Abstract
Salt stress is an environmental factor that limits plant growth and crop production. With the rapid expansion of salinized arable land worldwide, investigating the molecular mechanisms underlying the salt stress response in plants is urgently needed. Here, we report that GROWTH REGULATING FACTOR 7 (OsGRF7) promotes salt tolerance by regulating arbutin (hydroquinone-β-D-glucopyranoside) metabolism in rice (Oryza sativa). Overexpression of OsGRF7 increased arbutin content, and exogenous arbutin application rescued the salt-sensitive phenotype of OsGRF7 knockdown and knockout plants. OsGRF7 directly promoted the expression of the arbutin biosynthesis genes URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE 1 (OsUGT1) and OsUGT5, and knockout of OsUGT1 or OsUGT5 reduced rice arbutin content, salt tolerance, and grain size. Furthermore, OsGRF7 degradation through its interaction with F-BOX AND OTHER DOMAINS CONTAINING PROTEIN 13 reduced rice salinity tolerance and grain size. These findings highlight an underexplored role of OsGRF7 in modulating rice arbutin metabolism, salt stress response, and grain size, as well as its broad potential use in rice breeding.
Transcription factor control of arbutin biosynthesis gene expression regulates grain size and salt stress response in rice (Oryza sativa).
Introduction
As sessile organisms, plants are under constant threat due to different environmental stresses, such as extreme temperature, drought, and salinity, which limit plant development and have adverse effects on the biomass or yield of crops. The environmental impact of salt damage, which is currently underestimated, poses a major threat to food security in multiple regions of the world (Ismail and Horie 2017). Due to industrial pollution, the excessive use of chemical fertilizers, and irrational irrigation, soil salinization is becoming more common in many arable fields. Soil salinization, which has dramatically reduced available land resources and caused great losses in agricultural production, has become an ecological problem worldwide (Gong et al. 2020).
According to previous studies, several metabolites are associated with salinity tolerance in plants. The content of L-arabinose, a plant-specific monosaccharide present in cell wall polysaccharides or cell wall-localized glycoproteins, was altered after salt treatment. Plants with disrupted L-arabinose biosynthesis were hypersensitive to high salinity, suggesting that maintaining L-arabinose metabolism for cell wall components is critical for the adaptation of plants to salt stress (Zhao et al. 2019). A soluble sugar, trehalose, has been the focus of research on abiotic stress tolerance. TREHALOSE-6-PHOSPHATE SYNTHASE 8 (OsTPS8), a TPS-encoding gene in rice (Oryza sativa), positively regulates the salinity stress response. The OsTPS8 overexpression lines exhibited enhanced salinity tolerance, and an ostps8 mutant exhibited a salt sensitivity phenotype (Vishal et al. 2019). Moreover, vitamin B6, which is an essential cofactor of numerous metabolic enzymes, is involved in plant development and abiotic stress responses. Ectopic expression of MALATE DEHYDROGENASE 1 alters the expression levels of genes encoding key enzymes involved in vitamin B6 biosynthesis and reduces the level of vitamin B6 to negatively regulate salt tolerance (Nan et al. 2020).
Arbutin, also named hydroquinone-β-D-glucopyranoside, is a natural glucoside hydroquinone from plants that acts as an antioxidant and protects membranes in plants (Oliver et al. 2001). Arbutin has abundant biological activities and is known as a skin-whitening or depigmenting agent (Lim et al. 2009). In cucumber (Cucumis sativus), arbutin suppresses angular leaf spot disease by promoting the expression of Pathogen-Related gene 1, which is a marker gene of the salicylic acid signaling pathway (Kuzniak et al. 2015). In addition to its role in disease resistance, it has been suggested that arbutin contributes to resistance to abiotic stresses through its ability to stabilize membranes depending on lipid composition (Oliver et al. 2001). Among the 107 metabolites detected in potato (Solanum tuberosum), arbutin was enriched in drought-tolerant cultivars (Sprenger et al. 2016).
GROWTH REGULATING FACTORs (GRFs), a conserved plant-specific transcription factor family and downstream targets of OsmiR396, are involved in multiple developmental processes, such as grain size determination, inflorescence development, plant architecture determination, blast disease resistance, and brown planthopper resistance (Gao et al. 2015; Li et al. 2018; Chandran et al. 2019; Dai et al. 2019; Chen et al. 2020a, 2020b). In rice, the 12 existing GRFs have conserved QLQ and WRC domains, but they perform diverse functions during plant growth and development. Under salt stress conditions, the expression of OsmiR396c dramatically changed, and the overexpression of OsmiR396c decreased salt tolerance, indicating that OsmiR396 negatively regulates salinity tolerance (Gao et al. 2010). Although the roles of OsmiR396 in plant development and abiotic stress responses have been reported, how OsGRFs coordinate plant development and abiotic stress responses remains unknown.
In this study, we found that OsGRF7 overexpression (GRF7-OE) plants exhibited enhanced salt tolerance and increased grain size. Untargeted metabolite profiling analysis revealed that the content of arbutin significantly increased in the GRF7-OE transgenic plants compared with that in wild-type plants. Arbutin improved the salt tolerance of both OsGRF7 knockdown (GRF7-Ri) and knockout (grf7) plants. Moreover, arbutin repressed the accumulation of reactive oxygen species (ROS) generated by salt stress. Furthermore, in vivo and in vitro assays indicated that OsGRF7 binds directly to the promoters of arbutin biosynthesis genes (URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE 1 [OsUGT1] and OsUGT5) to regulate the content of arbutin. Loss of function of OsUGT1 or OsUGT5 resulted in hypersensitivity to salt stress and reduced grain size. Additionally, F-BOX AND OTHER DOMAINS CONTAINING PROTEIN 13 (OsFBO13) directly interacted with and promoted the degradation of OsGRF7 via the 26S proteasome system. Loss-of-function mutants of OsFBO13 exhibited enhanced salt tolerance similar to that of the GRF7-OE plants. Taken together, the results revealed that the OsFBO13–OsGRF7–OsUGT1/OsUGT5–arbutin regulatory module plays a vital role in regulating the rice salt stress response and grain development.
Results
OsGRF7 positively regulates rice salt tolerance
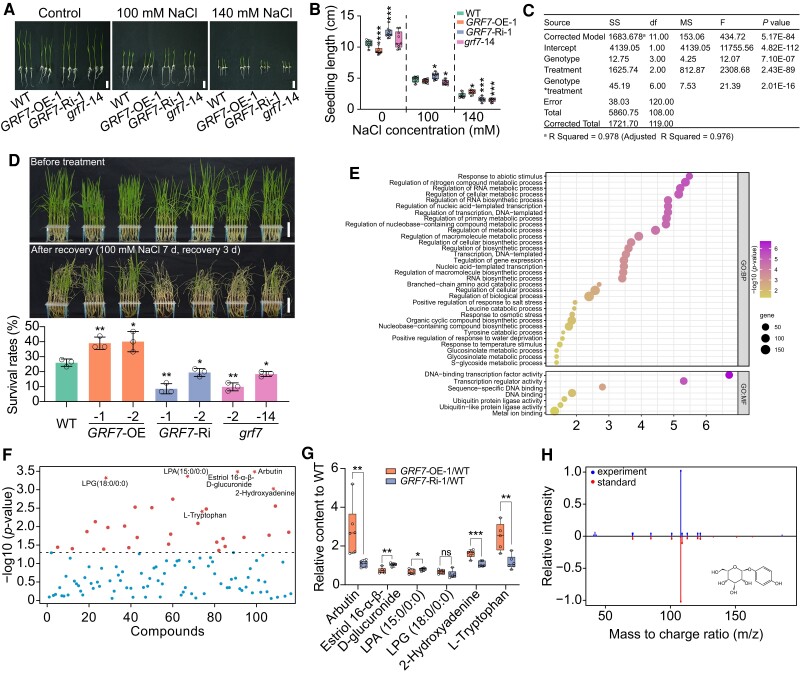
Previous studies have suggested that overexpression of OsmiR396c weakens salt tolerance and that repression of OsmiR396 enhances salt tolerance (Gao et al. 2010; Dai et al. 2019). However, the functions of the genes downstream of OsmiR396, namely, the GRFs, in salt tolerance remain unknown. To investigate whether OsGRF7 contributes to salt tolerance, we measured the seedling length of the OsGRF7 transgenic plants after treatment with different concentrations of NaCl. The seedling length of the GRF7-OE-1 plants under normal conditions was shorter than that of the wild type, which is consistent with the finding that OsGRF7 has a negative effect on plant height (Chen et al. 2020a). After a 5-d treatment with 140 mM NaCl, we found that the seedling lengths of the GRF7-Ri-1 and grf7-14 seedlings were significantly shorter than those of the wild type, while the opposite trend was observed for the GRF7-OE-1 seedlings (Fig. 1, A and B). We also found that genotype, treatment, and their interaction all contributed to seedling length (Fig. 1C). Moreover, the 15-d-old OsGRF7 transgenic plants were further treated with 100 mM NaCl, and the GRF7-OE plants exhibited higher salt-tolerance phenotypes and higher survival rates than did the wild-type plants (Fig. 1D; Supplementary Fig. S1), indicating that OsGRF7 positively regulates rice salt tolerance.
Figure 1.
Overexpression of OsGRF7 enhances tolerance to salinity stress with the involvement of arbutin. A) Phenotypes of WT and OsGRF7 transgenic seedlings treated with different concentrations of NaCl. Bars = 2 cm. B) Effects of different concentrations of NaCl on seedling length B) in the WT and OsGRF7 transgenic lines. Values are means ± SDs (n = 10 independent seedlings). C) Two-way analysis of variance of the contribution of genotype, treatment, and their interaction to seedling length. SS, type III sum of squares. df, degree of freedom. MS, mean square. F, freedom. D) Fifteen-day-old seedlings of WT and OsGRF7 transgenic lines were treated with 100 mM NaCl for 7 d and recovered for 3 d. Bars = 5 cm. Values are means ± SDs of 3 biological replicates. E) Gene ontology enrichment analyses of downregulated genes in grf7-14 after salt treatment. F) One-way analysis of variance of all identified metabolites in the WT and OsGRF7 transgenic lines. The six selected metabolites for further analysis are marked with red pentagrams. G) Relative levels of the six selected metabolites in F). H) Comparison of MS/MS spectra between experimental samples and the arbutin standard. Values are means ± SDs of 3 biological replicates. For B) and G), the line in the middle of each box represents the 50th percentile. The bottom and top lines represent the 25th and 75th percentiles, respectively. The whiskers are the minimum and maximum values. WT, wild type. GRF7-OE, OsGRF7 overexpression lines. GRF7-Ri, OsGRF7 RNAi lines. grf7, OsGRF7 knockout lines. ns, no significant difference. For this figure, asterisks indicate significant differences between the wild type and the other genotypes by a two-tailed Student's t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
To investigate the mechanism of OsGRF7 under salt stress, transcriptomic analysis was subsequently performed using 15-d-old seedlings of grf7-14 and wild-type plants after salt treatment. In total, 2,737 differentially expressed genes (DEGs) were detected (P < 0.001, fold change >1; Supplementary Data Set 1), 1,696 of which were upregulated and 1,041 of which were downregulated in the grf7-14 seedlings compared with the wild type (Supplementary Fig. S2). Gene Ontology analysis revealed that the 1,041 downregulated DEGs were enriched in “Response to abiotic stimulus” and “Regulation of metabolic process” (Fig. 1E), which was consistent with the positive role of OsGRF7 in salinity signaling.
To study the metabolites involved in salt stress, an untargeted metabolite profiling analysis was conducted with 15-d-old seedlings, and a total of 114 metabolites, including amino acids, pyridoxines, benzenediols, and steroidal glycosides, were identified from 31 subclasses (Supplementary Figs. S3 and S4; Supplementary Data Set 2). In total, 30 metabolites exhibited significant differences among the wild-type, GRF7-Ri, and GRF7-OE transgenic plants (Fig. 1F; Supplementary Table S1; P < 0.05). L-tryptophan, the precursor for auxin biosynthesis, showed significantly lower content in the wild-type and GRF7-Ri plants than in the GRF7-OE transgenic plants (Fig. 1G), which is consistent with the high auxin content in the GRF7-OE transgenic lines (Chen et al. 2020a). Notably, compared with that in the wild type, the content of arbutin in the GRF7-OE transgenic plants was significantly increased (Fig. 1, G and H). We thus speculated that arbutin might participate in the salt stress response.
