Abstract
Human enteroviruses consist of more than 60 serotypes, reflecting a wide range of evolutionary divergence. They have been genetically classified into four clusters on the basis of sequence homology in the coding region of the single-stranded RNA genome. To explore further the genetic relationships between human enteroviruses and to characterize the evolutionary mechanisms responsible for variation, previously sequenced genomes were subjected to detailed comparison. Bootstrap and genetic similarity analyses were used to systematically scan the alignments of complete genomic sequences. Bootstrap analysis provided evidence from an early recombination event at the junction of the 5′ noncoding and coding regions of the progenitors of the current clusters. Analysis within the genetic clusters indicated that enterovirus prototype strains include intraspecies recombinants. Recombination breakpoints were detected in all genomic regions except the capsid protein coding region. Our results suggest that recombination is a significant and relatively frequent mechanism in the evolution of enterovirus genomes.
Human enteroviruses have been subgrouped into polioviruses (PVs) (3 serotypes), coxsackie A viruses (CAVs) (23 serotypes), coxsackie B viruses (CBVs) (6 serotypes), echoviruses (EVs) (28 serotypes), and enteroviruses 68 to 71, mainly on the basis of pathogenicity in experimental animals. Recent studies have indicated that human enterovirus genomes, approximately 7,500-nucleotide (nt) single-stranded RNA molecules of positive polarity, can be phylogenetically divided into two distinct groups in the 5′ noncoding region (NCR) (nt 1 to 750); PVs, CAV21, CAV24, and enterovirus 70 belong to group I, while all sequenced EVs, CBVs, CAV9, CAV16, and enterovirus 71 form group II (13, 28, 29). In the coding region and the 3′ NCR, group I viruses divide further into clusters C and D and group II viruses divide into clusters A and B. Partial sequence analysis has shown that all enterovirus prototype strains fall into these clades (12, 27, 30). A proposed new species classification for human enteroviruses is based on the four clusters (A to D) (18). PVs, although genetically representatives of cluster C, have been separated as their own species on the basis of unique clinical features and receptor usage.
The spectrum of clinical manifestations of enterovirus infection varies from asymptomatic infections and the common cold to fatal cases of myocarditis and infections of the central nervous system. The high degree of enterovirus diversity is also reflected by the number of cell surface molecules they recognize during entry into the host cell. At least six different membrane proteins are known to interact with human enteroviruses (5). These include members of the immunoglobulin superfamily (poliovirus receptor, intercellular adhesion molecule 1, and coxsackievirus-adenovirus receptor), integrins, and decay accelerating factor, the normal function of which is to protect cells from the action of complement. Expression of virus receptors and other cellular factors interacting with viral macromolecules are important determinants in the pathogenesis of infection.
Since different parts of the enterovirus genome have distinct roles during the replication cycle, they may also evolve differently and possibly exhibit remarkable independence during evolution. The 5′ NCR has two functions: it contains the initiation site for synthesis of the genomic RNA strand and the internal ribosome entry site responsible for initiation of cap-independent translation. The capsid, encoded by the P1 region of the genome, mediates attachment and entry of the virus into target cells and is therefore essential for tissue and host tropism. The capsid is also an important target for host immune responses. The nonstructural (NS) (P2 and P3) region codes for proteins which function in RNA replication, and the 3′ NCR is involved in initiation of synthesis of the complementary RNA strand. The interplay between these elements includes processing of capsid proteins by NS proteases and recognition of replication initiation sites by the polymerase complex.
Mutation and recombination are the mechanistic alternatives for enterovirus evolution. Due to the absence of proofreading activity, the misinsertion rate of the viral polymerase is high, averaging up to one mutation per newly synthesized genome (4). Consequently, enteroviruses, like other RNA viruses, exist as quasispecies, diverse mixtures of virus mutants differing from each other at one or several sites (10). Recombination has been shown to occur between PVs of vaccine and wild-type origin (2, 8). The evidence supports a model of homologous recombination by strand switching (copy choice) (14, 19). For many other RNA viruses, recombination and reassortment have been shown to be important features of viral evolution. Acquisition of new genome segments by influenza A virus strains has been correlated with the initiation of new pandemics and therefore has a profound impact on the biology and evolution of the virus (16). Recently, evidence of recombination between hantaviruses has also been reported (39). Among retroviruses, such as the human immunodeficiency virus (HIV), recombination is a frequent phenomenon and contributes significantly to viral evolution (3, 9, 32, 36). HIV type 1 (HIV-1) strains are assigned to genetic subtypes (A to J), which are defined in a way similar to that of enterovirus clusters (25). It has been estimated that up to 10% of all characterized HIV-1 strains are intersubtype recombinants (32).
To obtain a detailed picture of the genetic relationships and molecular evolution of human enteroviruses, previously sequenced strains were selected for systematic analysis of all genomic regions.
MATERIALS AND METHODS
Bootscanning analysis.
