Abstract
Leaf litter microbes collectively degrade plant polysaccharides, influencing land–atmosphere carbon exchange. An open question is how substrate complexity—defined as the structure of the saccharide and the amount of external processing by extracellular enzymes—influences species interactions. We tested the hypothesis that monosaccharides (i.e. xylose) promote negative interactions through resource competition, and polysaccharides (i.e. xylan) promote neutral or positive interactions through resource partitioning or synergism among extracellular enzymes. We assembled a three-species community of leaf litter-degrading bacteria isolated from a grassland site in Southern California. In the polysaccharide xylan, pairs of species stably coexisted and grew equally in coculture and in monoculture. Conversely, in the monosaccharide xylose, competitive exclusion and negative interactions prevailed. These pairwise dynamics remained consistent in a three-species community: all three species coexisted in xylan, while only two species coexisted in xylose, with one species capable of using peptone. A mathematical model showed that in xylose these dynamics could be explained by resource competition. Instead, the model could not predict the coexistence patterns in xylan, suggesting other interactions exist during biopolymer degradation. Overall, our study shows that substrate complexity influences species interactions and patterns of coexistence in a synthetic microbial community of leaf litter degraders.
Keywords: biopolymer degradation, bottom-up approach, Curtobacterium, extracellular enzymes, interspecific interactions, leaf litter microbiome
Growth on a plant polymer minimizes negative interactions and promotes species coexistence in a simplified microbial community relevant to climate change studies.
Introduction
Microbial communities regulate biogeochemical processes, including the cycling of organic materials such as carbon through decomposition of recalcitrant plant materials (e.g. cellulose, lignin, and xylan). Bacteria and fungi associated with the top layer of soil—i.e. the leaf litter microbiome—break down plant saccharides, releasing carbon dioxide through respiration, which influences land–atmosphere carbon exchange (Coûteaux et al. 1995). Therefore, studying saccharide degradation is key for predicting climate change feedbacks and the rate of carbon remineralization (Bardgett et al. 2008, Arnosti et al. 2021).
During leaf litter decomposition, microbes incrementally decompose plant-derived saccharides and generate a shared pool of saccharides varying in substrate complexity. On the one end of the substrate complexity gradient are complex saccharides, defined as polymeric saccharides requiring external processing for microbial consumption (Lindemann 2020). An example of complex saccharides is xylan, the second most abundant plant cell wall polysaccharide after cellulose. Xylan can vary in structural complexity from a linear chain of β-1,4-linked xylose (homoxylan) to a decorated xylose backbone with branching sugar residues other than xylose (heteroxylan). Xylans are degraded by carbohydrate-active enzymes (CAZymes), which hydrolyze specific bonds within the polysaccharide chain. For example, endo-1,4,-β-xylanase hydrolyzes the xylan backbone and converts xylan into an accessible form of xylose. On the other end of the substrate complexity gradient are simple saccharides, including the monomeric forms of the polysaccharides (e.g. xylose), that are easily transported inside the cell without external processing. While a broad range of species have the transporters to import mono- and oligosaccharides, fewer species produce CAZymes that degrade polysaccharides (here referred to as “degraders”). Furthermore, degraders vary in the composition and abundance of their CAZymes, with the copy and type of genes varying widely among species.
Given that the degradation of complex saccharides involves CAZymes that act as public goods and any microbe in the vicinity can directly profit from the hydrolyzed products, the process of polysaccharide degradation has the potential to shape microbial behaviors and interactions (D’Souza et al. 2021, Pontrelli et al. 2022). For example, Caulobacter crescentus displayed different behaviors depending on its growth on the polysaccharide xylan or the monosaccharide xylose (D’Souza et al. 2021). In xylan, cells interact positively by coaggregating to localize extracellular enzymes that would otherwise be lost to diffusion. In contrast, in xylose, cells interact negatively and disperse to avoid resource competition (D’Souza et al. 2021). Thus, substrate complexity influences interactions among closely related individuals.
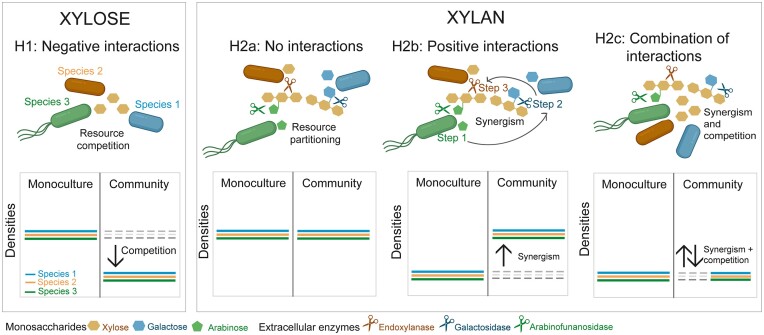
Yet, its effect on interspecific interactions remains less understood (but see Deng and Wang 2016, 2017). It is well-established that (1) simple carbohydrates promote negative interspecies interactions through resource competition (Tilman 1977) and (2) complex carbohydrates promote positive interactions between specialized degraders and downstream consumers, the latter of which lack the CAZymes necessary to degrade polysaccharides and, subsequently, cross-feed on simple saccharides released by the degraders (Datta et al. 2016, Enke et al. 2019, Pontrelli et al. 2022). Beyond these observations, it is unknown how substrate complexity influences interspecific interactions among degraders. Here, we hypothesized three outcomes (Fig. 1). In the no interactions hypothesis, degraders differentially hydrolyze and consume variable breakdown products from the polysaccharide that other degraders do not use (i.e. resource partitioning; Fig. 1, H2a). Alternatively, positive interactions among degraders are expected when synergisms emerge from the division of labor between species through their CAZymes (Lindemann 2020). For example, some CAZymes may initially degrade the branching sugar residues, facilitating other CAZymes to target other oligosaccharides (Fig. 1, H2b). Finally, a combination of interactions is plausible (Fig. 1, H2c).
Figure 1.
Expected interspecific interactions on simple and complex carbohydrates. In xylose, negative interactions among species are expected due to increased resource competition for simpler substrates (H1). Under this scenario, species’ densities in a community (represented by solid-colored lines) are expected to be lower than observed in monoculture. In xylan, several interactions may occur. First, no interactions are expected if species specialize in hydrolyzing and consuming part of the polysaccharide through differential production of various extracellular enzymes (represented by scissors) that differ in their hydrolytic enzyme activity. (H2a). Under this scenario, species’ densities are expected to be similar in the community than in monoculture. Second, positive interactions are expected if species cooperate to degrade the polysaccharide. Synergistic interactions can occur if the polysaccharide is degraded sequentially (H2b). Specialized CAZymes hydrolyze parts of the polysaccharide, and only after the hydrolysis products are removed the next hydrolysis step can proceed. Due to this synergism, species densities in the community are expected to be higher than in monoculture because species on their own cannot fully hydrolyze the polysaccharide. Finally, several interactions could be occurring simultaneously (H2c), which could be a combination of negative (H1), neutral (H2a), and/or positive interactions (H2b). In one possible scenario (of many), species may act synergistically to degrade the polymer but compete for the breakdown products released. Under this scenario, positive and negative effects may cancel each other out so that species’ densities are similar in the community than in monoculture.
Understanding species interactions during polysaccharide degradation can also provide insights into how nutrient complexity affects patterns of species coexistence. In theory, an environment with one resource can sustain one species only due to competition (Hardin 1960). However, recent models and experiments have shown that multiple species can coexist in a single resource (Goldford et al. 2018, Manhart et al. 2018). Thus, even a simple scenario is more nuanced than expected. Similarly, predicting coexistence patterns in polysaccharides is not trivial. One could expect that polysaccharides promote species coexistence through niche partitioning (Tilman 1982), or because species’ growth is limited due to increased energetic investment in extracellular enzyme production to degrade polysaccharides. These expectations have rarely been directly tested.