Arbutin improves the salt tolerance of rice
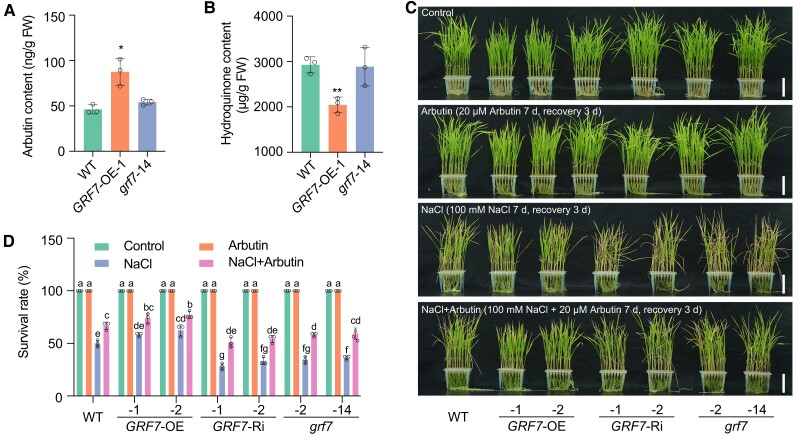
We subsequently measured the endogenous content of arbutin using the targeted metabolite profiling method and found that the GRF7-OE transgenic plants had higher arbutin concentrations in both the seedlings and roots than did the wild-type (Fig. 2A; Supplementary Fig. S5, A and C). Meanwhile, the content of hydroquinone was lower in GRF7-OE transgenic plants than that in the wild-type plants (Fig. 2B; Supplementary Fig. S5, B and D). To further determine the role of arbutin in salt tolerance, we applied exogenous treatment with arbutin to the wild type. The survival rates improved as the concentration of arbutin increased (Supplementary Fig. S6).
Figure 2.
Arbutin improves rice salt tolerance. A) and B) The in vivo contents of arbutin A) and hydroquinone B) in OsGRF7 transgenic seedlings. Values are means ± SDs of 3 biological replicates. C) Phenotypes of WT and OsGRF7 transgenic lines treated with 20 µM arbutin, 100 mM NaCl, and 100 mM NaCl plus 20 µM arbutin. Bars = 5 cm. D) Survival rates with different treatments in C). Different letters represent significant differences at P < 0.05 determined by Tukey's multiple comparisons test. WT, wild type. GRF7-OE, OsGRF7 overexpression lines. GRF7-Ri, OsGRF7 RNAi lines. grf7, OsGRF7 knockout lines. FW, fresh weight. For this figure, asterisks indicate significant differences between the wild type and the other genotypes by a two-tailed Student's t-test (*P < 0.05).
We then fixed 20 µM as a suitable concentration to determine whether arbutin could rescue the salt-sensitive phenotypes of the GRF7-Ri and grf7 transgenic plants. Without salt stress, the addition of arbutin to the culture solution resulted in no phenotypic difference among the OsGRF7 transgenic plants. However, in the presence of 100 mM NaCl, the addition of arbutin rescued the salt sensitivity of both the GRF7-Ri and grf7 transgenic plants (Fig. 2, C and D), indicating that arbutin enhanced rice salt tolerance.
Malondialdehyde (MDA), proline, soluble sugar, and chlorophyll levels are physiological and biochemical indicators of plants under stress in adverse environments (Chen and Murata 2002; Kavi Kishor and Sreenivasulu 2014). Biochemical analysis revealed that under saline conditions, the MDA content was significantly elevated in the GRF7-Ri-1 and grf7-14 lines compared with that in the wild type (Supplementary Fig. S7A). The proline content was significantly higher in the GRF7-OE-1 plants than in the wild-type plants, GRF7-Ri-1, and grf7-14 lines (Supplementary Fig. S7B). The soluble sugar content was markedly reduced in the GRF7-Ri-1 and grf7-14 plants and significantly increased in the GRF7-OE-1 plants (Supplementary Fig. S7C). The total chlorophyll content decreased in the GRF7-Ri-1 and grf7-14 plants and increased in the GRF7-OE-1 plants. When arbutin was applied, the physiological differences among the GRF7-OE, GRF7-Ri, grf7, and wild-type plants were decreased (Supplementary Fig. S7, D to F). These results indicated that arbutin effectively increased the salt stress tolerance of rice.
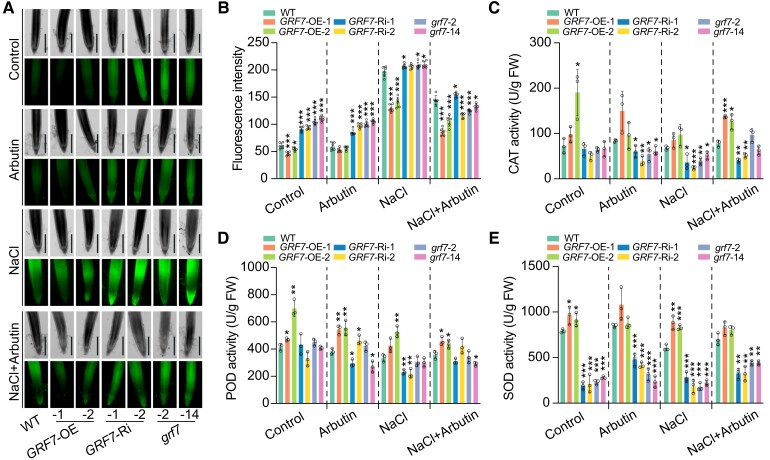
Arbutin is speculated to confer plant stress tolerance through the antioxidative properties of the hydroquinone moiety or by stabilizing the membranes depending on the lipid composition (Oliver et al. 2001; Kuzniak et al. 2015; Skłodowska et al. 2015). Therefore, arbutin might suppress the accumulation of ROS in rice. We thus examined the ROS content in OsGRF7 transgenic plants under salt stress using the ROS-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA). The results showed that the H2DCF-DA fluorescence signal was enriched in the roots of the GRF7-Ri or grf7 transgenic plants under salinity stress. Exogenous application of arbutin reduced the fluorescent signal in both the GRF7-Ri and grf7 transgenic plants (Fig. 3, A and B), which was consistent with the change in MDA content (Supplementary Fig. S7A). Consistent with these findings, the activities of the ROS-scavenging enzymes, catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD), were significantly reduced in both the GRF7-Ri and grf7 transgenic plants (Fig. 3, C to E). Hence, OsGRF7 appeared to promote the scavenging of ROS caused by salt stress by modulating arbutin metabolism.
Figure 3.
ROS accumulated in the root tips of the GRF7-Ri and grf7 lines under salinity stress. A) H2O2 detected by H2DCF-DA staining in the roots of WT and OsGRF7 transgenic lines. Bars = 500 µm. B) Quantitative analysis of H2O2 in the roots of WT and OsGRF7 transgenic lines. Values are means ± SDs (n = 6 independent root tips). C) to E) Enzyme activities of catalase (CAT; C), peroxidase (POD; D), and superoxide dismutase (SOD; E) in the WT and OsGRF7 transgenic lines. Values are means ± SDs of 3 biological replicates. WT, wild type. GRF7-OE, OsGRF7 overexpression lines. GRF7-Ri, OsGRF7 RNAi lines. grf7, OsGRF7 knockout lines. FW, fresh weight. For this figure, asterisks indicate significant differences between the wild type and the other genotypes by a two-tailed Student's t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
OsGRF7 directly modulates the biosynthesis of arbutin
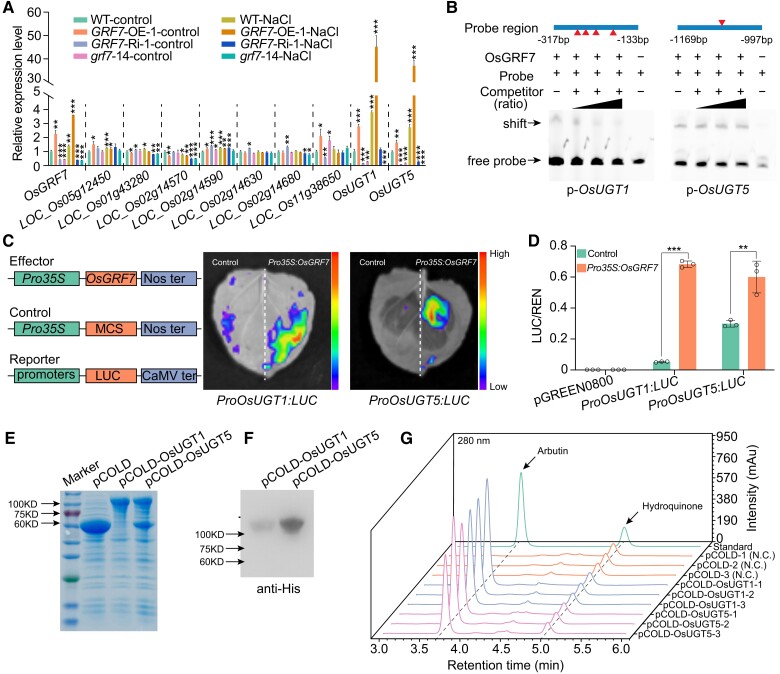
To investigate the relationship between OsGRF7 and arbutin, we dissected the biosynthesis pathway of arbutin via the Kyoto Encyclopedia of Genes and Genomes database (https://www.kegg.jp). UGTs catalyze the transfer of activated sugars linked with uridine diphosphate (UDP) to hydroquinone, directly regulating the biosynthesis of arbutin (Supplementary Fig. S8A). Bioinformatics analysis revealed that 9 genes encoding UGTs, among which the amino acid sequence of LOC_Os01g53330 was highly similar to that of LOC_Os05g08480 (Supplementary Fig. S8B). Expression analysis showed that the expression levels of only 3 genes, LOC_Os11g38650, LOC_Os01g53330, and LOC_Os05g08480, were significantly induced by salt treatment (Supplementary Fig. S8, C to K). Further expression analysis showed that only LOC_Os01g53330 (OsUGT1) and LOC_Os05g08480 (OsUGT5) were highly expressed in the GRF7-OE transgenic lines, suggesting that they might be directly modulated by OsGRF7. Moreover, the expression levels of OsUGT1 and OsUGT5 were upregulated in the GRF7-OE-1 transgenic plants and downregulated in the GRF7-Ri-1 and grf7-14 lines both with and without salinity stress (Fig. 4A). Time-series experiments further confirmed that both OsUGT1 and OsUGT5 were induced by salt treatment (Supplementary Fig. S9, A and B). Thus, we hypothesized that OsUGT1 and OsUGT5 participate in the biosynthesis of arbutin under salinity stress conditions.
Figure 4.
OsGRF7 directly regulates the biosynthesis of arbutin. A) Relative expression levels of arbutin biosynthesis-related genes in the WT and OsGRF7 transgenic lines with or without NaCl treatment. B) EMSA of the binding between OsGRF7 and the promoters of OsUGT1 and OsUGT5. Two-fold, 10-fold, and 100-fold unmodified probes were used as competitors. The presence (+) or absence (−) of components in the reaction mixture is indicated. Red triangles indicate the sites of ACRGDA motifs in the promoter region. C) and D) Transient expression assay in the infiltrated leaves of N. benthamianaC) and rice protoplast cells D). The color of the scale indicates increasing intensity from bottom to top. E) CBB staining of the expressed OsUGT1 and OsUGT5 proteins in E. coli. CBB, Coomassie Brilliant Blue. F) Immunoblot analysis of the OsUGT1 and OsUGT5 proteins using anti-His antibody. G) The in vitro glycosyltransferase activity of OsUGT1 and OsUGT5 determined by HPLC analyses. N.C., negative control. Values are means ± SDs of 3 biological replicates. WT, wild type. GRF7-OE, OsGRF7 overexpression lines. GRF7-Ri, OsGRF7 RNAi lines. grf7, OsGRF7 knockout lines. LUC, Luciferase. REN, Renilla luciferase. For this figure, asterisks indicate significant differences by a two-tailed Student's t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Subcellular localization analysis showed that OsUGT1 and OsUGT5 were localized in the nucleus in rice protoplasts (Supplementary Fig. S10). UDP-glucosyltransferases have been identified as cytoplasmic and nuclear regulators of metabolic homeostasis in mammals and may play roles in controlling the homeostasis of the nuclear receptor ligands and protecting the nuclear components from toxins (Hart et al. 1989; Husar et al. 2011). In plants, UGTs have been found in the nucleus, including linalyl-β-D-glucosyltransferase PpUGT85A2 (Wu et al. 2019), zeatin O-glucosyltransferase UGT85A1 (Jin et al. 2013), and UGT87A2 (Wang et al. 2012), suggesting that OsUGT1 and OsUGT5 might have other cellular functions in the nucleus.