Enteroviral sequences were analyzed systematically for phylogenetic clustering patterns in different genome regions. Full-length genomic sequences of 27 human enteroviruses, 3 swine vesicular disease viruses (SVDVs), and 4 human rhinoviruses (HRVs) available in the databases were analyzed (Table 1). SVDV strains were selected for analysis because of their antigenic and genetic similarity to CBV5 (43), while HRV sequences were included to serve as an outgroup, since they have previously been shown to represent the closest genetic relatives of enteroviruses (11, 13). Sequences were aligned with Clustal W software (41), and the resulting alignment was further optimized manually with Genetic Data Environment (GDE), version 2.2 (40). Regions where gaps had to be introduced in the alignment were deleted from all phylogenetic analyses by the masking feature of GDE. Bootscanning of the aligned sequences was performed as previously described (35). The method makes use of the bootstrapping procedure, which estimates the support for the clustering pattern of the phylogenetic tree by resampling the input sequence data (6). Bootstrap scores of >70% are usually thought to indicate significant support. During the bootscanning procedure, the alignment was divided into sequential segments of 300 nt, overlapping every previous segment by 250 nt. A bootstrapped phylogenetic analysis with 100 replicates was applied to each segment by using the neighbor-joining algorithm. For each segment, bootstrap values for the studied virus clusters were plotted along the genome. The PHYLIP package, version 3.572c, was used for phylogenetic analysis (7). PHYLIP outfiles give bootstrap values for all taxon clusters found in the bootstrap replicates, which permits estimation of support for clusters which are not represented in the consensus tree. The bootscanning package consists of subprograms for extraction of segments from the full-length alignment (chop) and for extraction of bootstrap values from the PHYLIP outfiles generated from each genome segment (analyse and collect). A series of shell scripts was used to perform phylogenetic analyses on each segment. The bootscanning package is available in ANSI-C source code, as well as in Sun and Linux binary formats (36a).
TABLE 1.
Enterovirus and rhinovirus strains used in analysis
| Serotype (strain) | Abbreviation | Speciesa | 5′ NCR groupb | Accession no.c |
|---|---|---|---|---|
| Coxsackievirus A9 (Griggs) | CAV9 | B | II | D00627 |
| Coxsackievirus A16 (G-10) | CAV16 | A | II | U05876 |
| Coxsackievirus A21 (Coe) | CAV21 | C | I | D00538 |
| Coxsackievirus A24 (EH24/70) | CAV24 | C | I | D90457 |
| Coxsackievirus B1 (Japan) | CBV1 | B | II | M16560 |
| Coxsackievirus B2 (Ohio-2) | CBV2 | B | II | AF081485 |
| Coxsackievirus B3 (Nancy) | CBV3N | B | II | M33854 |
| Coxsackievirus B3 (Woodruff) | CBV3W | B | II | U57056 |
| Coxsackievirus B4 (E2) | CBV4E | B | II | S76772 |
| Coxsackievirus B4 (JVB) | CBV4J | B | II | X05690 |
| Coxsackievirus B5 (1954/UK/85) | CBV5 | B | II | X67706 |
| Coxsackievirus B6 (Schmitt) | CBV6 | B | II | AF039205 |
| Echovirus 6 (Charles) | EV6 | B | II | U16283 |
| Echovirus 9 (Barty) | EV9B | B | II | X92886 |
| Echovirus 9 (Hill) | EV9H | B | II | X84981 |
| Echovirus 11 (Gregory) | EV11 | B | II | X80059 |
| Echovirus 12 (Travis) | EV12 | B | II | X79047 |
| Enterovirus 70 (J670/71) | Entero70 | D | I | D00820 |
| Enterovirus 71 (BrCr) | Entero71B | A | II | U22521 |
| Enterovirus 71 (MS7423/87) | Entero71M | A | II | U22522 |
| Poliovirus 1 (Mahoney) | PV1M | PV | I | J02281 |
| Poliovirus 1 (Sabin) | PV1S | PV | I | V01150 |
| Poliovirus 2 (Lansing) | PV2L | PV | I | M12197 |
| Poliovirus 2 (Sabin) | PV2S | PV | I | X00595 |
| Poliovirus 3 (23127/Finland/84) | PV3F | PV | I | X04468 |
| Poliovirus 3 (Leon) | PV3L | PV | I | K01392 |
| Poliovirus 3 (Sabin) | PV3S | PV | I | K00043 |
| Rhinovirus 1B (B632) | HRV1B | D00239 | ||
| Rhinovirus 2 (HGP) | HRV2 | X02316 | ||
| Rhinovirus 14 (1059) | HRV14 | K02121 | ||
| Rhinovirus 89 (41467-Gallo) | HRV89 | M16248 | ||
| Swine vesicular disease virus (J1′73) | SVDVJ | B | II | D16364 |
| Swine vesicular disease virus (H/3′76) | SVDVH | B | II | D00435 |
| Swine vesicular disease virus (P1) | SVDVP | B | II | X54521 |
A to D, human enterovirus A to D; PV, human poliovirus.
Genetic grouping in the 5′ NCR of the genome.
GenBank database.
Similarity analysis.
To analyze genetic relationships and recombination within enterovirus species, similarity analysis was employed separately for each cluster by using the SimPlot program (23, 31). A segmented analysis equivalent to the Bootscanning procedure was performed, but instead of bootstrap values, pairwise genetic similarities of a query sequence against other sequences of the alignment were calculated for each segment (a 400-nt window which was moved 20 nt at a time). Similarity values were then plotted along the genome.
