot analysis of the same panel of samples revealed a very similar pattern of binding with respect to infectious virion particles (Fig. 3B). Since the vast majority of envelope protein particles present in the duck serum are devoid of viral genomic DNA and hence noninfectious, detection of DHBV DNA indicates the attachment of infectious virion particles to the DCPD-transfected cells. Quantitative analysis revealed that about 10 to 20 DNA-containing particles were retained to each cell reconstituted with the full-length DCPD, assuming that one-half of the Bosc cell population expressed the DCPD protein and all of those cells subsequently became infected. A parallel study of PDH and transiently transfected Bosc cells revealed that Bosc cells overexpressing the DCPD protein compared to PDH will bind more DHBV particles (25a).
FIG. 3.
Binding of DHBV particles to DCPD-reconstituted Bosc cells as revealed by Western and Southern blot analyses. Transfected cells in six-well plates were incubated at 37°C for 8 h with a 1:10 dilution of viremic duck serum. Following a washing step, the cells were mechanically removed. (A) Western blot analysis of viral large envelope protein. Lanes: S, 0.05 μl of the viremic duck serum used for the binding experiment; L, lysate of DHBV-infected duck liver; pZeo, pcDNA3.1/Zeo(−); adhesin, sialoadhesin. Positions of the full-length 36 kDa large envelope protein and the 28-kDa truncated form are indicated. (B) Southern blot analysis of DHBV DNA. Lanes S, DNA corresponding to 0.3 μl of viremic duck serum; L, linear DHBV DNA (20 pg); non-infect, cells not incubated with virus. Positions of the relaxed circular (RC) and linear DHBV DNA forms are indicated.
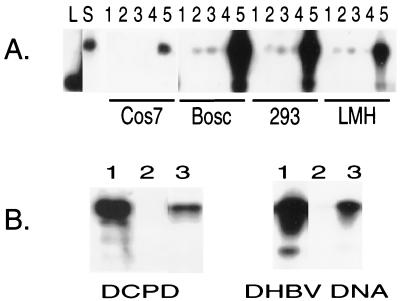
Bosc is a specialized retrovirus packaging cell line. To test the general applicability of DCPD as a DHBV receptor, we attempted reconstitution experiments in several other liver and kidney derived cell lines such as 293, COS (monkey kidney cells), and LMH (chicken hepatoma cells). Transient transfection with the full-length DCPD cDNA conferred highly efficient virus attachment to 293 cells and moderated virus binding to LMH and COS cells (Fig. 4A). Very little virus binding occurred in nontransfected cells and in cells transfected with the plasmid vector or the C-81 deletion mutant construct of DCPD. Therefore, of the liver- and kidney-derived cell lines analyzed thus far, the transfection of a full-length DCPD construct promoted binding of DHBV particles.
FIG. 4.
Binding of DHBV virions to several DCPD-reconstituted cell lines. (A) Transiently transfected cells. The transfection/infection profiles of the samples shown in lanes 1 to 5: 1, nontransfected/not infected; 2, nontransfected/infected; 3, pcDNA3.1/Zeo(−) transfected/infected; 4, C-81 mutant transfected/infected; 5, full-length DCPD transfected/infected. Lanes L and S denote 3-kb linear DHBV DNA and duck serum-derived viral DNA, respectively. (B) 293 cells with stable expression of the DCPD protein. Left, expression of DCPD protein in 293 cells by Western blot analysis; right, viral DNA of bound DHBV particles. Virus binding experiment was carried out at 37°C for 6 h, using a 1:5 dilution of viremic duck serum. For both panels, lanes 1 to 3 represent transiently transfected, nontransfected, and stable transfected 293-4 cells, respectively.
DHBV binding to DCPD-reconstituted cells is inhibited by a virus-neutralizing antibody recognizing pre-S amino acid residues 98 to 104 and by pre-S peptide 80-104.