Previous ChIP-seq results have shown that OsGRF7 directly binds to ACRGDA motifs to regulate downstream genes (Chen et al. 2020a). We analyzed the promoter regions of OsUGT1 and OsUGT5 and found several ACRGDA motifs. An electrophoretic mobility shift assay suggested the binding of OsGRF7 to ACRGDA-containing promoter fragments from OsUGT1 and OsUGT5 (Fig. 4B). Furthermore, transactivation assays in both Nicotiana benthamiana leaves and rice protoplasts showed that OsGRF7 activated the transcription of OsUGT1 and OsUGT5 via their promoters (Fig. 4, C and D).
To investigate whether OsUGT1 and OsUGT5 catalyze the biosynthesis of arbutin, we heterologously expressed OsUGT1 and OsUGT5 in Escherichia coli to explore the ability of the recombinant proteins to glycosylate hydroquinone (Fig. 4, E and F). High-performance liquid chromatography-mass spectrometry analysis of the reaction products showed that hydroquinone was glycosylated by both OsUGT1 and OsUGT5 in vitro (Fig. 4G; Supplementary Fig. S11; Supplementary Data Set 3). These results demonstrated that OsGRF7 directly regulated the expression of OsUGT1 and OsUGT5 to modulate arbutin biosynthesis in rice.
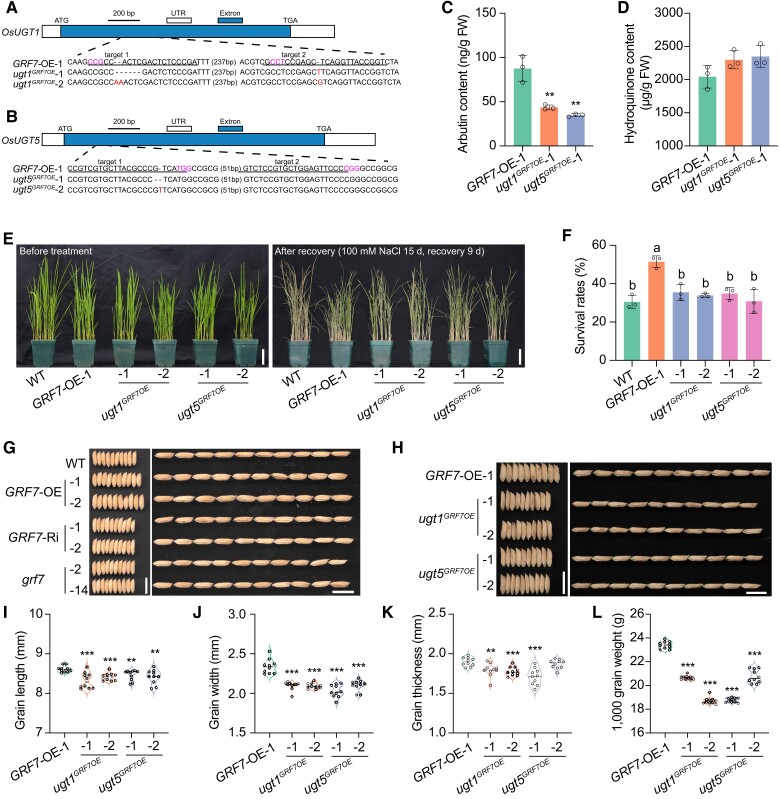
OsUGT1 and OsUGT5 positively regulate the salt tolerance and grain size of rice
To further understand the roles of OsUGT1 and OsUGT5 in the rice salt stress response, we generated OsUGT1 and OsUGT5 knockout lines in the GRF7-OE background (ugt1GRF7OE and ugt5GRF7OE). After sequence identification, we obtained two homozygous ugt1GRF7OE and ugt5GRF7OE lines without the Cas9 protein, in which OsUGT1 and OsUGT5 were prematurely terminated for each genotype (Fig. 5, A and B). After the content of arbutin was measured, we found that the levels of arbutin were reduced in both the seedlings and roots of the ugt1GRF7OE and ugt5GRF7OE mutants (Fig. 5C; Supplementary Fig. S12A). And the content of hydroquinone was higher in ugt1GRF7OE and ugt5GRF7OE mutants than that in the GRF7-OE transgenic plants (Fig. 5D; Supplementary Fig. S12B), confirming that OsGRF7 modulates arbutin metabolism by directly regulating the expression of OsUGT1 and OsUGT5.
Figure 5.
OsUGT1 and OsUGT5 positively participate in the rice salinity stress response and grain size development. A) and B) CRISPR/Cas9-mediated target mutagenesis of OsUGT1A) and OsUGT5B) in the GRF7-OE background. The exons and UTRs (untranslated region) are represented by blue and white boxes, respectively. Each alignment between GRF7-OE-1 and mutated sequences containing the target sites is shown below the schematic. The red font is the mutant sequence and the purple font is the NGG site. Bar = 200 bp. C) and D) The in vivo contents of arbutin C) and hydroquinone D) in GRF7-OE-1, ugt1GRF7OE, and ugt5GRF7OE seedlings. Values are means ± SDs of 3 biological replicates. E) Phenotypes of WT, GRF7-OE-1, ugt1GRF7OE, and ugt5GRF7OE plants before salt treatment and after recovery. Bars = 5 cm. F) Survival rates of WT, GRF7-OE-1, ugt1GRF7OE, and ugt5GRF7OE plants after recovery. Values are means ± SDs with 3 biological replicates. Different letters represent significant differences at P < 0.05 determined by Tukey's multiple comparisons test. G) Grain morphology of WT and OsGRF7 transgenic lines. Bars = 1 cm. H) Grain morphology of GRF7-OE-1, ugt1GRF7OE, and ugt5GRF7OE plants. Bars = 1 cm. I) to L) Statistical analysis of the grain length I), grain width J), grain thickness K), and 1,000-grain weight L). Values are means ± SDs (n = 10 independent grains). WT, wild type. GRF7-OE, OsGRF7 overexpression lines. ugt1GRF7OE, OsUGT1 knockout lines. ugt5GRF7OE, OsUGT5 knockout lines. FW, fresh weight. For this figure, asterisks indicate significant differences by a two-tailed Student's t-test (**P < 0.01 and ***P < 0.001).
We then treated 15-d-old GRF7-OE-1, ugt1GRF7OE and ugt5GRF7OE seedlings with 100 mM NaCl. The survival rate of GRF7-OE-1 was significantly higher than that of the wild-type plants, while that of ugt1GRF7OE and ugt5GRF7OE was reduced compared with that of the GRF7-OE-1 transgenic plants (Fig. 5, E and F; Supplementary Fig. S13). These results further confirmed that OsUGT1 and OsUGT5 were the direct targets of OsGRF7, which positively regulates arbutin metabolism and salt tolerance of rice.
Notably, the ugt1GRF7OE and ugt5GRF7OE mutants showed decreased grain size, similar to that of the GRF7-Ri and grf7 transgenic lines (Fig. 5, G and H). The grain width significantly increased by 10.7% and 9.8% in the GRF7-OE-1 and GRF7-OE-2 lines, respectively. The GRF7-Ri and grf7 transgenic lines exhibited significantly decreased grain width (Supplementary Fig. S14B). Moreover, the grain thickness of the GRF7-OE-1 and GRF7-OE-2 transgenic plants significantly increased by 4.6% and 3.9%, respectively (Supplementary Fig. S14C). However, the grain length was not significantly different among these transgenic lines (Supplementary Fig. S14A). As a result, compared with those of the wild type, the 1,000-grain weight of the GRF7-OE plants significantly increased, and those of the GRF7-Ri and grf7 lines significantly decreased (Supplementary Fig. S14D), indicating that OsGRF7 might positively regulate grain size through OsUGT1 and OsUGT5.
As demonstrated by scanning electron microscopy, histological section analysis of the grain lemma showed that the GRF7-OE-1 transgenic plants produced larger cells in the husk than did the wild-type and GRF7-Ri-1 plants (Supplementary Fig. S14E). However, the cell length had no significant change (Supplementary Fig. S14F). In contrast to the normal cell arrangements of the wild-type and GRF7-Ri-1 plants, the cell arrangement of the GRF7-OE-1 transgenic plants was disordered (Supplementary Fig. S14, E and F). These results demonstrated that OsGRF7 regulated grain size by changing cell size and arrangement.
Furthermore, we found that grain length, width, and thickness were lower in the ugt1GRF7OE and ugt5GRF7OE mutants than in the GRF7-OE-1 plants (Fig. 5, H to K). As a result, the 1,000-grain weight was significantly reduced in the ugt1GRF7OE and ugt5GRF7OE mutants compared with the GRF7-OE-1 transgenic plants (Fig. 5L), suggesting that OsGRF7 might regulate the biosynthesis of arbutin to participate in grain development.
OsGRF7 is ubiquitinated and degraded by OsFBO13
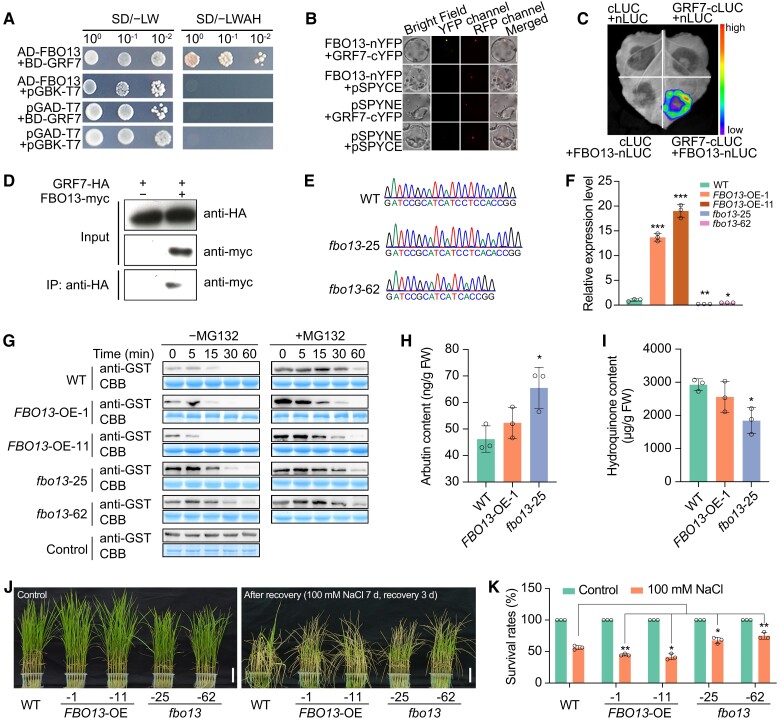
We subsequently performed a yeast two-hybrid screening assay to identify proteins that interact with OsGRF7. Sequence analysis of several clones revealed that the 23 putative interactors of OsGRF7 included an F-box-containing protein (here named rice F-BOX O 13, OsFBO13; Supplementary Table S2), which is particularly important due to its potential role in regulating protein stabilization, suggesting that this protein might be involved in the degradation of OsGRF7. Further yeast two-hybrid assays (Fig. 6A), transient bimolecular fluorescence complementation (Fig. 6B), firefly luciferase complementation imaging (Fig. 6C), and coimmunoprecipitation experiments in rice protoplast cells (Fig. 6D) confirmed that OsGRF7 interacted with OsFBO13 both in vitro and in vivo.
