Abstract
In December 2023, we observed a notable shift in the COVID-19 landscape, when JN.1 omicron emerged as the predominant SARS-CoV-2 variant with a 95% incidence. We characterized the clinical profile, and genetic changes in JN.1, an emerging SARS-CoV-2 variant of interest. Whole genome sequencing was performed on SARS-CoV-2 positive clinical specimens, followed by sequence analysis. Mutations within the spike protein sequences were analysed and compared with the previously reported lineages and sub-lineages, to identify the potential impact of the unique mutations on protein structure and possible alterations in the functionality. Several unique and dynamic mutations were identified herein. Molecular docking analysis showed changes in the binding affinity, and key interacting residues of wild-type and mutated structures with key host cell receptors of SARS-CoV-2 entry viz., ACE2, CD147, CD209L and AXL. Our data provides key insights on the emergence of newer variants and highlights the necessity for robust and sustained global genomic surveillance of SARS-CoV-2.
Keywords: Omicron variant, JN.1 variant, SARS-CoV-2, Whole genome sequencing, Mutation, Spike protein
Subject terms: Computational biology and bioinformatics, Evolution, Microbiology, Medical research
Introduction
The emergence of the novel coronavirus SARS-CoV-2 prompted an urgent need to investigate the global evolutionary dynamics of the virus. Since its initial identification in December 2019, the virus has evolved a slew of mutations, leading to the emergence of several ‘variants of concern’ due to evolutionary dynamics1. Viral mutations inherent to the replication process have led to the emergence of diverse variants characterized by distinct transmissibility and resistance profiles. Since the SARS-CoV-2 pandemic, the virus has evolved predominantly with five variants of concern (alpha, beta, gamma, delta, and omicron). Among these, the omicron variant (B.1.1.529) exhibited more than 50 characteristic mutations in different motifs of the spike protein2. The virus along with its subsequent concerning sub-lineages and variants, evolved with enhanced transmissibility, infectivity and immune evasion mechanism. Notable sub-variants, including BA.2, BA.5, and XBB were of particular interest due to their rapid spread across the globe and evasion from neutralizing and monoclonal antibodies3,4. Specific genetic changes within the virus have conferred selective advantages, augmenting its ability to propagate and evade host immune responses, thereby presenting formidable challenges to containment and development of treatment strategies5.
These dynamic and ever-evolving viral mutations underscore the imperative necessity for consistent monitoring and use of adaptive methodologies, such as whole-genome sequencing. The ongoing surveillance of viral genomes facilitates the prompt identification of emerging variants, playing a pivotal role in devising global public health measures and targeted interventions. Furthermore, a profound understanding of the genetic alterations steering the viral behavior becomes instrumental in developing effective treatments modalities and vaccines, ensuring a resilient response to the constantly evolving viral threats6. The impact of mutations on the protein function is now studied through in silico methods7,8. The difference in protein structures induced by mutations results in different lineages during the viral evolutionary process9. Here, we surveyed the population for SARS-CoV-2 omicron subvariant JN.1 between November 2023 and December 2023 as a part of the state’s public health genomic surveillance investigation, an ongoing program of the Directorate of Public Health and Preventive Medicine, Chennai, Tamil Nadu, India, since September 2021.
Results
Clinico-demographic characteristics of the JN.1 cohort
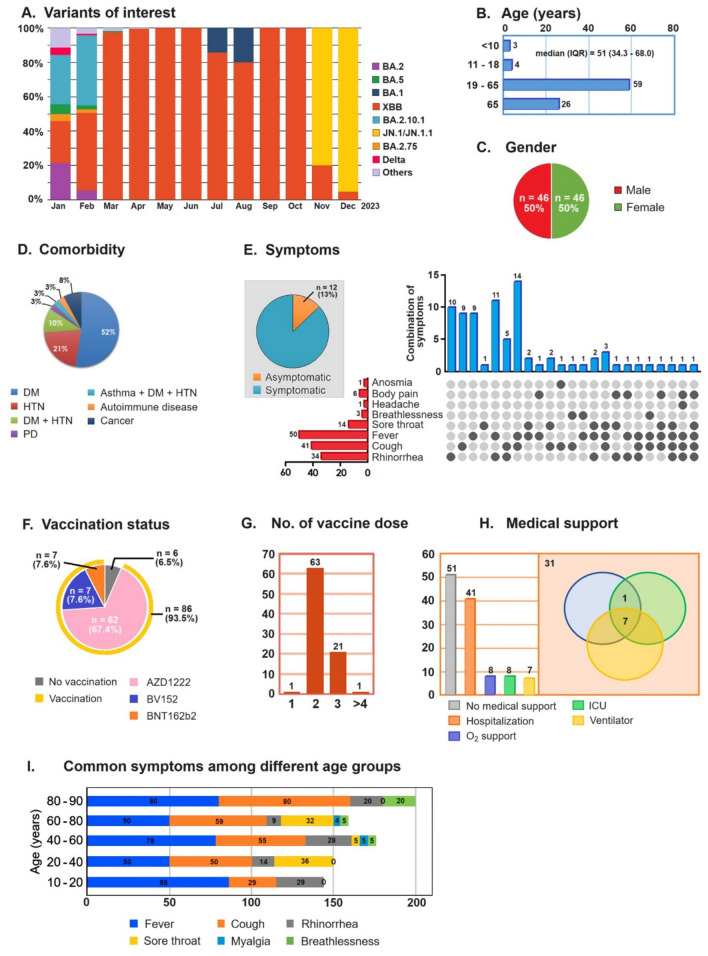
The ongoing SARS-CoV-2 genomic surveillance activities at SPHL have identified the dramatic emergence of the JN.1 variant of omicron by replacing the XBB variant between November 2023 and December 2023 (Fig. 1A). The age of the JN.1 positive patients ranged from 1 to 89 years, with the median age during the study period being 51 years (Fig. 1B) and with an equal proportion of male-to-female ratio (Fig. 1C). More than 50% of the JN.1 patients had underlying diabetes mellitus, followed by 21% of the cases with hypertension, and 10% of the cases had both diabetes mellitus and hypertension (Fig. 1D). About 87% of the patients were presenting with fever, cold, cough and sore throat (pharyngitis) (Fig. 1E,I). On analysing the vaccination status of the JN.1 positive patients, 93.5% had received COVID-19 vaccinations, indicating a high proportion of vaccine breakthrough infections (Fig. 1F). In total, 63 (73.2%) patients had received two doses while 21 (24.4%) patients had received at least 3 doses of vaccines against COVID-19. Among the vaccinated, 98% had received two or more doses of the COVID-19 vaccines (Fig. 1G). About 45% of the JN.1 patients were hospitalized, of which 20% had severe illnesses requiring oxygen support and intensive care unit (ICU) or high dependency unit (HDU) care with ventilator support although no deaths were recorded (Fig. 1H). Among the different symptoms/signs presented, fever (pyrexia) was the most common presentation (69%) and was predominant among the 10–20 years (86%) and 80–90 years (80%) of age. The other common symptoms found were cough (55%) which was predominant in older age groups, rhinorrhea (20%) in younger age groups and sore throat predominant in 20–40 years (36%) and 60–80 years (32%) age groups. Breathlessness (6%) and myalgia (1.8%) were less prevalent, and were mostly noticed in the older age groups only (Fig. 1I). Among the vaccinated, 42% of patients required hospitalization, 12% required oxygen support and ICU/HDU support and 10% required ventilation support.
Figure 1.