The previous results establish that DCPD expression in reconstituted cells mediates DHBV binding to the cell surface, but the question remains as to whether DHBV binding to PDH is also mediated by the DCPD protein. Four types of murine monoclonal antibodies raised against the pre-S domain have been previously shown to neutralize DHBV infection of PDH. Of these, three recognize adjacent sequences (pre-S residues 83 to 90, 91 to 99, and 100 to 107 [4, 28]), suggesting that residues 83 to 107 may constitute the receptor contact site. Indeed, pre-S peptide 80-104, which covers the clustered neutralizing epitopes, was found to efficiently inhibit DHBV infection of PDH in a dose-dependent manner (18). Since the minimal p170 (DCPD) binding site was mapped to a similar region (residues 87 to 102 [25]), it is a formal possibility that DCPD is the receptor molecule that mediates DHBV binding in PDH as well.
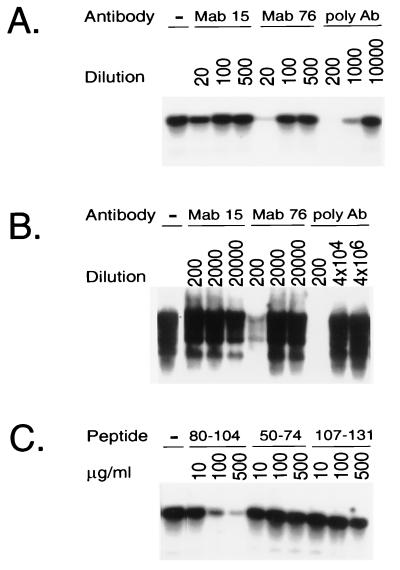
To directly prove that the neutralizing antibodies and the pre-S peptide 80-104 inhibit the DHBV-DCPD interaction, a 293 cell line stably expressing the DCPD protein was obtained by selection with zeocin (200 μg/ml)-containing medium. The 293-4 cell line expresses a level of DCPD protein similar to that found in PDH and actively binds DHBV particles (Fig. 4B). The cells were incubated with DHBV particles in the presence of a rabbit polyclonal pre-S antiserum (as a positive control) or two MAbs, 15 and 76. MAb 15 recognizes a pre-S epitope (residues 112 to 126) outside the DCPD contact site (negative control), whereas MAb 76 recognizes pre-S residues 98 to 104, a part of the putative DCPD contact site (residues 87 to 102). Virus binding was completely blocked by the polyclonal antiserum and nearly abolished by incubation with MAb 76 but not MAb 15 (Fig. 5A). In accordance with this finding, the polyclonal pre-S antibody and, to a lesser extent, MAb 76 inhibited DHBV infection of PDH (Fig. 5B). Thus, there was a good correlation between the antibody ability to inhibit DHBV binding to DCPD-reconstituted cells and the ability to block DHBV infection of PDH.
FIG. 5.
Inhibition of DHBV binding to the stable DCPD expressing 293-4 cell line by either pre-S antibodies or peptides. (A) Effect of anti-pre-S antibodies (Ab) on virus binding. Cells in six-well plates were incubated with a 1:30 dilution of viremic duck serum for 1 h in the presence of various dilution of pre-S antibodies. After a washing step, cells were scraped off the plates and lysed for Southern blot analysis. (B) Effect of anti-pre-S antibodies on DHBV infection of PDH. Virus infection was carried out in the presence of antibodies and viral replication was measured 7 days later (see Materials and Methods). (C) Effect of pre-S peptides on DHBV binding to 293-4 cell line. The pre-S peptides were expressed as GST fusion protein and purified from the GST moiety following thrombin cleavage (18). A 1:50 dilution of viremic duck serum was used, and binding was at 37°C for 1 h.
Additional support for the concept that DCPD is the major DHBV receptor in vivo was provided by the binding experiment carried out in the presence of pre-S peptides. As shown in Fig. 5C, virus binding to DCPD-reconstituted 293-4 cells was significantly inhibited by pre-S peptide 80-104, which is known to block DHBV infection of PDH (18). No inhibition of virus binding was observed with peptides 50-74 and 107-131 (Fig. 5C), which lie outside the critical DCPD binding site.
Internalization of DHBV particles.
