Abstract
Land plants (embryophytes) came about in a momentous evolutionary singularity: plant terrestrialization. This event marks not only the conquest of land by plants but also the massive radiation of embryophytes into a diverse array of novel forms and functions. The unique suite of traits present in the earliest land plants is thought to have been ushered in by a burst in genomic novelty. Here, we asked the question of how these bursts were possible. For this, we explored: (i) the initial emergence and (ii) the reshuffling of domains to give rise to hallmark environmental response genes of land plants. We pinpoint that a quarter of the embryophytic genes for stress physiology are specific to the lineage, yet a significant portion of this novelty arises not de novo but from reshuffling and recombining of pre-existing domains. Our data suggest that novel combinations of old genomic substrate shaped the plant terrestrialization toolkit, including hallmark processes in signalling, biotic interactions and specialized metabolism.
Keywords: streptophyte algae, mosaic evolution, reductive evolution, plant terrestrialization, protein domains, plant evolution
1. Introduction
Photosynthetic eukaryotes have a history of more than 1 billion years [1]. Many algal lineages have successfully made the wet-to-dry transition [2–4]. But only Embryophyta (land plants) rose above the substrate, evolved complex multicellular bodies and globally conquered land, constituting an evolutionary singularity [5–8]. Embryophytes belong to the Streptophyta, which encompass: (i) the Embryophyta (land plants), recovered by all molecular phylogenetic and phylogenomic analyses as a monophylum and united by synapomorphies that include the name-giving embryo [9], an alternation of generations [10] and likely the symbiotic interaction with mycorrhizal fungi [11,12]; and (ii) the paraphylum of streptophyte algae. Among the algae in this paraphylum, Charophyceae, Coleochaetophyceae and Zygnematophyceae are close relatives to land plants; together with land plants, they form a monophylum called Phragmoplastophyta (figure 1a). Therein, Zygnematophyceae are the closest algal relatives to land plants [17–19].
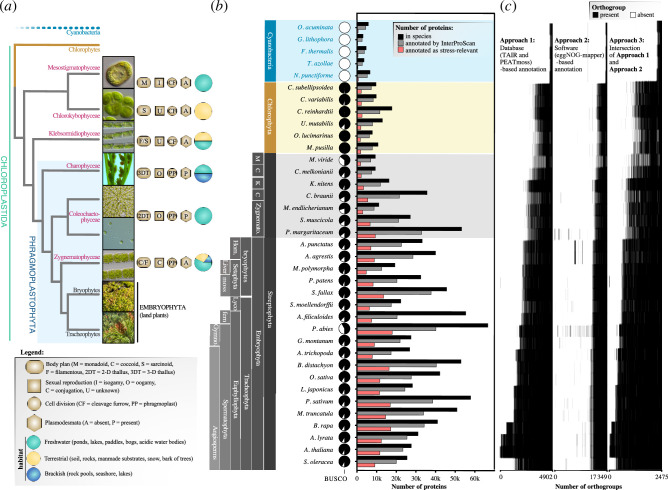
Figure 1.
Stress-relevant orthogroups across the green lineage. (a) Summary of the phylogeny, morphology, life history and ecology in cyanobacteria (blue) and the green lineage that includes chlorophyte algae (ochre) and the focus of this study: streptophytes, encompassing streptophyte algae (purple) and land plants (grey). (b) A phylodiverse set of land plants and algae assembled from the predicted proteomes of 37 species. Pie charts show BUSCO proteome completeness. Bar graphs show the total number of proteins per proteome (black), the subset annotated by InterProScan [13] (grey) and the subset that were stress-relevant from approach 3 in figure 1b (red). (c) Number of stress-annotated orthogroups (homologous protein group) were compiled using three approaches, building on: (1) the databases TAIR (v10) [14] + PEATmoss (4902 stress-relevant orthogroups) [15], (2) eggNOG-mapper [16] (17 349 orthogroups) and (3) an intersection of both approaches (Wilcox test p‐value = 2.2 × 10−16), yielding 2475 orthogroups that are considered the final set of stress-relevant orthogroups. Presence of orthogroups in species is indicated by black (and absence by white).
The independent wet-to-dry transitions in non-embryophytic plants incites the question: What was special about the biology of embryophytes that enabled them to dominate the land environment? There is a standing concept that the water- and/or land-dwelling streptophyte algal progenitor of land plants bore a mixture of exaptations and adaptations [6,7,20]. The genome sequences of extant members of the green lineage (Chloroplastida) provide a window through which to infer genetic potential in the ancestors along the trajectory of streptophyte evolution.
Comparative genomic analyses reveal that a substantial fraction of molecular physiological tools that land plants use to cope with terrestrial stressors can also be found in streptophyte algae, their last common ancestors (LCAs), and the LCA of streptophyte algae and land plants; all these ancestors had iterations of the complex genetic networks of land plants [21–27]. Several aspects have been investigated in the past decade through comparative analyses. One of the foremost are the deep evolutionary roots of phytohormone signalling cascades. For example, a complete chassis of homologues for the signalling cascade of the stress hormone abscisic acid (ABA) was found in several Zygnematophyceae [26,28,29] but functional analyses revealed that the protein homologues, albeit inhibiting downstream phosphatases, act ABA-independently [30]. Another interesting evolutionary pattern was recovered for the phytohormone auxin, whose polar distribution determines diverse aspects of growth and development in land plants [31–33]. Relevant homologues for the nuclear auxin response can be found in streptophyte algae [34–36] but a clear nuclear auxin response remains elusive in streptophyte algae. Where we do find a clear case of an auxin response conserved between land plants and streptophyte algae is the one that funnels through fast phosphorylation [37]. Next to phytohormones are several other specialized metabolites with important adaptive functions for terrestrial life, including phenylpropanoid-derived compounds [38], especially flavonoids [39] and unique algal compounds [40–42]. And of course, it has been pinpointed that the aforementioned adaptive symbiosis often requires genes to have had a deep evolutionary origin [11,43,44].
It has been proposed that the genes for these complex traits and networks arose in bursts of genetic novelty [45]; importantly, these burst are also recovered when corrected for homology detection failure [46]. But what made these bursts possible? Here, we explored the genetic substrate that underlies the complex stress-associated genetic networks now acting in land plants. From 37 species, we assembled a dataset of 57 795 orthogroups, extracted 2475 stress-associated orthogroups and traced their evolutionary assembly from protein domains. We infer that a quarter are land plant innovations, of which 15% emerged through novel domain shuffling. Our data explain that land plant-specific bursts of novelty for stress responses can arise from pre-existing protein domains, which are newly combined to create genetic novelty. New genes should allow for new network connections and fine-tuned responsiveness during the dawn of embryophytes.