Distribution dynamics and clinico-demographic characteristics of the study population. (A) Analysis of the proportions of different SARS-CoV-2 variants circulating in Tamil Nadu over time. The x-axis represents the timeline, the y-axis represents the proportion, and different colors represent distinct variants. (B)–(H) Clinico-demographic characteristics, where (B) median age, (C) gender, (D) symptoms, (E) comorbidity, (F) vaccination status, (G) number of doses received, (H) medical support, and (I) Common symptoms among different age groups. Note: All patients recovered from COVID-19.
Mutational analysis of spike protein of JN.1 omicron SARS-CoV-2 variant
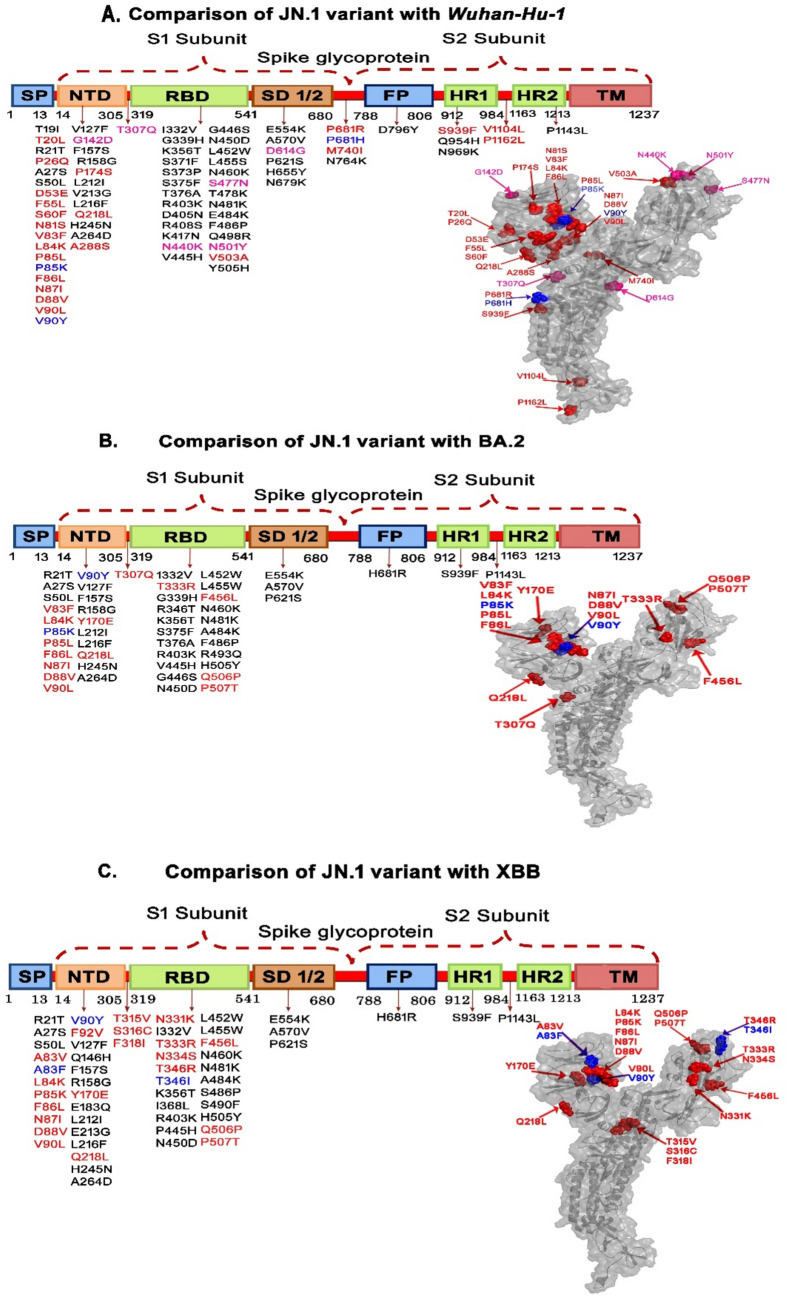
The S gene sequences of the 66 JN.1 variants were analysed with different reference genomes, and revealed a slew of unique mutations in the different domains of the S protein including the receptor binding domain (RBD), signal peptide (SP) and N-terminal domain (NTD) (Fig. 2 and Table 1).
Figure 2.
Amino acid changes in the different domains of JN.1 in comparison with Wuhan-Hu-1, BA.2, and XBB omicron variants. (Red color indicates the unique mutations, the blue color indicates the dynamic mutations, the black color indicates random mutations and pale red indicates universal mutations). Footnotes: FP Fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SP signal peptide, SD spike subdomain, TM transmembrane domain. The aforementioned are the different motifs of the SARS-CoV-2 spike glycoprotein.
Table 1.
Unique and dynamic mutational pattern in spike protein among different lineages.
| S. no | Comparison | Unique mutations (n) | Unique mutations | Dynamic mutations (n) | Dynamic mutations |
|---|---|---|---|---|---|
| 1 | JN.1 variants with Wuhan-Hu-1 | 22 | T20L, P26Q, D53E, F55L, S60F, N81S, V83F, L84K, P85L, F86L, N87I, D88V, V90L, P174S, Q218L, A288S, V503A, P681R, M740I, S939F, V1104L, P1162L | 3 | P85K, V90Y, P681R |
| 2 | JN.1 variants with BA.2 | 14 | V83F, L84K, P85L, F86L, N87I, D88V, V90L, Y170E, Q218L, T307Q, T333R, F456L, Q506P, P507T | 2 | P85L, V90Y |
| 3 | JN.1 variants with XBB | 20 | A83V, L84K, P85K, F86L, N87I, D88V, V90L, F92V, Y170E, Q218L, T315V, S316C, F318I, N331K, T333R, N334S, T346R, F456L, Q506P, P507T | 3 | A83F, V90Y, T346I |
Comparison of spike protein sequences of JN.1 with Wuhan-Hu-1
While comparing the JN.1 isolates with the reference sequence of the Wuhan-Hu-1 strain, we identified 22 unique mutations and three dynamic mutations. Of the unique mutations, the majority were found in the NTD (n = 16), followed by heptad repeat (HR1/2) (n = 3) and spike subdomain (SD) (n = 2), whereas RBD showed only one mutation (V503A). Among the dynamic mutations, two were seen in NTD (P85K and V90Y) and one in the signal domain (P681H). Further, JN.1 showed six universal mutations (three in NTD, two in RBD-N440K and N501Y and one in SD-D614G). This also encompassed certain universal mutations, such as G142D, which have been reported in several countries10,11. Upon analysis of random mutations, we found that JN.1 variants showed 40 mutations across the spike protein as compared to that from Wuhan-Hu-1 with a maximum number of mutations located in the RBD (23) followed by the NTD (12) regions (Table 2 and Fig. 2A).
Table 2.
