Abstract
Senescent cells accumulate in various organs with ageing, and its accumulation induces chronic inflammation and age-related physiological dysfunctions. Several remodelling of intracellular environments have been identified in senescent cells, including enlargement of cell/nuclear size and intracellular acidification. Although these alterations of intracellular environments were reported to be involved in the unique characteristics of senescent cells, the contribution of intracellular acidification to senescence-associated cellular phenotypes is poorly understood. Here, we identified that the upregulation of TXNIP and its paralog ARRDC4 as a hallmark of intracellular acidification in addition to KGA-type GLS1. These genes were also upregulated in response to senescence-associated intracellular acidification. Neutralization of the intracellular acidic environment ameliorated not only senescence-related upregulation of TXNIP, ARRDC4 and KGA but also inflammation-related genes, possibly through suppression of PDK-dependent anaerobic glycolysis. Furthermore, we found that expression of the intracellular acidification-induced genes, TXNIP and ARRDC4, correlated with inflammatory gene expression in heterogeneous senescent cell population in vitro and even in vivo, implying that the contribution of intracellular pH to senescence-associated cellular features, such as SASP.
Keywords: glycolysis, intracellular acidification, SASP, senescence
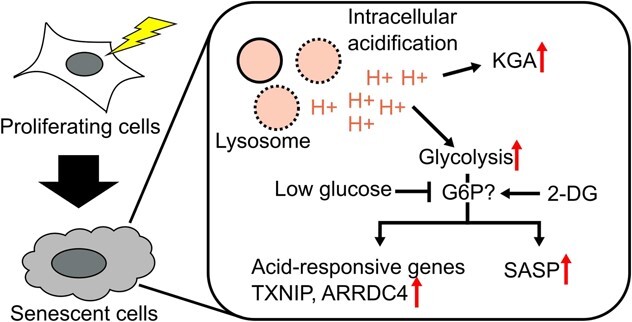
Graphical Abstract
Graphical Abstract.
Abbreviations
- MEF
mouse embryonic fibroblast
- PDK
pyruvate dehydrogenase kinase
- PT
proximal tubular
- ROS
reactive oxygen species
- SASP
senescence-associated secretory phenotype
- TXNIP
thioredoxin-interacting protein
When proliferating cells are exposed to various insults, such as DNA damage or oxidative stress, cells enter an irreversible cell cycle arrest state and become senescent cells. Senescent cells exhibit several unique characteristics such as cellular morphological changes (enlarged cell body and nucleus), expression of CDK inhibitors p16 and p21, abnormal β-galactosidase activity at pH 6 (senescence-associated β-galactosidase, SA-βGal) and secretion of various inflammatory molecules, termed senescence-associated secretory phenotype, SASP (1). Several mouse models have been generated to identify senescent cells and analyse their contributions to physiological ageing and disease in vivo. For example, in INK-ATTAC mice, an engineered caspase-8-FKBP fusion protein and GFP were expressed under the p16 promoter (2). When AP20187 was administered to naturally aged INK-ATTAC mice in order to induce cell death specifically in p16 expressing senescent cells, various ageing phenotypes in multiple organs were ameliorated, and healthy lifespan was prolonged. Previously, we generated p16-CreERT2 mouse model that allows the visualization and elimination of p16-high expressing senescent cells in vivo and found that the elimination of senescent cells prevented the progression of non-alcoholic steatohepatitis (3). Based on these findings, senescent cells are thought to contribute to age-related functional decline.
Several molecular pathways have been proposed as mechanisms by which a small number of senescent cells affect the function of entire organs. SASP is thought to play an important role in senescent cell-induced development of organ dysfunction (4). Mechanistically, the transcription factor NF-κB is activated in senescent cells, leading to the transcription of downstream inflammatory genes (5). The innate immune pathway cGAS-STING also contributes to the development of SASP by sensing cytoplasmic dsDNA generated by genomic instability (6), micronuclei rupture, transposable element activation (7,8) and mitochondrial DNA leakage (9). SASP is also regulated by senescence-associated metabolic signaling, such as the nutrition-sensing pathway mTOR signaling pathways (10,11) and the NAMPT-mediated NAD+ metabolism (12). Recently, it has been reported that glycolysis and lactate production are enhanced in senescent cells and that secreted lactate induces intracellular/extracellular acidification and inflammatory milieu, leading to cancer malignancy (13). Furthermore, we previously revealed that glutaminolysis mediated by GLS1 was essential for senescent cell survival through ammonia production and neutralization of acidic intracellular environment to prevent acidosis (14). Although these previous studies indicate the close relationship between metabolism and SASP, it is not fully understood how each metabolic change regulates inflammatory signaling in senescent cells.
Cellular acidosis occurs in various physiological conditions, such as exercise (15), metabolic dysfunctions (16,17) and cancer (18). In addition to GLS1-mediated ammoniagenesis, intracellular acidification induces a wide variety of metabolic changes, including glycolysis (19), fatty acid metabolism (20) and pentose phosphate pathway (19) to adapt to the environmental changes. Furthermore, it has been reported that intracellular acidification also modulates inflammatory pathways, especially in immune cells and other stromal cells through NLRP3 inflammasome and MAPK pathways (21–23). Although senescence-induced intracellular acidification has been reported in several cell types and conditions (13,14,24), the relationships between the intracellular acidification in senescent cells and their cellular features are still unknown.
Here, we show that intracellular acidification regulates several transcriptomic signatures in senescent cells, including glycolysis and SASP. Neutralization of intracellular acid down-regulated inflammation, which is possibly mediated by suppression of glycolysis. Furthermore, analysis of scRNA-seq datasets revealed that the expression of intracellular acidification-responsive genes may be positively correlated with glycolysis and inflammation even in senescent cell populations in vitro and in vivo. The present study provides a novel insight into the importance of intracellular acidification for senescence-associated traits.
Results
Acidification-induced transcriptome programme is activated in senescent cells
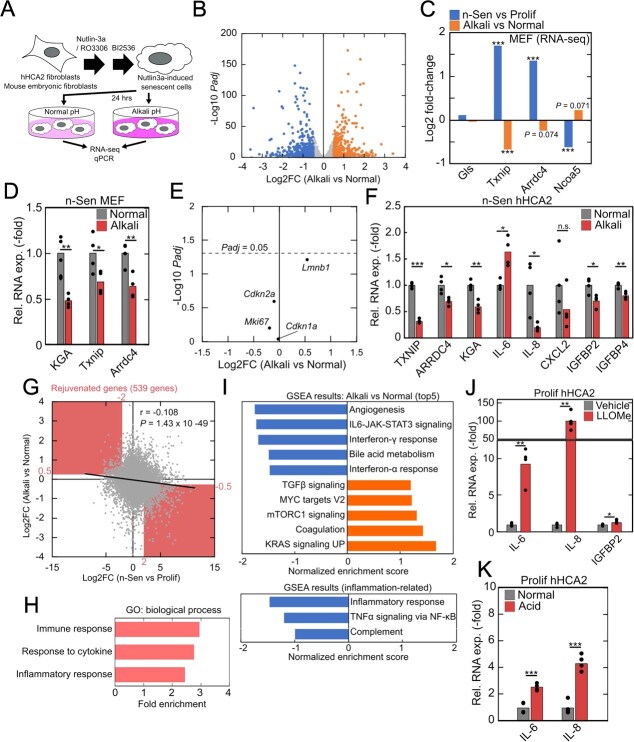
To evaluate how intracellular acidification affects transcriptional properties in senescent cells, we first analysed a public RNA-seq dataset of acidic environment-induced transcriptional reprogramming in human fibroblasts and smooth muscle cells (25) (Fig. 1A). We examined well-expressed, acid-responsive genes in both cell types, focusing on three upregulated genes, GLS, TXNIP and ARRDC4 and a single downregulated gene, NCOA5 (Fig. 1B). GLS (GLS1), especially a kidney-type isoform KGA, is reported to be upregulated by HuC stabilization of its mRNA in acidic environment and neutralized by ammonia production in senescent cells (14). TXNIP and its paralog ARRDC4 belong to the α-arrestin family and have been reported to be induced by acidic conditions such as lactate acidosis (26,27). NCOA5 has been reported as a well-conserved acid-responding gene in rat and human cell lines (25). We validated the acid-induced differential expression of these genes in hHCA2 primary human fibroblasts (28) (Fig. 1C and D) and confirmed that both extracellular acidification and lysosomal damage-induced proton leakage led to similar expression changes in marker gene expression.