RESULTS
Enterovirus clades along the genome.
By bootscanning of the full-length genome alignment, the occurrence and stability of the previously described genetic groups, I and II, and clusters A to C were systematically tested in different parts of the genome. Groups I and II were highly supported in the 5′ NCR (>70%) (Fig. 1). At positions 600 (group I) and 750 (group II) of the alignment, the bootstrap values of the groups fell sharply and stayed low along the rest of the genome, except for a few scattered positions (Fig. 1). Separate analysis of the genome areas corresponding to such peaks in the group II bootscan revealed that they were due to the relative proximity of the cluster A and B viruses, which, however, remained clearly separated in the coding region and the 3′ NCR (data not shown). Similarly, in some regions, enterovirus 70 appeared momentarily somewhat closer to cluster C viruses but still remained clearly distinct.
FIG. 1.
(A) Phylogenetic tree illustrating genetic relationships between enterovirus and rhinovirus genomes in the 5′ NCR. Enterovirus 5′ NCR groups I and II are indicated. (B) Bootscanning analysis of groups I (upper panel) and II (lower panel) along the genome was performed by the neighbor-joining method as implemented in the PHYLIP package (see text for details). PHYLIP reports bootstrap frequencies for all clusters found in the bootstrap replicates, which permits plotting of values for clusters which are not found in the consensus tree (areas of low bootstrap values). Bootstrap values are shown at the midpoint of each window.
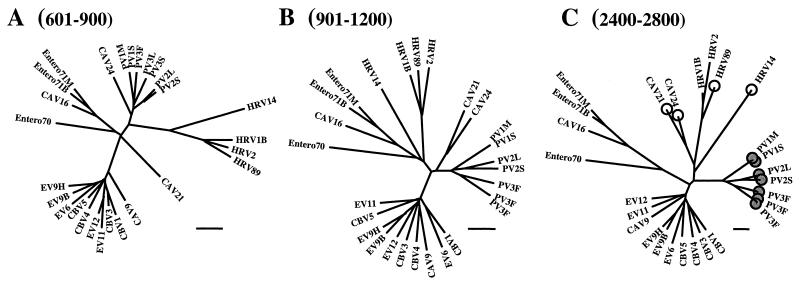
High bootstrap values were obtained for enterovirus clusters A to C in the coding region and in the 3′ NCR (Fig. 2). Since enterovirus 70 is the only completely sequenced member of proposed species D, its occurrence as an independent monophyletic lineage could not be directly tested by bootstrap analysis. However, it always remained as an outlier in trees derived from the coding region and the 3′ NCR, which supports its classification as a separate enteroviral species. Low bootstrap values for clusters A, B, and C in the 5′ NCR suggest that this clustering does not exist as a substructure of groups I and II. Two depressions in the cluster C bootscan, corresponding to the genome alignment from position 601 to 1200 and from position 2400 to 2800, were found to be due to separation of two viruses (CAV21 and CAV24) from the cluster into independent lineages. Separate phylogenetic analysis showed that between positions 601 and 900 of the alignment, CAV21 appears as a single outlier equidistant from the other clades (Fig. 3A). In the adjacent genome region (position 901 to 1200), CAV21 is joined by CAV24 (Fig. 3B). Again, in the VP1 capsid protein coding region (position 2400 to 2800), these two viruses separate from the rest of cluster C to form an independent clade which is not directly associated with any of the other clusters (Fig. 3C).
FIG. 2.
(A) Grouping of enteroviruses and rhinoviruses in the coding region and the 3′ NCR of the genome. The four genetic clusters (A to D) of enteroviruses are shown. (B) Bootscanning of clusters A, B, and C (performed as described in the legend to Fig. 1).
FIG. 3.
Alterations in clustering of enteroviruses and rhinoviruses in the 5′-terminal region of the genome. Trees correspond to nt 601 to 900 (A), 901 to 1200 (B), and 2400 to 2800 (C) of the alignment. Viruses interacting with poliovirus receptor are marked with shaded circles, and those recognizing intercellular adhesion molecule 1 are marked with open circles (C). Bars indicate 10% divergence.
Recombination may explain the phylogenetic division of groups I and II.
Bootscanning analysis extends the previous results by showing that the abrupt change in phylogenetic grouping which occurs between the 5′ NCR and the coding region-3′ NCR (approximately position 700 of the alignment) is a consistent feature of the enteroviral genome (Fig. 1 and 2). Groups I and II are highly supported only in the region preceding the initiation codon, and there is no indication of substructure within the two groups (Fig. 1B and 2B). After position 700, groups I and II split, and high support is seen only for the species-defining clusters A to C (Fig. 1B and 2B) (except for the regions where divergence of the CAV21 and CAV24 sequences lowers the bootstrap values of cluster C). Such a marked difference in clustering strongly supports the alternative that the mechanism resulting in the change may be recombination rather than conservation. Indirect evidence for this possibility could be drawn from the analysis of variation within clusters A to C, which indicates that intraspecies recombination frequently occurs. This was especially evident within cluster B (see below).
Variation within enterovirus clusters.