Unique, dynamic, universal, and random mutations in the spike protein of JN.1 compared to the Wuhan-Hu-1 sequence.
| Unique mutation | Dynamic mutation | Universal mutation | Random mutation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTD | RBD | SD | HR1/HR2 | NTD | RBD | SD | NTD | RBD | SD | NTD | RBD | SD | FP | HR1/HR2 |
| V83F | V503A | P681R | S939F | P85K | – | P681H | G142D | N440K | D614G | T19I | G339H | N764K | D796Y | Q954H |
| L84K | – | M740I | V1104L | V90Y | – | – | T307Q | S477N | – | R21T | K356T | – | – | N969K |
| P85L | – | – | P1162L | – | – | – | – | N501Y | – | A27S | S371F | – | – | P1143L |
| F86L | – | – | – | – | – | – | – | – | – | S50L | S373P | – | – | – |
| N87I | – | – | – | – | – | – | – | – | – | V127F | S375F | – | – | – |
| D88V | – | – | – | – | – | – | – | – | – | F157S | T376A | – | – | – |
| V90L | – | – | – | – | – | – | – | – | – | R158G | R403K | – | – | – |
| Q218L | – | – | – | – | – | – | – | – | – | L212I | D405N | – | – | – |
| T20L | – | – | – | – | – | – | – | – | – | V213G | R408S | – | – | – |
| P26Q | – | – | – | – | – | – | – | – | – | L216F | K417N | – | – | – |
| D53E | – | – | – | – | – | – | – | – | – | H245N | V445H | – | – | – |
| F55L | – | – | – | – | – | – | – | – | – | A264D | G446S | – | – | – |
| S60F | – | – | – | – | – | – | – | – | – | – | N450D | – | – | – |
| N81S | – | – | – | – | – | – | – | – | – | – | L452W | – | – | – |
| P174S | – | – | – | – | – | – | – | – | – | – | L455S | – | – | – |
| A288S | – | – | – | – | – | – | – | – | – | – | N460K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | T478K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | N481K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | E484K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | F486P | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | Q498R | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | Y505H | – | – | – |
FP fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SD spike subdomain, TM transmembrane domain.
Comparison of spike protein sequences of JN.1 with BA.2 omicron variant
Our results from the comparison of JN.1 with the BA.2 variant showed unique mutations, including V83F, L84K, P85L, F86L, N87I, D88V, V90L, Y170E, Q218L, and T307R and dynamic mutations such as P85L, V90Y in the NTD domain, and the following unique mutations T333R, F456L, Q506P, and P507T within the RBD domain. There was no evidence of universal mutations within any of the aforesaid domains. This shows that the amino acid changes as compared to BA.2 were identified individually (Table 3 and Fig. 2B). We also found a total of 34 random mutations when comparing JN.1 and BA.2 with 10 mutations found in the NTD and 18 in the RBD regions. The fusion peptide (FP) domain showed one mutation viz., H681R.
Table 3.
Unique, dynamic, universal, and random mutations in the spike protein of JN.1 as compared to the BA.2 sequence.
| Unique mutation | Dynamic mutation | Universal mutation | Random mutation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTD | RBD | SD | HR1/HR2 | NTD | RBD | SD | NTD | RBD | SD | NTD | RBD | SD | FP | HR1/HR2 |
| V83F | T33R | – | – | P85K | – | – | – | – | – | R21T | I332V | E554K | H681R | S939F |
| L84K | F456L | – | – | V90Y | – | – | – | – | – | A27S | G339H | A570V | – | P1143L |
| P85L | Q506P | – | – | – | – | – | – | – | – | S50L | R346T | P621S | – | |
| F86L | P507T | – | – | – | – | – | – | – | – | V127F | K356T | – | – | – |
| N87I | – | – | – | – | – | – | – | – | – | F157S | S375F | – | – | – |
| D88V | – | – | – | – | – | – | – | – | – | R158G | T376A | – | – | – |
| V90L | – | – | – | – | – | – | – | – | – | L212I | R403K | – | – | – |
| Q218L | – | – | – | – | – | – | – | – | – | L216F | V445H | – | – | – |
| Y170E | – | – | – | – | – | – | – | – | – | H245N | G446S | – | – | – |
| T307Q | – | – | – | – | – | – | – | – | – | A264D | N450D | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | L452W | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | L455W | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | N460K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | N481K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | A484K | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | F486P | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | R493Q | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | H505Y | – | – | – |
FP fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SD spike subdomain, TM transmembrane domain.
Comparison of spike protein sequences of JN.1 with the omicron variant XBB
Comparison of JN.1 with the XBB variant showed no trace of universal mutations, and hosted certain unique, dynamic, and random mutations. The unique mutations in the NTD domain were A83V, L84K, P85K, F86L, N87I, D88V, V90L, F92V, Y170E, and Q218L, besides two dynamic mutations viz., A83F and, V90Y. The unique mutations observed in the RBD domain included N331K, T333R, N334S, T346R, F456L, Q507P, and P507T, besides a dynamic mutation, T346I. The other unique mutations, T315V, S316C and F318I, were spotted in the intermediary region between the NTD and RBD domains (Table 4 and Fig. 2C). Our study showed 33 random mutations (13 in NTD, 14 in RBD, three in SD1/2, two in HR1-2 and one in the FP).
Table 4.
Unique, dynamic, universal, and random mutations in the spike protein of JN.1 as compared to the XBB sequence.
| Unique mutation | Dynamic mutation | Universal mutation | Random mutation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTD | RBD | SD | HR1/HR2 | NTD | RBD | SD | NTD | RBD | SD | NTD | RBD | SD | FP | HR1/HR2 |
| A83V | N331K | – | – | A83F | T346I | – | – | – | – | R21T | I332V | E554K | H681R | S939F |
| L84K | T333R | – | – | V90Y | – | – | – | – | – | A27S | K356T | A570V | – | P1143L |
| P85K | N334S | – | – | – | – | – | – | – | – | S50L | I368L | P621S | – | – |
| F86L | T346R | – | – | – | – | – | – | – | – | V127F | R403K | – | – | – |
| N87I | F456L | – | – | – | – | – | – | – | – | Q146H | P445H | – | – | – |
| D88V | Q506P | – | – | – | – | – | – | – | – | F157S | N450D | – | – | – |
| V90L | P507T | – | – | – | – | – | – | – | – | R158G | L452W | – | – | – |
| F92V | – | – | – | – | – | – | – | – | – | E183Q | L455W | – | – | – |
| Y170E | – | – | – | – | – | – | – | – | – | L212I | N460K | – | – | – |
| Q218L | – | – | – | – | – | – | – | – | – | E213G | N481K | – | – | – |
| T315V | – | – | – | – | – | – | – | – | – | L216F | A484K | – | – | – |
| S316C | – | – | – | – | – | – | – | – | – | H245N | S486P | – | – | – |
| F318I | – | – | – | – | – | – | – | – | – | A264D | S490F | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | H505Y | – | – | – |
FP fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SD spike subdomain, TM transmembrane domain.
Evolutionary mutation patterns in spike protein sequences
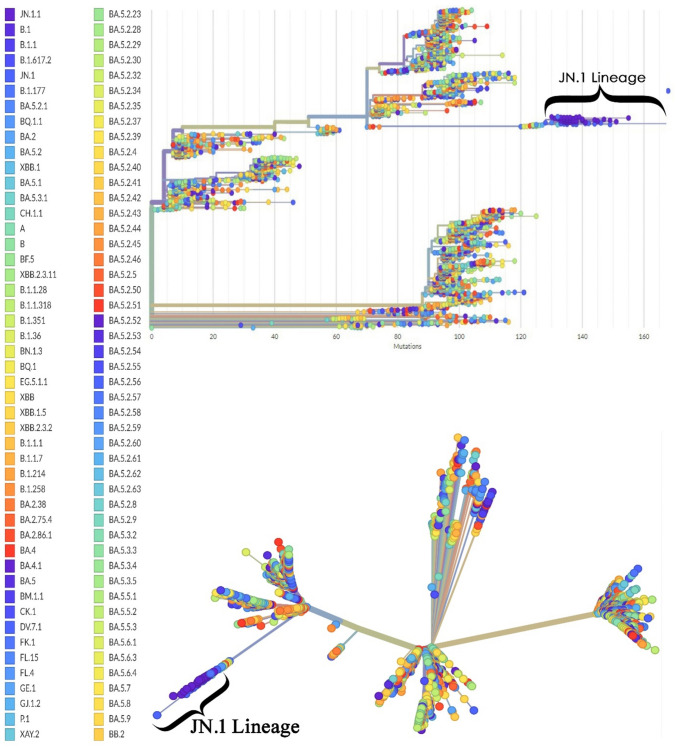
The evolutionary relatedness of JN.1 is presented in a phylogenetic tree that depicts the hierarchy of the different lineages of the selected strain, and the position of the JN.1 lineage in the tree. The JN.1 lineage showed an extended branch length in both rooted and unrooted trees (Fig. 3).