2. Results and discussion
(a). Assembling a genetic set for plant stress response
To trace the evolution of the elaborate molecular chassis for the response to stressors, we clustered all homologous proteins (OrthoFinder [47]) from predicted proteomes of five cyanobacteria, six chlorophyte algae, seven streptophyte algae and 19 land plants (figure 1a) into 57 795 orthogroups. Our dataset is highly diverse in terms of represented phylogenetic lineages, functional capacities and genome sizes. Species were selected to counterbalance existing biases in genome sequencing efforts by selecting representatives of all main branches in the green lineage. This species selection should cover a major share of the functional genomic diversity that is relevant for understanding the deep evolution of molecular functions of streptophytes. Thus, our analysis should not be influenced by the redundancy (or lack thereof) of protein families and variation of genome sizes. However, it is likely that additional genomic sequencing of species in key phylogenetic positions will in the future help to identify more complex evolutionary patterns. To identify stress-associated orthogroups, we used complementary approaches (figure 1b; §3 and electronic supplementary material, figure S1a,b), yielding 2475 orthogroups (figure 1b). While annotations here derive from land plants, the proteins often have deeper evolutionary origins. To scrutinize this, we searched for more distant homologues across the phylodiverse set of eukaryote proteins in the EukProt database [48]. The homology searches of these 2475 orthogroups include very few distant homologues in non-chloroplastidal species (electronic supplementary material, figure S1c); we thus focused on 32 Chloroplastida species. Our results suggest that while most stress response-related proteins occur throughout the green lineage (Chloroplastida), bursts in numbers of proteins per orthogroup with these domains have occurred at several points during evolution. We scrutinized this in the following.
(b). Bursts in protein domains across the evolution of Chloroplastida
To understand the emergence of a protein toolkit for stress response, we traced how these proteins emerged from domain building blocks. We used InterProScan [13] to predict the domain composition of all 57 234 proteins within 2475 orthogroups (figure 2a). We identified up to 5403 different domains (figure 2a and electronic supplementary material, figure S2). Most proteins are from tracheophytes, followed by bryophytes, streptophyte algae, chlorophyte algae and cyanobacteria (electronic supplementary material, figure S2). Yet, the majority of stress-associated orthogroups contained representatives across the green lineage, followed by tracheophyte-specific orthogroups and orthogroups shared across the entire dataset (i.e. also including cyanobacteria; electronic supplementary material, figure S2b). Among all 31 stress gene ontology (GO)-terms used as seed, most proteins were assigned to the category 'response to topologically incorrect protein' followed by 'cellular response to stress', both pertaining to common responses to various stresses. These were followed by several categories specifically related to environmental stresses. This captures well on a protein level that stress response derives from the requirement to maintain cell homeostasis.
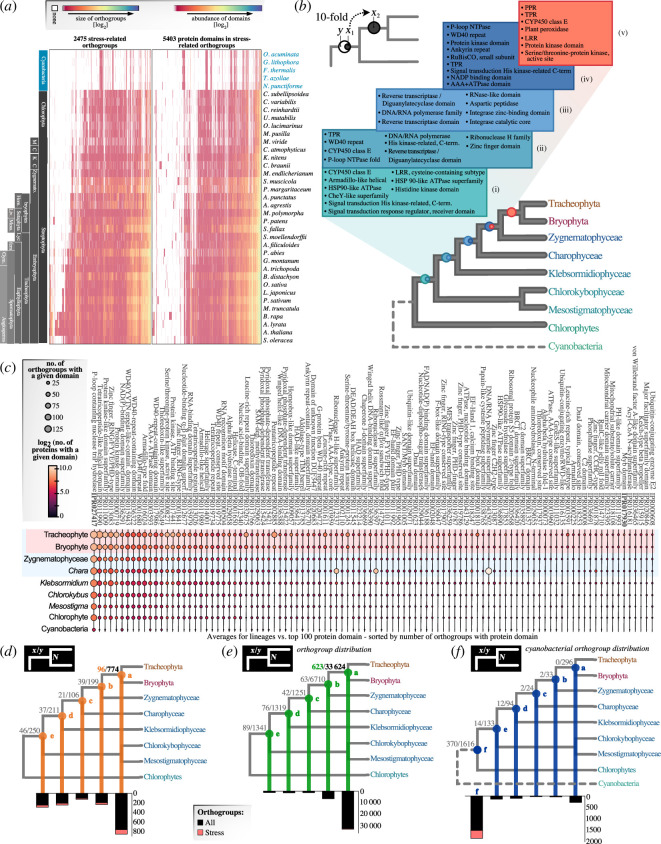
Figure 2.
Evolution of stress-associated domains and orthogroups across streptophytes. (a) Heatmaps on the number of proteins in the 2475 orthogroups (left) and the abundance of 5403 protein domains from stress-relevant orthogroups (right; both measures are log2-transformed; absences are in white). The x-axes are sorted by hierarchical clustering. (b) Bursts in protein domain expansions across the backbone of the streptophyte phylogeny. Named protein domains in the coloured boxes are those that expanded by ≥10-fold between adjacent LCAs (coloured nodes). Bottom-up, the LCAs correspond to: (i) streptophytes, (ii) Phragmoplastophyta + Klebsormidiophyceae, (iii) Phragmoplastophyta, (iv) Zygnematophyceae + land plants, (v) land plants. (c) Frequency of protein domains across the orthogroups in the dataset, averaged and collapsed into major lineages (individual values can be found in the electronic supplementary material, figure S3). The top 100 most frequent protein domains (x-axis) are sorted by their frequency of occurrence in stress-relevant orthogroups. Bubble colours indicate the number of proteins in which a given domain occurs; values are log2-transformed (number of proteins) and plotted as a gradient from black (lowest), to purple, red and white (highest). Bubble size indicates the number of orthogroups that bear a given domain; background colours: land plants (red) and Phragmoplastophyta (blue). (d–f) Inference of the: first occurrence of domains in stress-relevant orthogroups and their assemblage into the domain order in extant species, analysing the contribution of orthogroups from previous lineage/s to LGP of the next lineages. The x in x/y represents the number of orthogroups at node N that have protein domains pertaining to the orthogroups (embryophytic domain) present at all previous nodes (N − 1, and N − 2, and N − 3, etc.) but that did not occur in a single protein in the same order before node N. The y in x/y represents the number of orthogroups at node N that have key protein domains pertaining to the orthogroups (embrophytic domain) immediately at N − 1. (d) Distribution of stress-annotated and overall orthogroups with LGP in terms of conserved embryophytic domain set across the successive LCAs (represented as x/y). (e) Distribution of any stress-annotated and overall orthogroups (x and y represented as x/y) across successive LCAs. (f) Distribution of cyanobacterial stress-annotated and overall orthogroups across the successive LCAs (similarly represented as x/y). Abbreviations: PPR = pentatricopeptide repeat; TPR = tetratricopeptide repeat; LRR = leucine-rich repeat; HAD = haloacid dehydrogenase; TIM = triose-phosphate isomerase; FYVE/PHD = Fab 1 (yeast orthologue of PIKfyve), YOTB, Vac 1 (vesicle transport protein), and EEA1/plant homeodomain; SANT = Swi3, Ada2, N-Cor, and TFIIIB; RING = Really Interesting New Gene; MFS = Major Facilitator Superfamily; BRCT = Breast Cancer Gene 1 C terminus. Note that the DEAD/DEAH box family is named after the conserved Walker B motif (D-E-A-D sequence) and Myb after the Myeloblastosis genes.