Fig. 1.
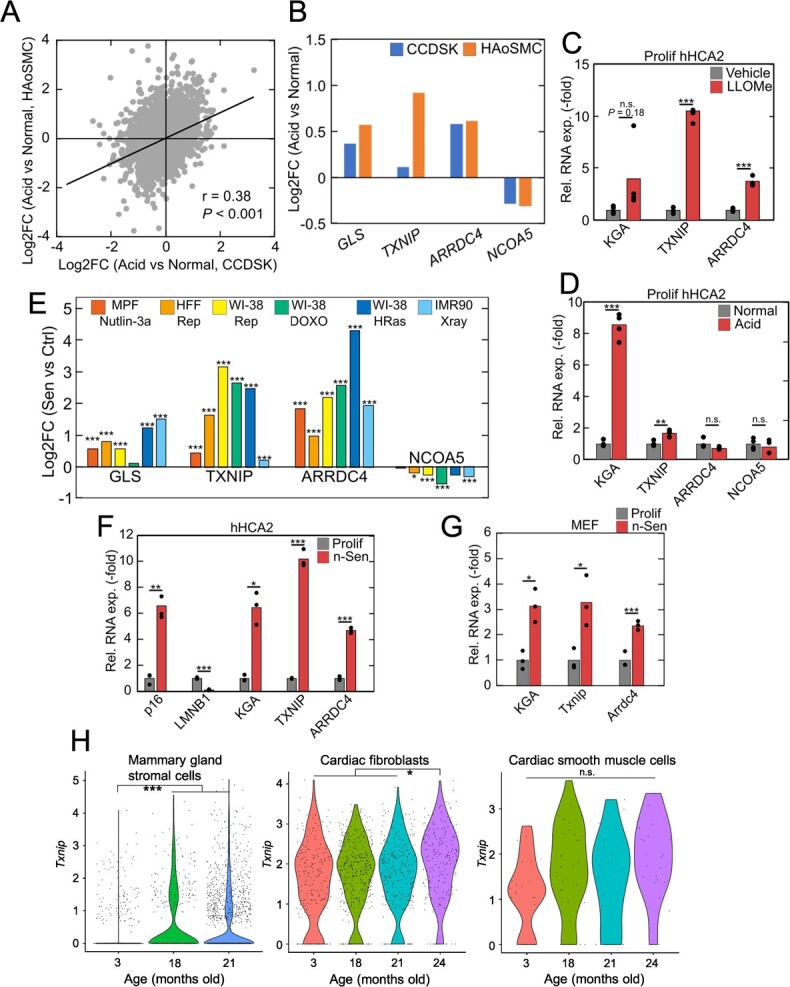
Activation of intracellular acidification-induced gene program in senescent cells. (A) Acid-induced expression changes of protein-coding genes in human fibroblasts (CCDSK) and human smooth muscle cells (HAoSMC) in GSE220788. The significant positive correlation between the two cell lines indicated the similar response to acidic stimulus. (B) Acid-induced expression changes of GLS, TXNIP, ARRDC4 and NCOA5 in the two cell lines of GSE220788. (C, D) Gene expression changes in LLOMe-treated (C) and acid-stimulated (D) hHCA2. (E) Senescence-induced expression changes of GLS, TXNIP, ARRDC4 and NCOA5 in several RNA-seq datasets of senescent cells. The datasets used in this figure were listed in Materials and Methods section. MPF, HFF and Rep indicate mouse primary fibroblasts, human foreskin fibroblasts, and replicative senescence, respectively. (F, G) qPCR validation of the gene expression changes in (E) using Nutlin3a-induced senescent hHCA2 cells and MEFs. (H) Age-related upregulation of Txnip in stromal cells in Tabula Muris Senis datasets. For qPCR analysis, Student’s t test was used, *P < 0.05; **P < 0.01; ***P < 0.001. For RNA-seq analysis, Benjamini–Hochberg method was used, *Padj < 0.05; **Padj < 0.01; ***Padj < 0.001.
Next, we obtained several public RNA-seq datasets of senescent cells in vitro and found that intracellular acidification-induced transcriptional changes were also observed in almost all datasets containing several types of senescent cells, such as nutlin3a-induced senescent cells (29), replicative senescent cells (30), doxorubicin-induced senescent cells and oncogene-induced senescent cells (31) (Fig. 1E). In addition, we prepared nutlin3a-induced senescent cells (n-Sen) from hHCA2 and mouse embryonic fibroblasts (MEFs), and the consistent gene expression changes were observed by qPCR (Fig. 1F and G). Senescence induction was confirmed by the induction of p16 expression and suppression of LMNB1 expression, well-known markers of senescence. We further validated the age-dependent induction of acid-induced transcriptional programme in publicly available single-cell RNA-seq datasets. We analysed mouse stromal cells in the Tabula Muris Senis dataset (32) and found that Txnip was well expressed and showed age-related upregulation in several tissue-derived stromal cells (Fig. 1H). The difference in cardiac smooth muscle cells was not significant probably due to small cell number, but an apparent upregulation in aged population was observed (18 m, 21 m, 24 m vs 3 m: logFC = 0.31, raw P = 0.012).
Together with previous reports that intracellular acidification was detected in senescent cells of various cell types (13,14,24), these results suggest that intracellular acidification-induced transcriptional reprogramming is generally activated in senescent cells.
Intracellular acidification contributes to SASP in senescent cells
The report that acid-induced upregulation of a glutaminase GLS1 is necessary for senescent cell survival suggests that intracellular acidification-induced transcriptional reprogramming may also govern other unique characteristics of senescent cells. To investigate the contribution of intracellular acidification to senescence-associated transcriptional remodelling, we cultured senescent cells in an alkaline environment to neutralize intracellular acid (Fig. 2A, Supplementary Fig. S1). RNA-seq analysis of senescent MEFs revealed global transcriptomic changes with senescence and intracellular neutralization (Fig. 2B). We also obtained transcriptome data of n-Sen and proliferating MEFs and found that senescence-induced expression changes of Txnip and other marker genes were ameliorated by extracellular alkaline environment (Fig. 2C and D), while the expression of Cdkn2a and Cdkn1a and of proliferation-related genes such as Mki67 remained unchanged (Fig. 2E) in our RNA-seq dataset, suggesting that intracellular acidification is not involved in the maintenance of cessation of cell proliferation in senescent cells. We also confirmed similar gene expression changes in n-Sen hHCA2 cells (Fig. 2F). Senescence-associated and alkali-induced expression changes of Gls gene were not drastic in our RNA-seq data of MEFs, probably because the contamination of the other isoform, GAC. Significant expression changes of KGA isoform were observed in response to senescence and alkali treatment (Fig. 2D and F) as described in (14). A weak but significant negative correlation was found between senescence-induced and alkali-induced gene expression changes (Fig. 2G), indicating the transcriptional dependence on intracellular acidification in senescent cells. Genes whose expression changes were normalized in alkali treatment were referred to as ‘rejuvenated genes’, and those genes were enriched in inflammatory pathways (Fig. 2G and H). In addition, in response to the neutralization of acidic intracellular environment, genes related to mTORC1 signaling and Myc signaling were upregulated (Fig. 2I). Previous studies have shown that mTORC1 signaling is involved in lysosome quality control and is inactivated in response to intracellular acidification (33). In contrast, a subset of inflammation-related genes was downregulated in alkali-treated cells (Fig. 2F and I). Upregulation of inflammatory genes was also observed in normal proliferating cells in response to lysosomal proton leak (Fig. 2J) and acidic environment (Fig. 2K), suggesting that intracellular acidification may contribute to the induction or maintenance of inflammation and SASP.
Fig. 2.