Here, we study the influence of carbohydrate complexity on the interactions and coexistence among three xylan-degraders isolated from the leaf litter microbiome of a Mediterranean grassland site in Southern California (Potts et al. 2012). The leaf litter microbiome has been used as a model system to study microbial responses to climate change as it performs an important ecosystem function—the decomposition of plant polysaccharides—and because it is less complex compared to bulk soil (Martiny et al. 2017, Glassman et al. 2018). While the leaf litter microbiome is less complex than bulk soil, it is still composed of hundreds of “species” (Glassman et al. 2018). Therefore, studying species interactions within leaf litter communities remains challenging. To tackle this challenge, we simplified the leaf litter microbiome by assembling a three-species synthetic community, with strains isolated from surface litter collected on the same field season, that is easy to manipulate and trackable. The three species—Curtobacterium sp., Flavobacterium sp., and Erwinia sp.—were selected for two reasons. First, the species belong to the most diverse and abundant phyla (Actinobacteria, Bacteroidetes, and Proteobacteria, respectively) from the grass litter communities comprising 95% of total bacterial abundance (Berlemont et al. 2014, Matulich et al. 2015, Chase et al. 2018). At the genus level, Curtobacterium is the most abundant in the grassland and represents 8%–12% of the bacterial cells on grass litter (Matulich et al. 2015, Chase et al. 2018). Second, the three species capture the metabolic diversity of the leaf litter microbiome. While most of the species from the leaf litter microbiome degrade complex polymers, species within each phylum share unique carbon use patterns (Dolan et al. 2017). For instance, species from the genus Curtobacterium are excellent “degraders” and breakdown complex polymers such as cellulose, hemicellulose (xylan), and other plant polysaccharides (Berlemont and Martiny 2015, Chase et al. 2016, 2017); whereas, γ-Proteobacterium species (e.g. Erwinia) are considered “consumers” defined as fast growers that specialize on oligosaccharides and monosaccharides such as glucose and xylose (Dolan et al. 2017). Finally, Bacteroidetes (e.g. Flavobacterium) are considered “generalists” having the genomic potential to use a broad set of polysaccharides and oligosaccharides (Berlemont and Martiny 2015).
Using this simplified system, we showed that substrate complexity influenced physiological responses (i.e. growth kinetics over 2 days), mediated interspecific interactions, and promoted the coexistence of leaf litter degraders over short ecological timescales (10 days or ∼50 generations).
Materials and methods
Bacterial isolates
To assemble a simple community representative of the grass litter microbiome, we followed a bottom-up approach and selected three bacterial strains isolated from the Loma Ridge site (Table 1). The grassland site from the Loma Ridge Global Change Experiment (Irvine, California, USA, 33° 44′ N, 117° 42′ W), is dominated by the annual grasses from the genera Avena, Bromus, and Lolium, and the perennial grass Nassella pulchra (Potts et al. 2012, Dolan et al. 2017). Strains were collected from surface litter sampled the same field season and isolated as previously described (Dolan et al. 2017). Briefly, grounded grass litter was resuspended on DI water and passed through a 100-µm filter. The filtered litter solution (i.e. the slur) was then plated on Luria broth agar plates or leaf litter tea agar plates and incubated at 21ºC for up to 2 weeks. Single colonies were picked and restreaked at least three time to ensure purity. Finally, sample were stored at −80°C in glycerol stocks (32.5%) for genomic and culturing analyses.
Table 1.
Bacterial isolates used in this study.
| Phylum (average relative abundance of phylum) | Family | Genus (relative abundance of genus) | Species (strain) | Accession number4 | Previous studies that have characterized the isolate |
|---|---|---|---|---|---|
| Actinobacteria | Microbacteriaceae | Curtobacterium | Curtobacterium sp. (MMLR14002) | PRJNA524191 | Chase et al. (2016, 2017, 2018, 2021), Ramin and Allison (2019) |
| (48.0%1–51.5%2) | (10.4%3–18.64%2) | MMLR14_002 | |||
| NZ_MJGT00000000 | |||||
| Bacteroidetes | Flavobacteriaceae | Flavobacterium | Flavobacterium sp. (LR40) | PRJNA391502 | Ramin and Allison (2019) |
| (6.8%1–26.5%2) | (0.78%2–5.5%3) | MMLR14_040 | |||
| JAWVWO000000000 | |||||
| Proteobacteria | Erwiniaceae | Erwinia | Erwinia sp. | PRJNA391502 | Dolan et al. (2017), Ramin and Allison (2019) |
| (25%2–35.4%1) | (<0.01% 2) | (LR17) | MMLR14_017 | ||
| JAWVWP000000000 |
Average from metagenomic sequencing data (Chase et al. 2018)
From natural litter pyrosequencing data (Matulich et al. 2015)
From 16S sequencing of the grassland microbial community (Finks et al. 2021)
Gene Bank accession number: BioProject, Organism, Accession number (this and previous studies).
The genome of Curtobacterium MMLR14002 was previously sequenced (GeneBank’s accession NZ_MJGT00000000; Chase et al. 2021). We sequenced the genomes of Flavobacterium sp. and Erwinia sp. (accession numbers JAWVWO000000000 and JAWVWP000000000, respectively). Bacterial isolates were revived from glycerol stocks and streaked on lysogeny broth (LB) agar plates. A colony was grown in 10 ml of LB media for 48 h at 26°C with shaking (100 rotations per minute, rpm). We used 1 ml of cultures and isolated the genomic DNA using the Wizard Genomic DNA Purification Kit (Promega; Madison, WI). We used the DNA with the Illumina DNA Prep kit (Illumina; San Diego, CA, USA) with the low-volume protocol (dx.doi.org/10.17504/protocols.io.bvv8n69w) for library preparation that was quality assessed using a Bioanalyzer, prior to sequencing on the NovaSeq6000 S4 flow cell.
Raw reads were processed for quality control (trimq=32 qtrim=rl) and adapter removal (ktrim=r k=25) with bbduk.sh (Bushnell 2014) and used as input for de novo assembly with SPAdes (Bankevich et al. 2012) by increasing k-mers from 31 to 111 under the “careful” h-path removal strategy to reduce mismatches and indels. Draft assemblies were screened for quality by constructing taxon-annotated GC-coverage plots using blastn (Chen et al. 2015) and bowtie2 (Langmead and Salzberg 2012), respectively. Based on the resulting metrics, we removed all contigs <5000 bp and coverage <30 for further analyses. Genome completeness and contamination were calculated checkM (Parks et al. 2015) and taxonomy was verified using the gtdbtk classifier (Chaumeil et al. 2019). To maintain consistency across genomes, all genomes were assigned open reading frames (ORFs) using prodigal (Hyatt et al. 2010) and gene annotations with prokka (Seemann 2014).
Whole genome analysis
To identify strains’ genomic potential for carbohydrate degradation and characterize the diversity of CAZymes present in each species, we screened all ORFs against the protein family database, Pfam-A (Finn et al. 2014), using hidden Markov models, hmmsearch (Finn et al. 2011). Resulting protein domains were filtered for their top match based on e-value and bitscore. Using a curated list, we queried the top domain hits for the presence of glycoside hydrolase (GH) and carbohydrate binding module (CBM) domains in each strain (Chase et al. 2017). This approach identified GH/CBM families with their associated enzymatic activity and target substrate (Berlemont and Martiny 2013). Potential xylan degraders were defined as bacteria having at least one GH/CBM gene targeting xylan (Table S1).