Figure 6.
OsFBO13 interacts with and degrades OsGRF7 to negatively regulate the salinity stress response. A) to D) The interaction between OsGRF7 and OsFBO13 was confirmed by a yeast two-hybrid assay A), bimolecular fluorescence complementation assay B), split luciferase complementation imaging assay C), and coimmunoprecipitation assay D). The color of the scale indicates increasing intensity from bottom to top. SD/–LW, Do Supplement/–Leu–Trp; SD/–LWAH, Do Supplement/–Leu–Trp–Ade–His. YFP, yellow fluorescent protein. LUC, luciferase. IP, immunoprecipitation. Bars = 200 µm. E) Sanger sequencing chromatography showed the mutation of OsFBO13 in the fbo13-25 and fbo13-62 mutants. F) Relative expression level of OsFBO13 in the seedlings of the WT and OsFBO13 transgenic lines. OsUBI was used as an internal control. Values are means ± SD of 3 biological replicates. G) Cell-free degradation assay of OsGRF7 in the WT, FBO13-OE-1, FBO13-OE-11, fbo13-25, and fbo13-62 cells. CBB, Coomassie Brilliant Blue. H) and I) The in vivo contents of arbutin H) and hydroquinone I) in OsFBO13 transgenic seedlings. J) and K) Phenotypes and survival rates of the WT and OsFBO13 transgenic lines under 100 mM NaCl treatment. Bars = 5 cm. Values are means ± SD of 3 biological replicates. WT, wild type. FBO13-OE, OsFBO13 overexpression lines. fbo13, OsFBO13 knockout lines. FW, fresh weight. For this figure, asterisks indicate significant differences by a two-tailed Student's t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
OsFBO13 is an F-box domain-containing protein, and OSKs (SKP1 homologs) act as bridges between Cullin and F-box-containing proteins to form the functional SCF complex (Kahloul et al. 2013). A yeast two-hybrid assay showed that OsFBO13 interacted with OSK4, OSK6, and OSK16 to form heterodimers (Supplementary Fig. S15A), indicating that they could form a functional SCF complex and that OsFBO13 was an integral part of the SCFOsFBO13 E3 ligase in rice.
Protein structure analysis with the Simple Modular Architecture Research Tool (http://smart.embl.de) revealed that OsFBO13 has a UBQ domain and an F-box domain and that both domains are necessary for the interaction between OsFBO13 and OSKs (Supplementary Fig. S15B). To establish whether OsGRF7 is a substrate of OsFBO13, a cell-free protein degradation system was used. The results showed that OsFBO13 promoted the degradation of OsGRF7, and this degradation was repressed by the addition of MG132 (a specific inhibitor of the 26S proteasome) (Supplementary Fig. S15C), indicating that OsFBO13 promoted the degradation of OsGRF7 via the 26S proteasome system.
OsFBO13 negatively regulates the rice salinity stress response and grain size
OsFBO13 interacted with and degraded OsGRF7, suggesting that OsFBO13 might function as a negative regulator of salt tolerance. Thus, OsFBO13 was overexpressed by the ubiquitin promoter and knocked out using the CRISPR/Cas9 system. Seventeen OsFBO13 overexpression (FBO13-OE) lines and 6 homozygous OsFBO13 knockout (fbo13) lines were obtained, respectively (Fig. 6, E and F). Immunoblot analysis using an anti-GST antibody showed that GST-OsGRF7 degraded when it was incubated with total protein extracts from the wild type. However, after incubating with the protein extracts from the FBO13-OE-1 and FBO13-OE-11 plants, the degradation of GST-OsGRF7 was faster than that of the wild type. In contrast to those of the wild-type and FBO13-OE plants, incubation with protein extracts from fbo13-25 and fbo13-62 plants retarded the degradation of GST-OsGRF7 (Fig. 6G). We thus concluded that OsFBO13 promoted the degradation of OsGRF7.
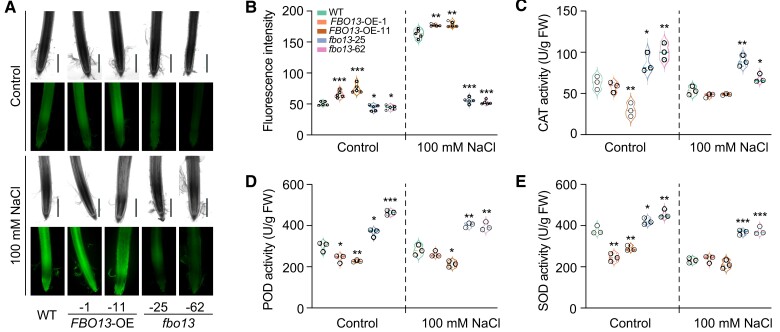
To investigate the function of OsFBO13 in arbutin biosynthesis, we measured the endogenous content of arbutin and hydroquinone. The fbo13-25 plants had a slightly higher level of arbutin than the wild-type, and the content of hydroquinone was lower in fbo13-25 transgenic plants than that in the wild-type (Fig. 6, H and I; Supplementary Fig. S16), suggesting that OsFBO13 may regulate the stability of OsGRF7 to modulate the biosynthesis of arbutin. Subsequently, we treated the OsFBO13 transgenic plants with 100 mM NaCl. After 3 d of recovery, the survival rates of the FBO13-OE-1 and FBO13-OE-11 transgenic lines were significantly lower than those of the wild type, while those of the fbo13-25 and fbo13-62 plants were significantly higher (Fig. 6, J and K). The MDA content was lower in the fbo13 mutants than in the wild type under normal conditions or NaCl treatment (Supplementary Fig. S17A). The fbo13 plants also accumulated more proline than the wild-type plants did (Supplementary Fig. S17B). These results were consistent with the lower ROS content and significantly increased CAT, POD, and SOD activities in the fbo13 plants compared with those in the wild type under salt stress (Fig. 7, A to E), confirming the negative role of OsFBO13 in the salt stress response in rice.
Figure 7.
OsFBO13 negatively regulates rice salinity stress response. A) Concentration of H2O2 detected by H2DCF-DA staining in roots of the WT and OsFBO13 transgenic lines with or without 100 mM NaCl treatment. Bars = 500 µm. B) Quantitative analysis of H2O2 in the roots of WT and OsFBO13 transgenic lines with or without 100 mM NaCl treatment. Values are means ± SDs (n = 5 independent root tips). C) to E) Enzyme activities of catalase (CAT; C), peroxidase (POD; D), and superoxide dismutase (SOD; E) in the WT and OsFBO13 transgenic lines with or without 100 mM NaCl treatment. WT, wild type. FBO13-OE, OsFBO13 overexpression lines. fbo13, OsFBO13 knockout lines. Values are means ± SDs of 3 biological replicates. For this figure, asterisks indicate significant differences between the wild-type and the other genotypes by a two-tailed Student's t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Phenotypic analysis of the grain size showed that, compared with those of the wild-type plants, the grain length, width, and thickness of the OsFBO13 overexpression lines were decreased, and the 1,000-grain weights of the FBO13-OE-1 and FBO13-OE-11 transgenic lines were decreased by 18.3% and 11.1%, respectively (Supplementary Fig. S18, A to D). Moreover, the 1,000-grain weight of the fbo13 mutants was significantly increased (Supplementary Fig. S18E). These results demonstrated that OsFBO13 interacted with and degraded OsGRF7 to participate in the salt stress response and grain size determination.
Given that OsFBO13 directly interacts with OsGRF7 and degrades it both in vitro and in vivo and that OsGRF7 regulates the expression of OsUGT1 and OsUGT5, the transcript status of OsUGT1 and OsUGT5 might change in OsFBO13 transgenic plants. Expression analysis showed that the OsUGT1 and OsUGT5 transcript levels were decreased in the FBO13-OE plants (Supplementary Fig. S19), suggesting that the role of OsGRF7 in regulating the salt stress response and grain size might be mediated by OsUGT1 and OsUGT5 in rice.
Lysine residue K365 is the primary ubiquitination site in OsGRF7
To identify the ubiquitination sites in OsGRF7, we analyzed the amino acid sequence of OsGRF7 and found 15 lysine residues (Supplementary Fig. S20A). According to the Predictor of Protein Ubiquitination Sites (http://www.ubpred.org), K315, K363, and K365 were the most likely ubiquitination sites in OsGRF7 (Supplementary Table S3). To determine whether OsFBO13 promoted ubiquitination at these sites, we performed a cell-free degradation assay of wild-type and mutated versions of OsGRF7. Compared to that of the control, the wild-type version of OsGRF7 was ubiquitinated by OsFBO13 in both the wild-type and FBO13-OE-1 plants. However, when the lysine residue K365 was mutated to arginine (K365R), the degree of degradation of OsGRF7K365R was lower than that of OsGRF7, OsGRF7K208RK315R, or OsGRF7K363R in both the wild-type and FBO13-OE-1 transgenic seedlings (Supplementary Fig. S20B), highlighting the crucial role of K365 in the degradation of OsGRF7.
We further performed a dual-luciferase activity assay to analyze whether the expression levels of OsUGTs were determined by the ubiquitination of OsGRF7. The results showed that OsGRF7 promoted the LUC activity of ProOsUGT5:LUC, and OsFBO13 repressed this effect. However, when OsGRF7K365R and OsFBO13 were cotransformed with ProOsUGT5:LUC, the repression was released (Supplementary Fig. S20, C and D). Taken together, these results indicated that the K365 residue was the primary ubiquitination site in OsGRF7.
Discussion
GRFs are conserved plant transcription factors that play important roles in plant growth and development. OsGRF1 was suggested to be the first identified GRF member identified in rice and to play a crucial role in leaf growth and early internode elongation (Luo et al. 2005). OsGRF3 and OsGRF10 play roles in rice tillering determination and leaf blade development (Kuijt et al. 2014). Moreover, OsGRF4, OsGRF6, and OsGRF10 affect inflorescence development, grain size, and grain yield through different signaling pathways (Liu et al. 2014; Che et al. 2015; Gao et al. 2015). Moreover, overexpression of OsGRF6, OsGRF7, OsGRF8, or OsGRF9 results in enhanced resistance to rice blast fungal infection (Chandran et al. 2019), and OsGRF8 promotes brown planthopper resistance by directly regulating the biosynthesis of flavonoids (Dai et al. 2019). These studies indicate that OsGRFs play pivotal roles in rice development and biotic stress responses. In Arabidopsis (Arabidopsis thaliana), GRFs are also involved in the biotic stress response (Hewezi et al. 2012), but little is known about how GRFs modulate the abiotic stress response. There are many abscisic acid-responsive element motifs in the promoters of GRF members in crops and Arabidopsis (Qin et al. 2022), suggesting that these genes may be involved in abiotic stress responses under adverse conditions.
A previous study has demonstrated that OsmiR396 directly cleaves the mRNA of OsGRF7 at the site within the OsmiR396 pairing region (Chen et al. 2020a). When OsmiR396s, nine members with different expression profiles and varying functions in rice, were expressed in the Pro35S:GRF7-GFP transgenic rice protoplasts, the content of the OsGRF7 protein was dramatically decreased in the OsmiR396e background. In addition, OsmiR396e exhibited the opposite expression pattern as OsGRF7 in different tissues, indicating that OsmiR396e is mainly responsible for the cleavage of OsGRF7 (Chen et al. 2020a).
Furthermore, OsGRF7 directly regulates the biosynthesis and signaling of auxin and gibberellic acid to modulate plant architecture. Phytohormones, including auxin, gibberellic acid, cytokinin, brassinosteroid, abscisic acid, jasmonic acid, and ethylene, play important roles in plant growth and development as well as biotic and abiotic stress responses (Waadt et al. 2022). A high concentration of NaCl causes overaccumulation of abscisic acid (ABA) in maize, which alters the polar localization of ZmPIN1 and disrupts the distribution of auxin (Lu et al. 2019). ABA also inhibits the function of gibberellic acid (GA) by regulating its biosynthesis. GA counteracts ABA signaling by enhancing ABA receptor degradation (Vanstraelen and Benkova 2012; Kawa 2020). Here, we found that OsGRF7 participates in the salinity stress response by directly regulating arbutin biosynthesis. Although the relationship between plant hormones and arbutin is unclear yet, we think that these metabolites may function together under different environmental conditions.