From which protein domains were stress-relevant proteins built during evolution (figure 2a)? The two main mechanisms are the emergence of new domains and the reassembly of existing ones. To quantify this, we asked which biological functions are associated with bursts of domain emergence and changes in proteins’ domain architectures. Our focus was on protein domains that emerged in the LCAs of: (i) streptophytes, (ii) various streptophyte algal ancestors including (iii) Phragmoplastophyta, (iv) Zygnematophyceae + land plants, and (v) land plants. We measured the differential increments in protein domains from one ancestral tree node to the next by assessing the differences in the mean number of protein domains between the extant lineages that derived from the respective ancestral nodes, henceforth simply referred to as fold change. Here, we had a particular look at the top 10-fold changes from one ancestral node to the next (figure 2b). Particularly noteworthy were recurring expansions salient to: (i) gene editing in phragmoplastophytes (PPR, TPR) [49,50], (ii) CYP450 class E family within streptophytes, coherent with embryophyte chemodiversity [51,52] and (iii) signal transduction across streptophyte evolution.
(c). Changes in domain usage recapitulate the evolution of signalling and regulation
An increase of proteomic complexity might follow two main scenarios: (i) an increase in the number of orthogroups with stress-associated domains, which suggests a diversified context for use of the same domains in different orthogroups; (ii) an increase in the number of proteins with stress-associated domains within orthogroups, which suggests duplications of genes coding for proteins with the stress-associated domain. A combination of (i) and (ii) can be most parsimoniously explained by sub- and neofunctionalization driven either by genetic divergence and/or domain-shuffling. Further, a less likely (non-parsimonious) scenario is duplication of just the domain and integration into another protein concomitant with the loss of the original orthogroup.
We quantified the contribution of 5403 protein domains to the 2475 stress-relevant orthogroups (figure 2c; electronic supplementary material, figures S3, S4; electronic supplementary material, table S3). While the 2475 orthogroups consist of proteins that are largely specific to the green lineage (electronic supplementary material, figure S1c) and therein particularly vascular plants, the protein domains are more ancient: the most frequent stress domains occurred either: (i) in a deep ancestor with prokaryotes or (ii) upon the emergence of the green lineage (figure 2c); none of the top contributors emerged exactly at the origin of either streptophytes or embryophytes. Some of the most pronounced changes occurred in the usage of domains salient to signalling, such as IPR000719 and IPR01109 (protein kinase(-like) domain) among the top five hits. Both are prominent across all lineages and showcase an increase in first abundance of proteins followed by orthogroup expansions upon the emergence of Phragmoplastophyta and of land plants (figure 2c). This speaks of a fine-tuning of environmental input and—the likely ancestral—multicellular growth of Phragmoplastophyta [26,53]. Indeed, among the top 100 changes in orthogroup numbers with stress domain-containing proteins, 90.2% of the relevant LCAs show a stronger increase in protein numbers than in orthogroup numbers. This highlights the propensity of gene duplications in gene families salient to stress responses, similar to morphogenic processes [54].
Next to small-scale duplications, we recover several bursts ascribed to major streptophyte lineages (see more on this below in the following paragraphs). The most used domain in stress-relevant orthogroups in land plants was IPR027417 (P-loop containing nuclease triphosphate hydrolase), with 27 379 occurrences in the 19 land plant genomes. The most common domain exclusive to land plants is IPR002902 (Gnk2-homologous domain), with 4359 occurrences; the superfamily of proteins with an Gnk2-homologous domain is highly diversified and associated with the protection of the seed from biotic and abiotic environmental threats [55,56].
In addition to the above observed pattern, some domains showed an increase in the number of proteins yet only small or no effects on the number of orthogroups (figure 2c). It is conceivable that an increase of proteins within the same orthogroup reflects less-divergent gene duplicates that are subfunctionalized, whereas an increase of proteins and of orthogroups indicates higher sequence divergence or domain composition, speaking of neofunctionalization. Yet, it is difficult to use protein and orthogroup patterns to derive biological functions, which are anyway defined loosely and not directly comparable among gene families. It is nonetheless interesting to speculate that under neofunctionalization we can expect more orthogroups, whereas under subfunctionalization the duplicates often will be clustered within the same original orthogroups and thus without an increase in orthogroup numbers. Examples include regulatory and signalling protein domains such as the transcription factor-associated Myb domain (IPR017930; figure 2c and electronic supplementary material, figure S3) and the signal transduction domains of the CheY-like superfamily (IPR011006; figure 2b). A similar pattern is observed with conserved domain (‘site’) of CYP450 (IPR017972; figure 2b), corroborating the high degree of subfunctionalization and metabolic versatility observed in enzyme families with these domains [57], and the ABC transporter-like ATP binding domain (IPR003439; figure 2b) and ATPase AAA core (IPR003959) domains (figure 2b and figure 3a). Some changes recapitulate signature expansions, e.g. PPRs [58], with IPR002885 (PPR) having undergone an increase of protein domains in land plants by a fold change of 2000 (i.e. the difference between the average number of IPR002885 domains in the LCA of land plants and the LCA of Phragmoplastophyta) (figure 2b,c). Similarly, leucine-rich repeat (LRR) domains IPR032675 and IPR003591 both increased upon the emergence of seed plants (electronic supplementary material, figure S3), concomitantly with a burst of nucleotide-binding site–leucine-rich repeat (NBS-LRR) resistance genes [59]—a burst that brought forth a new regulatory mechanism via microRNA family miR482/2118, regulating both stress responses and reproductive development, originating first in the LCA of seed plants [59,60].
Figure 3.
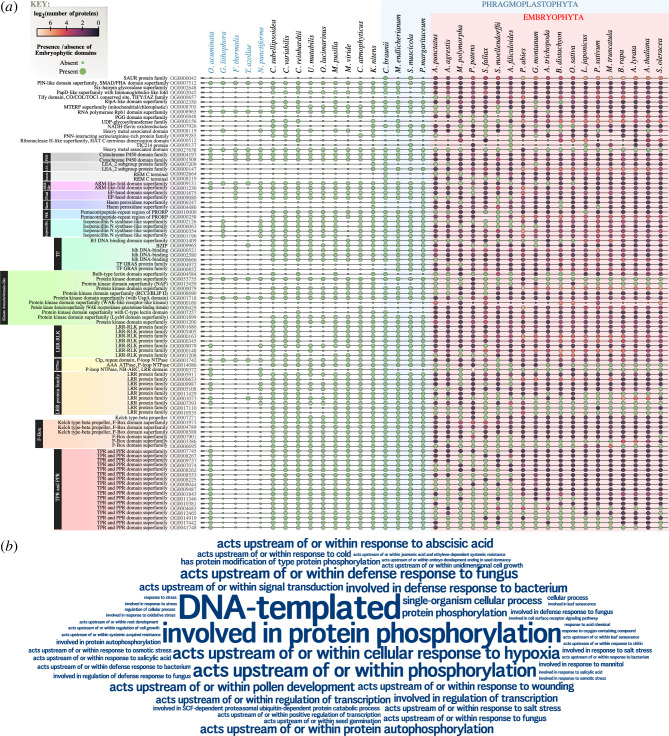
Latent genetic potential (LGP) in the protein building blocks for plant terrestrialization. (a) Phylogenetic distribution of key protein domains contributing to 96 stress-relevant orthogroups described in land plants. The bubble plot shows: (i) the 37 species/datasets on the x-axis; (ii) the 96 orthogroups and their corresponding functional annotations on the y-axis; (iii) presence (big circle) or absence (small circle) of embryophytic domains pertaining each orthogroup, and (iv) the count (log2-transformed) of proteins within each orthogroup, from black (absent) to white (highest). (b) Word cloud showing the top 50 most frequent Gene Ontology (GO) annotations (‘Biological Function’ via eggNOG-mapper) for proteins from the 96 orthogroups at the LCA of land plants node (node a in figure 2d). Word size represents GO term frequency.