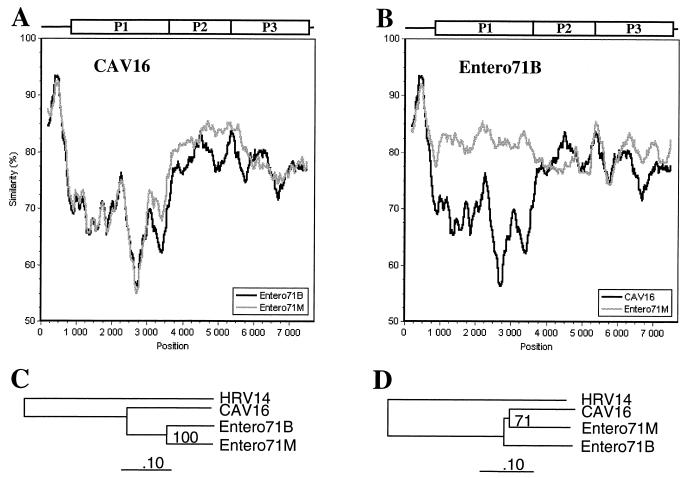
Sequences within each cluster were separately subjected to genetic similarity analyses, in which each strain was compared to all other strains of the cluster. For cluster A, three complete genome sequences were available (Table 1). Comparison of CAV16 with the two enteroviruses 71 strains resulted in similarity curves showing more variation in the P1 region than in the 5′ NCR or the NS region (Fig. 4A). The genetic similarity between the two enterovirus 71 strains in the P1 region (nt 1000 to 3500 in the alignment) was significantly higher than that observed between enterovirus 71 and CAV16 (Fig. 4B). In contrast, all three viruses appeared equidistant in the NS region (nt 3500 to 7500).
FIG. 4.
Genetic distances between cluster A enteroviruses along the genome as determined by similarity analysis (window, 400 nt; step, 20 nt). Query sequences are noted. Phylogenetic trees correspond to the capsid (P1 [C]) and NS (P2 and P3 [D]) protein coding regions.
Results of the similarity plots can be explained by the occurrence of intraspecies recombination. By this interpretation, the low divergence between the two enterovirus 71 strains in the P1 region could represent a recent recombination event between equidistantly evolved members of enterovirus species A which have recombined at the P1-P2 junction to form intraspecies chimeras. Findings of the similarity analysis were supported by phylogenetic analysis of the two regions (Fig. 4C and D).
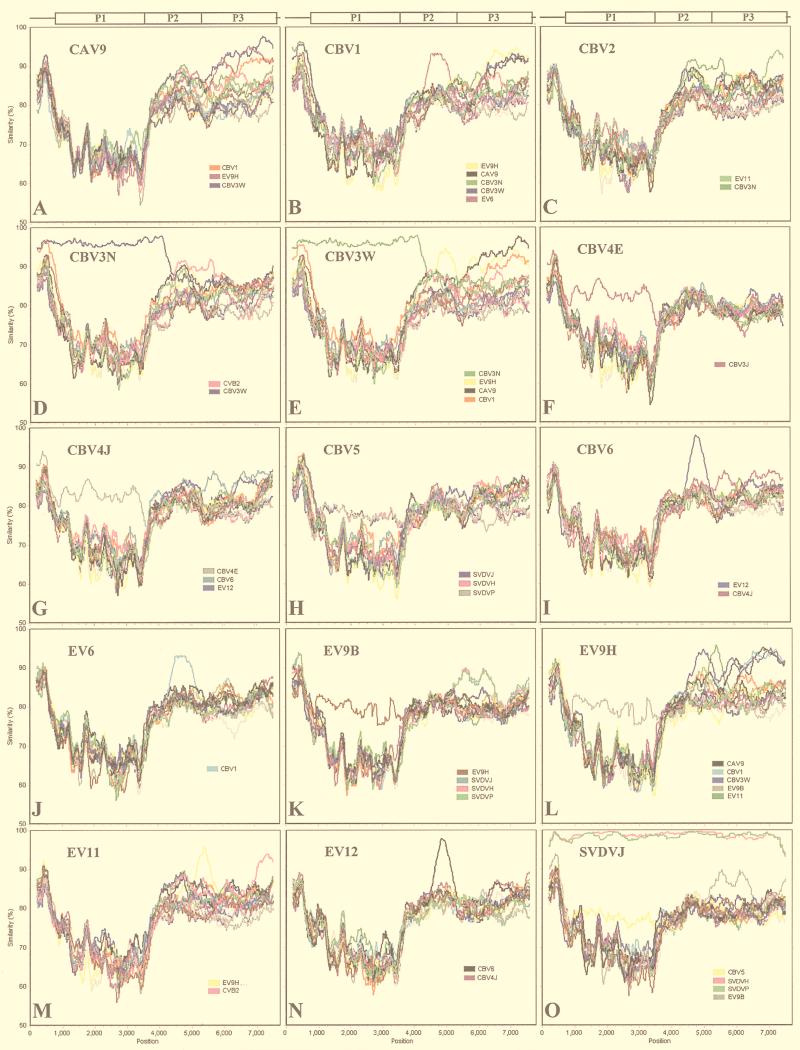
Seventeen cluster B genomic sequences were available (Table 1). SimPlot analysis resulted in a pattern of interstrain variation generally similar to that seen for cluster A (Fig. 5). High average sequence similarity was seen in the 5′ NCR (80 to 95%), lower similarity was seen in the P1 region (60 to 80%), and, again, higher similarity was seen in the region coding for NS proteins and the 3′ NCR (80 to 90%). However, in all viruses examined, and in all genome areas, regions of higher-than-average intersequence similarity were detected, suggesting a role for intraspecies recombination in the evolution of cluster B viruses.