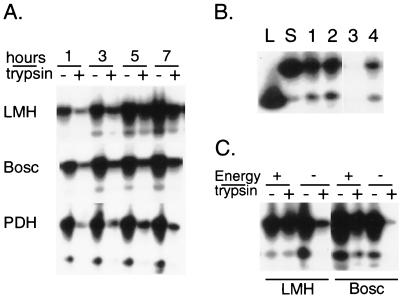
To verify whether bound DHBV particles are internalized, DCPD-reconstituted LMH and Bosc cells were preincubated with the DHBV inoculum for various times at 37°C. Freshly plated PDH were similarly infected in parallel experiments. Cells were washed and removed from the plates by scraping. Half of the sample was used directly for proteinase K digestion and DNA extraction, while the other half was pretreated with trypsin so as to remove viral particles exposed on the cell surface. A gradual increase in the proportion of trypsin-resistant DHBV DNA was observed in DCPD-transfected cells over the 7-h study period (Fig. 6A). Indeed, viral internalization was most efficient in LMH cells. Similar experiments carried out at 4°C, a nonpermissive temperature for virus internalization, resulted in efficient virus binding but lack of viral entry (Fig. 6B). Furthermore, DHBV entry into reconstituted Bosc or LMH cells was inhibited by sodium azide and 2-deoxy-d-glucose (Fig. 6C), the two energy-depleting agents that have been shown to block DHBV internalization into PDH (14).
FIG. 6.
Internalization of DHBV particles into DCPD-reconstituted cell lines. (A) Total and trypsin-resistant fractions of DHBV DNA at different time points of virus incubation. DCPD-reconstituted Bosc and LMH cells as well as freshly plated PDH were incubated at 37°C with a 1:10 dilution of 60 μl viremic duck serum for 1 to 7 h. Half of the cell pellet was pretreated with trypsin before DNA extraction. (B) DHBV entry at 4°C and at 37°C. Two wells of DCPD-transfected LMH cells were incubated with a 1:10 dilution of viremic duck serum at 4°C for 2 h. After washing, cells from one well (lanes 1 and 3) were removed, and half of the cell pellet (lane 3) was treated with trypsin. Cells in the other well (lanes 2 and 4) were further incubated at 37°C for 2 h and half of the cell pellet (lane 4) was treated with trypsin. Lanes L and S are as in Fig. 4. (C) Energy depletion inhibits DHBV entry. DCPD-reconstituted LMH cells were preincubated for 1 h with 600 μl of medium in the presence (energy −) or absence (energy +) of sodium azide (0.1%) and 2-deoxy-d-glucose (50 mM) and incubated for 3 h following the addition of 60 μl of viremic duck serum. The DCPD-transfected Bosc cells were preincubated with energy depleting agents for 3 h and incubated for 12 h with DHBV. Both total and trypsin-resistant fractions of DHBV DNA were measured.
DISCUSSION
The gp180/p170 protein was originally identified as a binding partner for the pre-S region of DHBV large envelope protein (15, 25). Since the pre-S region is believed to be the ligand for the cell surface receptor protein (11, 12, 22), p170/gp180 represents a potential candidate molecule in this regard. The role of p170/gp180 as a DHBV receptor has been strengthened by the following observations. First, the p170/gp180–pre-S interaction is species specific since the chicken homologue binds the pre-S protein poorly if at all (16, 18a). Furthermore, DHBV infection of hepatocytes is also species specific. Second, we have previously found that the interaction between GST–pre-S fusion protein and p170 was competitively inhibited by incubation with DHBV particles, suggesting that the binding site was present on native large envelope protein present on intact viral particles (25). Third, many DHBV mutants with linker substitutions in the pre-S region are no longer infectious when inoculated into PDH cultures (17). Indeed, these pre-S mutant proteins have also been shown to lose binding activity to gp180 (10). Fourth, the critical binding site for p170 has been mapped to pre-S residues 87 to 102, and a pre-S peptide composed of residues 80 to 104 has been found capable of binding p170 molecule (though quite weakly [25]). Of the four types of murine anti-pre-S MAbs that neutralize viral infectivity in cell culture, three antibodies have been shown to bind to this region of the pre-S protein (83 to 90, 91 to 99, and 100 to 107; [4, 28]). The clustering of several neutralizing epitopes in this pre-S domain is consistent with the concept of the location of a receptor contact site. Further support is provided by the observation that a synthetic peptide covering pre-S residues 80 to 104 inhibited DHBV infection of PDH (18).