Hundreds of F-box proteins occur in vascular plant genomes, where they recruit specific substrates to ubiquitin proteasome-based regulation [61]. We observed an increase of F-box protein family domains (IPR036047 and IPR001810; figure 2c and electronic supplementary material, figure S3) in land plants, and more so in angiosperms. F-Box proteins have apparently diversified in tracheophytes; concomitantly, both domains are reduced in liverworts and mosses, speaking to a combination of multiple events of duplication among tracheophytes and the reductive sweep in bryophyte genome evolution [62]. A similar trend occurs in the protein kinase domain (IPR000719; figure 2c and electronic supplementary material, figure S3) and the serine/threonine protein kinase domain (‘active site’; IPR008271; figure 2c and electronic supplementary material, figure S3). Taken together, we conclude from our data that domains integral to high-level signalling processes were seeded before plant terrestrialization but actualized through (often bursts in) usage of these domains in novel biological commitments.
(d). Genetic potential gleaned from protein domain emergence and assembly
The first land plants bore adaptations and exaptations to alleviate the stresses of the terrestrial habitat [6,7,20]. Exaptations are traits co-opted for a new function that either had a different ancestral (adaptive) function or were non-adaptive [63]. At the molecular level, protein domains represent the raw material from which new proteins can arise through new domain combinations. By integrating domains into new contexts—forming new protein architecture—or by being expressed in a different biological program, novelty in form and function can arise. This speaks of latent genetic potential (LGP) in the ancestors of land plants for assembling molecular programmes to respond to terrestrial stressors. Some of this should bear out from our data. We therefore explored the evolutionary time lapses between the emergence of bona fide stress-related proteins and the deeper origins of their individual protein domains.
Of the 2475 orthogroups, 623 had representatives in at least one angiosperm and one bryophyte, but in no other lineage outside of land plants (figure 2e), thus emerging in the LCA of land plants. Hence, one-quarter of stress-related orthogroups emerged with land plants. We determined the LCAs in which each protein domain first occurred for all 2475 stress-associated orthogroups (figure 2d).
We focused on the emergence of the 2475 stress-annotated orthogroups (x) and the total 57 795 orthogroups (y) (for comparison, see x/y representations in figure 2e). Here, our data capture the genetic bursts in the LCAs: (i) of land plants and Zygnematophyceae and (ii) of land plants (figure 2e, nodes a and b). Chloroplastida and cyanobacterial proteins are included in 2196 orthogroups, potentially capturing a signal from endosymbiotic gene transfers from plastids to nuclear genomes [64] (figure 2f). Out of these 2196, 400 are annotated as stress-related. Our data suggest that none of them contributed to stress-related genetic novelties that emerged with embryophytes, but instead were deployed earlier during evolution. We further validated this effect by searching for cyanobacterial homologues in the EukProt dataset (electronic supplementary material, table S1).
Do stress-annotated orthogroups emerge de novo or by combining pre-existing protein domains? We investigated how many land plant-specific orthogroups (figure 2d) are formed by domains with older evolutionary origins (figure 3a). For example, Remorin (OG0002664 and OG0008219; for all orthogroups, see electronic supplementary material, table S2) appear exclusively in embryophytes (figure 3a) but Remorin domains (‘C terminus’) are also present in Chara braunii, and thus they were likely present in their LCA. In total, 96 out of 623 (about 15%) are stress-annotated compared to 774 out of 33 624 (about 2%) overall embryophyte-specific novelty emerging from LGP (compare figure 2d,e).
Based on the most frequently predicted functions within these 96 orthogroups, we infer that the genetic potential for birth and expansion of homologues could have aided in signalling, molecular transport, microbe interaction, phenolic polymer formation, transcriptional regulation and RNA editing during terrestrialization (figure 3b). The embryophytic domains for JAZ proteins (OG0000657) occur across streptophyte algae, suggesting that its domains were present in the streptophyte LCAs. That said, JAZ proteins (OG0000657; electronic supplementary material, figure S3) belong to the 96 embryophyte-specific proteins, aligning with previous data indicating that functional JAZ and the co-occurrence of all the required domains are limited to embryophytes [65,66]. Yet, our data show that the LGP for a key component of the jasmonate signalling pathway, JAZ, was dwelling already in streptophyte algal ancestors and that functional JAZ derive from reshuffling and/or new domain combinations.
Streptophyte algae have orthologues for the genetic toolkit that in land plants establishes interactions with symbiotic fungi [43], especially genes involved in upstream signalling processes. Further, C. braunii has an expanded repertoire of LysM receptors [67]. Our data captured the RLK and LysMs orthogroups and proteins homologous to CERK1 in OG0001699 (electronic supplementary material, figure S3) and show that this orthogroup appeared in the LCA of land plants. Here, too, all its domains—the embryophytic domain assembly—were already present before the LCA of Chloroplastida. While chitin recognition via CERK1 homologues likely first emerged in the LCA of land plants [68], the LGP (i.e. its building blocks) is ancient.
Evolution of amino acid sequences in domains has shaped the functions that they fulfil in the genetic, protein and cellular context in which they reside. Thus, certain domain variants can have such divergent sequences that they are categorized as different domains. This has consequences for the insights garnered here. Identified changes can refer to the appearance of a new domain or a new domain variant. Thus, it is likely that certain domains are even more ancient than we currently estimate here. Overall, we suggest that the embryophyte-specific stress response toolkit (96 orthogroups) has a much earlier evolutionary origin and radiation if we consider the deep origin of the protein domains (figures 2c,d and 3a).