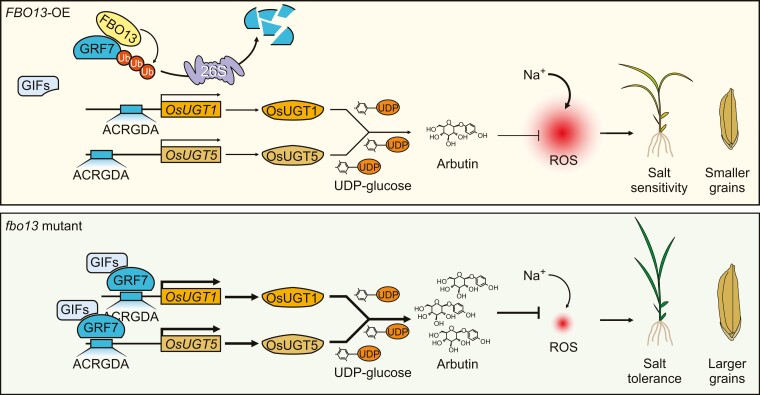
GRF-INTERACTING FACTORs (OsGIFs) have been proven to act as coactivators of OsGRF7 to modulate plant architecture (Chen et al. 2020a), but few GRF interactors have been identified in plants. Here, in addition to OsGIFs, OsFBO13 also interacted with OsGRF7 (Fig. 6, A to D; Supplementary Fig. S21, B to D). After direct interaction between OsFBO13 and OsGIFs was excluded (Supplementary Fig. S21A), competition analysis revealed that OsGIF1 interacted with OsGRF7 to promote the transactivation of ProOsUGT1:LUC, but this effect was diminished by the addition of OsFBO13 (Supplementary Fig. S21, E to G). These findings indicated that OsFBO13 competes with OsGIFs to interact with OsGRF7 and regulate its degradation via the 26S proteasome system, correspondingly slowing the expression of arbutin biosynthesis-related genes (OsUGT1 and OsUGT5) and reducing the content of arbutin. Hence, the downregulation of OsGRF7 promoted ROS accumulation and inhibited plant growth and development under salt stress (Fig. 8), highlighting the important role of OsGRF7 in resistance to abiotic stress.
Figure 8.
A proposed model for OsGRF7 coordinating salinity tolerance and grain size in rice. OsFBO13 interacts with and promotes the degradation of OsGRF7 via the 26S proteasome system. In fbo13 mutant, OsGRF7 bound directly to and activated the expression of OsUGT1/5, and the arbutin content was correspondingly increased. Under salt stress conditions, arbutin enhanced the activities of ROS-scavenging enzymes to eliminate ROS and ultimately induce a salt-tolerance phenotype. At the same time, arbutin also increased the grain size under normal conditions.
With the improvements in living standards, people have become more concerned about rice nutrients for health reasons. Thus, functional rice has been widely studied for its ability to improve the levels of specific nutrients and trace elements. Golden Rice was engineered for high β-carotene content to combat vitamin A deficiency (Ye et al. 2000; Paine et al. 2005), and resistant starch-containing rice can be used to effectively control diabetes by lowering blood glucose levels after meals in diabetic patients (Schulz et al. 1993; Raigond et al. 2015). Gamma-aminobutyric acid-enriched rice is also known to have various pharmaceutical properties and beneficial effects on health (Ngo et al. 2022). Arbutin is a natural product present in diverse plants. Due to its antimicrobial, antioxidant, and anti-inflammatory activities, arbutin has been used as a folk medicine for thousands of years, especially for wound healing and the treatment of urinary tract infections (Lim et al. 2009).
In this study, we found that OsGRF7 directly targets arbutin biosynthesis-related genes (OsUGT1 and OsUGT5) to activate their expression (Fig. 4) and increase the arbutin content (Fig. 2A). Both the grain size and arbutin content of the ugt1GRF7OE and ugt5GRF7OE mutants were decreased compared with those of the GRF7-OE transgenic plants, suggesting that arbutin may play a role in rice grain development. Although the detailed relationships between arbutin and grain size require further investigation, our results, including the finding that arbutin enhances salinity stress tolerance without affecting plant growth (Fig. 2, C and D), suggest that the exogenous application of arbutin in the field may facilitate the breeding of salt stress-tolerant rice. However, the stress conditions in the field are more complicated than that in the laboratory, so the persistent and mixed effects from multiple stress factors should be considered in field applications. Finally, the content of arbutin can be increased by engineering OsGRF7, and GRF is a conserved gene family in both monocotyledons and dicotyledons (Qin et al. 2022). The conserved GRF-arbutin module may provide an applicable strategy for functional crop breeding or even in medicinal plant research.
Materials and methods
Plant materials and growth conditions
The rice variety (O. sativa ssp. indica) Yuetai B (YB) was used as the wild-type background in this study. GRF7-OE-1, GRF7-OE-2, GRF7-Ri-1, GRF7-Ri-2, grf7-2, and grf7-14 transgenic lines were obtained from a previous study (Chen et al. 2020a). To generate the ugt1GRF7OE and ugt5GRF7OE mutants, two single guide RNAs (sgRNAs) were designed in the exons of OsUGT1 and OsUGT5. Then, the CRISPR/Cas9 knockout plasmids for ugt1GRF7OE and ugt5GRF7OE were transformed into calli of the GRF7-OE-1 transgenic lines using G418 as the selection marker. The mutants were identified by Sanger sequencing of the target sites, and the primers used are listed in Supplementary Data Set 4.
For the OsFBO13 overexpression plants, the full-length cDNA of OsFBO13 was amplified and inserted into pCAMBIA1301-Ubi via the BamHI site. The T4 generation of FBO13-OE lines was used for the experiments. The CRISPR/Cas9 knockout plasmid for OsFBO13 was constructed as previously reported (Ma et al. 2015). We generated two sgRNA constructs (designed in exons), in which the sgRNA was driven by the rice U6 promoter and the plant-optimized Cas9 was driven by the Ubi promoter. The integrated sgRNA expression cassettes were amplified and inserted into the CRISPR/Cas9 vector pYLCRISPR/Cas9Pubi-H (Ma et al. 2015). The construct was subsequently introduced into the wild-type accession YB. Seventy independent transgenic lines were subsequently subjected to PCR amplification and sequencing analysis. The T4 generation of fbo13 lines was used for the experiments. The primers used in this study are listed in Supplementary Data Set 4. All the rice plants used in this study were grown in Wuhan, Hubei, China. For stress treatment, all the rice plants were grown in a 28°C plant growth chamber. Nicotiana benthamiana plants were grown in a 22°C plant growth chamber. The light is provided by eight 16 W LED bulbs.
Salinity stress treatment
To measure the seedling length, sterilized seeds of the wild-type and OsGRF7 transgenic lines were grown on half Murashige and Skoog agar medium supplemented with or without NaCl for 5 d. The seedling length of each genotype was measured using 10 independent seedlings of each genotype. To measure the survival rates, the germinated seeds were sown and cultured in a bottomless 96-well plate with Yoshida culture solution. Fifteen-day-old seedlings were subjected to 100 mM NaCl, 20 µM arbutin, or 100 mM NaCl plus 20 µM arbutin treatment for 7 d. Subsequently, the rice seedlings were transferred to NaCl-free Yoshida culture solution and allowed to recover for 3, 7, 10, or 14 d. To analyze the sensitivity of ugt1GRF7OE and ugt5GRF7OE plants to salt stress in vermiculite, 15-d-old seedlings were treated with 100 mM NaCl for 15 d and allowed to recover for another 9 d. Photographs were taken before treatment and after recovery, and survival rates were recorded to determine plant sensitivity to salinity stress. Seedlings were regarded as survivors if healthy and still green. Three biological replicates were performed.
RNA-seq analysis
Seedlings were cultured in a bottomless 96-well plate with Yoshida culture solution. Fifteen-day-old wild-type and grf7-14 plants were cultured in Yoshida culture solution supplemented with 100 mM NaCl for 24 h. Subsequently, the seedlings were harvested and frozen in liquid nitrogen. Ten seedlings were sampled and pooled as one biological replicate, and 3 biological replicates were used for the wild-type and grf7-14 lines. After total RNA extraction, libraries were constructed using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. The DNA libraries were subsequently sequenced on the Illumina sequencing platform by Novogene Co., Ltd (Beijing, China), and the raw reads were filtered and mapped to the reference genome by Hisat2 v2.0.5. The significantly DEGs between the grf7-14 and wild-type lines were analyzed using ExpressAnalyst (P < 0.001, fold change >1) (https://www.expressanalyst.ca/ExpressAnalyst/home.xhtml). Then, the Gene Ontology analysis of the downregulated DEGs in the grf7 seedlings was performed using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost).
Untargeted metabolite profiling and analysis of metabolic data
Fifteen-day-old GRF7-OE, GRF7-Ri, and wild-type seedlings were bulk-harvested and frozen in liquid nitrogen. Ten seedlings were sampled and pooled as one biological replicate. Six replicates were used for the GRF7-OE and GRF7-Ri transgenic lines, and 3 replicates were used for the wild type. Untargeted metabolite profiling analysis was performed with a previously described method (Dan et al. 2020). Briefly, the seedling tissues of each biological replicate were first ground into fine powders in liquid nitrogen. After 80 mg of powder was transferred into 2 mL tubes, 1 mL of precooled extraction reagent (methanol/acetonitrile/water, 2/2/1, v/v/v) was added, and the mixture was vortexed for 60 s. A quality control sample was generated by pooling 10 μL of all the experimental samples. The samples were then subjected to 2 (30 min each time) ultrasonic treatments and placed in a freezer at −20°C for 60 min. Subsequently, the samples were centrifuged at 14,000 × g for 15 min, after which the supernatants were transferred and dried in a vacuum concentrator. After 100 μL of acetonitrile (acetonitrile/water, 1/1, v/v) was added to the sample tubes, the samples were vortexed and centrifuged at 14,000 × g for 15 min at 4°C.
Next, the supernatants were separated using an Agilent 1290 Infinity LC system equipped with a Waters reversed-phase T3 column (ACQUITY UPLC HSS T3; particle size, 1.8 μm; 2.1 mm × 100 mm) at 25°C. The injection volume of each sample was 2 μL and the flow rate was 0.3 mL/min. Mobile Phases A and B were 0.5 mM ammonium fluoride (Sigma Aldrich, United States) and acetonitrile (Merck), respectively. The gradient was set as follows: 0 to 1 min: 1% B; 1 to 8 min: 1% B to 100% B; 8 to 10 min: 100% B; 10 to 10.1 min: 100% B to 1% B; and 10.1 to 12 min: 1% B. The experimental samples were injected in random order and were combined with the same batch of seedling samples in a previous study (Dan et al. 2020). The quality control sample was injected into every 9 experimental samples and its distribution in a 3-dimensional principal component analysis score plot confirmed the stability of the LC-MS system (Dan et al. 2020). MS data were subsequently collected by an Agilent 6550 iFunnel quadruple time-of-flight mass spectrometer (Agilent Technologies) in negative electron spray ionization mode. The temperatures of the drying gas and sheath gas were 250°C and 400°C, respectively. The flow rates were 16 and 12 L/min, respectively. The capillary, nozzle, and fragment voltages were 3,000, 0, and 175 V, respectively. The parameters of the TOF mass scan were as follows: mass range, 60 to 1,200 Da; acquisition rate, 4 spectra/s; and cycle time, 250 ms.