FIG. 5.
Genetic distances between cluster B enteroviruses (SimPlot analysis) (window, 400 nt; step, 20 nt). Query sequences are noted.
As for species A, many examples of higher-than-average similarity in the P1 region were detected in comparisons between different strains of the same serotype. These included the two CBV3 (Fig. 5D and E), CBV4 (Fig. 5F and G), and EV9 strains (Fig. 5K and L), as well as the three strains of SVDV and CBV5 (Fig. 5H and O), which represent the same serotype despite different hosts. The three SVDV strains showed high similarity (>95%) to each other in all genome regions (Fig. 5O). However, in contrast to species A, many regions of higher-than-average similarity were also seen in the NS region: CAV9 showed >90% similarity to EV9H and CBV1 at nt 6210 to 7520 and to CBV3W at nt 5500 to 7520 (Fig. 5A). CBV1 also showed >90% similarity to EV6 from position 4470 to 5050 and >95% similarity to the two CBV3 strains in the 5′ NCR (Fig. 5B). CBV2 showed higher-than-average similarity to two other serotypes: CBV3N from position 4500 to 5500 and EV11 from position 7000 to the end of the genome (Fig. 5C). For the two CBV3 strains, positions 1 to 4000 showed high similarity (>95%), indicating recent divergence between the strains in this region (Fig. 5D and E). The viruses differed from each other in the NS region, where the CBV3N strain showed homology to CBV2 (Fig. 5D) whereas the CBV3W strain displayed high similarity to EV9H, CBV1, and CAV9 (Fig. 5E). For the CBV3W strain, the longest region of similarity in the NS region was to EV9H (Fig. 5E). Clear areas of high similarity in the NS region were also detected between CBV6 and EV12 (Fig. 5I and N), as well as between EV9H and EV11 (Fig. 5L and M). Increased similarity between EV9B and SVDVs, which Zhang et al. (43) detected in a partial sequence (615 nt), was seen at nt 5200 to 6500 (Fig. 5K and O).
The results of intraspecies comparisons for cluster B provide strong evidence that multiple recombination events, both within and between serotypes, have shaped the evolution of this cluster. It would be difficult to envision any other evolutionary process which would lead to the patterns observed, especially in the case of the CBV3 strains included in the analysis. The results for cluster B also make the recombination interpretation of the patterns seen in cluster A more likely.
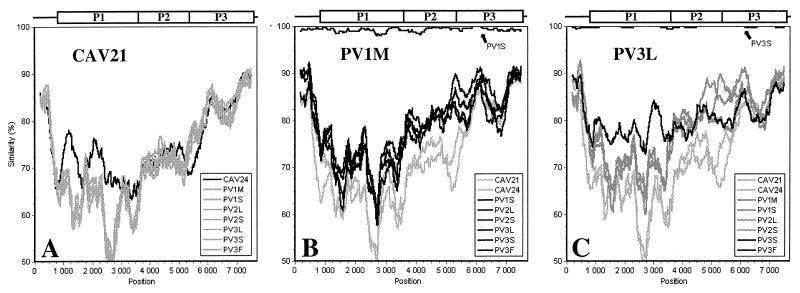
Results of similarity analysis for enterovirus cluster C agree with the findings observed in the bootscanning procedure. However, the analysis did not find suggestions of intraspecies recombination as clear as for clusters A and B. Comparison of each strain against the other viruses of cluster C indicates that CAV21 and CAV24 diverge from the PVs in the P1 region but also around nt 3000 to 5300 (Fig. 6A and B). In the P1 region, CAV21 and CAV24 are more closely related to each other than to PVs. However, in the P2 region, the two CAVs are as distant from each other as from the PVs, which form a more coherent group. These patterns of similarity suggest that the viruses have evolved through complex evolutionary pathways which the methods employed here are not able to resolve. The serotype classification seems to follow the degree of similarity between the strains in the P1 region but is not necessarily reflected in the NS region. This is illustrated particularly well by the PV3 isolate from Finland (PV3F), which shows increased homology to the other PV3 strains only in the P1 region (Fig. 6C) and actually seems to be closest to PV2 in the NS region (data not shown).
FIG. 6.
SimPlot analysis of cluster C enteroviruses (window, 400 nt; step, 20 nt). Query sequences are noted.
The patterns of similarity between members of enterovirus cluster C are more difficult to interpret than are those of clusters A and B. Although some suggestions for possible interstrain recombination were seen, especially in comparisons of the CAV21 and CAV24 strains, we cannot rule out the possibility that the evolution of cluster C viruses follows pathways different from those of clusters A and B. However, the number of analyzed full-length sequences that belong to cluster C is currently small, representing mostly PV vaccine and vaccine-related strains, and may therefore not be representative of wild-type viruses.