Growth conditions
Strains were grown in a modified M63 medium supplemented with either xylan or xylose. Xylan was selected as a focal polysaccharide instead of cellulose (the most abundant plant cell wall polysaccharide) because all three strains grew poorly on cellulose. The chemical composition of this modified M63 medium was: 47 mM K2HPO4, 39 mM KH2PO4, 15 mM (NH4)2SO4, 1.8 µM FeSO4, 1 mM MgSO4, 34 nM CaCl2, 92 nM Fe III Chloride, 0.73 nM ZnCl2, 6.3 nM CuSO4 5H20, 8.4 nM CoCl2 6H20, 4.1 nM Na2MoO4 2H20, 5.9 µM thiamine, and 0.02% (w/v) peptone. Peptone (Sigma Aldrich, USA) was added to the medium as a source of amino acids. We added d-(+)-xylose (Sigma Aldrich) or xylan from corn core (Tokyo Chemical Industry, Japan), composed of at least 76% xylose after hydrolysis, to the medium as source of carbon. All solutions were sterilized using 0.22-µm PES filters. Growth curve experiments were performed in 96-well plates and batch culture experiments were performed in 40 ml glass vials with screw caps containing TFE-lined silicone septa (Rodríguez-Verdugo et al. 2019).
Growth curves experiments
To determine if xylose and xylan were limiting growth factors, we performed growth curves experiments at different concentrations of xylan and xylose (from 0% to 0.36% w/v). We revived the bacteria by streaking 10 µl of glycerol stock onto LB agar plates and incubating the plates at room temperature. We then picked a colony from each species and grew them in 10 ml of fresh M63 medium supplemented with 0.06% xylose. Cultures were incubated for 48 h at 26°C with shaking (100 rpm). We then transferred 2 µl of the saturated culture into 198 µl of fresh media dispensed in each of the wells of the 96-wells plate. An additional washing step was performed for Erwinia sp. which grew poorly in xylan. This washing step was performed to remove any excess of xylose that could be carry over in the fresh medium. We washed the saturated culture by spinning down the cells and washing them three times with 1% MgSO4 solution. We then used 2 µl of washed cells to inoculate the plates with fresh medium. To avoid evaporation, we covered the plates with a lid, which we sealed with silicon grease. We incubated the plates at 30°C with shaking inside a photospectrometer plate reader (Epoch2TM, AgilentTM), which lower detection limit is 0.005. Optical density (OD600) measurements were acquired every 10 min for 72 h (48 h for Erwinia sp.). Three technical replicates per strain and condition were obtained. From each growth curve, we estimated three parameters as in Rodriguez-Verdugo et al. (2019). These parameters were: (1) the maximum growth rate µ max, defined as the maximum growth rate during the exponential growth phase (h−1); (2) the maximum population size ODmax, defined as the maximum OD after saturation minus the minimum OD at the start of the experiment (unitless OD); (3) the yield Y, defined as the amount of biomass produced per unit of resource (unitless OD mM−1). The fitting of parameters was done using Matlab (version R2018b, Mathworks) and R (version 4.1.3).
Finally, we did an in-depth characterization of the growth kinetics for a selected concentration of xylan/xylose which was 0.06% (w/v). We used the same protocol, except that we incubated the plate at 26ºC and used mineral oil instead of a lid to avoid evaporation and condensation. These adjustment were made to match the conditions between growth curve experiments and the batch culture experiments (full details in Supplementary material and methods).
Coculture invasion experiments
To determine if species could coexist in pairs regardless of their initial frequency (e.g. if one species could establish itself in a coculture despite starting from rare), we did coculture invasion experiments in batch culture. We coinoculated species at varying initial fractions so each species was underrepresented, equally represented, and overrepresented. To reach these initial fractions, we first determined the relationship between the OD and the cell density estimated by colony forming units (CFU) counts. To do so, we grew each species in M63 medium with 0.25% of xylose to reach high bacterial densities. We then used the saturated cultures and diluted them to obtain 2-, 4-, and 8-fold dilutions. We measured the OD from these dilutions in the plate reader and plated them in LB agar to estimate their CFU. We replicated the experiment two times and fitted a linear relationship between the OD and the cell density for each species (Fig. S1).
To start the invasion experiments, we acclimated cultures by inoculating a colony from each species into 10 ml fresh M63 medium with 0.25% xylose and incubated them for 48 h. We used the OD/CFU relationship (Fig. S1) to calculate the volumes needed from each acclimated culture to reach the targeted cell densities. We used these adjusted volumes and mixed the competing species to reach the targeted ratios of 10:90%, 50:50%, and 90:10%. We launched two cocultures targeting each initial ratio but sometimes failed to achieve the targeted ratios. Ultimately, we obtained six cocultures (independent observations) per species pair. The cocultures were then incubated at 26°C with shaking (100 rpm). After 48 h, we transferred 0.1% of the population into a fresh medium and repeated this transfer again to obtain three growth-dilution cycles (i.e. 6 days). After each cycle, we estimated the species densities by plating cultures on LB plates and determining the CFU for each species. This was possible given that each species has a unique colony size and morphology on LB plates (Fig. S2). Species densities were used to estimate relative abundances for pairs of species estimated as fractions; e.g. fraction species A = cell density species A/(cell density species A + cell density species B). Initial fractions were 0.1 ± 0.15 (rare), 0.5 ± 0.15 (equal), and 0.9 ± 0.15 (common). Changes in relative abundances over time were used to determine the type of competition outcome (Chang et al. 2023). Coexistence occurred when one species increased in frequency when rare and decreased in frequency when common until it reached a final fraction statistically different from 0 or 1 (one sample t-test, n = 5–6). Competitive exclusion was observed when one species increased or decreased in frequency regardless of its initial frequency and reached a final fraction nondifferent from 0 or 1 (one sample t-test, n = 5–6).
Pairwise species interactions
To quantify the ecological interactions between pairs of species in each carbon source (xylose or xylan), we compared the densities of species grown alone (monocultures) or with a second species (pairwise cocultures). We used the same data from the invasion experiments previously described. We compared the densities (CFU ml−1) at day 6 in monoculture and coculture, performing a two sample t-test over the means (n = 3–6). If the densities were not significantly different in both monocultures and cocultures, we concluded that species had no interaction. If the densities in cocultures were significantly lower than in monocultures, we concluded that species had a negative interaction.
Three-species community experiments
To determine if all three species coexisted in a community, we did batch culture experiments over 10 days. We also determined the effect that other species had on growth by comparing species densities in monoculture and in a community. Monocultures were started by diluting the acclimated cultures 1000-fold into 10 ml M63 medium with xylose or xylan. In communities, the three species were mixed at an equal ratio by adjusting the volumes of each species to reach an initial target population density of 105 cells ml−1 per species. Cultures were incubated at 26°C with shaking (100 rpm) for 48 h. Monocultures and communities were propagated by transferring 0.01 ml of the culture into 9.99 ml of fresh medium every 48 h for a total of 10 days. We repeated these experiments two more times to obtain three temporal replicates.
To define the effect that other species have on growth, we compared the densities over 10 days for each species in monoculture and in the community and performed a two sample t-test over the means (n = 3). If the densities were not significantly different between a species grown in monoculture and in the community, we concluded that species in the community had no effect on the species of interest. If the densities in monocultures were higher than in communities, we concluded that species in the community had a negative effect on the species of interest.
Statistical analyses
For all the density data (CFU ml−1) obtained from pairwise and community experiments, we tested that the data followed the assumption of normality and equal variance with a Shapiro–Wilk normality test and an F test, respectively. When the assumption of equality of variance was not met, we conducted a Welch two sample t-test. For the few cases in which the data did not follow the assumption of normality, we performed a Wilcox test instead of a t-test. Finally, when temporal replicates were used, we controlled for the effect of the weeks in which the experiments were conducted and the grouped data structure by using a linear mixed model. We used the lmer function in the R package “lme4,” using the formula lmer (density ∼ treatment + (1|replicate)), where treatment (fixed effects) refers to the comparison between monocultures and communities, and replicate (random effects) refers to the weeks in which the experiment was conducted. All statistical analyses were done using R (version 4.1.3).