(e). Conclusion
Extant streptophytes display a mosaic of traits, brought about by processes like parallelism, convergence and rampant gene loss of key traits, concomitant with genomic streamlining in the Zygnematophyceae [20,26,62,69,70]. Yet, it is difficult to infer deep evolutionary processes that underlie genomic novelty by looking only at extant species: properties salient to a terrestrial lifestyle shared by land plants and streptophyte algae can be interpreted as exaptations [6], or adaptations under the assumption that the LCA of streptophytes was a terrestrial organism [71]—but there have been multiple habitat transitions [72]. Here, we reconciled trait mosaicism, bursts in genetic novelty and the deep evolutionary origins of protein domains underpinning stress response in land plants. We propose that domain co-option driven by integrations into new biological contexts was an important force in streptophyte evolution: in our analyses, domain co-option explains at least 15% of the apparent genetic novelty in land plants—likely an underestimate given the conservative nature of our analyses. Molecular traits emerging from LGP include the moulding of hallmark signalling, biotic interactions and specialized metabolic processes. This is likely no coincidence, consistent with signalling network remodelling [26,73], biotic stress-driven co-evolution [74] and the promiscuity of specialized metabolism [75], and as such a hotbed for biological novelty. Overall, our data suggest that novel combinations of old genomic substrate shaped the plant terrestrialization toolkit.
3. Methods
(a). Assembly of a protein database across the green lineage
We downloaded predicted proteins from genomes of: (i) cholorophyte algae [76–81], (ii) streptophyte algae [22,29,39,67,82] (following naming [83]), (iii) bryophytes [84–87], and (iv) tracheophytes [14,88–101]. Additionally, Cyanobacteria were included [102–106]. Completeness was assessed using BUSCO v. 4.1.4 [107] with the ‘Viridiplantae odb10’ dataset (parameters: e-value threshold 0.001, three candidate regions considered per BUSCO).
(b). Stress-annotation of protein data
We used OrthoFinder2 [47] on all proteomes and obtained 57 795 orthogroups (full analysis mode; default parameters). We searched for stress-related proteins in either Arabidopsis thaliana [14] or Physcomitrella patens PEATmoss [15] and used these proteins to annotate orthogroups. For A. thaliana, we extracted stress-relevant proteins based on 31 parent Gene Ontology (GO) terms (GO:0001666, GO:0002931, GO:0003299, GO:0006952, GO:0006970, GO:0006979, GO:0006991, GO:0009271, GO:0009408, GO:0009409, GO:0009413, GO:0009414, GO:0009611, GO:0009635, GO:0033554, GO:0033555, GO:0034059, GO:0034405, GO:0035900, GO:0035902, GO:0035966, GO:0042594, GO:0051409, GO:0051599, GO:0055093, GO:0061771, GO:0080134, GO:0090664, GO:0097501, GO:0097532, GO:1990911) and all their children (yielding 11 641 Arabidopsis proteins). For P. patens, we worked with the GO annotations provided by PEATmoss [15]. Among the total of 57 795 orthogroups, we identified 4902 as being relevant to ‘stress’ whenever they contained at least one stress-relevant protein each from Arabidopsis and P. patens. Independently, we performed de novo GO term assignment of orthogroups using eggNOG-mapper (e-value threshold 0.001, minimum bit score 60, 1 best HMM-hit reported, sequences >5000 bp ignored, database size 40 000 000 and diamond v. 0.9.24). This approach yielded 17 349 stress-related orthogroups after parsing de novo annotations for the occurrence of at least one of the aforementioned 31 GO terms or its children.
We intersected the two annotation methods, yielding 2475 stress-annotated orthogroups out of the initial 57 795 orthogroups see electronic supplementary material, figure S2 for details).
(c). Stress-annotated orthogroup distribution across EukProt
Homology of proteins from 2475 stress-annotated orthogroups was checked against the EukProt database using DIAMOND v. 0.9.24 [108,109] with e-value <1 × 10−22 and percentage identity as >25%. The EukProt database comprises annotated proteins from 993 species across eukaryotes, out of which 118 species belong to Chloroplastida; the remaining 875 belong to Glaucophyta, Rhodophyta, TSAR, Cryptista, Haptista, Amorphea, Metamonada, Discoba, CRuMs, Hemimastigophora, Malawimonadida and Ancyromonadida.
(d). Domain prediction
Protein domains were predicted for 37 proteomes using InterProScan 5 [13] using all databases (CDD + COILS + Gene3D + HAMAP + MOBIDB + PANTHER + Pfam + PIRSF + PRINTS + PROSITE + SFLD + SMART + SUPERFAMILY + NCBIFAM; parameters: default pre-calculated match lookup service using MD5 checksum, n = 8 longest ORFs taken as input for analysis). Protein domains predicted totalled 14 367, out of which 5403 were found in the 2475 stress-relevant orthogroups. To compute protein domain frequencies, the intervals of occurrences were merged given that the domain is predicted more than once on overlapping intervals. The count and domain order of proteins are finalized based on merged intervals and the total frequency of protein domain occurrence in all species and orthogroups is computed.
For fold changes in protein domains predicted in the 2475 stress-annotated orthogroups between sister lineages, the average count of a given domain in the LCA of a lineage was divided by the average count in its sister lineage. The top 10-fold changes of protein domains between adjacent LCAs (nodes) are reported in figure 2b.
(e). Embryophytic domain set and latent genetic potential
We define ‘embryophytic domain set’ as the domain configuration in a given orthogroup that originated in the land plant LCA, i.e. each of the domains in the set is predicted in at least one species of: (i) Tracheophyta and (ii) Bryophyta. For a given species, we define domain completion when all the domains in the embryophytic domain set are predicted for this species. We calculated (x/y) of orthogroups at each node (a–f). For example, the numbers at node b indicate the count of orthogroups in Tracheophyta + Bryophyta + Zygnemaotphyceae that have LGP (or key Embryophytic protein domains) in all the rest of the lineages (Charophyceae + Klebsormidiophyceae + Chlorokybophyceae + Mesostigmatophyceae + Chlorophytes). We define LGP as the case when the domains that make up a given protein were individually present at an earlier evolutionary time; thus, the required domains emerged earlier than the orthogroup. The origin of orthogroups is assessed parsimoniously assuming common ancestry of all the represented taxa.
(f). Data visualization
The heatmap plot was created using gplots (https://cran.r-project.org/web/packages/gplots), curl (https://cran.r-project.org/web/packages/curl), dendextend [110], colorspace (https://cran.r-project.org/web/packages/colorspace/index.html) and RColorBrewer (https://cran.r-project.org/web/packages/RColorBrewer). We used ggplot2 [111], viridis [112] and reshape2 [113] for bubble plots. For the respective R packages, R v. 4.0.3 [114] was used. Upset plots were created using pandas, matplotlib.pyplot and upsetplot packages in Python v. 3.9.5. The wordle was created using wordle.net.
Contributor Information
Amra Dhabalia Ashok, Email: amradhabalia.ashok@uni-goettingen.de.
Sophie de Vries, Email: sophie.devries@uni-goettingen.de.
Tatyana Darienko, Email: tdarien@gwdg.de.
Iker Irisarri, Email: iker.irisarriaedo@uni-goettingen.de.
Jan de Vries, Email: devries.jan@uni-goettingen.de.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
Code is available online [115]. All computational analyses were performed with published tools and are cited in §3. All intermediate files were deposited on Dryad [116].