DISCUSSION
Enterovirus recombination has previously been demonstrated with PVs, and artificial chimeras between different enteroviral species have been produced in vitro. However, direct evidence of recombination contributing to the evolution of enteroviruses other than PVs has not been presented. In PVs, it has been estimated that the intratypic recombination frequency for the entire genome is approximately 15% (17). Recombination between intertypic PV strains, which have 85% nucleotide identity, has been observed at a 100-fold-lower frequency than intratypic recombination between completely homologous parents (19). Viable recombinants have also been produced between members of different enterovirus species by exchanging various genomic regions in vitro with molecular cloning techniques (15, 24, 33, 34, 37, 42). While not all artificially produced interspecies chimeras were viable, these studies have demonstrated that some genome regions may be interchangeable between different enteroviruses.
The existence of only two enterovirus clades in the 5′ NCR but four elsewhere in the genome has previously been proposed to be a consequence of strong evolutionary restrictions in this genomic region (29) and to reflect conservation of the RNA secondary structures required for efficient replication and translation (34). We were interested in studying this phenomenon further by using novel phylogenetic approaches.
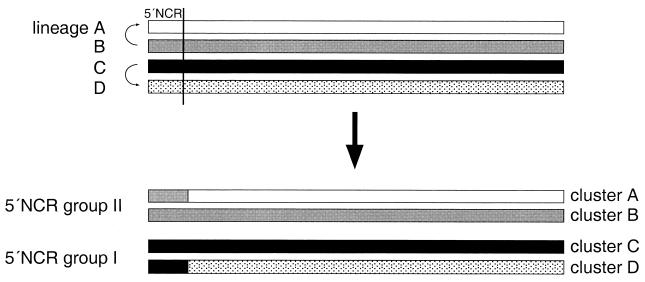
Results of bootscanning analyses of 30 complete enterovirus genomes were, in general, consistent with previously established clades for the proposed regions: groups I and II were supported by high bootstrap values in the 5′ NCR, while clusters A, B, and C were supported in the coding region and the 3′ NCR. However, our findings suggest that the reduction in the number of enterovirus clades observed in the 5′ NCR compared to the coding region and the 3′ NCR could be a consequence of recombination rather than structural conservation. In support of such a hypothesis, the low bootstrap values for groups I and II in the coding region and the 3′ NCR indicate that groups I and II do not represent earlier states of enterovirus evolution in these genome regions. Similarly, 5′ NCR groups I and II do not exhibit signs of separate subclades for cluster A, B, C, or D, which would be expected if the viruses had evolved only by accumulation of point mutations. Since the clusters are equidistantly separated in the different regions, a model in which four clusters have evolved from two is difficult to envisage. Instead, the different phylogenetic grouping of enteroviruses in the 5′ NCR compared to the rest of the genome could be explained by two recombination events, in which two of the progenitors of current clades A to D have replaced their original 5′ regions with those of the other two clades (Fig. 7), perhaps to gain more efficient translation initiation machinery. Although this model might be overly simplistic, it provides an approximation for a hypothesis based on recombination. The alternative possibility is that viruses originating from common progenitors have evolved to form two extra clusters in the coding region and the 3′ NCR, while in the 5′ NCR the same virus genomes have remained in the two original genetic groups. This would require strong conservation in the 5′ NCR, which is certainly possible, but also a large number of highly convergent point mutations in the rest of the genome to form the two additional clusters, which may be more difficult to generate during evolution. However, such a possibility cannot be formally excluded.
FIG. 7.
Schematic presentation of proposed recombination events between the 5′ NCR and the rest of the genome during enterovirus evolution which could explain the existence of currently known subgroups. First, four genetic lineages evolved from a common enterovirus progenitor by point mutations. During evolution, the lineage A virus acquired the lineage B virus 5′ NCR by recombination. Alternatively, the lineage B virus could have acquired the lineage A virus 5′ NCR. A similar recombination event may have occurred between lineage C and D viruses. Current species classification of human enteroviruses is based mainly on four genetic lineages (clusters A to D) observed in the coding region and the 3′ NCR. Due to the proposed recombination, only two genetic groups are seen in the 5′ NCR. Viruses containing the 5′ NCR from the other two lineages have disappeared or have not been sampled to date.
A highly speculative possibility concerning the origin of the two extra clades in the coding region-3′ NCR is that these genetic lineages originate from animal viruses which have acquired a more efficient 5′ NCR for human cells by recombination. Recent work on animal models showing that the 5′ NCR can include determinants that affect the host range supports this hypothesis (38). However, without direct evidence from animal enteroviruses, such a hypothesis cannot be verified. An alternative hypothesis is that the original 5′ NCRs were simply lost due to chance or poor fitness compared to the current 5′ NCRs.
More direct evidence for the role of recombination in enterovirus evolution was provided by sequence comparisons within the species-defining clusters. In enteroviruses, variation in the capsid protein coding sequences is known to be clearly more extensive than that in the NS regions, and this was also evident in the similarity analysis. Examination of sequence variation within enterovirus species revealed a general pattern of high sequence homology in the 5′ NCR, lower homology in the P1 region, and, again, higher homology in the region coding for the NS proteins and the 3′ NCR, resulting in the characteristic wavelike shape of the curve in similarity plots (Fig. 4 to 6). While the exact degree and localization of similarity was somewhat different between clusters, the basic shape of the similarity plot was repeated in the intraspecies comparisons of all clusters.