Another mass spectrometer (TripleTOF 6600, AB SCIEX) was used to collect tandem MS data for the quality control sample. The source parameters were as follows: ion source gas 1 (GS1), 40 psi; ion source gas 2 (GS2), 80 psi; curtain gas (CUR), 30 psi; source temperature, 650°C; ion spray voltage floating, −4,500 V in negative mode; and declustering potential, −60 V. The mass range in the TOF mass scan was 60 to 1,200 Da and the MS accumulation time was 200 ms/spectra. To increase the data acquisition rate, the quality control sample was injected twice with 4 sequential windows of mass range: 60 to 200, 190 to 400, 390 to 600, 590 to 1,200, 60 to 1,200, and 200 to 600, 590 to 750, 740 to 900, 890 to 1,200, and 200 to 1,200 Da. The data were acquired in high-sensitivity mode in an information-dependent manner. The parameters for product ion scanning were as follows: collision energy, 35 ± 15 eV; accumulation time, 50 ms; isotope isolation width, 4.0 Da; and number of candidate ions to monitor per cycle, 10.
The raw MS files were subsequently converted to mzXML format with ProteoWizard (Chambers et al. 2012) and processed with XCMS (Gowda et al. 2014), including feature detection, retention time correction, and alignment of features. The generated peak table, which included the mass-to-charge ratio and peak abundances, contained 3,746 peaks and was used for metabolite annotation. The MS/MS data of the quality control sample were matched against an in-house standard MS2 spectral library (Shen et al. 2019) and a lipid MS2 library (Tu et al. 2017). A total of 114 metabolites were annotated with a mass tolerance of 25 ppm. To align the MS peaks and MS/MS spectra, the errors in the mass-to-charge ratio and retention time were set to 15 ppm and 20 s, respectively. The intensity of the annotated metabolites was normalized with the MetaboAnalyst web-based system (Supplementary Fig. S3) (Xia et al. 2015). The parameters for sample normalization, data transformation, and data scaling were quantile normalization, cubic root transformation, and range scaling, respectively. One-way ANOVA (P < 0.05) with a post hoc test (Fisher's LSD) was performed to identify differentially abundant metabolites among the GRF7-OE, GRF7-Ri, and the wild-type plants with the “Statistical Analysis” module of MetaboAnalyst (www.metaboanalyst.ca) (Xia et al. 2015).
Targeted metabolite profiling
Fifteen-day-old seedlings and roots of the wild-type, GRF7-OE-1, grf7-14, ugt1GRF7OE-1, ugt5GRF7OE-1, FBO13-OE-1, and fbo13-25 transgenic lines were bulk harvested and frozen in liquid nitrogen. Five plants were sampled and pooled as 1 biological replicate, and 3 biological replicates were used for each genotype. Briefly, the tissues were first ground into fine powers using liquid nitrogen. After approximately 100 mg of power was transferred into 2 mL tubes, 1 mL of methanol/acetonitrile/water (2:2:1, v/v/v) was added, and the mixture was vortexed uniformly. The samples were then ultrasonically extracted for 30 min and centrifuged at 13,523 × g for 5 min. The supernatants were moved to new EP tubes and concentrated to dryness in a vacuum concentrator. After reconstitution with 200 µL of 50% (v/v) methanol solution, the supernatants were analyzed via LC-MS/MS.
Targeted analyses of arbutin were conducted using a Nexera X2 LC-30AD apparatus equipped with an ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 µm) at 40°C. The gradient elution method was applied, and the Mobile Phases A and B were water and acetonitrile, respectively, with a flow rate of 0.3 mL/min. The gradient was set as follows: 0 to 2 min: 15% B; 2 to 2.5 min: 15% B to 90% B; 2.5 to 4.5 min: 90% B; 4.5 to 5 min: 90% B to 15% B; and 5 to 8 min: 15% B. MS analysis was carried out on a 5500 QTRAP (AB SCIEX) in a negative ion mode. The source parameters were as follows: source temperature, 550°C; ion source gas 1 (GS1), 55; ion source gas 2 (GS2), 55; curtain gas (CUR), 35; and ion spray voltage (IS), 5500 V. Arbutin was detected in multiple-reaction monitoring mode with 271 > 108* (quantitative ion pair) and 271 > 161 (qualitative ion pair) ion pairs. Analyst 1.6.3 was used to determine the chromatographic peak area and retention time. Quantitative analyses of hydroquinone were conducted using a Q Exactive Combined Quadrupole Orbitrap Mass Spectrometer (ThermoFisher Scientific) equipped with a Hypersil Gold column (2.1 mm × 100 mm, 3 µm) at 25°C. The mobile phases A and B for gradient elution were water and acetonitrile, respectively. The gradient was set as follows: 0 min: 5% B; 10 min: 20% B; and 35 min: 90% B. The flow rate was 0.35 mL/min. Based on the retention times and peak shapes of the standards, all the samples were qualitatively and quantitatively analyzed (Supplementary Fig. S5, A and B).
H2DCF-DA staining
To detect H2O2 produced in the roots of the OsGRF7 and OsFBO13 transgenic lines under normal and salinity stress conditions, 7-d-old plants were cultured in a bottomless 96-well plate with Yoshida culture solution supplemented with or without 100 mM NaCl for 24 h. Then, the root tips were cut and immersed in 2 mL of H2DCF-DA staining buffer. The incubation was conducted at room temperature in the dark for 10 min with shaking at 100 rpm. The reactions were stopped by transferring the roots to deionized water. The fluorescence signals were detected under a DMi8 microscope system (Leica). The root tips of 6 OsGRF7 transgenic plants and 5 OsFBO13 transgenic plants were measured.
Measurements of SOD, POD, and CAT enzyme activities
To analyze the enzyme activities in the OsGRF7 and OsFBO13 transgenic plants, the seedlings were grown under normal conditions for 15 d, and sampled after 24 h of 100 mM NaCl treatment. Ten seedlings were sampled and pooled as one biological replicate, and 3 biological replicates were performed for each analysis. Approximately 0.1 g of rice seedlings was ground in 10 mM potassium phosphate buffer. The homogenate was centrifuged at 1,150 × g for 10 min at 4°C. The supernatant was used for crude enzyme extraction. SOD enzyme activity was measured using a previously reported protocol. The POD and CAT enzyme activities were measured using the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute).
Measurements of the MDA, proline, soluble sugar, and chlorophyll levels
To analyze the physiological and biochemical indicators of the OsGRF7 transgenic plants, the wild-type and OsGRF7 transgenic plants were grown under normal conditions for 15 d and sampled after 24 h of 100 mM NaCl treatment. Ten seedlings were sampled and pooled as one biological replicate, and 3 biological replicates were performed for each analysis. To measure the MDA content, approximately 0.5 g of rice seedlings was homogenized in 5 mL of 5% (w/v) trichloroacetic acid and centrifuged at 845 × g for 10 min at 4°C. The supernatant (2 mL) was reacted with 2 mL of 0.67% (w/v) thiobarbituric acid. The mixture was boiled for 30 min and centrifuged at 13,523 × g at room temperature. The absorbance of the supernatant was subsequently read at 450, 532, and 600 nm, respectively. The MDA content was estimated using the formula MDA (µmol/g FW) = [6.45*(A532 to A600)−0.56*A450]*Vt/Vs/W, where Vt indicates the total volume of the extraction buffer, Vs indicates the volume of the measured extraction buffer, and W indicates the fresh weight of the sample.
To generate the standard curve for proline, 10 mg of proline was dissolved in 100 mL of ddH2O to prepare a 100 µg/mL stock buffer. The stock buffer was then diluted to different concentrations (1, 2, 3, 4, 5, and 6 µg/mL). The mixture of 2 mL of proline, 2 mL of acetic acid, and 2 mL of acid ninhydrin was boiled for 30 min. Then, 4 mL of methylbenzene was added, and the absorbance of the supernatant was measured at 520 nm. To determine the proline content, approximately 0.5 g of rice seedlings was pulverized in liquid nitrogen and homogenized in 5 mL of salicylic acid. After the samples were boiled for 30 min to extract free proline, the proline content was estimated using the formula proline (µg/g FW) = X*5/W, where X indicates the concentration of proline and W indicates the fresh weight of the sample.
To generate the standard curve for sugar, 1 g of sucrose was dissolved in 100 mL of ddH2O to prepare a 1% (w/v) stock buffer. The stock buffer was then diluted to different concentrations. To measure the soluble sugar content, approximately 0.5 g of rice seedlings was homogenized in 5 mL of ddH2O. The supernatant was mixed with 5% (v/v) phenol and 98% (v/v) sulfuric acid for 30 min at room temperature. Then, the absorbance at 485 nm was subsequently determined with a spectrophotometer. The soluble sugar content was estimated using the formula soluble sugar (mg/g FW) = (C*D/W)*100, where C indicates the concentration of sugar, D indicates the dilution ratio, and W indicates the fresh weight of the sample.
To measure the chlorophyll content, 0.1 g of rice seedlings was pulverized in liquid nitrogen and homogenized in 80% (v/v) ethyl alcohol for 1 h in the dark. After centrifugation at 13,523 × g for 3 min at 4°C, the absorbance of the supernatant was measured at 663 and 645 nm. The chlorophyll content was estimated using the formula Chlorophyll a (µg/g FW) = (12.72*A663 to 2.59*A645)*V/W*1000, Chlorophyll b (µg/g FW) = (22.88*A645–4.67*A663)*V/W*1000, and Chlorophyll (µg/g FW) = (20.29*A645 + 8.05*A663)*V/W, where V indicates the volume of the sample and W indicates the fresh weight of the sample.
Total RNA isolation and RT-qPCR
To analyze the expression levels of the arbutin biosynthesis genes in the OsGRF7 transgenic plants, the wild-type and OsGRF7 transgenic plants were grown under normal conditions for 15 d and subsequently transferred to a solution with or without 100 mM NaCl. After 24 h, the wild-type and OsGRF7 transgenic seedlings were sampled. To analyze the expression levels of the genes involved in arbutin biosynthesis under different abiotic stress conditions, wild-type plants (YB) were grown under normal conditions for 15 d and sampled after 24 h of treatment with 150 mM mannitol, 20% (w/v) PEG, 4°C, 37°C, or 100 mM NaCl, respectively. To analyze the expression levels of OsUGT1 and OsUGT5 at different time points, wild-type plants (YB) were grown under normal conditions for 15 d and sampled after 0, 0.5, 1, and 4 h of 100 mM NaCl treatment. To analyze the expression levels of OsFBO13, OsUGT1, and OsUGT5 in the OsFBO13 transgenic plants, 15-d-old seedlings of wild-type and OsFBO13 transgenic plants under normal growth conditions were sampled. Three seedlings were sampled and pooled as one biological replicate, and 3 biological replicates were performed for each analysis. Total RNA was extracted and used to synthesize cDNA with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) according to the instruction manual. RT-qPCR was performed with a Light Cycler 480 II (Roche) with SYBR Green PCR Mixture (YEASEN) according to the manufacturer's instructions. The OsUBI gene was used as the internal control. Relative expression levels were calculated according to the 2−ΔΔCt method. Gene expression levels were normalized to the expression level of OsUBI.
Electrophoretic mobility shift assay
After labeling with FAM, the promoter regions of OsUGT1 and OsUGT5 were amplified and purified as probes. Then, a reaction mixture containing 200 ng of DNA probe, 2 µg of protein (His-GRF7), 2 µL of 10× binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT, pH 7.5), 1 µL of 1 M KCl, 1 µL of 50% (v/v) glycerol, 1 µL of 0.5 M EDTA, 1 µL of 1% (v/v) Nonidet P-40, 1 µL of 1 mg mL−1 poly (dI-dC), and double-distilled water was prepared to a final volume of 20 µL was prepared. After 20 min of reaction at 25°C, the products were electrophoresed on 3.5% (w/v) native polyacrylamide gels and then scanned by an Amersham Typhoon 2000 (GE Healthcare).
Transient transactivation assay
Approximately 2-kb promoter regions from OsUGT1 and OsUGT5 with restriction sites KpnI and BamHI restriction sites were amplified from the genomic DNA of YB and subsequently cloned and inserted into a pGreenII-0800-Luc (EK-Bioscience) vector. Transient transactivation assays were performed using rice protoplast cells, and the Dual-Luciferase Reporter Assay System (Promega) was used to detect luciferase activity, with the Renilla luciferase serving as the internal control. Independent transformations were considered as biological replicates, and 3 biological replicates were performed for each analysis.