Resource-explicit model
To evaluate if we could predict community dynamics from single-species behaviors, we build a mathematical model with parameters estimated from monocultures. A similar resource-explicit model has been shown to recapitulate community dynamics in a two-species system (Rodriguez-Verdugo et al. 2019). The full details on how we estimated the parameters from growth curve experiments are found in the Supplementary materials and methods.
The model explicitly modeled the changes in resources (xylose, xylan, and peptone) and the bacterial growth of the three species grown in isolation in each carbon source. The model assumes the resources are readily used, regardless of whether they are in polymeric or monomeric form. Bacteria and resource concentrations were modeled with the following differential equations:
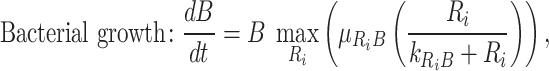 |
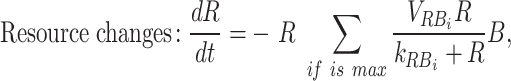 |
in which B is the density of bacteria (unitless OD), R is the concentration of resources in the culture (mM), µ R,B is the maximum growth rate of bacteria B on resource R (h−1), kR,B is the half-saturation constant of bacteria B on resource R (mM), and VR, B the maximum uptake rate of bacteria B on resource R (mM unitless OD−1 h−1). Each resource is used only if bacteria grow on it (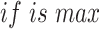 ).
).
The growth of each species was dependent on the concentration of resources, which was modeled explicitly using Matlab’s ODE45 solver. We incorporated the daily dilution that resulted from transferring 1/1000 of the culture into fresh media every 48 h and added an extinction threshold in which extinction occurred when less than one bacterium was transferred to the next cycle. Finally, to compare the results generated by the mathematical model to the experimental data, we converted the numerical values generated by the model (in OD units) to cell densities (in CFU) using the slope of the linear relationship between the OD and the cell density as the conversion rate (Fig. S1).
Metabolic analyses
To assess the degree to which resource competition and niche partitioning influenced the community dynamics, we performed genome-level metabolic profiling. We used dbCAN3 (Version 12) to identify CAZymes and potential substrates and METABOLIC-G (version 4.0) to build on dbCAN3 and predict biogeochemical and metabolic functional traits. The three genomes were independently fed into each respective pipeline using the defaults settings. Within the output files generated, we focused on the carbon cycles and the type and frequency of CAZymes. To validate some of the substrate use predictions and the potential to use the branched parts of the xylan, we did growth curve experiments in different carbon sources at a concentration of 4 mM. We tested growth on d-(-)-arabinose, d-(+)-galactose, d-(+)-glucose monohydrate, d-glucuronic acid, and d-(+)-mannose (all from Sigma Aldrich). These monosaccharides have been previously identified as part of the branched sugar residues of xylan from corn cobs (Melo-Silveira et al. 2019). We performed growth curve experiments with the same protocol previously described with some minor modifications. We acclimated the strains by growing them in M63 medium supplemented with a mixture of xylose (0.25%), xylan (0.25%), and glucose (0.25%) to reach high bacterial densities. We then washed the cells three times with 1% MgSO4 solution to remove any excess carbon that could be carried over in the fresh medium. Finally, instead of using a lid to avoid evaporation, we added 50 µl of mineral oil to each well containing 200 µl culture. OD600 measurements were acquired every 10 min for 48 h.
Results
Species in monoculture are capable of degrading xylan but utilize it less efficiently than xylose
First, we assessed the genomic potential for xylan degradation of the three isolates by characterizing the abundance and composition of CBM and GH protein families associated with xylanase activity. The abundance of CBM/GH genes involved in xylan degradation varied among genomes, with Flavobacterium encoding the most xylanases (34 CBM/GH xylanases copies), followed by Curtobacterium (6 CBM/GH xylanases copies) and Erwinia (2 CBM/GH xylanases copies; Table S1). All strains had at least two copies of endo-1–4-xylanases (“true xylanases”), which degrade the xylan backbone (Vandenberghe et al. 2020). Based on the presence of xylanase genes, all three species were classified as potential xylan degraders.
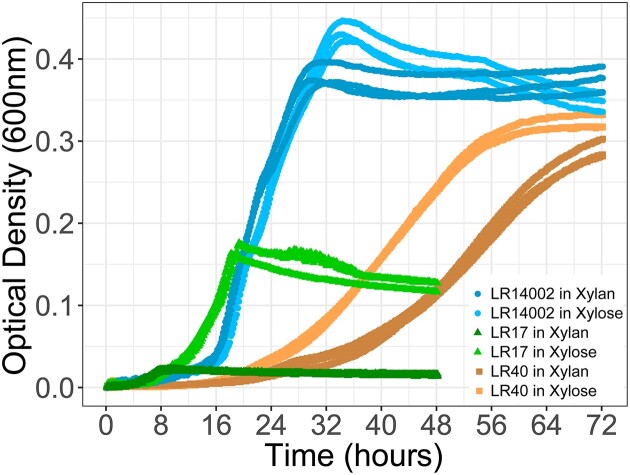
To determine if genomic potential translated into polymer degradation, we grew the species in monoculture with different concentrations of xylan (Fig. S3). The maximum population size increased with the concentration of xylan supplied, indicating that xylan was a limiting growth factor in our monocultures (Fig. S3). While all three species utilized and degraded xylan as their main source of carbon and energy (Monod 1949), there was high variability in growth among species (Table 2). At 0.06% of xylan, the maximum growth rate and population size significantly varied among species (one-way ANOVA, F2,6 = 19.294, P = .002 and F2,6 = 840.41, P = 4.5 × 10−8, respectively). Curtobacterium and Flavobacterium were efficient xylan degraders and achieved a higher maximum population size than Erwinia (Fig. 2; Fig. S3). Unexpectedly and unlike the other strains, Curtobacterium also grew in the control treatment without xylan (Fig. S3). That is, Curtobacterium utilized peptone as a carbon source (Fig. S4). Curtobacterium’s ability to use peptone as a source of carbon resulted in a diauxic shift when grown in xylan and peptone (Fig. S3), with the growth on peptone accounting for approximately half of its total growth. Therefore, after we normalized Curtobacterium growth by accounting for peptone, Curtobacterium and Flavobacterium achieved similar maximum population sizes. Curtobacterium had a higher maximum growth rate than Flavobacterium in xylan, but this difference was partially significant (0.526 h−1 for Curtobacterium, 0.207 h−1 for Flavobacterium; two sample t-test, d.f. = 2, and P = .052). In sum, even though all three species utilized xylan—i.e. they are all xylan degraders—they each had unique growth behaviors. Curtobacterium grew fast and achieved a high final population size, Flavobacterium grew slow and achieved a high final population size, and Erwinia grew fast but achieved a low final population size.
Table 2.