This study was designed to directly assess the ability of DCPD to mediate virus binding to reconstituted intact cells. By performing comparative studies with PDH, we attempted to verify whether DCPD was the major DHBV receptor in vivo. A significant amount of the DCPD protein expressed in transiently transfected Bosc cells was available on the cell surface, as revealed by IF staining (Fig. 2a) and by Western blot analysis (data not shown). Binding of DHBV particles to DCPD-transfected Bosc cells was independently demonstrated by three techniques: (i) IF staining of the large envelope protein (Fig. 2), (ii) Western blot analysis of the large envelope protein (Fig. 3A), and (iii) Southern blot analysis of viral DNA (Fig. 3B). The specificity of virus binding was established by trace background binding found in nontransfected or mock-transfected Bosc cells and in cells transfected with vector backbone, plasmids encoding nonrelevant cell surface proteins, or DCPD deletion mutants that have lost affinity for the pre-S protein.
Reconstitution of DHBV binding is not restricted to Bosc cells but may also take place in other liver- or kidney-derived cell lines, including 293, COS, and LMH (Fig. 4A). The quantitative difference in virus binding (Bosc > 293 > LMH) may be explained, in part, by a higher level of DCPD expression in Bosc cells due to the presence of simian virus 40 large T antigen as well as by higher transfection efficiency (about 50%) in Bosc and 293 cells than in LMH cells (<10%). In fact, transiently transfected Bosc and 293 cells have higher virus binding capacities than native PDH (25a). However, binding of DHBV particles does not require overexpression of the DCPD receptor protein in reconstituted cells, since a 293-4 cell line that stably expresses low levels of the DCPD protein binds DHBV particles quite well (Fig. 4B).
While our previous study (25) revealed an overlap between the critical p170 binding site on the pre-S domain (amino acids 87 to 102) and the binding sites of three classes of neutralizing MAbs (amino acids 83 to 107), the present study demonstrates directly that such neutralizing monoclonal antibodies strikingly inhibit DHBV binding to DCPD reconstituted cells (Fig. 5A). Furthermore, pre-S peptide 80-104, which covers the clustered neutralizing antibody binding sites, was found to significantly inhibit DHBV infection of PDH (18) as well as block DHBV binding to DCPD reconstituted cells (Fig. 5C). These results together with the ability of energy-depleting reagents to inhibit DHBV entry into both PDH (14) and DCPD-reconstituted cells (Fig. 6C) strongly suggest that DCPD is the major viral receptor in duck hepatocytes.
Trypsin pretreatment of reconstituted cells revealed the presence of internalized DHBV DNA (Fig. 6A). We confirmed that the trypsin-resistant signal represents internalized intracellular DHBV DNA by the following experiments. First, no trypsin-resistant signal was detectable in DCPD-transfected LMH cells preincubated with virus inoculum at a nonpermissive temperature of 4°C, which prevents capping and internalization of viral particles (Fig. 6B and data not shown). Second, the trypsin-resistant signals were greatly reduced or eliminated by performing the viral infection experiments in the presence of energy-depleting agents, as shown by the studies presented in Fig. 6C.
In addition to the observations reported here, several lines of indirect evidence also suggest that DCPD may serve as a DHBV receptor. For example, Sunyach et al. (23) found that pre-S residues 88 to 90, an essential region of the DCPD binding site, were critical for viral binding to PDH and also for viral infectivity. In addition, Bruns and colleagues (3) reported that at a low multiplicity of infection, the pre-S domain enhances DHBV infection. Although the mechanism(s) of this phenomenon remains to be elucidated, the region responsible for such enhancement corresponds to the binding site of p170. Moreover, other investigators have directly tested role of DCPD as the primary DHBV receptor (2, 26). Based on the ability of truncation mutants of the pre-S protein to compete for the interaction between DCPD and immobilized pre-S protein, Breiner et al. (2) proposed that DCPD binding site is composed of a main binding region located at residues 85 to 115 and an auxiliary binding in the N terminus. This finding is in accord with our observation that a small pre-S peptide composed of residues 80 to 104 is capable of binding to p170, although much less efficiently than full-length pre-S protein (25). Finally, it has been determined that soluble DCPD inhibits DHBV infection of PDH in a dose-dependent manner (2). It remains to be determined if pretreatment of PDH with antibodies against DCPD will block subsequent DHBV infection.