To detect the transactivation of these genes in N. benthamiana leaves, the constructs were introduced into the Agrobacterium tumefaciens strain GV3101 (pSoup-p19) and infiltrated into N. benthamiana leaves. After 3 d, the leaves were infiltrated with luciferin buffer, and the LUC signals were detected with an LB985 NightSHADE system (Berthold). Independent transformations were considered as biological replicates, and 3 biological replicates were performed for each analysis.
Protein expression in E. coli and glycosyltransferase activity assay
The full-length cDNA sequences of OsUGT1 and OsUGT5 were amplified with the restriction sites KpnI and BamHI and fused into the expression vector pCOLD-TF. The constructs were subsequently introduced into the E. coli strain Rosetta (DE3), after which the transformants were grown in 200 mL of LB buffer at 37°C until the OD600 reached 0.6. Then, 0.2 mM isopropyl-β-D-thiogalactopyranoside was added, and the cultures were grown for 17 h at 16°C. Cultured cells were harvested by centrifugation (1,902 × g, 10 min) at 4°C and resuspended in 5 mL of PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.5). The samples were lysed and centrifuged at 13,523 × g at 4°C, and the supernatants were used as the crude enzyme extract. The glycosyltransferase activity assay was performed as described previously (Hou et al. 2004; Dong et al. 2020; Ma et al. 2021) with some modifications. Briefly, a 200 µL aliquot of the reaction mixture contained 5 mM PBS buffer (pH 7.5), 0.5 mM hydroquinone, 1.5 mM UDP-glucose, 10 mM phenylmethylsulfonyl fluoride, and 50 µL of crude enzyme extract. The reaction mixtures were incubated at 30°C for 3 h, after which 10 µL of trichloroacetic acid (240 mg/mL) was added to quench the reaction. The reaction products were quickly frozen and stored at −80°C before HPLC analysis. The empty vector of pCOLD-TF was used as the negative control. Three independent reactions were performed for each genotype.
Qualitative analyses of arbutin and hydroquinone were conducted using an Ultimate 3000 apparatus with Hypersil GOLD C18 column (4.6 mm × 250 mm, 5 µm) thermostated at 30°C. The isocratic method was applied, and the mobile phase was methanol/water (15:85, v/v) with a flow rate of 1 mL/min. Methanol, arbutin, and hydroquinone (HPLC grade) were purchased from Merck (Darmstadt, Germany). The water was purified by a Milli-Q gradient (Millipore, Vimidrone, Italy). Arbutin and hydroquinone were detected at 280 nm (retention times: 3.7 and 5.0 min, respectively). The chromatographic data are listed in Supplementary Data Set 3.
Histological analysis
Plant tissues were fixed in FAA solution (3.7% [v/v] formaldehyde, 50% [v/v] ethanol, and 5% [v/v] acetic acid) and embedded in Paraplast Plus (Sigma Aldrich). Eight-micron-thick sections were stained with toluidine blue for light microscopic analysis (Leica). For scanning electron microscopy, spikelet husks were fixed in 4% paraformaldehyde (Sigma Aldrich) for 12 h and then processed according to the instruction manual for the scanning electron microscope (Hitachi S-3000N, Japan).
Yeast two-hybrid assay
The full-length cDNAs of OsGIF1, OsGIF2, OsGIF3, and OSKs were cloned and inserted into the prey vector pGAD-T7, and the full-length or truncated cDNAs of OsGRF7 and OsFBO13 were cloned and inserted into pGBK-T7. The prey and bait plasmids were subsequently cotransformed into yeast (Saccharomyces cerevisiae) AH109 strains and plated on SD/–Leu–Trp medium for 3 d at 30°C. Interactions between bait and prey were further tested on SD/–Trp–His–Leu–Ade medium. Empty vectors of the yeast two-hybrid constructs pGAD-T7 and pGBK-T7 were used as the negative controls.
Subcellular localization and bimolecular fluorescence complementation assay
For subcellular localization, the full-length OsUGT1 and OsUGT5 cDNAs were cloned and inserted into the pSAT-RFP vector (at the KpnI and BamHI sites) to generate the pSAT-UGT1/5-RFP construct. The empty vector pSAT-RFP was used as the negative control, and the Pro35S:GRF7-GFP was used as the nuclear marker. For the bimolecular fluorescence complementation assay, the full-length OsGRF7 cDNA was amplified and inserted into pSPYCE (C-terminal end of YFP) via the XbaI and ClaI sites, and the full-length OsFBO13 cDNA was amplified and inserted into pSPYNE (N-terminal end of YFP) via the XbaI and ClaI sites, respectively. Rice protoplasts were isolated as described previously (Yoo et al. 2007). For protoplast transformation, 2 μg of plasmids harboring UGT1/5-RFP, GRF7-GFP, FBO13-nYFP, GRF7-cYFP, nYFP, or cYFP; 100 μL of protoplasts; and 110 μL of PEG4000 solution (40% [w/v] PEG4000, 0.6 M mannitol, and 100 mM CaCl2) were mixed gently and incubated for 20 min at room temperature. The cells were then washed twice with 440 μL of W5 solution (154 mM NaCl, 5 mM KCl, 125 mM CaCl2, 5 mM glucose, and 2 mM MES, pH 5.8) and resuspended in 1 mL of W5 solution. After 16 h of incubation in the dark, fluorescence signals and bright-field images of the protoplasts were obtained with a DMi8 confocal system (Leica) with the excitation at 488 nm/emission at 500 nm for GFP, excitation at 532 nm/emission at 588 nm for RFP, and excitation at 514 nm/emission at 527 nm for YFP. Empty vectors of bimolecular fluorescence complementation constructs were used as negative controls.
Split firefly luciferase complementation imaging assay
The full-length cDNA of OsGRF7 was amplified and inserted into the cLUC vector with BamHI and SalI sites, while the full-length cDNAs of OsFBO13 and OsGIF1 were amplified and inserted into the nLUC vector with BamHI and SalI sites. These vectors were subsequently introduced into the A. tumefaciens strain GV3101 and coinfiltrated into N. benthamiana leaves. After 3 d, the leaves were infiltrated with luciferin buffer and incubated in the dark for 5 min. Finally, the LUC signals were detected with an LB985 NightSHADE system (Berthold). Empty vectors of split firefly luciferase complementation constructs were used as the negative controls.
Coimmunoprecipitation and immunoblotting
Full-length cDNAs of OsFBO13 and OsGRF7 were amplified and inserted into pSPYNE (myc tag fused) and pSPYCE (HA tag fused), respectively. Rice protoplasts were transfected with 30 μg of plasmid and incubated for 24 h at 28°C. Total protein was extracted from harvested protoplasts. The lysates were incubated with agarose beads conjugated with the anti-HA antibody (E12-003-3, EnoGene) at 4°C for 5 h. The beads were washed 3 times with protein extraction buffer. Immunoprecipitates were separated by SDS-PAGE and detected by immunoblotting using anti-HA (E12-003-3, EnoGene, 1:5,000 dilution) and anti-myc (E12-002-3, EnoGene, 1:5,000 dilution) antibodies.
Cell-free degradation assay
The full-length OsFBO13 cDNA was amplified and inserted into the pSPYNE vector, which was fused with the myc tag. Rice protoplasts were transfected with 30 μg of plasmid and then incubated for 48 h at 28°C. Total protein was extracted from the protoplasts in the extraction buffer. The purified bacterially expressed OsGRF7-His protein was incubated at 30°C in total protein extracts with or without 50 μM MG132 supplementation for the requisite duration. Immunoblotting was performed using an anti-His antibody (E12-004-2, EnoGene, 1:4,000 dilution).
The total protein content of the OsFBO13 transgenic plants was extracted in the degradation buffer (25 mM Tris-HCl [pH 7.5], 10 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol, 10 mM ATP, and 10 mM phenylmethylsulfonyl fluoride). Five micrograms of recombinant GST-OsGRF7 protein were incubated with 30 μg of various protein extracts with or without 50 µM MG132 at 30°C for 0, 5, 15, 30, or 60 min. The GST-OsGRF7 protein was detected by an anti-GST antibody (E12-007-2, EnoGene, 1:10,000 dilution). Coomassie brilliant blue staining indicated that equal amounts of proteins were loaded. Degradation buffer was used as a negative control, and the GST-OsGRF7 protein was incubated with a degradation buffer to exclude the effect of the degradation buffer.
Phylogenetic analysis
For phylogenetic analysis of arbutin biosynthesis-related genes in rice, 9 rice arbutin biosynthesis-related protein sequences were obtained from the Rice Genome Annotation Project database (http://rice.uga.edu/index.shtml). The amino acid sequences were aligned using the ClustalX program. The phylogenetic tree was constructed using MEGAX based on the neighbor-joining method with 1,000 bootstrap replicates. The alignment file is provided in Supplementary File 1. The phylogenetic tree in Newick format is provided in Supplementary File 2.
Statistical analysis
Unpaired Student's t-test (two-tailed) and Tukey’s multiple comparisons test were performed using GraphPad Prism 8. Student's t-test (two-tailed) was used for significant difference analysis between the 2 samples. Tukey's multiple comparisons test was used for significant difference analysis among multiple samples. P value <0.05 was considered to indicate statistical significance. Two-way ANOVA of the contributions of genotype, treatment, and their interaction to the phenotype were performed using IBM SPSS statistics, version 20.0. The detailed statistical analysis data are shown in Supplementary Data Set 5.
Accession numbers
Sequences from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsGRF7, Os12g0484900; OsGIF1, Os03g0733600; OsGIF2, Os11g0615200; OsGIF3, Os12g0496900; OsFBO13, Os03g0232000; OsUGT1, Os01g0734600; OsUGT5, Os05g0177500; OsUBI, Os03g0234200.
Supplementary Material
Acknowledgments
We thank Dr Huanran Yuan (Wuhan University) for providing assistance in the yeast two-hybrid screening. We thank Dr Mingqiang Zhu (Wuhan University) for providing assistance in the detection of hydroquinone.
Contributor Information
Yunping Chen, State Key Laboratory of Hybrid Rice, Key Laboratory for Research and Utilization of Heterosis in Indica Rice of Ministry of Agriculture, Engineering Research Center for Plant Biotechnology and Germplasm Utilization of Ministry of Education, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Zhiwu Dan, State Key Laboratory of Hybrid Rice, Key Laboratory for Research and Utilization of Heterosis in Indica Rice of Ministry of Agriculture, Engineering Research Center for Plant Biotechnology and Germplasm Utilization of Ministry of Education, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Shaoqing Li, State Key Laboratory of Hybrid Rice, Key Laboratory for Research and Utilization of Heterosis in Indica Rice of Ministry of Agriculture, Engineering Research Center for Plant Biotechnology and Germplasm Utilization of Ministry of Education, College of Life Sciences, Wuhan University, Wuhan 430072, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Author contributions
S.L. and Y.C. designed the experiments. Y.C. and Z.D. performed the experiments and analyzed the data. Y.C., Z.D., and S.L. wrote and revised the manuscript.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. OsGRF7 positively regulates rice salt stress response.
Supplementary Figure S2. RNA-seq analysis of salt-treated grf7-14 and WT.
Supplementary Figure S3. Metabolomic analysis of OsGRF7 transgenic seedlings.
Supplementary Figure S4. Classification of 114 metabolites.
Supplementary Figure S5. The in vivo contents of arbutin and hydroquinone in OsGRF7 transgenic roots.
Supplementary Figure S6. Quantification of arbutin concentration.
Supplementary Figure S7. Physiological and biochemical indicators of WT and OsGRF7 transgenic seedlings under different treatments.
Supplementary Figure S8. Phylogenetic analysis and expression patterns of arbutin biosynthesis-related genes under different stress conditions.
Supplementary Figure S9. Expression of OsUGT1 and OsUGT5 is induced by salt stress.
Supplementary Figure S10. Subcellular localization of OsUGT1 and OsUGT5 in rice protoplast cells.
Supplementary Figure S11. Mass spectra of the arbutin and hydroquinone standards.
Supplementary Figure S12. The in vivo contents of arbutin and hydroquinone in wild type (GRF7-OE-1), ugt1GRF7OE, and ugt5GRF7OE roots.