However, exceptions to the basic shape were also detected within each cluster. Especially in cluster B, many regions of higher-than-average similarity in pairwise comparisons were seen between individual strains (Fig. 5). Perhaps the most striking example of this was between the two CBV3 strains (Nancy and Woodruff 20–22]). From the 5’ end of the genome up to position 4000, similarity values in excess of 95% were observed. In contrast, similarity in the NS gene region was much lower and did not correlate with the serotype classification. Instead, several regions in the 3’ half of the genome exhibited higher-than-average sequence homology to other strains. Versions of this pattern were also repeated between viruses of different serotypes. We interpret the patterns of higher-than-average similarity as evidence of recombination events between viruses of the same species.
In intraspecies comparisons, none of the putative recombination breakpoints were detected within the P1 region, suggesting that the virus capsid is a relatively stable unit. In contrast, several breakpoints occurred within the P2 and P3 regions. The fact that the NS genes are more homologous than the structural genes between members of genetic clusters may explain the occurrence of frequent recombination events in the NS protein coding region. Notably, only a few of the putative recombination breakpoints observed in the NS region were situated at the cleavage sites of individual viral proteins. Instead, most of the regions of higher-than-average sequence homology between different members of a species either covered only a part of one individual viral gene or spanned several neighboring genes. In general, crossover sites in the NS region seemed to be relatively randomly distributed rather than being concentrated on some specific genome region. Based on the results obtained by similarity analyses of the cluster A and B viruses, it seems likely that enterovirus genomes contain regions that are interchangeable and that viable virus chimeras arise frequently between the members of a species.
Results of the bootstrap as well as the similarity analysis of cluster C indicated that CAV21 and CAV24 carry sequences in the capsid protein coding region which clearly separate them from the PVs. However, in other genomic regions these strains were no more related to each other than to the PVs. It is known that cluster C coxsackieviruses utilize intercellular adhesion molecule 1 (1, 30) for cell entry while polioviruses recognize another member of the immunoglobulin superfamily, the poliovirus receptor (26). The differences seen between CAVs and PVs in the capsid protein coding region could be responsible for the differences seen in receptor utilization and could therefore affect the clinical outcomes of diseases caused by these viruses. Based on the results of our analyses, we were not able to conclude how the regions of the capsid shared by CAV21 and CAV24 and unrelated to the PVs have evolved.
In conclusion, the complex patterns of regions of higher-than-average similarity in pairwise comparisons across serotypes, as observed for enterovirus clusters A, B, and C, are not easily explained with a model of evolution based only on gradual accumulation of mutations and subsequent starlike divergence from a common ancestor. Instead, we propose that they could have been generated through multiple homologous recombination events, leading to shuffling of gene segments between various strains. All of the observed relationships of high homology between different serotypes could be verified with high (>90%) bootstrap values (data not shown). The results indicate that there may be several potential recombination hot spots in the enterovirus genome, including the junction between the 5′ NCR and the P1 region and around the region coding for the C terminus of VP1. The NS region may also contain locations where recombination events accumulate. Our observations support the possibility that exchange of different genomic regions by recombination, in addition to the great adaptability provided by the high mutation rate, is an important mechanism in enterovirus evolution. This would allow independent evolution of elements with different functions needed during the viral replication cycle and would permit rapid alterations in host and tissue tropism. Our observations also suggest that enterovirus serotypes accurately reflect the capsid type of the virus but not necessarily other parts of the genome.
ACKNOWLEDGMENTS
This study was supported by grants from the Turku University Foundation, the Finnish Cultural Foundation, the Finnish Medical Foundation, the Academy of Finland, and the Sigrid Juselius Foundation.
We thank Tapani Hovi, Alexander Plyusnin, and Glyn Stanway for helpful comments.