Breiner et al. found that DCPD is expressed only in the Golgi apparatus of Huh7 cells transfected with DCPD cDNA (2). They also reported that transfection of DCPD cDNA into Huh7 human hepatocellular carcinoma cells mediated binding of a fusion construct of pre-S–green fluorescent protein as well as fluorescence-labeled viral particles (2). In this regard, DHBV may bind nonspecifically to human hepatocellular carcinoma cell lines such as HepG2 (12, 13, 25a) and Huh7 (25a). In our studies, DCPD transfection failed to increase DHBV binding to Huh7 cells (25a). In the Bosc cells, we found that the DCPD molecule is available on the cell surface (Fig. 2a), and this was confirmed by confocal microscopy (data not shown). Cell surface expression was still detectable if cells were fixed by nonpermeabilizing agents such as formaldehyde. By Western blot analysis, it was found that trypsin treatment could remove approximately one-half of the DCPD signal (25a), which also suggests cell surface expression. Discrepancies related to cellular location may be due in part on the high level of expression of DCPD achieved in transfected Bosc cells.
In conclusion, cumulative evidence from different groups, including the direct transfection experiments, strongly suggest that DCPD serves as an avian hepatitis B virus receptor. It is of interest that another protease, aminopeptidase N, has been found to serve as a coronavirus receptor (7, 27). Whether the enzymatic function of the DCPD protein is required for its biologic function as a viral receptor remains to be established. With the recent cloning of the human homologue of DCPD (24), it will be important to determine whether this molecule can serve as a hepadnavirus receptor for HBV as well. Identification of DCPD as a DHBV receptor has several implications. For example, the availability of transfected cell lines stably expressing the DCPD molecule will provide a useful system for study of the early events of the viral life cycle as well as permit study of antiviral agents that may block binding of virus to its receptor.
Although DCPD-reconstituted cells were capable of binding and internalizing viral particles, no viral replication was observed even in LMH cells, which support DHBV DNA replication when transfected with cloned DHBV DNA (5). Thus, additional factors are required for productive viral replication in established cell lines. Such factors, for example, may be responsible for fusion event(s), nuclear translocation of viral genome, or the repair of viral genome prior to formation of covalently closed circular DNA. In this regard, we have previously identified p120 as a binding partner for truncated pre-S peptide (18). Molecular cloning has revealed p120 to be duck glycine decarboxylase (18a). It will be interesting to test if this molecule can serve as a cofactor for DHBV infection in combination with DCPD.
ACKNOWLEDGMENTS
We are grateful to D. Sgroi for contributing the sialoadhesin cDNA and antibody, A. Luster for the CD4 cDNA, J. Zhao for the CD20 cDNA and antibody, and W. Pear for the Bosc 23 cell line. We thank S. de la Monte for suggestions on IF techniques, R. I. Carlson for preparation of the figures, and members of the Molecular Hepatology Laboratory and the MGH Cancer Center for helpful discussions.
This work was supported in part by National Institutes of Health grants CA-35711, AA-02169, and AA-02666 and grants from the American Cancer Society and the Tan Yan Kee Foundation.