Figure 3.
The phylogenetic analysis of the JN.1 variant as compared with the wild-type Wuhan-Hu-1 genome. Phylogenetic analysis using a rooted tree (top panel) and an unrooted tree (bottom panel) illustrates the evolutionary relatedness among the sequences of JN.1 (n = 66) compared with the reference genome of SARS-CoV-2 (Wuhan-Hu-1).
Mutational and conformational changes in the spike protein of JN.1 omicron variant
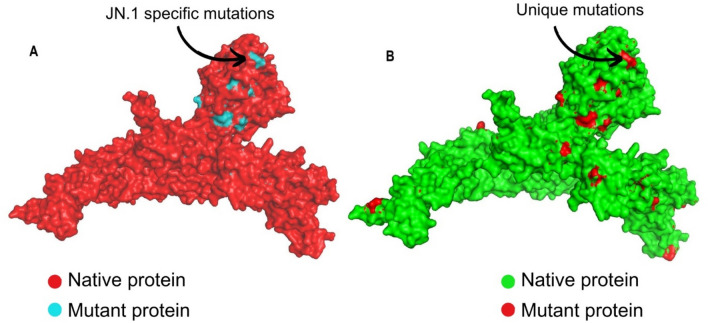
When our study sequences were compared with the reference genomes of different variants, 19 mutations were observed in our strains and the previously reported JN.1 variants (but not seen in other lineages). This indicated that the mutations were unique to the JN.1 variant. The list of mutations and the specific motifs is given in Table 5, and the frequency of these mutations is given in Supplementary Table S1. The other 24 mutations that were observed in a few lineages/sub-lineages but not found in our 66 sequences were identified and listed in Table 6. Intrigued by these mutations, we next elucidated the resulting protein structural changes with the reference protein sequences. The 19 JN.1-specific mutations were created in the model and superimposed with the reference protein. The superimposed structure showed a root mean square deviation (RMSD) value of 0.071 Å (Fig. 4A). The 17 study-specific mutations were created and superimposed with the same model, which showed an RMSD value of 0.081 Å (Fig. 4B).
Table 5.
List of mutations observed in our strains and previously reported JN.1 variants.
| S. no | Mutations | Motif | Subunit |
|---|---|---|---|
| 1 | R21T | NTD | S1 |
| 2 | S50L | NTD | |
| 3 | V127F | NTD | |
| 4 | F157S | NTD | |
| 5 | R158G | NTD | |
| 6 | L216F | NTD | |
| 7 | H245N | NTD | |
| 8 | A270D | NTD | |
| 9 | I332V | RBD | |
| 10 | K356T | RBD | |
| 11 | R403K | RBD | |
| 12 | N450D | RBD | |
| 13 | L455S | RBD | |
| 14 | N481K | RBD | |
| 15 | E554K | SD1/2 | |
| 16 | A570V | SD1/2 | |
| 17 | P621S | SD1/2 | |
| 18 | S939F | HR1 | S2 |
| 19 | P1143L | HR2 |
FP fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SD spike subdomain.
Table 6.
List of mutations observed in a few lineages/sub-lineages but not in our current investigation.
| S. no | Mutations | Motif | Variants bearing the mutation |
|---|---|---|---|
| 1 | L24S | NTD | BA.2; BA.2.75; BA.2.10; BA.4; BA.5 |
| 2 | A67V | NTD | BA.1 |
| 3 | H146Q | NTD | XBB.1 |
| 4 | K147E | NTD | BA.2.75 |
| 5 | W152R | NTD | BA.2.75 |
| 6 | F157L | NTD | BA.2.75 |
| 7 | Q183E | NTD | XBB1 |
| 8 | I210V | NTD | BA.2.75 |
| 9 | L212V | NTD | BA.1 |
| 10 | R214K | NTD | BA.1 |
| 11 | G252V | NTD | BA.1 |
| 12 | G257S | NTD | BA.2.75 |
| 13 | L368I | RBD | XBB1 |
| 14 | Y449S | RBD | BA.1 |
| 15 | F490L | RBD | BA.1 |
| 16 | Q493R | RBD | BA.2;BA.1.1.529;BA2.10 |
| 17 | G496N | RBD | BA.1 |
| 18 | T500A | RBD | BA.1 |
| 19 | G502S | RBD | XBB1 |
| 20 | N658S | SD1/2 | BA.4 |
| 21 | Q755R | SD1/2-FP | BA.2.75 |
| 22 | D950N | HR1 | BA.1.617.2 |
| 23 | L981F | HR1 | BA.1 |
| 24 | S982A | HR1 | BA.1.1 |
| 25 | V1228L | TM | BA.1 |
FP fusion peptide, HR1/2 heptad repeat, NTD N-terminal domain, RBD receptor binding domain, SD spike subdomain, TM transmembrane domain.
Figure 4.
Superimposition of protein models of SARS-CoV-2. (A) Superimposed protein model of Wuhan Hu-1 reference model (red) with JN.1 specific mutations (blue). (B) Superimposed protein model of JN.1 variant (green) with the study-specific unique mutation model identified in our study participants (red).
Molecular docking revealed high binding affinity of the mutated structures of JN.1 compared with Wuhan Hu-1 reference SARS-CoV-2
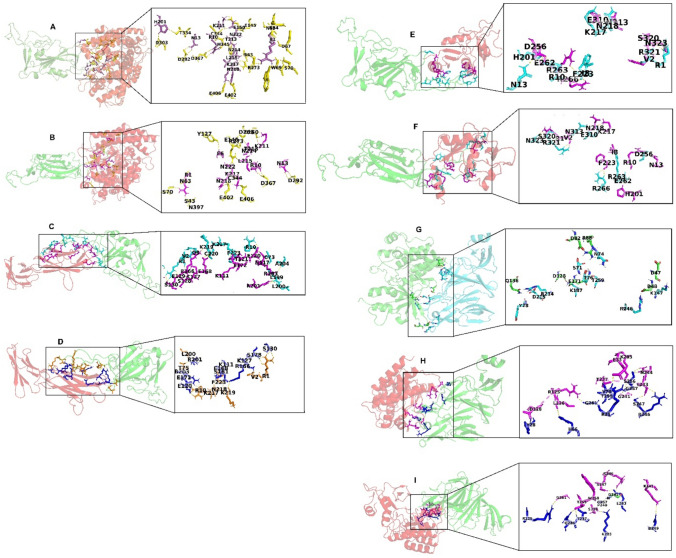
The results of docking different receptors (ACE2, CD147, CD209L and AXL) with wild-type and mutated structures of RBD and NTD are summarized in Table 7. The study indicated a high negative energy value of mutated RBD structures with ACE2, CD147 and CD209L compared to the wild-type. Of these, N481K and R403K showed a significantly low energy score (− 1011.9 kcal/mol, − 945 kcal/mol and − 1183.9 kcal/mol, respectively) demonstrating a strong binding affinity. In contrast, the energy value of mutated NTD structures with AXL was higher compared to the wild-type implying a weak binding affinity. The individual molecular interactions with different receptors are shown in Fig. 5. The key interacting residues of the selected mutated structures with strong binding affinity were analysed and listed in the Supplementary Table S1. The results signify high binding affinity with the mutated structures of JN.1 as compared with the Wuhan Hu-1 wild-type strain.