REFERENCES
- 1.Abraham G, Colonno R J. Many rhinovirus serotypes share the same cellular receptor. J Virol. 1984;51:340–345. doi: 10.1128/jvi.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cammack N, Phillips A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology. 1988;167:507–514. [PubMed] [Google Scholar]
- 3.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 4.Drake J W. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans D J, Almond J W. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 1998;6:198–202. doi: 10.1016/s0966-842x(98)01263-3. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP: phylogeny inference package, version 3.572c. Seattle: University of Washington; 1996. [Google Scholar]
- 8.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:199–208. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland J, Domingo E. Origin and evolution of viruses. Virus Genes. 1998;16:13–21. doi: 10.1023/a:1007989407305. [DOI] [PubMed] [Google Scholar]
- 11.Horsnell C, Gama R E, Hughes P J, Stanway G. Molecular relationships between 21 human rhinovirus serotypes. J Gen Virol. 1995;76:2549–2555. doi: 10.1099/0022-1317-76-10-2549. [DOI] [PubMed] [Google Scholar]
- 12.Huttunen P, Santti J, Pulli T, Hyypiä T. The major echovirus group is genetically coherent and related to coxsackie B viruses. J Gen Virol. 1996;77:715–725. doi: 10.1099/0022-1317-77-4-715. [DOI] [PubMed] [Google Scholar]
- 13.Hyypiä T, Hovi T, Knowles N J, Stanway G. Classification of enteroviruses based on molecular and biological properties. J Gen Virol. 1997;78:1–11. doi: 10.1099/0022-1317-78-1-1. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis T C, Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson V H, Semler B L. Defined recombinants of poliovirus and coxsackievirus: sequence-specific deletions and functional substitutions in the 5′-noncoding regions of viral RNAs. Virology. 1988;162:47–57. doi: 10.1016/0042-6822(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 16.Kendal A P. Epidemiologic implications of changes in the influenza virus genome. Am J Med. 1987;82:4–14. doi: 10.1016/0002-9343(87)90554-7. [DOI] [PubMed] [Google Scholar]
- 17.King A M Q. Recombination in positive strand RNA viruses. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. Vol. 2. Boca Raton, Fla: CRC; 1988. pp. 149–165. [Google Scholar]
- 18.King A M Q, Brown F, Christian P, Hovi T, Hyypiä T, Knowles N J, Lemon S M, Minor P D, Palmenberg A C, Skern T, Stanway G. Picornaviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 1999. p. 996. [Google Scholar]
- 19.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klump W M, Bergmann I, Müller B C, Ameis D, Kandolf R. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–1583. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowlton K U, Jeon E-S, Berkley N, Wessely R, Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindberg A M, Stålhandske P O K, Pettersson U. Genome of coxsackievirus B3. Virology. 1987;156:50–63. doi: 10.1016/0042-6822(87)90435-1. [DOI] [PubMed] [Google Scholar]
- 23.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu H-H, Li X, Cuconati A, Wimmer E. Analysis of picornavirus 2Apro proteins: separation of proteinase from translation and replication functions. J Virol. 1995;69:7445–7452. doi: 10.1128/jvi.69.12.7445-7452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCutchan F E, Salminen M O, Carr J K, Burke D S. HIV-1 genetic diversity. AIDS. 1996;10:S13–S20. [PubMed] [Google Scholar]
- 26.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 27.Oberste M S, Maher K, Pallansch M A. Molecular phylogeny of all human enterovirus serotypes based on comparison of sequences at the 5′ end of the region encoding VP2. Virus Res. 1998;58:35–43. doi: 10.1016/s0168-1702(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 28.Pöyry T, Hyypiä T, Horsnell C, Kinnunen L, Hovi T, Stanway G. Molecular analysis of coxsackievirus A16 reveals a new genetic group of enteroviruses. Virology. 1994;202:982–987. doi: 10.1006/viro.1994.1423. [DOI] [PubMed] [Google Scholar]
- 29.Pöyry T, Kinnunen L, Hyypiä T, Brown B, Horsnell C, Hovi T, Stanway G. Genetic and phylogenetic clustering of enteroviruses. J Gen Virol. 1996;77:1699–1717. doi: 10.1099/0022-1317-77-8-1699. [DOI] [PubMed] [Google Scholar]
- 30.Pulli T, Koskimies P, Hyypiä T. Molecular comparison of coxsackie A virus serotypes. Virology. 1995;212:30–38. doi: 10.1006/viro.1995.1450. [DOI] [PubMed] [Google Scholar]
- 31.Ray, S. C. 1997. SimPlot for Windows 95/NT, version 1.2.2. [Online.] Baltimore, Md. (Distributed by author.) http://www.welch.jhu.edu/∼sray/download.
- 32.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 33.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohll J B, Percy N, Ley R, Evans D J, Almond J W, Barclay W S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 36.Salminen M O, Carr J K, Robertson D L, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P M, Hahn B H, Burke D S, McCutchan F E. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Salminen, M. O., and W. Cobb. 11 September 1998, revision date. [Online.] Bootscanning package for Unix/Linux, version 1. National Public Health Institute, Helsinki, Finland. http://www.ktl.fi/hiv.
- 37.Semler B L, Johnson V H, Tracy S. A chimeric plasmid from cDNA clones of poliovirus and coxsackievirus produces a recombinant virus that is temperature-sensitive. Proc Natl Acad Sci USA. 1986;83:1777–1781. doi: 10.1073/pnas.83.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiroki K, Ishii T, Aoki T, Ota Y, Yang W X, Komatsu T, Ami Y, Arita M, Abe S, Hashizume S, Nomoto A. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J Virol. 1997;71:1–8. doi: 10.1128/jvi.71.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sibold C, Meisel H, Krüger D H, Labuda M, Lysy J, Kozuch O, Pejcoch M, Vaheri A, Pluysnin A. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The Genetic Data Environment: an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kuppeveld F J, van den Hurk P J, van der Vliet W, Galama J M, Melchers W J. Chimeric coxsackie B3 virus genomes that express hybrid coxsackievirus-poliovirus 2B proteins: functional dissection of structural domains involved in RNA replication. J Gen Virol. 1997;78:1833–1840. doi: 10.1099/0022-1317-78-8-1833. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Wilsden G, Knowles N J, McCauley J W. Complete nucleotide sequence of a coxsackie B5 virus and its relationship to swine vesicular disease virus. J Gen Virol. 1993;74:845–853. doi: 10.1099/0022-1317-74-5-845. [DOI] [PubMed] [Google Scholar]