REFERENCES
- 1.Beasley R P, Hwang L Y, Lin C C, Chien C S. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22702 men in Taiwan. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 2.Breiner K M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns M, Miska S, Chassot S, Will H. Enhancement of hepatitis B virus infection by noninfectious subviral particles. J Virol. 1998;72:1462–1468. doi: 10.1128/jvi.72.2.1462-1468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chassot S, Lambert V, Kay A, Godinot A, Roux C, Trepo C, Cova L. Fine mapping of neutralization epitopes on duck hepatitis B virus (DHBV) pre-S protein using monoclonal antibodies and overlapping peptides. Virology. 1993;192:217–223. doi: 10.1006/viro.1993.1024. [DOI] [PubMed] [Google Scholar]
- 5.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crocker P R, Mucklow S, Bouckson V, McWilliam A, Wills A C, Gordon S, Milon G, Kelm S, Bradfield P. Sialoadhesin, a macrophage sialic acid binding receptor for haematopoietic cells with 17 immunoglobulin-like domains. EMBO J. 1994;13:4490–4503. doi: 10.1002/j.1460-2075.1994.tb06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmas B, Gelfi J, L’Haridon R, Vogel L K, Sjostrom H, Noren O, Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Einfeld D A, Brown J P, Valentine M A, Clark E A, Ledbetter J A. Molecular cloning of the human B cell CD20 receptor predicts a hydrophobic protein with multiple transmembrane domains. EMBO J. 1988;7:711–717. doi: 10.1002/j.1460-2075.1988.tb02867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa T, Kuroki K, Lenhoff R, Summers J, Ganem D. Analysis of binding of a host cell surface glycoprotein to the pre-S protein of duck hepatitis B virus. Virology. 1994;202:1061–1064. doi: 10.1006/viro.1994.1440. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa T, Ganem D. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B virus. Proc Natl Acad Sci USA. 1995;92:6259–6263. doi: 10.1073/pnas.92.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klingmuller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kock J, Schlicht H-J. Analysis of the earliest steps of hepadnavirus replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J Virol. 1993;67:4867–4874. doi: 10.1128/jvi.67.8.4867-4874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kock J, Borst E M, Schlicht H J. Uptake of duck hepatitis B virus into hepatocytes occurs by endocytosis but does not require passage of the virus through an acidic intracellular compartment. J Virol. 1996;70:5827–5831. doi: 10.1128/jvi.70.9.5827-5831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroki K, Cheung R, Marion P L, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 17.Lenhoff R J, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Tong S, Wands J R. Characterization of a 120-kilodalton pre-S binding protein as a candidate duck hepatitis B virus receptor. J Virol. 1996;70:6029–6035. doi: 10.1128/jvi.70.9.6029-6035.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Li, J. S., S. P. Tong, and J. R. Wands. Identification and expression of glycine decarboxylase (P120) as a duck hepatitis B virus pre-S envelope binding protein. J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Moradpour D, Wands J R. Hepatic oncogenesis. In: Zakim D, Boyer T, editors. Hepatology, 3rd (W. B. Philadelphia, Pa: Saunders; 1996. pp. 1490–1512. [Google Scholar]
- 20.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamenkovic I, Seed B. The B-cell antigen CD22 mediates monocyte and erythrocyte adhesion. Nature. 1990;345:74–77. doi: 10.1038/345074a0. [DOI] [PubMed] [Google Scholar]
- 22.Summers J, Smith P M, Huang M J, Yu M S. Morphogenic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunyach C, Rollier C, Robaczewska M, Borel C, Barraud L, Kay A, Trepo C, Will H, Cova L. Residues critical for duck hepatitis B virus neutralization are involved in host cell interaction. J Virol. 1999;73:2569–2575. doi: 10.1093/gao/9781884446054.article.t048360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan F, Rehli M, Krause S W, Skidgel R A. Sequence of human carboxypeptidase D reveals it to be a member of the regulatory carboxypeptidase family with three tandem active site domains. Biochem J. 1997;327:81–87. doi: 10.1042/bj3270081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong S, Li J, Wands J R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Tong, S., et al. Unpublished data.
- 26.Urban S, Breiner K, Fehler F, Klingmuller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J Virol. 1998;72:8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeager C L, Ashmun R A, Williams R K, Cardellichio C B, Shapiro L H, Look A T, Holmes K V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuasa S R, Cheung R C, Pham Q, Robinson W S, Marion P L. Peptide mapping of neutralizing and nonneutralizing epitopes of duck hepatitis B virus pre-S polypeptide. Virology. 1991;181:14–21. doi: 10.1016/0042-6822(91)90465-n. [DOI] [PubMed] [Google Scholar]