Intracellular acidification-dependent gene expression in senescent cells. (A) Schematic illustration of senescence induction and intracellular acid neutralization. (B) Differential gene expression analysis of alkali-treated senescent MEFs. Genes with |Log2FC| > 0.5 and Padj < 0.01 were highlighted. (C-E) Alkali-induced expression changes of pH-responsive genes Gls, Txnip, Arrdc4, and Ncoa5 in n-Sen MEFs (RNA-seq: C, qPCR: D) and senescence-related genes Cdkn2a, Cdkn1a, Mki67, and Lmnb1 (E). (F) qPCR analysis of alkali-induced expression changes of pH-responsive genes in n-Sen hHCA2. (G) Senescence-induced and alkali-induced gene expression changes in MEFs. The significant negative correlation indicated the intracellular acidification dependency of senescence transcriptome. Highlighted area indicates ‘rejuvenated genes’. (H) GO analysis of ‘rejuvenated genes’. (I) GSEA results of differentially expressed genes in response to alkali. Positive values of normalized enrichment score indicate upregulation in alkali-treated senescent MEFs. (J, K) qPCR analysis of gene expression changes in LLOMe-treated (H) and acid-stimulated (I) hHCA2. For qPCR analysis, Student’s t test was used, *P < 0.05; **P < 0.01; ***P < 0.001. For RNA-seq analysis, Benjamini–Hochberg method was used, *Padj < 0.05; **Padj < 0.01; ***Padj < 0.001.
Glycolysis contributes to intracellular acidification-induced inflammation in senescent cells
Increased glucose uptake and glycolysis is one of the major metabolic changes in senescent cells (13,34,35), but contribution of these metabolic changes to SASP remains unknown. Consistent with previous studies, several glycolysis-related genes were upregulated in senescent MEFs in our RNA-seq dataset (Fig. 3A). In general, glycolysis is upregulated by hypoxia and other factors, leading to lactate accumulation, and intracellular acidification, especially in cancer cells (18). Consistent with a previous report (13), we found that expression of pyruvate dehydrogenase kinases (PDKs) and phosphorylation of pyruvate dehydrogenase (PDH) increased in senescent cells, which are a hallmark of anaerobic glycolysis and promote lactate production instead of acetyl-CoA from pyruvate (Fig. 3B). The decline of PDH activity was also observed (Fig. 3C), indicating the enhancement of anaerobic glycolysis in senescent cells. Interestingly, several studies have shown that intracellular acidosis induces the accumulation of glucose-6-phosphate (G6P) to activate the transcription factor MondoA and the transcription of TXNIP and ARRDC4 (27). In addition, the hexokinase2 (HK2)-G6P axis modulates the transcriptional activity of NF-kB and the activation (36) of the inflammasome (37,38). Thus, we speculated that enhanced glycolysis may positively affect intracellular acidification-induced inflammation in senescent cells.
Fig. 3.
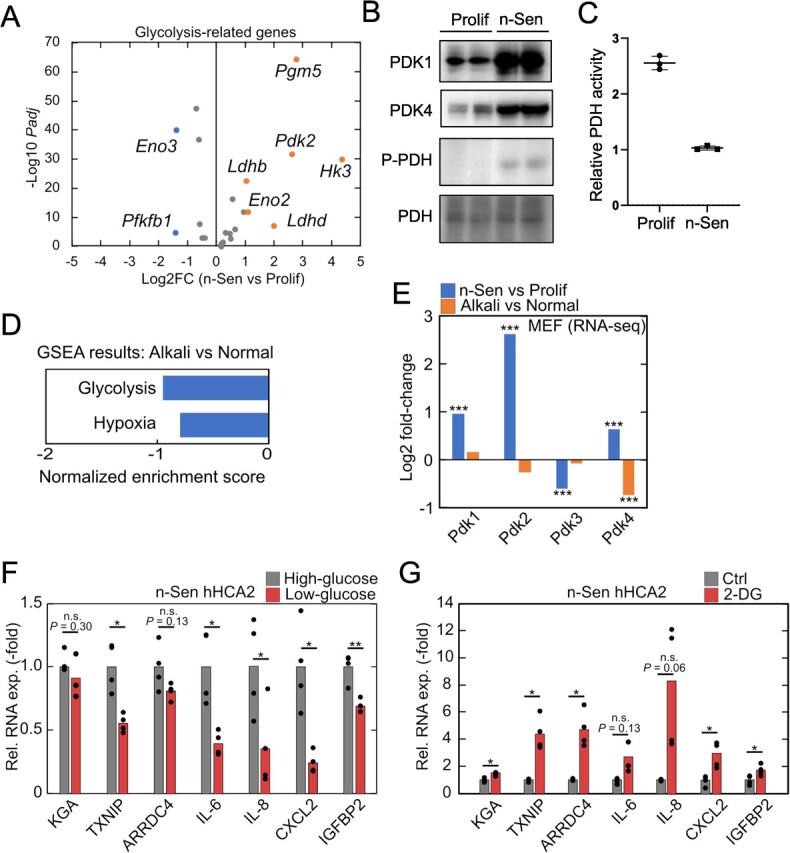
Senescence-associated anaerobic glycolysis-dependent regulation of TXNIP/ARRDC4 and SASP. (A) Senescence-induced expression changes of glycolysis-related genes in our RNA-seq data of MEFs. Genes with |Log2FC| > 0.5 and Padj < 0.01 were highlighted. (B) Immunoblotting of proliferating and n-Sen hHCA2 cells. (C) PDH activity measurement of proliferating and n-Sen hHCA2 cells. (D) GSEA results indicated that glycolysis-related genes were enriched in downregulated genes in response to alkali treatment of senescent MEFs. (E) Senescence-induced and alkali-induced expression changes of pyruvate dehydrogenase kinase genes in our RNA-seq data of MEFs. (F, G) qPCR analysis of gene expression changes in low glucose media-treated (D) and 2-DG-treated (E) n-Sen hHCA2. For qPCR analysis, Student’s t test was used, *P < 0.05; **P < 0.01; ***P < 0.001. For RNA-seq analysis, Benjamini–Hochberg method was used, *Padj < 0.05; **Padj < 0.01; ***Padj < 0.001.
We then examined our RNA-seq data and found that glycolysis-related genes were downregulated in alkali-treated senescent cells (Fig. 3D). Consistent with this, the expression of Pdk4 was significantly downregulated in response to an alkali environment (Fig. 3E), indicating that glycolysis responded to changes in intracellular pH in senescent cells. To investigate whether glycolysis activity contributes to intracellular acidification-induced transcriptional changes and SASP in senescent cells, we cultured senescent cells in low glucose-containing media and found that TXNIP/ARRDC4 and SASP-related genes were downregulated in the glucose-restricted condition (Fig. 3F). In contrast, administration of 2-deoxyglucose (2-DG), a glucose analogue used to block glycolysis, induced opposite expression changes in TXNIP/ARRDC4 and inflammatory genes (Fig. 3G). 2-DG is normally imported into cells and metabolized to 2-deoxyglucose 6-phosphate (2-DG6P) but cannot be further metabolized. The upregulation of inflammation by 2-DG treatment suggests that glucose or G6P, rather than other downstream glycolytic metabolites, may regulate intracellular acidification-induced TXNIP/ARRDC4 expression and inflammation in senescent cells. Taken together, these results suggest that the intracellular acidification-related glycolysis and G6P production may contribute to the strong expression of SASP-related genes in senescent cells.
Expression of intracellular acidification marker genes TXNIP and ARRDC4 predicts increased glycolysis and inflammation in senescent cells
Recent studies have proposed that senescent cells exhibit heterogeneous characteristics, such as immune surveillance and inflammation (3,29,39). Based on our findings that glycolysis and SASP were upregulated in response to intracellular acidification in senescent cells, we hypothesized that the heterogeneity of intracellular acidification may lead to heterogeneous inflammatory states in senescent cells.