Growth parameters estimated from growth curves in xylan (0.06%, 4 mM) and xylose (0.06%, 4 mM).
| Growth on Xylan (4 mM) | Growth on Xylose (4 mM) | ||
|---|---|---|---|
| Species | Parameter (unit) | Parameters estimated (SE)1 | Parameters estimated (SE)1 |
| Curtobacterium sp. | (1) Maximum growth rate† | µ maxC = 0.526 (0.077) | µ maxC = 0.501 (0.013) |
| (MMLR14002) | (2) Maximum population size | ODmaxC = 0.374 (0.009) | ODmaxC = 0.427 (0.007) |
| (3) Yield | YC = 0.094 (0.002) | YC = 1.107 (0.002) | |
| (4) Half-saturation constant† | kXnC = 2.335 (0.842) | k XoC = 2.479 (0.539) | |
| (5) Maximum uptake rate† | VXnC = 3.727 (0.788) | V XoC = 2.876 (0.400) | |
| Flavobacterium sp. | (1) Maximum growth rate† | µ maxF = 0.207 (0.006) | µ maxF = 0.252(0.005) |
| (LR40) | (2) Maximum population size | ODmaxF = 0.285 (0.007) | ODmaxF = 0.317 (0.005) |
| (3) Yield | YF = 0.071 (0.002) | YF = 0.079 (0.001) | |
| (4) Half-saturation constant† | kXnF = 1.843 (0.362) | k XoF = 2.401 (0.201) | |
| (5) Maximum uptake rate† | VXnF = 1.640 (0.166) | V XoF = 1.782 (0.169) | |
| Erwinia sp. | (1) Maximum growth rate† | µ maxE = 0.568 (0.012) | µ maxE = 0.424 (0.032) |
| (LR17) | (2) Maximum population size | ODmaxE = 0.016 (0.002) | ODmaxE = 0.212 (0.006) |
| (3) Yield | YE = 0.004 (4.6*10−4) | YE = 0.053 (0.002) | |
| (4) Half-saturation constant† | kXnE = 3.111 (0.538) | kXoE = 1.057 (0.525) | |
| (5) Maximum uptake rate† | VXnE = 213.958 (22.796) | VXoE = 4.701 (1.228) |
Parameters: (1) Maximum growth rate in h−1, (2) Maximum population size in unitless OD, (3) Yield in unitless OD mM−1, (4) Half-saturation constant in mM, and (5) Maximum uptake rate in mM OD−1 h−1.
The mean and standard error (SE) were calculated from three replicate growth curves.
Parameters used in the mathematical model.
Figure 2.
Species have unique growth profiles in xylan and xylose. Growth curves of species grown in isolation on 0.06% (w/v) of the polymer xylan and on 0.06% (w/v) of its constituent monomer xylose. A close-up of Erwinia’s growth on xylan is available in Fig. S3. The optical density measured at 600 nm over 72 h (48 h for Erwinia sp.) is plotted for three technical replicates per species.
Next, we grew the species in monomer xylose at the same concentrations (ranging from 0% to 0.36% w/v) previously used for the polymer xylan. Overall, the three species grew better in xylose than in xylan (Fig. S3). For example, at 0.06% (w/v) of xlyan or xylose, Erwinia sp. achieved a 13-fold higher maximum population size in xylose than in xylan (0.212 unitless OD in xylose; 0.016 unitless OD in xylan; two sample t-test, d.f. = 2, and P = 5.041 × 10−4; Fig. 2 and Table 2). But, it grew slower in xylose (0.424 h−1 in xylose, 0.568 h−1 in xylan; two sample t-test, d.f. = 2, and P = .035). The growth limitation in xylan was less severe but significant for Curtobacterium with a 1.1-fold lower maximum population size in 0.06% xylan than in 0.06% xylose (0.427 unitless OD in xylose; 0.374 unitless OD in xylan; two sample t-test, d.f. = 2, P = .01; Fig. 2). Finally, Flavobacterium grew slower in xylan than in xylose (maximum growth rate of 0.207 h−1 in xylan, and of 0.252 h-1 in xylose; two sample t-test, d.f. = 2, and P = .004).
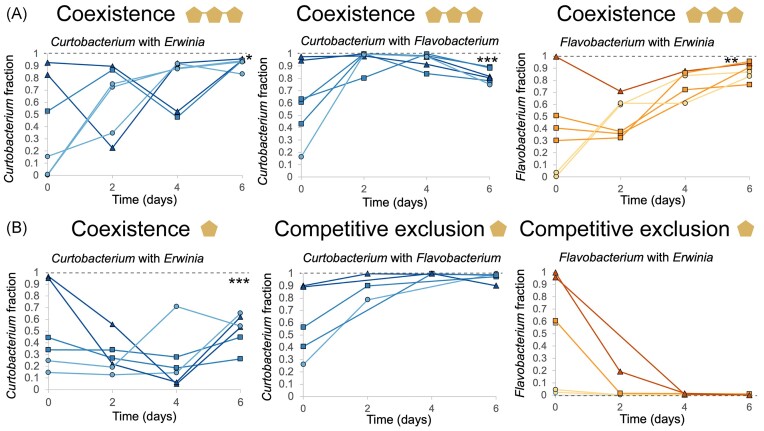
All pairs of species coexist in xylan, but two pairs outcompete each other in xylose
Next, we assessed if species could coexist when grown in coculture in xylan or xylose. We used 0.06% of xylan and xylose, which is a concentration below the saturation threshold for all three species (Fig. S3). All pairs coexisted in xylan (Fig. 3, Fig. S5). For each pair, both species invaded each other when starting at low initial frequencies. On day 6, species converged into a final fraction regardless of the starting fractions. For all pairs, the final fraction was skewed toward the dominance of one species. For example, Curtobacterium was overrepresented when growing in xylan and reached a final fraction of 0.93 ± 0.02 in pair with Erwinia and 0.82 ± 0.02 in pair with Flavobacterium (Fig. 3). For the Flavobacterium–Erwinia pair, the coexistence equilibrium was shifted toward Flavobacterium being overrepresented (final fraction of 0.87 ± 0.03). Erwinia was always underrepresented (Fig. 3, Fig. S5). In contrast to xylan, coexistence patterns differed in xylose, with two pairs displaying competitive exclusion. In both cases, Flavobacterium was outcompeted. Only Curtobacterium and Erwinia coexisted in xylose (Fig. 3).
Figure 3.
Pairwise competitions in xylan result in coexistence, whereas, in xylose, they result in competitive exclusion or coexistence. Pairwise competitions in xylan (panel A) and xylose (panel B) for the Curtobacterium–Erwinia pair, Curtobacterium–Flavobacterium pair, and Flavobacterium–Erwinia pair. Graphs represent the fraction (values between 0 and 1) of each competing species over 6 days. The circles, squares, and triangles correspond to low, equal, and common initial fractions, respectively. In coexistence, a species’ fraction increases when started from rare and decreases when started from common until an equilibrium fraction statistically different from 1 is reached (one sample t-test, d.f. = 5, P = .011, P = 6.13 × 10−4, P = .009, P = 3.67 × 10−4 for the Curtobacterium–Erwinia pair in xylan, Curtobacterium–Flavobacterium pair in xylan, Flavobacterium–Erwinia pair in xylan, and Curtobacterium–Erwinia pair in xylose, respectively). In competitive exclusion, the fraction of the strongest competitor increases until it reaches a fraction of 1 regardless of whether it started from rare or common.
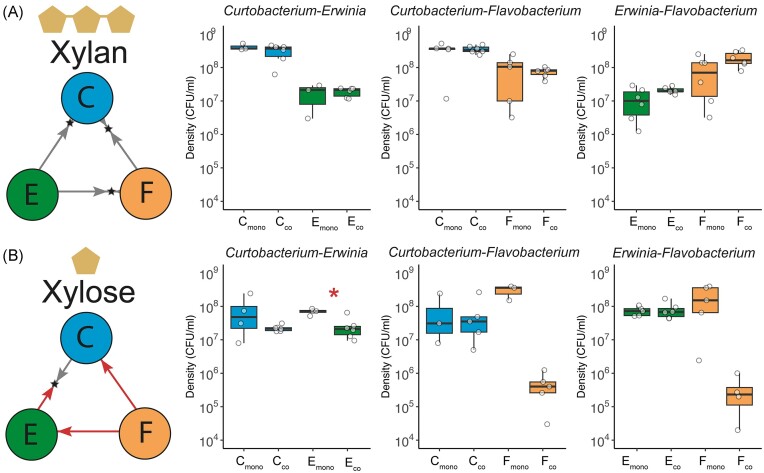
Pairwise interactions are negative in xylose and not significant in xylan
We further characterized the network of pairwise interactions by comparing the growth of species in monoculture to its growth in pairwise coculture. For all pairs in xylan, there were no significant differences in the final densities in monoculture and coculture (Fig. 4A). In contrast, in xylose, negative pairwise interactions prevailed. Erwinia had significantly lower densities in coculture with Curtobacterium than when growing in monoculture (2.7-fold lower in the coculture than in monoculture, two sample t-test, d.f. = 8, P = .003). Flavobacterium also reached lower densities in coculture with Erwinia and Curtobacterium than in monoculture (Fig. 4B), but these differences were marginally significant (two sample t-test, d.f. = 4, P = .068, and d.f. = 2, P = .0576, respectively). This lack of significance is likely due to a large variability among replicates and because we did not include the replicates for which the final densities dropped below the detection limit. Neither Erwinia nor Curtobacterium were affected by the presence of other species. Thus, in xylose, interactions between pairs of species were amensalistic.