Supplementary material is available online [117].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors’ contributions
A.D.A.: data curation, formal analysis, investigation, methodology, software, visualization, writing—original draft, writing —review and editing; S.d.V.: conceptualization, formal analysis, supervision, writing—original draft, writing—review and editing; T.D.: resources, visualization; I.I.: conceptualization, resources, software, supervision, writing—original draft, writing—review and editing; J.d.V.: conceptualization, funding acquisition, investigation, project administration, supervision, visualization, writing—original draft, writing— review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
J.d.V. thanks the European Research Council for funding under the European Union’s Horizon 2020 research and innovation programme (Grant Agreement No. 852725; ERC-StG “TerreStriAL”) the DFG (grant 440231723, VR 132/4-1, SPP2237 “MAdLand”). A.D.A. thanks the IMPRS Genome Science.
References
- 1. Strassert JFH, Irisarri I, Williams TA, Burki F. 2021. A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids. Nat. Commun. 12, 1879. ( 10.1038/s41467-021-22044-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffmann L. 1989. Algae of terrestrial habitats. Bot. Rev. 55, 77–105. ( 10.1007/BF02858529) [DOI] [Google Scholar]
- 3. Terlova EF, Holzinger A, Lewis LA. 2021. Terrestrial green algae show higher tolerance to dehydration than do their aquatic sister-species. Microb. Ecol. 82, 770–782. ( 10.1007/s00248-020-01679-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCourt RM, Lewis LA, Strother PK, Delwiche CF, Wickett NJ, de Vries J, Bowman JL. 2023. Green land: Multiple perspectives on green algal evolution and the earliest land plants. Am. J. Bot. 110, e16175. ( 10.1002/ajb2.16175) [DOI] [PubMed] [Google Scholar]
- 5. Bierenbroodspot M, Pröschold T, Fürst-Jansen JMR, de Vries S, Irisarri I, Darienko T, de Vries J. 2024. Phylogeny and evolution of streptophyte algae. Ann. Bot. ( 10.1093/aob/mcae091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Becker B, Marin B. 2009. Streptophyte algae and the origin of embryophytes. Ann. Bot. 103, 999–1004. ( 10.1093/aob/mcp044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fürst-Jansen JMR, de Vries S, de Vries J. 2020. Evo-physio: on stress responses and the earliest land plants. J. Exp. Bot. 71, 3254–3269. ( 10.1093/jxb/eraa007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donoghue PCJ, Harrison CJ, Paps J, Schneider H. 2021. The evolutionary emergence of land plants. Curr. Biol. 31, R1281–R1298. ( 10.1016/j.cub.2021.07.038) [DOI] [PubMed] [Google Scholar]
- 9. Rensing SA. 2016. (Why) does evolution favour embryogenesis? Trends Plant Sci. 21, 0562–573. ( 10.1016/j.tplants.2016.02.004) [DOI] [PubMed] [Google Scholar]
- 10. Bowman JL, Sakakibara K, Furumizu C, Dierschke T. 2016. Evolution in the cycles of life. Annu. Rev. Genet. 50, 133–154. ( 10.1146/annurev-genet-120215-035227) [DOI] [PubMed] [Google Scholar]
- 11. Rich MK, et al. 2021. Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science 372, 864–868. ( 10.1126/science.abg0929) [DOI] [PubMed] [Google Scholar]
- 12. Delaux PM, Schornack S. 2021. Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 371, eaba6605. ( 10.1126/science.aba6605) [DOI] [PubMed] [Google Scholar]
- 13. Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. ( 10.1093/bioinformatics/btu031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamesch P, et al. 2012. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–10. ( 10.1093/nar/gkr1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandez-Pozo N, et al. 2020. PEATmoss (Physcomitrella expression Atlas tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J. 102, 165–177. ( 10.1111/tpj.14607) [DOI] [PubMed] [Google Scholar]
- 16. Huerta-Cepas J, et al. 2019. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. ( 10.1093/nar/gky1085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wickett NJ, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA 111, E4859–E4868. ( 10.1073/pnas.1323926111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Puttick MN, et al. 2018. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr. Biol. 28, 733–745.( 10.1016/j.cub.2018.01.063) [DOI] [PubMed] [Google Scholar]
- 19. One Thousand Plant Transcriptomes Initiative. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. ( 10.1038/s41586-019-1693-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Vries J, Archibald JM. 2018. Plant evolution: landmarks on the path to terrestrial life. New Phytol. 217, 1428–1434. ( 10.1111/nph.14975) [DOI] [PubMed] [Google Scholar]
- 21. Delaux PM, et al. 2012. Origin of strigolactones in the green lineage. New Phytol. 195, 857–871. ( 10.1111/j.1469-8137.2012.04209.x) [DOI] [PubMed] [Google Scholar]
- 22. Hori K, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978. ( 10.1038/ncomms4978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ju C, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C. 2015. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 1, 14004. ( 10.1038/nplants.2014.4) [DOI] [PubMed] [Google Scholar]
- 24. Van de Poel B, Cooper ED, Van Der Straeten D, Chang C, Delwiche CF. 2016. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 172, 533–545. ( 10.1104/pp.16.00299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohtaka K, Hori K, Kanno Y, Seo M, Ohta H. 2017. Primitive auxin response without Tir1 and Aux/IAA in the Charophyte alga Klebsormidium nitens Plant Physiol. 174, 1621–1632. ( 10.1104/pp.17.00274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng X, et al. 2024. Genomes of multicellular algal sisters to land plants illuminate signaling network evolution. Nat. Genet. 56, 1018–1031. ( 10.1038/s41588-024-01737-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dadras A, et al. 2023. Environmental gradients reveal stress hubs predating plant terrestrialization. Nat. Plants 9, 1419–1438. ( 10.1038/s41477-023-01491-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Vries J, Curtis BA, Gould SB, Archibald JM. 2018. Embryophyte stress signaling evolved in the algal progenitors of land plants. Proc. Natl Acad. Sci. USA 115, E3471–E3480. ( 10.1073/pnas.1719230115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng S, et al. 2019. Genomes of subaerial zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067.( 10.1016/j.cell.2019.10.019) [DOI] [PubMed] [Google Scholar]
- 30. Sun Y, et al. 2019. A ligand-independent origin of abscisic acid perception. Proc. Natl Acad. Sci. 116, 24892–24899. ( 10.1073/pnas.1914480116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153. ( 10.1038/nature02085) [DOI] [PubMed] [Google Scholar]