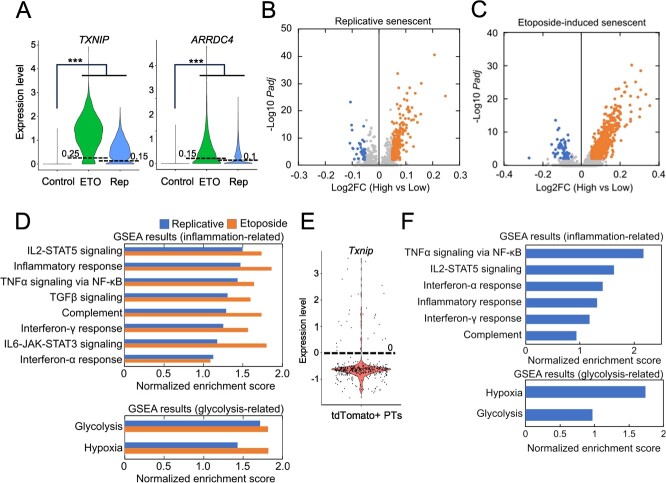
Single-cell RNA-seq techniques allow the analysis of transcriptomic heterogeneity at single-cell resolution and have recently been applied to the field of ageing and senescence. We obtained an scRNA-seq dataset of senescent WI-38 fibroblasts in vitro (40), and a significant upregulation of TXNIP and ARRDC4 was observed in both replicative senescent and etoposide-induced senescent populations (Fig. 4A). We extracted senescent cells highly expressing TXNIP and ARRDC4 (TXNIP/ARRDC4high senescent cells) and found that TXNIP/ARRDC4high senescent cells exhibited higher expression of inflammatory genes and glycolysis-related genes when compared to TXNIP/ARRDC4low senescent cells (Fig. 4B–D). We further analysed our previous scRNA-seq dataset of p16 high-expressing cells (p16high cells) in vivo (3). We previously performed scRNA-seq analysis of p16high senescent cells from adult p16-CreERT2-tdTomato mice and found the heterogeneous senescence-associated phenotypes including SASP. In the dataset, tdTomato+ cells represent p16high cells, which were collected from p16-CreERT2-tdTomato mice by FACS. We focused on proximal tubular (PTs) cells in the kidney, and upregulation of inflammatory genes and glycolysis-related genes was observed in Txnip high-expressing, p16high senescent PTs (Fig. 4E and F).
Fig. 4.
Transcriptomic signatures of TXNIP and/or ARRDC4 highly expressing cells in heterogenous senescent cell population. (A) Identification of TXNIP/ARRDC4high senescent WI-38 cells from scRNA-seq datasets. ETO and Rep indicate etoposide-induced and replicative senescent cells, respectively. Lines and values in the violin plots indicate expression thresholds to extract TXNIP/ARRDC4high and TXNIP/ARRDC4low cells. The definition of TXNIP/ARRDC4high cells was described in Materials and Methods part. (B, C) Gene expression changes in TXNIP/ARRDC4high replicative senescent cells (B) and etoposide-induced senescent cells (C). Genes with |Log2FC| > 0.05 and Padj < 0.01 were highlighted as differentially expressed genes. ‘High’ and ‘Low’ indicate TXNIP/ARRDC4high and TXNIP/ARRDC4low senescent cells, respectively. (D) GSEA results of differentially expressed genes in TXNIP/ARRDC4high senescent WI-38 cells. Positive values of normalized enrichment score indicate upregulation in TXNIP/ARRDC4high senescent WI-38 cells. (E) Identification of Txniphigh- p16high senescent proximal tubule cells from the scRNA-seq dataset of kidney. (F) GSEA results of differentially expressed genes in Txniphigh- p16high senescent proximal tubule cells. Positive values of normalized enrichment score indicate upregulation in Txniphigh- p16high senescent proximal tubule cells. For RNA-seq analysis, Benjamini–Hochberg method was used, *Padj < 0.05; **Padj < 0.01; ***Padj < 0.001.
In conclusion, the present study suggests that the intracellular acidification contributes to the upregulation of inflammatory genes in senescent cells. The acid-induced inflammation may be due to increased glycolysis and G6P production, which also activate the transcription of TXNIP and ARRDC4. In addition, the cells highly expressing TXNIP and/or ARRDC4 exhibit strong senescent characteristics, suggesting the importance of intracellular acidification for senescence-associated changes in cellular phenotypes.
Discussion
Intracellular pH is not only strictly maintained by various homeostatic mechanisms (41), but is also used as a mediator to respond to internal/external stimuli, such as metabolic dysregulation (42), infection (43,44) and tumour microenvironment (45). Changes in intracellular pH modulate cellular properties through a variety of gene expression regulation, including transcription (27,46), mRNA stabilization (14,47), phase separation (48), splicing (49) and RNA editing (50). In the present study, we found that intracellular acidification drives several pH-dependent transcriptional programmes in both human and mouse senescent cells (Fig. 1). Neutralization of the acidic intracellular environment partially reversed senescence-associated gene expression changes, including glycolysis-related pathways and inflammation (Fig. 2). Cellular experiments and analysis of scRNA-seq data suggested that glycolysis is tightly coupled with intracellular acidification and inflammation (Figs 3 and 4), although detailed experiments are needed to clarify the molecular links between intracellular acidification and senescence-associated inflammation.
Although our transcriptome analysis revealed that a proportion of senescence-associated gene expression changes depend on intracellular acidification, the molecular mechanisms of pH-dependent transcriptional reprogramming are still elusive. Our previous study showed that an RNA-binding factor, HuC, binds to the AU-rich motif of GLS1 and enhances its expression through RNA stabilization (14). Another RNA-binding factor, HuR, a homologue of HuC also plays an important role in transcriptomic changes in the acidic tumour microenvironment (47). It is possible that HuC stabilizes RNAs of the acid-responsive gene subset in senescent cells. In addition to RNA binding proteins, MondoA also responds to intracellular acidosis through HK2-mediated G6P production (26,27). Previous studies reported that MondoA not only modulates glucose metabolisms (51) but also promotes TFEB activation and autophagy (52,53), indicating that activation of MondoA in acidic environment in senescent cells is possibly a feedback response to maintain intracellular homeostasis. Further studies are needed to identify the factors responsible for intracellular acidification-induced transcriptional remodelling in senescent cells.
Increased glucose uptake and G6P production occur not only in acidic environments but also in response to infection (37), diabetes (54) and cardiac reperfusion injury (55,56). Under these conditions, it is reported that increased glucose uptake is reported to drive ‘unscheduled glycolysis’, resulting in massive accumulation of G6P rather than other glycolysis-related metabolites (57). Unscheduled glycolytic overload and G6P accumulation induce mitochondrial dysfunction, reactive oxygen species (ROSs) production and AGE accumulation, which are also well-known features of senescent cells. Consistent with this, G6PD deficiency, which can lead to G6P accumulation, is known to promote cellular senescence (58). G6P is produced by mitochondria-localized HK2, and G6P suppresses HK2 activity through its dissociation from mitochondria. HK2 dissociation induces mitochondrial dysfunction, mtDNA leakage, mPTP opening and NLRP3 inflammasome activation (37,38,57). Furthermore, dissociated HK2 phosphorylates IκBα, leading to its degradation and activation of NF-κB in cancer cells (36). Based on these previous findings, it is suggested that the HK2-G6P axis is involved in intracellular acidification-induced inflammation in senescent cells.
In addition to G6P and HK2, TXNIP is another candidate of molecular link between intracellular acidification and inflammation. Thioredoxin-interacting protein (TXNIP) not only interacts with thioredoxin and senses redox states but also regulates a wide variety of signaling pathways (59). TXNIP controls glucose metabolism through multiple molecular pathways including the direct binding to and inactivation of GLUT1/4 (60,61). TXNIP also responds to ROS, resulting in its subsequent dissociation from thioredoxin and binding to the NLRP3 inflammasome (62). Recently, it has been reported that thioredoxin binds to NLRP1 inflammasome and inhibits its activity, and that TXNIP is a potential activator of NLRP1 inflammasome (63). Together with our data showing a strong correlation between TXNIP expression and inflammation in senescent cells, it is suggested that TXNIP is involved in the induction of SASP in senescent cells in an intracellular acidification-dependent manner. In contrast, some reports showed that deficiency of TXNIP or its upstream regulator MondoA accelerated cellular senescence (52,64), while age-related upregulation of TXNIP and its contribution to diseases have been well described (59,65,66). It remains to be investigated how TXNIP upregulation contributes to the induction of cellular senescence and the establishment of unique characteristics of senescent cells.