Figure 4.
The pairwise interaction network differs in xylan and xylose. In xylan (panel A), pairwise competitions result in stable coexistence of Curtobacterium (C), Erwinia (E), and Flavobacterium (F), which is represented by two arrows pointing to a star representing the final fraction on day 6 (summary from Fig. 3). The growth in monoculture and in coculture at day 6 is represented by box plots (average of three to six replicates), with the data points shown with empty circles. For all three species, the growth in monoculture (Cmono, Emono, and Fmono) is not statistically different than the growth in coculture (Cco, Eco, and Fco). Thus, interactions between species were not significant in xylan. Instead, in xylose (panel B), pairs interact negatively. Flavobacterium (F) was outcompeted by both Curtobacterium (C) and Erwinia (E), which is represented by a single arrow (summary from Fig. 3). The growth of Erwinia at day 6 in coculture with Curtobacterium was significantly lower than its growth in monoculture (Wilcox Test, W = 23, P = .019). The growth of Flavobacteriu m at day 6 in coculture with the other species was lower, but these differences were only marginally significant (two sample t-tests, d.f. = 2, P = .058 and, d.f. = 4, P = .068).
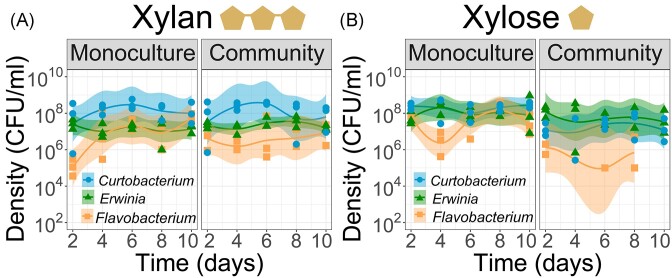
All three species coexist in xylan, whereas only two species coexist in xylose
Next, we assembled a three-species community and compared species’ growth in the community and in monoculture. In xylan, the three species grown in a community sustained a high population over 10 days (Fig. 5A, right panel). These population densities did not differ to those in monocultures (linear mixed model with n = 30 and groups = 3; t-values of −0.215, 1.575, and −1.055, for Curtobacterium sp., Flavobacterium sp., and Erwinia sp., respectively). This suggests that other species had no effect on a species’ net growth. In contrast, in xylose, growth was diminished in the three-species community compared to monocultures (Fig. 5B). Curtobacterium sustained significantly lower densities in the community than in monoculture (8-fold lower growth in the community than in monoculture; linear mixed model, t-value = 5.506). Erwinia also had a lower density in the community than in monoculture (1.6-fold lower growth in the community than in monoculture), even though the linear mixed model showed a nonsignificant trend (t-value = 1.141). Finally, Flavobacterium was the most impacted by the presence of other species, and its density dropped below the detection limit after the fourth serial transfer (Fig. 5B, right panel). These negative effects of growing in a community were also captured by analyzing the area under the curve over 10 days (Fig. S6). In sum, all three species coexisted in xylan, whereas only two of them coexisted in xylose over 10 days.
Figure 5.
Coexistence patterns in a three-species community change according to the substrate complexity. In xylan, all three species coexisted and grew equally well in a three-species community than alone (panel A). Instead, in xylose, only two species coexisted for 10 days, and species grew less well in the community than alone (panel B). For each panel, population density trajectories of monocultures are represented on the left side and of communities on the right side, estimated from CFU ml−1 during five serial transfers (1000-fold dilution every 2 days). Each line corresponds to a local polynomial regression fitting of three replicates with 95% confidence interval. Graphs were constructed with the function stat_smooth (R version 3.2.3).
Potential mechanisms undelying species coexistence in xylan
We previously observed that the growth of species in isolation is limited in xylan compared to xylose (Fig. 2). We wondered whether this growth limitation suffices to explain the coexistence of three species in xylan (Fig. 5); i.e. whether Curtobacterium and Erwinia’s lower growth in xylan buffers the negative impact on Flavobacterium. To assess this possibility, we used a mathematical model simulating the simplest case of resource competition, which was parameterized by single-species growth measurements (Table 2).
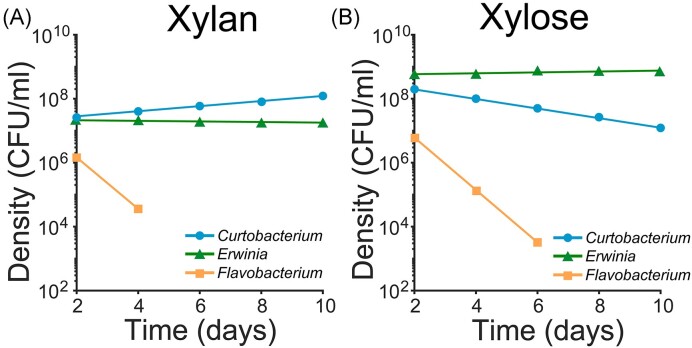
Having verified that our model captured the monocultures’ dynamics in batch culture based on the fitted parameters to growth curves (Fig. S7), we used the model to predict community dynamics. When simulating five serial passages of communities in xylose and peptone, Flavobacterium went extinct and only Curtobacterium and Erwinia coexisted over 10 days (Fig. 6B). This result validates our experimental findings, except for the time of extinction of Flavobacterium, which occurred after 6 days in the model (Fig. 6B) compared to after 8 days in the experiments (Fig. 5B). Thus, in xylose our model qualitatively recapitulated the community dynamics. Instead, in xylan, our model predicted the extinction of Flavobacterium after 4 days (Fig. 6A). This prediction contrasted with our experimental data, where we observed coexistence of all three species over 10 days (Fig. 5A). Thus, the resource model partially recapitulated community dynamics in xylan, suggesting the coexistence of the three species in the community cannot fully be explained by the growth behaviors of monocultures.
Figure 6.
The mathematical model recapitulates community dynamics in xylose but not in xylan. The results derived from the resource competition model are plotted when simulating growth in a three-species community during five serial transfers in xylan (panel A) and in xylose (panel B). Each line corresponds to the connection between the final densities of each species after a 48-h cycle of growth.
Next, we explored whether resource partitioning could explain the coexistence of three species in xylan. First, we conducted a genome-level metabolic profiling to assess whether species had the genes encoding enzymes involved in the debranching of sugar residues decorating the xylan. The three species varied in the type and number of enzymes involved in hemicellulose debranching (Table S2). The presence of α-l-arabinofuranosidase (arabinosidase) was particularly interesting, given that this enzyme is directly involved in debranching of heteroxylans (Malgas et al. 2019). Copy numbers for this enzyme were identified in Curtobacterium sp. (1 hit) and Flavobacterium sp. (4 hits), but not in Erwinia sp. (0 hits). Finally, in terms of CAZymes, Flavobacterium sp. was the only species that had one copy of a glucuronidase (GH67 family), which has been associated with the hydrolysis of the glycosidic bond releasing glucuronic acid residues (Table S2; Malgas et al. 2019). This suggest that Flavobacterium may specialize in using xylan breakdown products other than xylose.