- 32. Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32. ( 10.1105/tpc.114.134874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carrillo-Carrasco VP, Hernandez-Garcia J, Mutte SK, Weijers D. 2023. The birth of a giant: evolutionary insights into the origin of auxin responses in plants. EMBO J. 42, e113018. ( 10.15252/embj.2022113018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D. 2018. Origin and evolution of the nuclear auxin response system. Elife 7, e33399. ( 10.7554/eLife.33399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flores-Sandoval E, Romani F, Bowman JL. 2018. Co-expression and Transcriptome analysis of Marchantia polymorpha transcription factors supports class C ARFs as independent actors of an ancient auxin regulatory module. Front. Plant Sci. 9, 1345. ( 10.3389/fpls.2018.01345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin-Arevalillo R, Thévenon E, Jégu F, Vinos-Poyo T, Vernoux T, Parcy F, Dumas R. 2019. Evolution of the auxin response factors from charophyte ancestors. PLoS Genet. 15, e1008400. ( 10.1371/journal.pgen.1008400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuhn A, et al. 2024. RAF-like protein Kinases mediate a deeply conserved, rapid Auxin response. Cell 187, 130–148.( 10.1016/j.cell.2023.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Vries S, et al. 2021. The evolution of the phenylpropanoid pathway entailed pronounced radiations and divergences of enzyme families. Plant J. 107, 975–1002. ( 10.1111/tpj.15387) [DOI] [PubMed] [Google Scholar]
- 39. Jiao C, et al. 2020. The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell 181, 1097–1111.( 10.1016/j.cell.2020.04.019) [DOI] [PubMed] [Google Scholar]
- 40. Remias D, Schwaiger S, Aigner S, Leya T, Stuppner H, Lütz C. 2012. Characterization of an UV- and VIS-absorbing, purpurogallin-derived secondary pigment new to algae and highly abundant in Mesotaenium berggrenii (Zygnematophyceae, Chlorophyta), an extremophyte living on glaciers. FEMS Microbiol. Ecol. 79, 638–648. ( 10.1111/j.1574-6941.2011.01245.x) [DOI] [PubMed] [Google Scholar]
- 41. Busch A, Gerbracht JV, Davies K, Hoecker U, Hess S. 2024. Comparative transcriptomics Illuminates the cellular responses of an aeroterrestrial zygnematophyte to UV radiation. J. Exp. Bot. 75, 3624–3642. ( 10.1093/jxb/erae131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davies KM, et al. 2022. Evolution and function of red pigmentation in land plants. Ann. Bot. 130, 613–636. ( 10.1093/aob/mcac109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Delaux PM, et al. 2015. Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl Acad. Sci. USA 112, 13390–13395. ( 10.1073/pnas.1515426112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kodama K, et al. 2022. An ancestral function of strigolactones as symbiotic rhizosphere signals. Nat. Commun. 13, 3974. ( 10.1038/s41467-022-31708-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bowles AMC, Bechtold U, Paps J. 2020. The origin of land plants is rooted in two bursts of genomic novelty. Curr. Biol. 30, 530–536.( 10.1016/j.cub.2019.11.090) [DOI] [PubMed] [Google Scholar]
- 46. Barrera-Redondo J, Lotharukpong JS, Drost HG, Coelho SM. 2023. Uncovering gene-family founder events during major evolutionary transitions in animals, plants and fungi using genera. Genome Biol. 24, 54. ( 10.1186/s13059-023-02895-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. ( 10.1186/s13059-019-1832-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richter DJ, Berney C, Strassert JFH, Poh YP, Herman EK, Muñoz-Gómez SA, Wideman JG, Burki F, de Vargas C. 2022. EukProt:A database of genome-scale predicted proteins across the diversity of eukaryotes. Peer. Community. J. 2. ( 10.24072/pcjournal.173) [DOI] [Google Scholar]
- 49. Fujii S, Small I. 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. ( 10.1111/j.1469-8137.2011.03746.x) [DOI] [PubMed] [Google Scholar]
- 50. Small ID, Schallenberg-Rüdinger M, Takenaka M, Mireau H, Ostersetzer-Biran O. 2020. Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 101, 1040–1056. ( 10.1111/tpj.14578) [DOI] [PubMed] [Google Scholar]
- 51. Weng JK. 2014. The evolutionary paths towards complexity: a metabolic perspective. New Phytol. 201, 1141–1149. ( 10.1111/nph.12416) [DOI] [PubMed] [Google Scholar]
- 52. Rieseberg TP, Dadras A, Fürst-Jansen JMR, Dhabalia Ashok A, Darienko T, de Vries S, Irisarri I, de Vries J. 2023. Crossroads in the evolution of plant specialized metabolism. Semin. Cell Dev. Biol. 134, 37–58. ( 10.1016/j.semcdb.2022.03.004) [DOI] [PubMed] [Google Scholar]
- 53. Buschmann H. 2020. Into another dimension: how streptophyte algae gained morphological complexity. J. Exp. Bot. 71, 3279–3286. ( 10.1093/jxb/eraa181) [DOI] [PubMed] [Google Scholar]
- 54. Rensing SA. 2014. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 17, 43–48. ( 10.1016/j.pbi.2013.11.002) [DOI] [PubMed] [Google Scholar]
- 55. Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. 2009. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 149, 916–928. ( 10.1104/pp.108.131144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miyakawa T, Hatano K ichi, Miyauchi Y, Suwa Y ichi, Sawano Y, Tanokura M. 2014. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 166, 766–778. ( 10.1104/pp.114.242636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nelson D, Werck-Reichhart D. 2011. A P450-centric view of plant evolution. Plant J. 66, 194–211. ( 10.1111/j.1365-313X.2011.04529.x) [DOI] [PubMed] [Google Scholar]
- 58. Gutmann B, et al. 2020. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant 13, 215–230. ( 10.1016/j.molp.2019.11.002) [DOI] [PubMed] [Google Scholar]
- 59. Zhang Y, Xia R, Kuang H, Meyers BC. 2016. The diversification of plant NBS-LRR defense genes directs the evolution of micrornas that target them. Mol. Biol. Evol. 33, 2692–2705. ( 10.1093/molbev/msw154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Vries S, Kloesges T, Rose LE. 2015. Evolutionarily dynamic, but robust, targeting of resistance genes by the Mir482/2118 gene family in the Solanaceae. Genome Biol. Evol. 7, 3307–3321. ( 10.1093/gbe/evv225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. 2006. F-box proteins everywhere. Curr. Opin. Plant Biol. 9, 631–638. ( 10.1016/j.pbi.2006.09.003) [DOI] [PubMed] [Google Scholar]
- 62. Harris BJ, Clark JW, Schrempf D, Szöllősi GJ, Donoghue PCJ, Hetherington AM, Williams TA. 2022. Divergent evolutionary tajectories of bryophytes and tracheophytes from a complex common ancestor of land plants. Nat. Ecol. Evol. 6, 1634–1643. ( 10.1038/s41559-022-01885-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gould SJ, Vrba ES. 1982. Exaptation—a missing term in the science of form. Paleobiology 8, 4–15. ( 10.1017/S0094837300004310) [DOI] [Google Scholar]
- 64. Ku C, Nelson-Sathi S, Roettger M, Garg S, Hazkani-Covo E, Martin WF. 2015. Endosymbiotic gene transfer from prokaryotic pangenomes: inherited chimerism in eukaryotes. Proc. Natl Acad. Sci. USA 112, 10139–10146. ( 10.1073/pnas.1421385112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Monte I, Franco-Zorrilla JM, García-Casado G, Zamarreño AM, García-Mina JM, Nishihama R, Kohchi T, Solano R. 2019. A single JAZ repressor controls the Jasmonate pathway in Marchantia polymorpha. Mol. Plant 12, 185–198. ( 10.1016/j.molp.2018.12.017) [DOI] [PubMed] [Google Scholar]
- 66. Guo Q, Yoshida Y, Major IT, Wang K, Sugimoto K, Kapali G, Havko NE, Benning C, Howe GA. 2018. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl Acad. Sci. USA 115, E10768–E10777. ( 10.1073/pnas.1811828115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nishiyama T, et al. 2018. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174, 448–464.( 10.1016/j.cell.2018.06.033) [DOI] [PubMed] [Google Scholar]