Finally, we found that senescent cells highly expressing TXNIP and/or ARRDC4 exhibited a more inflammatory phenotype in a heterogeneous population of senescent cell in vitro and in vivo (Fig. 4). The acidic intracellular environment in senescent cells is due to lysosomal proton leakage induced by accumulation of protein aggregates (14). Lysosomal dysfunction is also observed in age-related immune activation inflammaging (67) and various age-associated diseases including non-alcoholic fatty liver disease (68) and Alzheimer’s disease (69). These lines of previous work suggest that age-related accumulation of senescent cells with dysfunctional lysosomes and acidic intracellular environment may contribute to chronic inflammation and disease progression. The senescent cells with intracellular acidification may be an attractive therapeutic target for age-associated physiological dysfunctions, e.g. by a TXNIP expression inhibitor SRI-37330 (70).
Materials and Methods
Cells and treatments
The primary MEFs is prepared as described in (71) with minor modifications. Briefly, mouse embryos at E12–14 were isolated, and differentiated parts were manually removed. The remaining fragments were digested in the 0.25% trypsin solution for 30 min at 37 °C after overnight incubation at 4 °C. The digested fragments were triturated with 10 mL pipette in the normal culture media [Dulbecco's Modified Eagle Medium (DMEM, high glucose, Nacalai Tesque) with 10% Fetal Bovine Serum (FBS, Sigma) and 1× penicillin/streptomycin/amphotericin B (PSA, Nacalai Tesque)]. Early passage hHCA2 (28), and MEFs were maintained in the normal culture media at 37 °C under 5% CO2. hHCA2 cells were maintained with normoxia, but MEFs were maintained with 3–5% O2 condition.
Nutlin3a-induced senescent cells (n-Sen) were prepared as described in our previous work (14) with minor modification. Cells were synchronized at G2 phase by treatment with 9 mM of RO3306 (Roche) for 16–20 h, treated with 9 mM of RO3306 and 5 mM of nutlin3a (Sigma-Aldrich) for 8 h and then with 5 mM of nutlin3a for 48 h. After induction of cellular senescence, cells were maintained in 100 nM of BI-2536-containing media for 14 days to eliminate residual-proliferating cells.
For pH-adjusted experiments, CO2 Independent Media (Gibco) with 1× GlutaMAX (Gibco) were used unless otherwise described. pH was adjusted to acidic (6.5) or alkali (8.5) by NaOH and HCl. Cells in culture dishes were cultured in pH-adjusted media for 24 h in a 37 °C incubator under atmospheric conditions before sampling. Neutralization of intracellular pH was measured using pHrodo Red AM (Thermo) under the 24-h treatment of BPTES (Selleck) and decylubiquinone (Cayman), as described previously (14).
For glucose metabolism-related experiments, culture media were replaced to DMEM (low glucose, Nacalai Tesque) with 10% FBS and 1× PSA, or 2-DG (5 mM, TCI chemicals) was added 24 h before sampling.
LLOMe (500 μM, Sigma) was added 24 h before sampling.
RNA extraction and qPCR
Total RNA from cultured cells was extracted using TRIzol (Thermo Fisher) and purified using RNeasy Mini kit (QIAGEN) according to the manufacturer’s instructions. For qPCR analysis, concentration-adjusted RNA samples were reverse-transcribed with ReverTra Ace qPCR RT Master Mix (TOYOBO). Real-time PCR amplifications were performed with THUNDERBIRD® SYBR® qPCR Master Mix (TOYOBO) and monitored with StepOnePlus (Applied Biosystems). The relative expression of each gene was determined by normalization to beta-actin expression for each sample. All the primers used in this study are shown in Supplementary Table S1.
RNA-seq and data processing
Proliferating MEFs, n-Sen MEFs and n-Sen MEFs under pH 7.4/8.5 media (HEPES-buffered normal culture media) were used for transcriptomic analysis. Sequencing of extracted RNA samples was performed at Novogene inc. (China). Briefly, PolyA-tailed mRNAs were isolated, and strand-specific RNA-seq libraries were prepared. Sequencing of 150-bp paired-end reads was performed on the NovaSeq 6000 Sequencing System (Illumina). Sequenced reads were quality-controlled using fastp (v0.23.2) and aligned to mouse 10 mm genome using HISAT2 (v2.1.0). TPM values were calculated with TPMcalculator (v0.0.4), using aligned bam files and a reference gene annotation file (Ensembl 95). Read counts for each gene features were calculated using featureCounts function in subread package (v2.0.2) and used for differential gene expression analysis using DESeq2 (v1.34.0). Genes with |log2FC| > 2 in senescence-associated changes and |log2FC| > 0.5 in alkali-associated changes were used to extract ‘rejuvenated genes’ in response to alkali stimulation (Fig. 2G and H). GO analysis was performed using ShinyGO (v0.77).
Measurement of PDH activity
PDH activity was determined using PDH Enzyme Activity Microplate Assay Kit (abcam, ab109902) according to the manufacturer’s instructions. Cell lysates were prepared from 1 × 107 cells and adjusted to 150 mg/ml. The relative activities were determined based on OD450 values at 30 min after the reaction initiation.
Immunoblotting analyses
Immunoblotting was performed, as described previously (14). Cells were lysed in TBSN buffer (20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.5% NP-40, 5 mM EGTA, 1.5 mM EDTA and 0.5 mM Na3VO4). The resulting lysates were clarified by centrifugation at 15,000× g for 20 min at 4 °C before immunoprecipitation with the specified antibody. For whole lysates, cells or tissues were directly lysed with Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.002% bromophenol blue and 62.5 mM Tris–HCl at pH 6.8). The whole lysates (20–50 μg) were separated by SDS-PAGE, transferred to a PVDF (Immobilon-P; Millipore) membrane and then subjected to immunoblotting with the indicated antibodies using the ECL detection system. Antibodies used in this study were follows: anti-PDK1 (CST, 3062), anti-PDK4 (abcam, ab110336), anti-P-PDHA1 (abcam, ab177461) and anti-PDHA1 (abcam, ab168379).
Data processing of publicly available bulk RNA-seq and scRNA-seq data
The bulk RNA-seq datasets of acid-stimulated human cell lines (GSE220788), n-Sen MPFs (GSE198397), replicative senescent HFF and WI-38 (GSE63577) and damage-induced senescent WI-38 and IMR90 (GSE130727) were obtained from the Gene Expression Omnibus (GEO). Downloaded raw fastq files were processed as described above. The raw matrix files of the single-cell RNA-seq dataset of replication-induced and etoposide-induced WI-38 cells (GSE226225) were obtained from GEO and processed with Seurat (v4.3.0) package in R. Cells with the following criteria were extracted (nFeature_RNA > 3000 & nFeature_RNA < 10,000 & percent.mt < 5 & nCount_RNA < 100,000). Expression levels of each gene were calculated by log-transformation (scale.factor = 10,000) and scaling. TXNIP/ARRDC4high cells were determined as follows: TXNIP > 0.25 and ARRDC4 > 0.15 for the etoposide-induced senescent cells, and TXNIP > 0.15 and ARRDC4 > 0.1 for the replicative senescent cells (Fig. 4A). These thresholds were calculated from the position of the neck of the violin plots. For the single-cell RNA-seq dataset of p16high cells in vivo (GSE155182), processed and quality-controlled data were obtained from GEO, and data of FACSorted tdTomato+ proximal tubule cells were extracted. Txniphigh cells were determined as Txnip > 0, and Arrdc4 was not used because the dataset failed to detect its expression. For the Tabula Muris Senis dataset, processed and quality-controlled data were obtained from the website.
Differential gene expression analysis was performed by FindMarkers function in Seurat, and genes that were expressed in less than 0.1% of cell populations were removed from the results. GSEA analysis was performed by fgsea package (v1.20.0) in R, using ‘Hallmark’ gene sets obtained from MsigDB using msigdbr package (v7.5.1).
Supplementary Material
Supplementary data
Supplementary data are available at JB Online.