Table 7.
The binding energies of the interaction of wild-type and mutated structures with different receptors in the molecular docking investigation.
| Mutations (RBD) | Docking score (ACE2) | Docking score (CD147) | Docking score (CD209L) | Mutations (NTD) | Docking score (AXL) |
|---|---|---|---|---|---|
| Wild | − 998.7 | − 922 | − 1170.3 | Wild | − 848.1 |
| I332V | 997.3 | − 910.7 | − 1171.4 | R21T | − 718.2 |
| K356T | − 992.2 | − 932.9 | − 1175.7 | S50L | − 740.0 |
| N450D | − 997.9 | − 942.5 | − 1172.4 | V127F | − 737.5 |
| L455S | − 995 | − 940.9 | − 1171.5 | F157S | − 720.0 |
| N481K | − 1011.9 | − 928.9 | − 1183.7 | R158G | − 716.5 |
| R403K | − 996.4 | − 945 | − 1183.9 | L216F | − 741.8 |
| All | − 959.2 | − 951.2 | − 1177.7 | H245N | − 726.0 |
| A264D | − 740.5 | ||||
| All | − 702.1 |
ACE2 angiotensin-converting enzyme-2, NTD N-terminal domain, RBD receptor binding domain.
Significant values are in bold.
Figure 5.
Molecular interactions of wild-type and mutated structures with host cell receptors. (A) Interaction of wild-type RBD with ACE2. (B) Interaction of N481K mutated RBD with ACE2. (C) Interaction of wild-type RBD with CD147. (D) Interaction of R403K mutated RBD with CD147. (E) Interaction of wild-type RBD with CD209L. (F) Interaction of R403K mutated RBD with CD209L. (G) Interaction of wild-type NTD with AXL. (H) Interaction of R158G mutated NTD with AXL. (I) Interaction of L216F mutated NTD with AXL. Footnotes: ACE2 angiotensin-converting enzyme-2, NTD N-terminal domain, RBD receptor binding domain. AXL is a tyrosine-protein kinase receptor, with potential oncogenic properties; CD147 is an alternate receptor for SARS-CoV-2 entry into host cells with low ACE2 expression; CD209L can also act as a receptor for SARS-CoV-2 entry into susceptible host cells.
Discussion
Our study describes the detection of SARS-CoV-2 omicron subvariant JN.1 during November 2023 and December 2023, as a part of the state public health genomic surveillance activity initiated since September 2021. In our recent investigation, we showed the mutational patterns of omicron variants and the emergence of the XBB as a dominant variant in January 2023 replacing BA.2, which continued till October 202312,13. In September 2023, JN.1 was first identified in the United States14 followed by Canada, France, Singapore and the United Kingdom reporting the expanding global emergence of JN.115–18 thereafter. In India, the JN.1 variant was first identified on 6th October 2023. In Tamil Nadu, the first case of JN.1 was reported on 27th December 2023, which subsequently replaced the XBB variant to evolve into the most predominant SARS-CoV-2 variant in Tamil Nadu, India.
Along with the L455S, a hallmark FLip mutation of JN.1 in the spike protein, the present study identified mutations in different domains of the protein, particularly in the NTD, RBD, SD1/2 and HR1/2 domains. Furthermore, the study identified several mutations when compared to Wuhan-Hu-1, BA.2 and XBB. This indicates the adaptive evolutionary trends of the virus possibly due to reduced neutralizing antibody responses19,20. Since the emergence of JN.1 in August 2023 as a descendant of the BA.2.86 lineage, it drew much attention due to mutation-induced interference in viral binding to angiotensin-binding enzyme (ACE2) receptors21,22. A significant rise in numbers of JN.1 variant, post-vaccinations indicate breakthrough infections with enhanced capacity of immune evasion23. The predecessor XBB variant had 82% of immune evasiveness among the vaccinated individuals12, whereas the current JN.1 variant by acquiring several mutations led to structural changes in the spike protein, conferring 95% of immune evasiveness24. This warranted a detailed investigation on other additional mutations that JN.1 harbored subsequently from its predecessors25. The present study analysed the mutational dynamics of the virus it inherited across the lineages and sub-lineages. Though the study does not represent the true prevalence of the JN.1 in India, the widespread distribution of this variant in the community warrants nationwide genomic surveillance of SARS-CoV-2 under the INSACOG network.
Although viruses are constantly known to evolve due to genetic diversity and evolutionary selection pressure, the clinical and immunological outcomes are always unpredictable. It is therefore important to carry out continuous surveillance on SARS-CoV-2 variants and their mutational analysis26. The mutational pattern leads to virus evolution resulting in the emergence of variants with enhanced transmissibility, severity and immune evasion27. The disappearance and re-appearance of certain mutations (dynamic mutations), during virus evolution with any possible clinical relevance is generally overlooked in evolutionary studies. The present study takes cognizance of such dynamic patterns during virus evolution. The analysis revealed dynamic mutations such as P85L to P85K, V90L to V90Y in the NTD domain and P681R to P681H in the intermediate region between SD1/2 and FP. These dynamic mutations were identified by comparing JN.1 with the Wuhan-Hu-1, BA.2 and XBB. The mutational spectrum in the NTD domain of the spike protein has been an important mechanism in driving antigenic variation and host adaptation28. Studies indicate that potent neutralizing antibodies to NTD interact with residues F140S, G142D, Y145D, K150E, W152R and R158S29. The study also showed that antibodies against both RBD and NTD could efficiently neutralize SARS-CoV-2, limiting the emergence of neutralization-escape mutants.
Our study indicated universal mutation-G142D and common mutations-Q146H and R158G identified in JN.1 variants in line with other studies29,30. These findings further augment the emergence of vaccine-escape mutants. Similarly, by comparing the JN.1 with XBB, we show two dynamic mutations—A83V to A83F, V90L to V90Y in the NTD domain and T346R to T346I in the RBD domain. This indicates that these mutations would possibly contribute to the ongoing evolution of viral lineages, and their overall fitness and adaptability31. Our study identified a dynamic mutation P85K that has gradually evolved from the Wuhan-Hu-1 and BA.2 strain and reshaped into a unique mutation in comparison with XBB12. This indicates the genetic drift within the SARS-CoV-2 infection particularly in the JN.1 variant. These results were further supported by the phylogenetic tree where the JN.1 lineage showed an increased branch length demonstrating both the evolutionary divergence and its relationships with the other lineages of SARS-CoV-232.
The impact of unique mutations identified in the spike protein of the study strains on its structure was analysed by superimposing the mutant model with the wild-type model. The structural alignment between the two models was analysed with the equivalent backbone atoms. Every single mutation causes concomitant changes in protein folding, conformation, physiochemical properties and in situ function33. The conformational changes upon superimposition are analysed with RMSD as a measurement. The predicted structural modifications on ab initio protein models are a known limitation on the robustness yet considered to be the best approach for comprehensive analysis of variants7. Protein function predictions are now studied increasingly through computational methods as the curated protein structures, along with experimentally determined host–pathogen protein interactions8. In the present study, as anticipated, the superimposed structure showed only a slight deviation (< 0.1 Å) compared to the reference strains. Nevertheless, these mutations may be the prelude for further mutations during the evolution of the new sub-lineage. Therefore, monitoring the periodical changes in the virus dynamics is important34. Through the combination of high-throughput sequencing technologies with phylogenetic analysis, it is possible to assess parallel patterns of evolution driving significant phenotypic shifts. These methods offer a framework to measure and predict future evolutionary prospects35.