Finally, we assessed species’ ability to grow on simple carbohydrates from the branched part of xylan. All three species were able to grow well on diverse monosaccharides—except for arabinose—with different degrees of growth (Fig. S8). Curtobacterium sp. was the only species able to grow on arabinose (after accounting for its growth on peptone). Thus, although there is resource use overlap among species, differences in growth abilities to use sugar residues suggest a potential for species to partition resources in xylan.
Discussion
There is a growing interest in understanding how interspecies interactions shape the degradation of biopolymers in nature. Recent studies have focused on studying the interactions among multiple microbial guilds; for example, the interaction among degraders, exploiters, and scavengers in marine polysaccharide-degrading bacteria (Enke et al. 2019, Pontrelli et al. 2022, D’Souza et al. 2023, Daniels et al. 2023). Here, we explored the interactions among three species belonging to the guild of xylan degraders from a natural leaf litter community that mediate the turnover of organic matter. We found that the complexity of the substrate influences the interactions and coexistence of species.
Substrate complexity influences patterns of coexistence with less coexistence in xylose than in xylan
The complexity of the carbon source influenced the coexistence patterns: three species coexisted in xylan for 10 days; instead, only two species coexisted in xylose for 10 days. According to a simple view of the coexistence theory, it is expected that, an environment with a single carbon source could only sustain one species. At first, it seemed surprising to see more coexistence than expected. Nevertheless, it is important to recognize that xylan/xylose may not act as the only carbon source, as Curtobacterium grew on peptone without an additional source of carbon. Thus, based on our system, we could expect that at least two species coexist, one of them being Curtobacterium. This is what we observed from experiments in pairs and communities in xylose. The observation that three species coexist in xylan (instead of two) could be explained either because xylan is a polymer or because the xylan in our study is composed of a chain of xylose decorated with branches of other carbon sources such as glucose and arabinose (Melo-Silveira et al. 2019). Even if these other carbon sources are the minority (24%), it is possible that these extra carbon sources sustained the coexistence of three species through resource partitioning. To disentangle these possibilities, future studies should compare the coexistence patterns in homoxylan and xylose. For instance, Dal Bello et al. (2021) observed that the number of species coexisting in cellulose (a homopolysaccharide of glucose) was roughly double that of species coexisting in glucose (Dal Bello et al. 2021). Thus, our results agree with the trend that complex substrates promote coexistence while simple substrates hinder it.
Substrate complexity influences species interactions with more negative interactions in xylose than in xylan
Our study shows that in the polysaccharide xylan, pairs of species grew equally well in the presence of another species than in monoculture. In comparison, in the monosaccharide xylose, one of the species grew worse in the presence of another species than on its own. These pairwise dynamics scaled up to three species communities. For example, in xylose, species grew worse in the community than in monoculture. Our mathematical model showed that in xylose, these dynamics could be explained by resource competition, whereas the resource competition model partially predicted the coexistence patterns in xylan.
One of the most interesting results was that, in xylan, all three species grew equally well in the community than alone. We posit that a combination of resource partitioning (hypothesis H2a) and resource competition may explain this observation. All three species encoded the genomic potential (endo-1–4-xylanases enzymes) to hydrolyze the xylose backbone to utilize the majority of the xylan polysaccharide (76% xylose). Given that we observed that species compete for the monomer xylose (Figs 4B, 5B, and 6B), we might expect species to compete and have reduced growth in the xylan community than in isolation, as predicted by the mathematical model (Fig. 6A). However, the negative effects of resource competition may have been alleviated by enzyme complementarity and resource partitioning. For example, Flavobacterium encodes unique copies of arabinosidases and CAZymes involved in heteroxylan debranching that the other species did not have (Table S2). Thus, Flavobacterium may specialize in degrading and consuming specific parts of the xylan (Malgas et al. 2019). In the community, these enzymes (not produced by other species) may facilitate the debranching of xylan, which could improve degradation efficiency. Previous studies using communities of cellulose-degrading bacteria have shown a positive relationship between species diversity and community productivity. This positive relationship has been associated with a subset of species being able to degrade recalcitrant compounds that other species cannot degrade (Wohl et al. 2004, Evans et al. 2017). In addition to complementarity, resource partitioning may explain coexistence of species in xylan. For example, Curtobacterium was capable of using carbon sources such as peptone and arabinose that the other species could not use. In conclusion, complex polysaccharides may buffer the effects of resource competition through complementarity and resource partitioning.
In conclusion, our study suggests that negative interactions may be buffered when growing in complex polysaccharides. This is relevant because substrates in nature often exist in the form of polysaccharides, which can promote species coexistence. Importantly, we have established a model community with three species from grass litter that coexist in the polysaccharide xylan for 10 days (∼50 generations). This community can be used as a starting point to study the evolution of species interactions under environmental change, which could be already occurring in our system (Lawrence et al. 2012). Although, one needs to be cautious about extrapolating the finding from these simple systems to natural ecosystem, which are inherently complex. Our hope is to isolate some of the variables (e.g. abiotic factors), while keeping some of the complexities of microbial communities (natural isolates growing on complex substrates) to generate general principles on how species evolve in response to environmental stressors and their consequences for ecosystems functions (Rodriguez-Verdugo et al. 2021, McClure et al. 2022, Martiny et al. 2023).
Supplementary Material
Acknowledgement
We thank Melissa Martens for her assistance with the experiments. Special thanks to Jennifer Martiny for providing the strains and for providing valuable feedback on earlier manuscript versions. We thank four anonymous reviewers for helpful comments that improved the paper. Finally, we thank Rodriguez-Verdugo lab members for providing useful feedback during this research.
Contributor Information
Parmis Abdoli, Department of Ecology and Evolutionary Biology, University of California Irvine, 321 Steinhaus Hall, Irvine, CA 92697, United States.
Clément Vulin, Department of Fundamental Microbiology, University of Lausanne, Biophore, CH-1015 Lausanne, Switzerland.
Miriam Lepiz, Department of Ecology and Evolutionary Biology, University of California Irvine, 321 Steinhaus Hall, Irvine, CA 92697, United States.
Alexander B Chase, Department of Earth Sciences, Southern Methodist University, 3225 Daniel Avenue, Suite 207, Heroy Hall, Dallas, TX 75205, United States.
Claudia Weihe, Department of Ecology and Evolutionary Biology, University of California Irvine, 321 Steinhaus Hall, Irvine, CA 92697, United States.
Alejandra Rodríguez-Verdugo, Department of Ecology and Evolutionary Biology, University of California Irvine, 321 Steinhaus Hall, Irvine, CA 92697, United States.
Author contributions
Parmis Abdoli (Conceptualization, Data curation, Investigation, Methodology, Writing – review & editing), Clément Vulin (Data curation, Formal analysis, Methodology, Writing – review & editing), Miriam Lepiz (Investigation, Writing – review & editing), Alexander B. Chase (Data curation, Formal analysis, Software, Writing – review & editing), Claudia Weihe (Investigation, Resources, Writing – review & editing), and Alejandra Rodríguez-Verdugo (Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing)
Conflict of interest
The authors declare no conflicts of interest.
Funding
We thank all the collaborators from the Loma Ridge Global Change Experiment, including Kathleen Treseder and Steve Allison, which are currently funded along with A.R.V. by the National Science Foundation (award number 2308342). P.A. was supported by UC Irvine’s Summer Undergraduate Research Fellowship (SURP). M.L. was supported by the NIH-Maximizing Access to Research Careers (MARC) grant. A.R.V. was supported by UC Irvine’s Charlie Dunlop School of Biological Sciences and two grants from the National Science Foundation (award numbers 2308342 and 2234627).