Author contributions
MN and YJ conceived the idea of the project. SK, YJ and MN planned the experiments. SK and YJ performed the experiments. SK, YJ and MN analysed the results, and MN and SK wrote the manuscript with editing by all the other authors.
Conflict of interest
M.N. is a Scientific Advisor and a shareholder of reverSASP Therapeutics.
Data availability
The bulk RNA-seq datasets performed in this study were uploaded to Gene Expression Omnibus (GSE252672). All other data needed to evaluate the conclusions in the paper are presented in the paper and/or provided by corresponding authors.
Acknowledgements
We are grateful to Dr. Tei-Wei Wang for the technical assistance. The super-computing resource was provided by the Human Genome Center (University of Tokyo). This study was supported by AMED under Grant Numbers 21zf0127003 (M.N.), 21 cm0106175 (M.N.) and 21gm5010001 (M.N.), 214600040 (Y.J.), and by MEXT/JSPS KAKENHI under Grant Numbers 20H00514 (M.N.), 19H05740 (M.N.), JP18H05026m (Y.J.), JP16H06148 (Y.J.), JP16K15238 (Y.J.), 22KJ0841 (S.K.) and by the Princess Takamatsu Cancer Research Fund (M.N.).
Contributor Information
Satoshi Kawakami, Division of Cancer Cell Biology, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; Department of Biological Sciences, School of Science, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Yoshikazu Johmura, Division of Cancer and Senescence Biology, Cancer Research Institute, Kanazawa University, Kakuma-machi, Kanazawa 920-1192 Japan.
Makoto Nakanishi, Division of Cancer Cell Biology, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan.
REFERENCES
- 1. Muñoz-Espín, D. and Serrano, M. (2014) Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 15, 482–496 [DOI] [PubMed] [Google Scholar]
- 2. Baker, D.J., Childs, B.G., Durik, M., Wijers, M.E., Sieben, C.J., Zhong, J., A. Saltness, R., Jeganathan, K.B., Verzosa, G.C., Pezeshki, A., Khazaie, K., Miller, J.D., and van Deursen, J.M. (2016) Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Omori, S., Wang, T.-W., Johmura, Y., Kanai, T., Nakano, Y., Kido, T., Susaki, E.A., Nakajima, T., Shichino, S., Ueha, S., Ozawa, M., Yokote, K., Kumamoto, S., Nishiyama, A., Sakamoto, T., Yamaguchi, K., Hatakeyama, S., Shimizu, E., Katayama, K., Yamada, Y., Yamazaki, S., Iwasaki, K., Miyoshi, C., Funato, H., Yanagisawa, M., Ueno, H., Imoto, S., Furukawa, Y., Yoshida, N., Matsushima, K., Ueda, H.R., Miyajima, A., and Nakanishi, M. (2020) Generation of a p16 reporter mouse and its use to characterize and target p16high cells in vivo. Cell Metab. 32, 814–828.e6 [DOI] [PubMed] [Google Scholar]
- 4. Birch, J. and Gil, J. (2020) Senescence and the SASP: many therapeutic avenues. Genes Dev. 34, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chien, Y., Scuoppo, C., Wang, X., Fang, X., Balgley, B., Bolden, J.E., Premsrirut, P., Luo, W., Chicas, A., Lee, C.S., Kogan, S.C., and Lowe, S.W. (2011) Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi, A., Loo, T.M., Okada, R., Kamachi, F., Watanabe, Y., Wakita, M., Watanabe, S., Kawamoto, S., Miyata, K., Barber, G.N., Ohtani, N., and Hara, E. (2018) Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 9, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Cecco, M., Ito, T., Petrashen, A.P., Elias, A.E., Skvir, N.J., Criscione, S.W., Caligiana, A., Brocculi, G., Adney, E.M., Boeke, J.D., Le, O., Beauséjour, C., Ambati, J., Ambati, K., Simon, M., Seluanov, A., Gorbunova, V., Slagboom, P.E., Helfand, S.L., Neretti, N., and Sedivy, J.M. (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu, X., Liu, Z., Wu, Z., Ren, J., Fan, Y., Sun, L., Cao, G., Niu, Y., Zhang, B., Ji, Q., Jiang, X., Wang, C., Wang, Q., Ji, Z., Li, L., Esteban, C.R., Yan, K., Li, W., Cai, Y., Wang, S., Zheng, A., Zhang, Y.E., Tan, S., Cai, Y., Song, M., Lu, F., Tang, F., Ji, W., Zhou, Q., Belmonte, J.C.I., Zhang, W., Qu, J., and Liu, G.H. (2023) Resurrection of endogenous retroviruses during aging reinforces senescence. Cell 186, 287–304.e26 [DOI] [PubMed] [Google Scholar]
- 9. Victorelli, S., Salmonowicz, H., Chapman, J., Martini, H., Vizioli, M.G., Riley, J.S., Cloix, C., Hall-Younger, E., Machado Espindola-Netto, J., Jurk, D., Lagnado, A.B., Sales Gomez, L., Farr, J.N., Saul, D., Reed, R., Kelly, G., Eppard, M., Greaves, L.C., Dou, Z., Pirius, N., Szczepanowska, K., Porritt, R.A., Huang, H., Huang, T.Y., Mann, D.A., Masuda, C.A., Khosla, S., Dai, H., Kaufmann, S.H., Zacharioudakis, E., Gavathiotis, E., LeBrasseur, N.K., Lei, X., Sainz, A.G., Korolchuk, V.I., Adams, P.D., Shadel, G.S., Tait, S.W.G., and Passos, J.F. (2023) Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature 622, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herranz, N., Gallage, S., Mellone, M., Wuestefeld, T., Klotz, S., Hanley, C.J., Raguz, S., Acosta, J.C., Innes, A.J., Banito, A., Georgilis, A., Montoya, A., Wolter, K., Dharmalingam, G., Faull, P., Carroll, T., Martínez-Barbera, J.P., Cutillas, P., Reisinger, F., Heikenwalder, M., Miller, R.A., Withers, D., Zender, L., Thomas, G.J., and Gil, J. (2015) mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 17, 1205–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laberge, R.-M., Sun, Y., Orjalo, A.V., Patil, C.K., Freund, A., Zhou, L., Curran, S.C., Davalos, A.R., Wilson-Edell, K.A., Liu, S., Limbad, C., Demaria, M., Li, P., Hubbard, G.B., Ikeno, Y., Javors, M., Desprez, P.Y., Benz, C.C., Kapahi, P., Nelson, P.S., and Campisi, J. (2015) MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 17, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nacarelli, T., Lau, L., Fukumoto, T., Zundell, J., Fatkhutdinov, N., Wu, S., Aird, K.M., Iwasaki, O., Kossenkov, A.V., Schultz, D., Noma, K.I., Baur, J.A., Schug, Z., Tang, H.Y., Speicher, D.W., David, G., and Zhang, R. (2019) NAD+ metabolism governs the proinflammatory senescence-associated secretome. Nat. Cell Biol. 21, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dou, X., Fu, Q., Long, Q., Liu, S., Zou, Y., Fu, D., Xu, Q., Jiang, Z., Ren, X., Zhang, G., Wei, X., Li, Q., Campisi, J., Zhao, Y., and Sun, Y. (2023) PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer malignancy. Nat. Metab. 5, 1887–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johmura, Y., Yamanaka, T., Omori, S., Wang, T.W., Sugiura, Y., Matsumoto, M., Suzuki, N., Kumamoto, S., Yamaguchi, K., Hatakeyama, S., Takami, T., Yamaguchi, R., Shimizu, E., Ikeda, K., Okahashi, N., Mikawa, R., Suematsu, M., Arita, M., Sugimoto, M., Nakayama, K.I., Furukawa, Y., Imoto, S., and Nakanishi, M. (2021) Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science 371, 265–270 [DOI] [PubMed] [Google Scholar]
- 15. Robergs, R.A., Ghiasvand, F., and Parker, D. (2004) Biochemistry of exercise-induced metabolic acidosis. Am. J. Phys. Regul. Integr. Comp. Phys. 287, R502–R516 [DOI] [PubMed] [Google Scholar]
- 16. Kraut, J.A. and Madias, N.E. (2010) Metabolic acidosis: pathophysiology, diagnosis and management. Nat. Rev. Nephrol. 6, 274–285 [DOI] [PubMed] [Google Scholar]