Supplementary Figure S13. OsUGT1 and OsUGT5 positively regulate the rice salinity stress response.
Supplementary Figure S14. OsGRF7 positively regulates rice grain size.
Supplementary Figure S15. OsFBO13 is an integral part of a SCFOsFBO13 E3 ligase in rice.
Supplementary Figure S16. The in vivo contents of arbutin and hydroquinone in OsFBO13 transgenic roots.
Supplementary Figure S17. The contents of MDA and proline in WT and OsFBO13 transgenic lines.
Supplementary Figure S18. OsFBO13 negatively regulates rice grain size.
Supplementary Figure S19. OsFBO13 negatively regulates the expression of OsUGT1 and OsUGT5.
Supplementary Figure S20. The lysine residue K365 is the primary ubiquitination site of OsGRF7.
Supplementary Figure S21. OsFBO13 competes with OsGIFs to interact with OsGRF7.
Supplementary Table S1. Differential metabolites between the wild type and OsGRF7 transgenic seedlings.
Supplementary Table S2. OsGRF7-interacting proteins identified by yeast two-hybrid screening.
Supplementary Table S3. Candidate ubiquitinated sites of OsGRF7.
Supplementary Data Set 1. DEGs in grf7-14 seedlings compared with wild type.
Supplementary Data Set 2. Untargeted metabolic data of OsGRF7 transgenic plants at the seedling stage.
Supplementary Data Set 3. Chromatogram data.
Supplementary Data Set 4. Primers used in this paper.
Supplementary Data Set 5. Statistical analyses.
Supplementary File 1. Multiple sequence alignment for Supplementary Fig. S8B.
Supplementary File 2. Newick format of the phylogenetic tree for Supplementary Fig. S8B.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (grant nos. 32101667 and U20A2023) and the China Postdoctoral Science Foundation (grant no. 2023T160497).
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012:30(10):918–920. 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Wang H, Gao F, Cao X, Chen Y, Li G, Zhu Y, Yang X, Zhang L, Zhao Z, et al. miR396-OsGRFs module balances growth and rice blast disease-resistance. Front Plant Sci. 2019:9:1999. 10.3389/fpls.2018.01999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015:2(1):15195. 10.1038/nplants.2015.195 [DOI] [PubMed] [Google Scholar]
- Chen TH, Murata N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol. 2002:5(3):250–257. 10.1016/S1369-5266(02)00255-8 [DOI] [PubMed] [Google Scholar]
- Chen Y, Dan Z, Gao F, Chen P, Fan F, Li S. Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and indole-3-acetic acid metabolism. Plant Physiol. 2020a:184(1):393–406. 10.1104/pp.20.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dan Z, Li S. Rice GROWTH-REGULATING FACTOR 7 controls tiller number by regulating strigolactone synthesis. Plant Signal Behav. 2020b:15(11):1804685. 10.1080/15592324.2020.1804685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Tan J, Zhou C, Yang X, Yang F, Zhang S, Sun S, Miao X, Shi Z. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol J. 2019:17(8):1657–1669. 10.1111/pbi.13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Z, Chen Y, Zhao W, Wang Q, Huang W. Metabolome-based prediction of yield heterosis contributes to the breeding of elite rice. Life Sci Alliance. 2020:3(1):e201900551. 10.26508/lsa.201900551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong NQ, Sun Y, Guo T, Shi CL, Zhang YM, Kan Y, Xiang YH, Zhang H, Yang YB, Li YC, et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun. 2020:11(1):2629. 10.1038/s41467-020-16403-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wang K, Liu Y, Chen Y, Chen P, Shi Z, Luo J, Jiang D, Fan F, Zhu Y, et al. Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat Plants. 2015:2(1):15196. 10.1038/nplants.2015.196 [DOI] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta. 2010:231(5):991–1001. 10.1007/s00425-010-1104-2 [DOI] [PubMed] [Google Scholar]
- Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, et al. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020:63(5):635–674. 10.1007/s11427-020-1683-x [DOI] [PubMed] [Google Scholar]
- Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B, et al. Interactive XCMS online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem. 2014:86(14):6931–6939. 10.1021/ac500734c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Haltiwanger RS, Holt GD, Kelly WG. Glycosylation in the nucleus and cytoplasm. Annu Rev Biochem. 1989:58(1):841–874. 10.1146/annurev.bi.58.070189.004205 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012:159(1):321–335. 10.1104/pp.112.193649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem. 2004:279(46):47822–47832. 10.1074/jbc.M409569200 [DOI] [PubMed] [Google Scholar]
- Husar S, Berthiller F, Fujioka S, Rozhon W, Khan M, Kalaivanan F, Elias L, Higgins GS, Li Y, Schuhmacher R, et al. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 2011:11(1):51. 10.1186/1471-2229-11-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol. 2017:68(1):405–434. 10.1146/annurev-arplant-042916-040936 [DOI] [PubMed] [Google Scholar]
- Jin SH, Ma XM, Kojima M, Sakakibara H, Wang YW, Hou BK. Overexpression of glucosyltransferase UGT85A1 influences trans-zeatin homeostasis and trans-zeatin responses likely through O-glucosylation. Planta. 2013:237(4):991–999. 10.1007/s00425-012-1818-4 [DOI] [PubMed] [Google Scholar]
- Kahloul S, HajSalah El Beji I, Boulaflous A, Ferchichi A, Kong H, Mouzeyar S, Bouzidi MF. Structural, expression and interaction analysis of rice SKP1-like genes. DNA Res. 2013:20(1):67–78. 10.1093/dnares/dss034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sreenivasulu N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2014:37(2):300–311. 10.1111/pce.12157 [DOI] [PubMed] [Google Scholar]
- Kawa D. APC/c(TE) shapes rice architecture from top to bottom. Plant Cell. 2020:32(6):1786–1787. 10.1105/tpc.20.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt S, Greco R, Agalou A, Shao J, t Hoen CCJ, Overnas E, Osnato M, Curiale S, Meynard D, van Gulik R, et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 2014:164(4):1952–1966.. 10.1104/pp.113.222836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniak E, Wielanek M, Chwatko G, Głowacki R, Libik-Konieczny M, Piatek M, Gajewska E, Skłodowska M. Salicylic acid and cysteine contribute to arbutin-induced alleviation of angular leaf spot disease development in cucumber. J Plant Physiol. 2015:181:9–13. 10.1016/j.jplph.2015.03.017 [DOI] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature. 2018:560(7720):595–600. 10.1038/s41586-018-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YJ, Lee EH, Kang TH, Ha SK, Oh MS, Kim SM, Yoon TJ, Kang C, Park JH, Kim SY. Inhibitory effects of arbutin on melanin biosynthesis of alpha-melanocyte stimulating hormone-induced hyperpigmentation in cultured Brownish Guinea pig skin tissues. Arch Pharm Res. 2009:32(3):367–373. 10.1007/s12272-009-1309-8 [DOI] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014:165(1):160–174. 10.1104/pp.114.235564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Chen MX, Liu R, Zhang L, Hou X, Liu S, Ding X, Jiang Y, Xu J, Zhang J, et al. Abscisic acid regulates auxin distribution to mediate maize lateral root development under salt stress. Front Plant Sci. 2019:10:716. 10.3389/fpls.2019.00716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, Liu L, Tang Z, Bai Z, Cao S, Chu C. Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. J Integr Plant Biol. 2005:47(6):745–752. 10.1111/j.1744-7909.2005.00071.x [DOI] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015:8(8):1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Ma Y, Cao J, Chen Q, He J, Liu Z, Wang J, Li X, Yang Y. Abscisic acid receptors maintain abscisic acid homeostasis by modulating UGT71C5 glycosylation activity. J Integr Plant Biol. 2021:63(3):543–552. 10.1111/jipb.13030 [DOI] [PubMed] [Google Scholar]
- Nan N, Wang J, Shi Y, Qian Y, Jiang L, Huang S, Liu Y, Wu Y, Liu B, Xu ZY. Rice plastidial NAD-dependent malate dehydrogenase 1 negatively regulates salt stress response by reducing the vitamin B6 content. Plant Biotechnol J. 2020:18(1):172–184. 10.1111/pbi.13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo DH, Tran QT, Kim YS, Hang NTN, Ngo DN, Vo TS. GABA-enriched rice bran inhibits inflammation in LPS-stimulated macrophages via suppression of TLR4-MAPK/NF-kappaB signaling cascades. J Food Biochem. 2022:46(12):e14421. 10.1111/jfbc.14421 [DOI] [PubMed] [Google Scholar]
- Oliver AE, Hincha DK, Tsvetkova NM, Vigh L, Crowe JH. The effect of arbutin on membrane integrity during drying is mediated by stabilization of the lamellar phase in the presence of nonbilayer-forming lipids. Chem Phys Lipids. 2001:111(1):37–57. 10.1016/S0009-3084(01)00141-4 [DOI] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol. 2005:23(4):482–487. 10.1038/nbt1082 [DOI] [PubMed] [Google Scholar]
- Qin L, Chen H, Wu Q, Wang X. Identification and exploration of the GRF and GIF families in maize and foxtail millet. Physiol Mol Biol Plants. 2022:28(9):1717–1735. 10.1007/s12298-022-01234-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raigond P, Ezekiel R, Raigond B. Resistant starch in food: a review. J Sci Food Agric. 2015:95(10):1968–1978. 10.1002/jsfa.6966 [DOI] [PubMed] [Google Scholar]
- Schulz AG, Van Amelsvoort JM, Beynen AC. Dietary native resistant starch but not retrograded resistant starch raises magnesium and calcium absorption in rats. J Nutr. 1993:123(10):1724–1731. 10.1093/jn/123.10.1724 [DOI] [PubMed] [Google Scholar]
- Shen X, Wang R, Xiong X, Yin Y, Cai Y, Ma Z, Liu N, Zhu ZJ. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat Commun. 2019:10(1):1516. 10.1038/s41467-019-09550-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skłodowska M, Naliwajski M, Wielanek M, Gajewska E, Kuźniak E. Arbutin- and benzotiadiazole-mediated cucumber response to Pseudomonas syringae pv. lachrymans infection in carbohydrate metabolism. Sci Hortic. 2015:192:200–210. 10.1016/j.scienta.2015.06.007 [DOI] [Google Scholar]
- Sprenger H, Kurowsky C, Horn R, Erban A, Seddig S, Rudack K, Fischer A, Walther D, Zuther E, Kohl K, et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016:39(11):2370–2389. 10.1111/pce.12780 [DOI] [PubMed] [Google Scholar]
- Tu J, Yin Y, Xu M, Wang R, ZJ Z. Absolute quantitative lipidomics reveals lipidome-wide alterations in aging brain. Metabolomics. 2017:14(1):5. 10.1007/s11306-017-1304-x [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Benkova E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012:28(1):463–487. 10.1146/annurev-cellbio-101011-155741 [DOI] [PubMed] [Google Scholar]
- Vishal B, Krishnamurthy P, Ramamoorthy R, Kumar PP. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol. 2019:221(3):1369–1386. 10.1111/nph.15464 [DOI] [PubMed] [Google Scholar]
- Waadt R, Seller CA, Hsu PK, Takahashi Y, Munemasa S, Schroeder JI. Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol. 2022:23(10):680–694. 10.1038/s41580-022-00479-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Jin SH, Hu HQ, Sun YG, Wang YW, Han P, Hou BK. UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol. 2012:194(3):666–675. 10.1111/j.1469-8137.2012.04107.x [DOI] [PubMed] [Google Scholar]
- Wu B, Cao X, Liu H, Zhu C, Klee H, Zhang B, Chen K. UDP-glucosyltransferase PpUGT85A2 controls volatile glycosylation in peach. J Exp Bot. 2019:70(3):925–936. 10.1093/jxb/ery419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015:43(W1):W251–W257. 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000:287(5451):303–305. 10.1126/science.287.5451.303 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007:2(7):1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhao C, Zayed O, Zeng F, Liu C, Zhang L, Zhu P, Hsu CC, Tuncil YE, Tao WA, Carpita NC, et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019:224(1):274–290. 10.1111/nph.15867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.