Data availability
The genome sequences generated during the current study are available in the DDBJ/ENA/GenBank repository under accession numbers JAWVWO000000000 and JAWVWP000000000. All the other data generated or analyzed during this study are included in this published article and its supplementary information files.
References
- Arnosti C, Wietz M, Brinkhoff T et al. The biogeochemistry of marine polysaccharides: sources, inventories, and bacterial drivers of the carbohydrate cycle. Annu Rev Mar Sci. 2021;13:81–108. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett RD, Freeman C, Ostle NJ. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008;2:805–14. [DOI] [PubMed] [Google Scholar]
- Berlemont R, Allison SD, Weihe C et al. Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol. 2014;5. 10.3389/fmicb.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microb. 2013;79:1545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont R, Martiny AC. Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microb. 2015;81:1513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. BBMap: A Fast, Accurate, Splice-Aware Aligner. Berkeley: Lawrence Berkeley National Lab, 2014. https://www.osti.gov/servlets/purl/1241166. [Google Scholar]
- Chang C-Y, Bajic D, Vila JCC et al. Emergent coexistence in multispecies microbial communities. Science. 2023;381:343–8. [DOI] [PubMed] [Google Scholar]
- Chase AB, Arevalo P, Polz MF et al. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front Microbiol. 2016;7. 10.3389/fmicb.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AB, Karaoz U, Brodie EL et al. Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio. 2017;8. 10.1128/mbio.01809-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AB, Gomez-Lunar Z, Lopez AE et al. Emergence of soil bacterial ecotypes along a climate gradient. Environ Microbiol. 2018;20:4112–26. [DOI] [PubMed] [Google Scholar]
- Chase AB, Weihe C, Martiny JBH. Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient. Proc Natl Acad Sci USA. 2021;118. 10.1073/pnas.2101254118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil PA, Mussig AJ, Hugenholtz P et al. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ye W, Zhang Y et al. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43:7762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coûteaux MM, Bottner P, Berg B. Litter decomposition, climate and liter quality. Trends Ecol Evol. 1995;10:63–6. [DOI] [PubMed] [Google Scholar]
- D'Souza GG, Povolo VR, Keegstra JM et al. Nutrient complexity triggers transitions between solitary and colonial growth in bacterial populations. ISME J. 2021;15:2614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza GG, Schwartzman J, Keegstra J et al. Interspecies interactions determine growth dynamics of biopolymer degrading populations in microbial communities. Proc Natl Acad Sci USA. 2023;120. 10.1073/pnas.2305198120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bello M, Lee H, Goyal A et al. Resource-diversity relationships in bacterial communities reflect the network structure of microbial metabolism. Nat Ecol Evol. 2021;5:1424–34. [DOI] [PubMed] [Google Scholar]
- Daniels M, van Vliet S, Ackermann M. Changes in interactions over ecological time scales influence single-cell growth dynamics in a metabolically coupled marine microbial community. ISME J. 2023;17:406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta MS, Sliwerska E, Gore J et al. Microbial interactions lead to rapid micro-scale succession on model marine particles. Nat Commun. 2016;7. 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YJ, Wang SY. Synergistic growth in bacteria depends on substrate complexity. J Microbiol. 2016;54:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YJ, Wang SY. Complex carbohydrates reduce the frequency of antagonistic interactions among bacteria degrading cellulose and xylan. FEMS Microbiol Lett. 2017;364:fnx019. 10.1093/femsle/fnx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan KL, Peña J, Allison SD et al. Phylogenetic conservation of substrate use specialization in leaf litter bacteria. PLoS One. 2017;12:e0174472. 10.1371/journal.pone.0174472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enke TN, Datta MS, Schwartzman J et al. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol. 2019;29:1528–1535.e6. [DOI] [PubMed] [Google Scholar]
- Evans R, Alessi AM, Bird S et al. Defining the functional traits that drive bacterial decomposer community productivity. ISME J. 2017;11:1680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finks SS, Weihe C, Kimball S et al. Microbial community responses to a decade of simulated global changes depends on the plant community. Element Sci Anthropocene. 2021;9. 10.1525/elementa.2021.00124. [DOI] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J et al. Pfam: the protein families database. Nucl Acids Res. 2014;42:D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman SI, Weihe C, Li J et al. Decomposition responses to climate depend on microbial community composition. Proc Natl Acad Sci USA. 2018;115:11994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldford JE, Lu N, Bajic D et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin G. The competitive exclusion principle. Science. 1960;131:1292–7. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11. 10.1186/1471-2015-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D, Fiegna F, Behrends V et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann SR. A piece of the pie: engineering microbiomes by exploiting division of labor in complex polysaccharide consumption. Curr Opin Chem Eng. 2020;30:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgas S, Mafa MS, Mkabayi L et al. A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J Microbiol Biotechnol. 2019;35. 10.1007/s11274-019-2765-z. [DOI] [PubMed] [Google Scholar]
- Manhart M, Shakhnovich EI. Growth tradeoffs produce complex microbial communities on a single limiting resource. Nat Commun. 2018;9. 10.1038/s41467-018-05703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JB, Martiny AC, Weihe C et al. Microbial legacies alter decomposition in response to simulated global change. ISME J. 2017;11:490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JB, Martiny AC, Brodie E et al. Investigating the eco-evolutionary response of microbiomes to environmental change. Ecol Lett. 2023;26:S81–90. [DOI] [PubMed] [Google Scholar]
- Matulich KL, Weihe C, Allison SD et al. Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J. 2015;9:2477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, Farris Y, Danczak R et al. Interaction networks are driven by community-responsive phenotypes in a chitin-degrading consortium of soil microbes. mSystems. 2022;7. 10.1128/msystems.00372-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Silveira RF, Viana RLS, Sabry DA et al. Antiproliferative xylan from corn cobs induces apoptosis in tumor cells. Carbohydr Polym. 2019;210:245–53. [DOI] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–94. [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontrelli S, Szabo R, Pollak S et al. Metabolic cross-feeding structures the assembly of polysaccharide degrading communities. Sci Adv. 2022;8. 10.1126/sciadv.abk3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts DL, Suding KN, Winston GC et al. Ecological effects of experimental drought and prescribed fire in a southern California coastal grassland. J Arid Environ. 2012;81:59–66. [Google Scholar]
- Ramin KI, Allison SD. Bacterial tradeoffs in growth rate and extracellular enzymes. Front Microbiol. 2019;10. 10.3386/fmicb.2019.02956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Vulin C, Ackermann M. The rate of environmental fluctuations shapes ecological dynamics in a two-species microbial system. Ecol Lett. 2019;22:838–46. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A. Evolving interactions and emergent functions in microbial consortia. mSystems. 2021;6. 10.1128/msystems.00774-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Tilman D. Resource competition between planktonic algae—experimental and theoretical approach. Ecology. 1977;58:338–48. [Google Scholar]
- Tilman D. Resource competition and community structure. Monogr Popul Biol. 1982;17:1–296. [PubMed] [Google Scholar]
- Vandenberghe LPS, SG Karp MGBP et al. Chapter 2–classification of enzymes and catalytic properties. In: Singh SP, Pandey A, Singhania RR et al. (eds.), Biomass, Biofuels, Biochemicals. Amsterdam: Elsevier Science, 2020, 11–30. [Google Scholar]
- Wohl DL, Arosa D, Gladstone JR. Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology. 2004;85:1534–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences generated during the current study are available in the DDBJ/ENA/GenBank repository under accession numbers JAWVWO000000000 and JAWVWP000000000. All the other data generated or analyzed during this study are included in this published article and its supplementary information files.