The present study identified 19 unique mutations among the study sequences—the majority in the NTD and RBD domains followed by the HR1/2 regions. The mutations and the following structural changes in the NTD appear to be implicated in reduced epitope recognition ensuing immune viral escape. The mutations of the RBD domain have been implicated significantly with infectability, transmissibility, and antibody resistance. Spike proteins are known to accumulate multiple mutations upon evolution, subsequently ranking up the virion spike density and infectivity. The present study identified novel mutations of the JN.1 variant, possibly contributing to high transmissibility and immune evasion. In addition, the structural conformation analysis performed by superimposing them with the reference protein structure indicated structural divergence. The considerable deviations measured in conventional RMSD could have been studied further to elucidate its functional variability. Our study also identified 24 mutations that were present in lineages described in the past, which nonetheless appears to have been lost in the JN.1 variant, indicating the active dynamicity of such mutations. These dynamic mutations might play a substantial role in viral evolution and pathogenicity mechanism, and therefore remains a grey area of investigation.
The molecular docking results indicated a high binding affinity of the mutated structures of JN.1 relative to the wild-type Wuhan Hu-1 strain against ACE2, CD147 and CD209L. ACE2 plays a pivotal role in viral entry into a susceptible and permissive host cell. Nonetheless, alternative receptors have also been proposed, including AXL, CD147 and CD209L that likely could modulate the host’s cellular and immune functions36. The variants are reported to use alternate receptors as an immune escape mechanism, resulting in vaccine breakthrough infections. AXL receptors have been shown to augment virus entry significantly, and promote infection of pulmonary airway and bronchial epithelial cells37. Our study revealed reduced binding affinity to AXL receptors as compared to the Wuhan Hu-1 strain indicating a possible immune-escape mechanism. The alterations in the receptor binding properties with different host cell receptors owing to mutations could provide key insights into the pathogenesis mechanisms involving the rapidly evolving SARS-CoV-2 variants. The identification of key interacting residues could lead to the discovery of potential drugs and an effective vaccine construct against emerging variants of SARS-CoV-2.
Conclusions
Recognizing the significance of mutations is pivotal to modify public health strategies and therapeutic interventions. This study aimed to characterize the clinical profiling and genetic modifications of the JN.1 variant relative to the previously reported variants of SARS-CoV-2, including the wild-type Wuhan-Hu-1, BA.2, and XBB. The current study showed that the prevalent mutations in JN.1 may attribute to their immune evasive properties, and acknowledged that the genetic landscape of SARS-CoV-2 is inconsistent. This capability to respond to alterations in the genetic composition of the virus is vital for effective pandemic governance at the global forefront.
Methods
Study design
The study was part of the SARS-CoV-2 genomic surveillance by the State Public Health Laboratory (SPHL), Directorate of Public Health and Preventive Medicine, Chennai, India. COVID‐19 diagnosis was based on clinical and laboratory tests using nasopharyngeal and oropharyngeal swabs as per the guidelines of the Centers for Disease Control and Prevention (CDC), Atlanta, USA. Written informed consent was obtained before sampling. Patients’ demographic details and clinical presentations were collected, including underlying comorbidities, vaccination history, disease progression, and outcome. Of the 471 COVID-19-positive samples reported in Tamil Nadu between November and December 2023, 92 samples with a cycle threshold (Ct) value of < 25 were chosen for whole genome sequencing. RNA extraction was carried out using MagMAX Viral/Pathogen II Nucleic Acid isolation kit (Thermo Fisher Scientific, USA) and tested for SARS-CoV-2 using TaqPath COVID-19 RT-PCR kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The RNA elutes were stored at − 80 °C until further testing.
Ethics approval
The study was approved (EC No. 03092021) by the Institutional Ethics Committee of the Madras Medical College and Hospital, Chennai. The clinical classification was based on the Clinical Guidance for Management of Adult COVID‐19 Patients by the Ministry of Health and Family Welfare, Government of India (January 2022).
Whole genome sequencing
Complementary DNA (cDNA) was prepared from RNA elutes using the SuperScript VILO cDNA Synthesis kit (Invitrogen, Thermo Fisher Scientific, USA) as per the manufacturer’s instructions. Among 471 SARS-CoV-2 positive samples reported during the study period, whole genome sequencing was carried out on 66 of 92 samples with cycle threshold (Ct) values < 25 as per the World Health Organization (WHO) criterion. Ion AmpliSeq library kit (ThermoFisher Scientific, Waltham, USA) was used for the library preparation, and the final library was adjusted to a final concentration of 75 pM using the low TE buffer and loaded onto Ion Chef instrument for emulsion PCR, enrichment, and subsequent placement onto an Ion 540 chip. Next-generation sequencing (NGS) was conducted using the Ion Torrent NGS System using the Ion GenStudio S5 Plus System (Thermo Fisher Scientific, Waltham, USA). Data analysis was performed using Torrent Suite™ software ver.5.18.1. The consensus sequence was analysed using IRMA report ver.1.3.0.2. Annotation was performed using the SnpEff program. The reads were aligned to the Wuhan Hu-1 strain as a reference genome (NCBI ID: NC_045512.2).
Phylogenetic and evolutionary analysis of spike protein
The phylogenetic analysis of SARS-CoV-2 was carried out to identify the mutations and relatedness of the JN.1 variant with other SARS-CoV-2 variants using the Nextclade online software v.3.2.0. (https://clades.nextstrain.org/)38. The nucleotide sequences coding for spike protein from the datasets available in the Nextclade as reference sequences and reference genomes of SARS-CoV-2 variants from the NCBI database were selected and used to detect the mutations. The Nextclade reference datasets, GenBank accession numbers and the list of mutations are provided in Supplementary Table S1. The mutations that were unique to the study sequences are called ‘unique mutations’, whereas those observed in the global JN.1 strains are called ‘universal mutations’. The dynamic mutations characteristically appearing and reappearing in the subsequent variants of SARS-CoV-2 are called ‘dynamic mutations’. Other mutations that were observed with no characteristic pattern were denoted as ‘random mutations’.
Mutations and superimposition of 3D spike protein structure of SARS-CoV-2
Mutations unique to the study sequences were compared with the Wuhan Hu-1 and other major lineages and sub-lineages, including XBB and the other reported JN.1 sequences. Mutations were introduced in the 3D protein structure using PyMol Molecular Graphics System. Using the mutagenesis function and best rotamer based on the frequency of its occurrence and clashes with neighbouring amino acids, the mutation was selected and introduced. A 3D structure model was created with JN.1 specific mutations using Wuhan-Hu-1 as the reference (model A), and a model with unique mutations was identified in our study using the reference JN.1 (model B). The above models were superimposed to identify any structural variations due to these mutations. A 3D protein model for the Wuhan Hu-1 spike protein was built using the Swiss-model protein structure homology modelling (www.swissmodel.expasy.org), upon which mutations were introduced using the Pymol program. The quality of the developed model was analysed using SAVES and Procheck online server programs. The root mean square deviation (RMSD) values were used to assess the mutation-induced structural effects on the protein, where a score of 0 indicates identical structures and a high value indicates higher dissimilarity33,39.
Molecular docking
The impact of the identified mutations on binding to the receptors was analysed using a molecular docking experiment. The PDB structures of the receptors and virus domains were obtained from the protein data bank (www.rcsb.org). The binding of the RBD domain to ACE2 (PDB ID: 6M0J), CD147 (PDB ID: 4U0Q) and CD209L (PDB ID: 1XAR) receptors and the NTD domain to AXL (PDB ID: 5U6B) receptors were analysed. The 3D structures of RBD and NTD domains lacked a few crucial amino acid positions, and hence a 3D model was developed using the Swiss-model protein modelling program for the two proteins. Wuhan-Hu-1 strain sequence served as wild-type and mutations identified in the RBD and NTD regions in our study were created in the structure using the Pymol program. The mutated structures and the wild-type structure were docked against the receptors and compared with each other. Following the energy minimization, docking was performed using the Cluspro 2.0 docking server program (https://cluspro.bu.edu/login.php) with default settings. The top-ordered protein–protein docked complex was selected and the binding scores were analysed. The complex that showed high deviation from the wild-type was analysed further for residue interactions and the number of hydrogen bonds. The interacting residues and hydrogen bonding were identified using Pymol program.