- 17. Wesson, D.E., Buysse, J.M., and Bushinsky, D.A. (2020) Mechanisms of metabolic acidosis–induced kidney injury in chronic kidney disease. J. Am. Soc. Nephrol. 31, 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li, X., Yang, Y., Zhang, B., Lin, X., Fu, X., An, Y., Zou, Y., Wang, J.X., Wang, Z., and Yu, T. (2022) Lactate metabolism in human health and disease. Sig. Transduct. Target Ther. 7, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. LaMonte, G., Tang, X., Chen, J.L.-Y., Wu, J., Ding, C.K.C., Keenan, M.M., Sangokoya, C., Kung, H.N., Ilkayeva, O., Boros, L.G., Newgard, C.B., and Chi, J.T. (2013) Acidosis induces reprogramming of cellular metabolism to mitigate oxidative stress. Cancer Metab. 1, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corbet, C., Pinto, A., Martherus, R., Santiago de Jesus, J.P., Polet, F., and Feron, O. (2016) Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metab. 24, 311–323 [DOI] [PubMed] [Google Scholar]
- 21. Riemann, A., Wußling, H., Loppnow, H., Fu, H., Reime, S., and Thews, O. (2016) Acidosis differently modulates the inflammatory program in monocytes and macrophages. Biochim. Biophys. Acta Mol. Basis Dis. 1862, 72–81 [DOI] [PubMed] [Google Scholar]
- 22. Thi Tran, U. and Kitami, T. (2019) Niclosamide activates the NLRP3 inflammasome by intracellular acidification and mitochondrial inhibition. Commun. Biol. 2, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riemann, A., Ihling, A., Thomas, J., Schneider, B., Thews, O., and Gekle, M. (2015) Acidic environment activates inflammatory programs in fibroblasts via a cAMP–MAPK pathway. Biochim. Biophys. Acta 1853, 299–307 [DOI] [PubMed] [Google Scholar]
- 24. Li, W., Kawaguchi, K., Tanaka, S., He, C., Maeshima, Y., Suzuki, E., and Toi, M. (2023) Cellular senescence triggers intracellular acidification and lysosomal pH alkalinized via ATP6AP2 attenuation in breast cancer cells. Commun. Biol. 6, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubourg, V., Schulz, M.-C., Terpe, P., Ruhs, S., Kopf, M., and Gekle, M. (2023) Hypothesis-generating analysis of the impact of non-damaging metabolic acidosis on the transcriptome of different cell types: Integrated stress response (ISR) modulation as general transcriptomic reaction to non-respiratory acidic stress? PLoS One 18, e0290373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen, J.L.-Y., Merl, D., Peterson, C.W., Wu, J., Liu, P.Y., Yin, H., Muoio, D.M., Ayer, D.E., West, M., and Chi, J.T. (2010) Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 6, e1001093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilde, B.R., Ye, Z., Lim, T.-Y., and Ayer, D.E. (2019) Cellular acidosis triggers human MondoA transcriptional activity by driving mitochondrial ATP production. elife 8, e40199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakanishi, M., Robetorye, R.S., Adami, G.R., Pereira-Smith, O.M., and Smith, J.R. (1995) Identification of the active region of the DNA synthesis inhibitory gene p21Sdi1/CIP1/WAF1. EMBO J. 14, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang, T.-W., Johmura, Y., Suzuki, N., Omori, S., Migita, T., Yamaguchi, K., Hatakeyama, S., Yamazaki, S., Shimizu, E., Imoto, S., Furukawa, Y., Yoshimura, A., and Nakanishi, M. (2022) Blocking PD-L1–PD-1 improves senescence surveillance and ageing phenotypes. Nature 611, 358–364 [DOI] [PubMed] [Google Scholar]
- 30. Marthandan, S., Baumgart, M., Priebe, S., Groth, M., Schaer, J., Kaether, C., Guthke, R., Cellerino, A., Platzer, M., Diekmann, S., and Hemmerich, P. (2016) Conserved senescence associated genes and pathways in primary human fibroblasts detected by RNA-Seq. PLoS One 11, e0154531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Casella, G., Munk, R., Kim, K.M., Piao, Y., de, S., Abdelmohsen, K., and Gorospe, M. (2019) Transcriptome signature of cellular senescence. Nucleic Acids Res. 47, 7294–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The Tabula Muris Consortium (2020) A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walton, Z.E., Brooks, R.C., and Dang, C.V. (2019) mTOR senses intracellular pH through lysosome dispersion from RHEB. BioEssays 41, e1800265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. James, E.L., Michalek, R.D., Pitiyage, G.N., de Castro, A.M., Vignola, K.S., Jones, J., Mohney, R.P., Karoly, E.D., Prime, S.S., and Parkinson, E.K. (2015) Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. J. Proteome Res. 14, 1854–1871 [DOI] [PubMed] [Google Scholar]
- 35. Dörr, J.R., Yu, Y., Milanovic, M., Beuster, G., Zasada, C., Däbritz, J.H.M., Lisec, J., Lenze, D., Gerhardt, A., Schleicher, K., Kratzat, S., Purfürst, B., Walenta, S., Mueller-Klieser, W., Gräler, M., Hummel, M., Keller, U., Buck, A.K., Dörken, B., Willmitzer, L., Reimann, M., Kempa, S., Lee, S., and Schmitt, C.A. (2013) Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 [DOI] [PubMed] [Google Scholar]
- 36. Guo, D., Tong, Y., Jiang, X., Meng, Y., Jiang, H., du, L., Wu, Q., Li, S., Luo, S., Li, M., Xiao, L., He, H., He, X., Yu, Q., Fang, J., and Lu, Z. (2022) Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 34, 1312–1324.e6 [DOI] [PubMed] [Google Scholar]
- 37. Wolf, A.J., Reyes, C.N., Liang, W., Becker, C., Shimada, K., Wheeler, M.L., Cho, H.C., Popescu, N.I., Coggeshall, K.M., Arditi, M., and Underhill, D.M. (2016) Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166, 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baik, S.H., Ramanujan, V.K., Becker, C., Fett, S., Underhill, D.M., and Wolf, A.J. (2023) Hexokinase dissociation from mitochondria promotes oligomerization of VDAC that facilitates NLRP3 inflammasome assembly and activation. Sci. Immunol. 8, eade7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohn, R.L., Gasek, N.S., Kuchel, G.A., and Xu, M. (2023) The heterogeneity of cellular senescence: insights at the single-cell level. Trends Cell Biol. 33, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wechter, N., Rossi, M., Anerillas, C., Tsitsipatis, D., Piao, Y., Fan, J., Martindale, J.L., de, S., Mazan-Mamczarz, K., and Gorospe, M. (2023) Single-cell transcriptomic analysis uncovers diverse and dynamic senescent cell populations. Aging 15, 2824–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Casey, J.R., Grinstein, S., and Orlowski, J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 42. Isom, D.G., Sridharan, V., Baker, R., Clement, S.T., Smalley, D.M., and Dohlman, H.G. (2013) Protons as second messenger regulators of G protein signaling. Mol. Cell 51, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu, Y., Zhou, P., Cheng, S., Lu, Q., Nowak, K., Hopp, A.K., Li, L., Shi, X., Zhou, Z., Gao, W., Li, D., He, H., Liu, X., Ding, J., Hottiger, M.O., and Shao, F. (2019) A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 178, 552–566.e20 [DOI] [PubMed] [Google Scholar]
- 44. Liu, B., Carlson, R.J., Pires, I.S., Gentili, M., Feng, E., Hellier, Q., Schwartz, M.A., Blainey, P.C., Irvine, D.J., and Hacohen, N. (2023) Human STING is a proton channel. Science 381, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kato, Y., Ozawa, S., Miyamoto, C., Maehata, Y., Suzuki, A., Maeda, T., and Baba, Y. (2013) Acidic extracellular microenvironment and cancer. Cancer Cell Int. 13, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Triandafillou, C.G., Katanski, C.D., Dinner, A.R., and Drummond, D.A. (2020) Transient intracellular acidification regulates the core transcriptional heat shock response. elife 9, e54880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohani, N., Hao, L., Alexis, M.S., Joughin, B.A., Krismer, K., Moufarrej, M.N., Soltis, A.R., Lauffenburger, D.A., Yaffe, M.B., Burge, C.B., Bhatia, S.N., and Gertler, F.B. (2019) Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 79, 1952–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aryan, F., Detrés, D., Luo, C.C., Kim, S.X., Shah, A.N., Bartusel, M., Flynn, R.A., and Calo, E. (2023) Nucleolus activity-dependent recruitment and biomolecular condensation by pH sensing. Mol. Cell 83, 4413–4423.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Borsi, L., Allemanni, G., Gaggero, B., and Zardi, L. (1996) Extracellular pH controls pre-mRNA alternative splicing of tenascin-C in normal, but not in malignantly transformed, cells. Int. J. Cancer 66, 632–635 [DOI] [PubMed] [Google Scholar]
- 50. Malik, T.N., Doherty, E.E., Gaded, V.M., Hill, T.M., Beal, P.A., and Emeson, R.B. (2021) Regulation of RNA editing by intracellular acidification. Nucleic Acids Res. 49, 4020–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stoltzman, C.A., Peterson, C.W., Breen, K.T., Muoio, D.M., Billin, A.N., and Ayer, D.E. (2008) Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc. Natl. Acad. Sci. 105, 6912–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamamoto-Imoto, H., Minami, S., Shioda, T., Yamashita, Y., Sakai, S., Maeda, S., Yamamoto, T., Oki, S., Takashima, M., Yamamuro, T., Yanagawa, K., Edahiro, R., Iwatani, M., So, M., Tokumura, A., Abe, T., Imamura, R., Nonomura, N., Okada, Y., Ayer, D.E., Ogawa, H., Hara, E., Takabatake, Y., Isaka, Y., Nakamura, S., and Yoshimori, T. (2022) Age-associated decline of MondoA drives cellular senescence through impaired autophagy and mitochondrial homeostasis. Cell Rep. 38, 110444. [DOI] [PubMed] [Google Scholar]
- 53. Nakamura, S., Karalay, Ö., Jäger, P.S., Horikawa, M., Klein, C., Nakamura, K., Latza, C., Templer, S.E., Dieterich, C., and Antebi, A. (2016) Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7, 10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brereton, M.F., Rohm, M., Shimomura, K., Holland, C., Tornovsky-Babeay, S., Dadon, D., Iberl, M., Chibalina, M.V., Lee, S., Glaser, B., Dor, Y., Rorsman, P., Clark, A., and Ashcroft, F.M. (2016) Hyperglycaemia induces metabolic dysfunction and glycogen accumulation in pancreatic β-cells. Nat. Commun. 7, 13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taegtmeyer, H., Roberts, A.F.C., and Raine, A.E.G. (1985) Energy metabolism in reperfused heart muscle: metabolic correlates to return of function. J. Am. Coll. Cardiol. 6, 864–870 [DOI] [PubMed] [Google Scholar]
- 56. Karlstaedt, A., Khanna, R., Thangam, M., and Taegtmeyer, H. (2020) Glucose 6-phosphate accumulates via phosphoglucose isomerase inhibition in heart muscle. Circ. Res. 126, 60–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rabbani, N. and Thornalley, P.J. (2019) Hexokinase-2 glycolytic overload in diabetes and ischemia–reperfusion injury. Trends Endocrinol. Metab. 30, 419–431 [DOI] [PubMed] [Google Scholar]
- 58. Cheng, M., Ho, H., Wu, Y., and Chiu, D.T.Y. (2004) Glucose-6-phosphate dehydrogenase-deficient cells show an increased propensity for oxidant-induced senescence. Free Radic. Biol. Med. 36, 580–591 [DOI] [PubMed] [Google Scholar]
- 59. Choi, E.-H. and Park, S.-J. (2023) TXNIP: A key protein in the cellular stress response pathway and a potential therapeutic target. Exp. Mol. Med. 55, 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoshihara, E. (2020) TXNIP/TBP-2: a master regulator for glucose homeostasis. Antioxidants 9, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sullivan, W.J., Mullen, P.J., Schmid, E.W., Flores, A., Momcilovic, M., Sharpley, M.S., Jelinek, D., Whiteley, A.E., Maxwell, M.B., Wilde, B.R., Banerjee, U., Coller, H.A., Shackelford, D.B., Braas, D., Ayer, D.E., de Aguiar Vallim, T.Q., Lowry, W.E., and Christofk, H.R. (2018) Extracellular matrix remodeling regulates glucose metabolism through TXNIP destabilization. Cell 175, 117–132.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou, R., Tardivel, A., Thorens, B., Choi, I., and Tschopp, J. (2010) Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 11, 136–140 [DOI] [PubMed] [Google Scholar]
- 63. Zhang, Z., Shibata, T., Fujimura, A., Kitaura, J., Miyake, K., Ohto, U., and Shimizu, T. (2023) Structural basis for thioredoxin-mediated suppression of NLRP1 inflammasome. Nature 622, 188–194 [DOI] [PubMed] [Google Scholar]
- 64. Huy, H., Song, H.Y., Kim, M.J., Kim, W.S., Kim, D.O., Byun, J.E., Lee, J., Park, Y.J., Kim, T.D., Yoon, S.R., Choi, E.J., Lee, C.H., Noh, J.Y., Jung, H., and Choi, I. (2018) TXNIP regulates AKT-mediated cellular senescence by direct interaction under glucose-mediated metabolic stress. Aging Cell 17, e12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ismael, S., Nasoohi, S., Li, L., Aslam, K.S., Khan, M.M., el-Remessy, A.B., McDonald, M.P., Liao, F.F., and Ishrat, T. (2021) Thioredoxin interacting protein regulates age-associated neuroinflammation. Neurobiol. Dis. 156, 105399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. He, Q., Li, Y., Zhang, W., Chen, J., Deng, W., Liu, Q., Liu, Y., and Liu, D. (2021) Role and mechanism of TXNIP in ageing-related renal fibrosis. Mech. Ageing Dev. 196, 111475 [DOI] [PubMed] [Google Scholar]
- 67. Jin, J., Mu, Y., Zhang, H., Sturmlechner, I., Wang, C., Jadhav, R.R., Xia, Q., Weyand, C.M., and Goronzy, J.J. (2023) CISH impairs lysosomal function in activated T cells resulting in mitochondrial DNA release and inflammaging. Nat. Aging 3, 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeng, J., Acin-Perez, R., Assali, E.A., Martin, A., Brownstein, A.J., Petcherski, A., Fernández-del-Rio, L., Xiao, R., Lo, C.H., Shum, M., Liesa, M., Han, X., Shirihai, O.S., and Grinstaff, M.W. (2023) Restoration of lysosomal acidification rescues autophagy and metabolic dysfunction in non-alcoholic fatty liver disease. Nat. Commun. 14, 2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee, J.-H., Yang, D.-S., Goulbourne, C.N., Im, E., Stavrides, P., Pensalfini, A., Chan, H., Bouchet-Marquis, C., Bleiwas, C., Berg, M.J., Huo, C., Peddy, J., Pawlik, M., Levy, E., Rao, M., Staufenbiel, M., and Nixon, R.A. (2022) Faulty autolysosome acidification in Alzheimer’s disease mouse models induces autophagic build-up of Aβ in neurons, yielding senile plaques. Nat. Neurosci. 25, 688–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thielen, L.A., Chen, J., Jing, G., Moukha-Chafiq, O., Xu, G., Jo, S.H., Grayson, T.B., Lu, B., Li, P., Augelli-Szafran, C.E., Suto, M.J., Kanke, M., Sethupathy, P., Kim, J.K., and Shalev, A. (2020) Identification of an anti-diabetic, orally available small molecule that regulates TXNIP expression and glucagon action. Cell Metab. 32, 353–365.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kawakami, S., Yoshitane, H., Morimura, T., Kimura, W., and Fukada, Y. (2022) Diurnal shift of mouse activity by the deficiency of an ageing-related gene Lmna. J. Biochem. 171, 509–518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.