Supplementary Information
Acknowledgements
S.T.S. and S.R. are funded by the National Health Mission, Tamil Nadu (680/NGS/NHMTNMSC/ENGG/2021) for the Directorate of Public Health and Preventive Medicine, WGS facility. M.L. is supported by grants through AI52731, the Swedish Research, the Swedish Physicians against AIDS Research Foundation, the Swedish International Development Cooperation Agency, SIDASARC, VINNMER for Vinnova, Linköping University Research Fund, CALF, and the Swedish Society of Medicine. V.V. is supported by the Office of Research Infrastructure Programs (ORIP/NIH) base grant P51 OD011132 to ENPRC. A.M. is supported by Grant No. 12020/04/2018‐HR, Department of Health Research, Government of India. The funders of this study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The authors thank the Indian SARS CoV‐2 Genomic Consortium (INSACOG), Department of Biotechnology, Ministry of Science and Technology, Government of India for their approval and inclusion of the State Public Health Laboratory (SPHL) as INSAGOG Genomic Sequencing Laboratory (IGSL) vide File No: RAD‐22017/28/2020‐KGDDBT‐Part (6) Dated 29th December 2021. The authors also thank all the national and international members of the Infectious Diseases Society of India (IDSI), Chennai for extending insightful discussions as well as technical and logistic support.
Abbreviations
- ACE2
Angiotensin‐converting enzyme-2
- CDC
Centers for Disease Control and Prevention
- cDNA
Complementary DNA
- COVID‐19
Coronavirus disease 2019
- Ct
Cycle threshold value
- FP
Fusion peptide
- HDU
High dependency unit
- HR
Heptad repeat
- ICU
Intensive care unit
- PCR
Polymerase chain reaction
- NGS
Next‐generation sequencing
- NTD
N‐terminal domain
- QC
Quality control
- RBD
Receptor binding domain
- RMSD
Root mean square deviation
- RT-PCR
Reverse transcription PCR
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SD
Spike subdomain
- SP
Signal peptide
- SPHL
State Public Health Laboratory
- TE
Tris–EDTA
- TM
Transmembrane domain
- VOC
Variant of concern
- WGS
Whole genome sequencing
Author contributions
S.T.S., M.S.K., Y.K.Y., S.Su., A.M., M.L., P.B., S.N.B., V.V., E.M.S., and S.R. designed the study and were responsible for conceptualization and data curation. S.T.S., N.C., K.H., S.Sa, Y.K.Y., H.Y.T., Y.Z., P.A., R.P.P., M.R., A.K., N.G., M.K., M.L., S.Sh, V.V., P.B., E.M.S., and S.R. conducted the analysis, and were responsible for methodology, formal analysis, validation, and visualization. S.Su., Y.K.Y., A.M., E.M.S and S.R. wrote the first draft of the manuscript. All authors reviewed the manuscript.
Funding
S.T.S. and S.R. are funded by the National Health Mission, Tamil Nadu (680/NGS/NHMTNMSC/ENGG/2021) for the Directorate of Public Health and Preventive Medicine, WGS facility. M.L. is supported by grants through AI52731, the Swedish Research Council, the Swedish, Physicians against AIDS Research Foundation, the Swedish International Development Cooperation Agency, SIDASARC, VINNMER for Vinnova, Linköping University Hospital Research Fund, CALF, and the Swedish Society of Medicine. V.V. is supported by the Office of Research Infrastructure Programs (ORIP/NIH) base grant P51 OD011132 to ENPRC. A.M. is supported by Grant No. 12020/04/2018‐HR, Department of Health Research, Government of India. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The authors thank the Indian SARS CoV‐2 Genomics Consortium (INSACOG), Department of Biotechnology, Ministry of Science and Technology, Government of India for their approval and inclusion of the State Public Health Laboratory (SPHL) as INSAGOG Genomic Sequencing Laboratory (IGSL) vide File No: RAD‐22017/28/2020‐KGDDBT‐Part (6) Dated 29th December 2021.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The identified clinical data of patients in the study and the ‘in-house’ bioinformatics pipeline in Python are available on request to the corresponding authors. The whole genome sequences of the study strains have been deposited in the NCBI database. The list of 66 accession numbers is provided in the supplementary table.
Competing interests
Esaki M. Shankar is an Editorial Board Member in the Scientific Reports. There are no conflicts of interest to disclose by other authors.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sivaprakasam T. Selvavinayagam, Sathish Sankar, Yean K. Yong and Suvaiyarasan Suvaithenamudhan.
Contributor Information
Esaki M. Shankar, Email: shankarem@cutn.ac.in
Sivadoss Raju, Email: sivraju@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68678-z.
References
- 1.Zaidi, A. K. & Singh, R. B. SARS-CoV-2 variant biology and immune evasion. Prog. Mol. Biol. Transl. Sci.202, 45–66. 10.1016/bs.pmbts.2023.11.007 (2024). 10.1016/bs.pmbts.2023.11.007 [DOI] [PubMed] [Google Scholar]
- 2.Fan, Y. et al. SARS-CoV-2 omicron variant: Recent progress and future perspectives. Signal Transduct. Target Ther.7, 141. 10.1038/s41392-022-00997-x (2022). 10.1038/s41392-022-00997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selvavinayagam, S. T. et al. Factors associated with the decay of anti-SARS-CoV-2 S1 IgG antibodies among recipients of an adenoviral vector-based AZD1222 and a whole-virion inactivated BBV152 vaccine. Front. Med. (Lausanne)9, 887974. 10.3389/fmed.2022.887974 (2022). 10.3389/fmed.2022.887974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selvavinayagam, S. T. et al. Low SARS-CoV-2 viral load among vaccinated individuals infected with Delta B.1.617.2 and omicron BA.1.1.529 but not with omicron BA.1.1 and BA.2 variants. Front. Public Health10, 1018399. 10.3389/fpubh.2022.1018399 (2022). 10.3389/fpubh.2022.1018399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahilkar, S. et al. SARS-CoV-2 variants: Impact on biological and clinical outcome. Front. Med. (Lausanne)9, 995960. 10.3389/fmed.2022.995960 (2022). 10.3389/fmed.2022.995960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari, M. et al. A critical overview of current progress for COVID-19: Development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci.29, 68. 10.1186/s12929-022-00852-9 (2022). 10.1186/s12929-022-00852-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, D., Redfern, O. & Orengo, C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol.8, 995–1005. 10.1038/nrm2281 (2007). 10.1038/nrm2281 [DOI] [PubMed] [Google Scholar]
- 8.Pál, C., Papp, B. & Lercher, M. J. An integrated view of protein evolution. Nat. Rev. Genet.7, 337–348. 10.1038/nrg1838 (2006). 10.1038/nrg1838 [DOI] [PubMed] [Google Scholar]
- 9.Lomoio, U., Puccio, B., Tradigo, G., Guzzi, P. H. & Veltri, P. SARS-CoV-2 protein structure and sequence mutations: Evolutionary analysis and effects on virus variants. PLoS ONE18, e0283400. 10.1371/journal.pone.0283400 (2023). 10.1371/journal.pone.0283400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, J. J. et al. Analysis of the ARTIC version 3 and version 4 SARS-CoV-2 primers and their impact on the detection of the G142D amino acid substitution in the spike protein. Microbiol. Spectr.9, e0180321. 10.1128/Spectrum.01803-21 (2021). 10.1128/Spectrum.01803-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh, P. et al. Genomic characterization unravelling the causative role of SARS-CoV-2 Delta variant of lineage B.1.617.2 in 2nd wave of COVID-19 pandemic in Chhattisgarh, India. Microb. Pathog.164, 105404. 10.1016/j.micpath.2022.105404 (2022). 10.1016/j.micpath.2022.105404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selvavinayagam, S. T. et al. Clinical characteristics and novel mutations of omicron subvariant XBB in Tamil Nadu, India—A cohort study. Lancet Reg. Health Southeast Asia19, 100272. 10.1016/j.lansea.2023.100272 (2023). 10.1016/j.lansea.2023.100272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selvavinayagam, S. T. et al. Genomic surveillance of omicron B.1.1.529 SARS-CoV-2 and its variants between December 2021 and March 2023 in Tamil Nadu, India—A state-wide prospective longitudinal study. J. Med. Virol.96, e29456. 10.1002/jmv.29456 (2024). 10.1002/jmv.29456 [DOI] [PubMed] [Google Scholar]
- 14.Looi, M. K. Covid-19: WHO adds JN.1 as new variant of interest. BMJ383, 2975. 10.1136/bmj.p2975 (2023). 10.1136/bmj.p2975 [DOI] [PubMed] [Google Scholar]
- 15.Planas, D. et al. Distinct evolution of SARS-CoV-2 omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun.15, 2254. 10.1038/s41467-024-46490-7 (2024). 10.1038/s41467-024-46490-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wannigama, D. L. et al. Wastewater-based epidemiological surveillance of SARS-CoV-2 new variants BA.2.86 and offspring JN.1 in south and Southeast Asia. J. Travel Med.31, taae040. 10.1093/jtm/taae040 (2024). 10.1093/jtm/taae040 [DOI] [PubMed] [Google Scholar]
- 17.Ou, G. et al. Evolving immune evasion and transmissibility of SARS-CoV-2: The emergence of JN.1 variant and its global impact. Drug Discov. Ther.18, 67–70. 10.5582/ddt.2024.01008 (2024). 10.5582/ddt.2024.01008 [DOI] [PubMed] [Google Scholar]
- 18.Kaku, Y. et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis.24, e82. 10.1016/S1473-3099(23)00813-7 (2024). 10.1016/S1473-3099(23)00813-7 [DOI] [PubMed] [Google Scholar]
- 19.Wang, Q. et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell186, 279-286.e278. 10.1016/j.cell.2022.12.018 (2023). 10.1016/j.cell.2022.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel, N. et al. XBB.1.5 spike protein COVID-19 vaccine induces broadly neutralizing and cellular immune responses against EG.5.1 and emerging XBB variants. Sci. Rep.13, 19176. 10.1038/s41598-023-46025-y (2023). 10.1038/s41598-023-46025-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eshraghi, R., Bahrami, A., Karimi Houyeh, M. & Nasr Azadani, M. JN1 and the ongoing battle: Unpacking the characteristics of a new dominant COVID-19 variant. Pathog. Glob. Health24, e82. 10.1080/20477724.2024.2369378 (2024). 10.1080/20477724.2024.2369378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uriu, K. et al. Transmissibility, infectivity, and immune evasion of the SARS-CoV-2 BA.2.86 variant. Lancet Infect. Dis.23, e460–e461. 10.1016/S1473-3099(23)00575-3 (2023). 10.1016/S1473-3099(23)00575-3 [DOI] [PubMed] [Google Scholar]
- 23.Yang, S. et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis.24, e70–e72. 10.1016/S1473-3099(23)00744-2 (2024). 10.1016/S1473-3099(23)00744-2 [DOI] [PubMed] [Google Scholar]
- 24.Karyakarte, R. P. et al. Appearance and prevalence of JN.1 SARS-CoV-2 variant in India and its clinical profile in the state of Maharashtra. Cureus16, e56718. 10.7759/cureus.56718 (2024). 10.7759/cureus.56718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosugi, Y. et al. Characteristics of the SARS-CoV-2 omicron HK.3 variant harbouring the FLip substitution. Lancet Microbe5, e313. 10.1016/S2666-5247(23)00373-7 (2024). 10.1016/S2666-5247(23)00373-7 [DOI] [PubMed] [Google Scholar]
- 26.Singh, J., Pandit, P., McArthur, A. G., Banerjee, A. & Mossman, K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J.18, 166. 10.1186/s12985-021-01633-w (2021). 10.1186/s12985-021-01633-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goga, A. et al. Breakthrough SARS-CoV-2 infections during periods of delta and omicron predominance, South Africa. The Lancet400, 269–271. 10.1016/S0140-6736(22)01190-4 (2022). 10.1016/S0140-6736(22)01190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantoni, D. et al. Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep.23, e54322. 10.15252/embr.202154322 (2022). 10.15252/embr.202154322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haslwanter, D. et al. A combination of receptor-binding domain and N-terminal domain neutralizing antibodies limits the generation of SARS-CoV-2 spike neutralization-escape mutants. mBio12, e0247321. 10.1128/mBio.02473-21 (2021). 10.1128/mBio.02473-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suryadevara, N. et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell184, 2316-2331.e15. 10.1016/j.cell.2021.03.029 (2021). 10.1016/j.cell.2021.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey, W. T. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol.19, 409–424. 10.1038/s41579-021-00573-0 (2021). 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negi, S. S., Schein, C. H. & Braun, W. Regional and temporal coordinated mutation patterns in SARS-CoV-2 spike protein revealed by a clustering and network analysis. Sci. Rep.12, 1128. 10.1038/s41598-022-04950-4 (2022). 10.1038/s41598-022-04950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravantti, J. J., Martinez-Castillo, A. & Abrescia, N. G. A. Superimposition of viral protein structures: A means to decipher the phylogenies of viruses. Viruses12, 1146. 10.3390/v12101146 (2020). 10.3390/v12101146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloom, J. D., Beichman, A. C., Neher, R. A. & Harris, K. Evolution of the SARS-CoV-2 mutational spectrum. Mol. Biol. Evol.40, msad085. 10.1093/molbev/msad085 (2023). 10.1093/molbev/msad085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolan, P. T., Whitfield, Z. J. & Andino, R. Mapping the evolutionary potential of RNA viruses. Cell Host Microbe23, 435–446. 10.1016/j.chom.2018.03.012 (2018). 10.1016/j.chom.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim, S., Zhang, M. & Chang, T. L. ACE2-independent alternative receptors for SARS-CoV-2. Viruses14, 2535. 10.3390/v14112535 (2022). 10.3390/v14112535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, S. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res.31, 126–140. 10.1038/s41422-020-00460-y (2021). 10.1038/s41422-020-00460-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aksamentov, I., Roemer, C., Hodcroft, E. & Neher, R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw.6, 3773. 10.21105/joss.03773 (2021). 10.21105/joss.03773 [DOI] [Google Scholar]
- 39.Carugo, O. & Pongor, S. A normalized root-mean-spuare distance for comparing protein three-dimensional structures. Protein Sci.10, 1470–1473. 10.1110/ps.690101 (2001). 10.1110/ps.690101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The identified clinical data of patients in the study and the ‘in-house’ bioinformatics pipeline in Python are available on request to the corresponding authors. The whole genome sequences of the study strains have been deposited in the NCBI database. The list of 66 accession numbers is provided in the supplementary table.