- 68. Bressendorff S, et al. 2016. An innate immunity pathway in the moss Physcomitrella patens. Plant Cell 28, 1328–1342. ( 10.1105/tpc.15.00774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Delwiche CF. 2016. The genomes of charophyte green algae. In Advances in botanical research (ed. Rensing SA), pp. 255–270, vol. 78. Cambridge, MA: Academic Press. ( 10.1016/bs.abr.2016.02.002) [DOI] [Google Scholar]
- 70. Hess S, et al. 2022. A phylogenomically informed five-order system for the closest relatives of land plants. Curr. Biol. 32, 4473–4482.( 10.1016/j.cub.2022.08.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Harholt J, Moestrup Ø, Ulvskov P. 2016. Why plants were terrestrial from the beginning. Trends Plant Sci. 21, 96–101. ( 10.1016/j.tplants.2015.11.010) [DOI] [PubMed] [Google Scholar]
- 72. Bierenbroodspot MJ, Darienko T, de Vries S, Fürst-Jansen JMR, Buschmann H, Pröschold T, Irisarri I, de Vries J. 2024. Phylogenomic insights into the first multicellular streptophyte. Curr. Biol. 34, 670–681.( 10.1016/j.cub.2023.12.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Romani F, Moreno JE. 2021. Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytol. 230, 1345–1353. ( 10.1111/nph.17161) [DOI] [PubMed] [Google Scholar]
- 74. de Vries S, Stukenbrock EH, Rose LE. 2020. Rapid evolution in plant–microbe interactions – an evolutionary genomics perspective. New Phytol. 226, 1256–1262. ( 10.1111/nph.16458) [DOI] [PubMed] [Google Scholar]
- 75. Dadras A, Rieseberg TP, Zegers JMS, Fürst-Jansen JMR, Irisarri I, de Vries J, de Vries S. 2023. Accessible versatility underpins the deep evolution of plant specialized metabolism. Phytochem. Rev. ( 10.1007/s11101-023-09863-2) [DOI] [Google Scholar]
- 76. Worden AZ, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. ( 10.1126/science.1167222) [DOI] [PubMed] [Google Scholar]
- 77. Palenik B, et al. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl Acad. Sci. 104, 7705–7710. ( 10.1073/pnas.0611046104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. De Clerck O, et al. 2018. Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol. 28, 2921–2933.( 10.1016/j.cub.2018.08.015) [DOI] [PubMed] [Google Scholar]
- 79. Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. ( 10.1126/science.1143609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Blanc G, et al. 2010. The Chlorella variabilis Nc64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943–2955. ( 10.1105/tpc.110.076406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Blanc G, et al. 2012. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13, 1–12. ( 10.1186/gb-2012-13-5-r39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang S, et al. 2020. Genomes of early-diverging streptophyte algae shed light on plant Terrestrialization. Nat. Plants. 6, 95–106. ( 10.1038/s41477-019-0560-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Irisarri I, Darienko T, Pröschold T, Fürst-Jansen JMR, Jamy M, de Vries J. 2021. Unexpected cryptic species among streptophyte algae most distant to land plants. Proc. R. Soc. B 288, 20212168. ( 10.1098/rspb.2021.2168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lang D, et al. 2018. The Physcomitrella patens chromosome‐scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533. ( 10.1111/tpj.13801) [DOI] [PubMed] [Google Scholar]
- 85. Healey AL, et al. 2023. Newly identified sex chromosomes in the Sphagnum (peat moss) genome alter carbon sequestration and ecosystem dynamics. Nat. Plants. 9, 238–254. ( 10.1038/s41477-022-01333-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bowman JL, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304. ( 10.1016/j.cell.2017.09.030) [DOI] [PubMed] [Google Scholar]
- 87. Li FW, et al. 2020. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants. 6, 259–272. ( 10.1038/s41477-020-0618-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. ( 10.1126/science.1203810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hulse-Kemp AM, et al. 2021. An anchored chromosome-scale genome assembly of spinach improves annotation and reveals extensive gene rearrangements in euasterids. Plant. Genome. 14, e20101. ( 10.1002/tpg2.20101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hu TT, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481. ( 10.1038/ng.807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rawat V, Abdelsamad A, Pietzenuk B, Seymour DK, Koenig D, Weigel D, Pecinka A, Schneeberger K. 2015. Improving the annotation of Arabidopsis lyrata using RNA-Seq data. PLoS One 10, e0137391. ( 10.1371/journal.pone.0137391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–86. ( 10.1093/nar/gkr944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tang H, et al. 2014. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15, 312. ( 10.1186/1471-2164-15-312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kreplak J, et al. 2019. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 51, 1411–1422. ( 10.1038/s41588-019-0480-1) [DOI] [PubMed] [Google Scholar]
- 95. Li H, Jiang F, Wu P, Wang K, Cao Y. 2020. A high-quality genome sequence of model legume Lotus japonicus (MG-20) provides insights into the evolution of root nodule symbiosis. Genes (Basel) 11, 483. ( 10.3390/genes11050483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kawahara Y, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. ( 10.1186/1939-8433-6-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Initiative IB. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. ( 10.1038/nature08747) [DOI] [PubMed] [Google Scholar]
- 98. Project AG, et al. 2013. The Amborella genome and the evolution of flowering plants. Science 342, 1241089. ( 10.1126/science.1241089) [DOI] [PubMed] [Google Scholar]
- 99. Wan T, et al. 2018. A genome for gnetophytes and early evolution of seed plants. Nat. Plants 4, 82–89. ( 10.1038/s41477-017-0097-2) [DOI] [PubMed] [Google Scholar]
- 100. Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. ( 10.1038/nature12211) [DOI] [PubMed] [Google Scholar]
- 101. Li FW, et al. 2018. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4, 460–472. ( 10.1038/s41477-018-0188-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosyn. Res. 70, 85–106. ( 10.1023/A:1013840025518) [DOI] [PubMed] [Google Scholar]
- 103. Ran L, et al. 2010. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 5, e11486. ( 10.1371/journal.pone.0011486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dagan T, et al. 2013. Genomes of Stigonematalean Cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44. ( 10.1093/gbe/evs117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ponce-Toledo RI, Deschamps P, López-García P, Zivanovic Y, Benzerara K, Moreira D. 2017. An early-branching freshwater Cyanobacterium at the origin of Plastids. Curr. Biol. 27, 386–391. ( 10.1016/j.cub.2016.11.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shih PM, et al. 2013. Improving the coverage of the Cyanobacterial Phylum using diversity-driven genome sequencing. Proc. Natl Acad. Sci. USA 110, 1053–1058. ( 10.1073/pnas.1217107110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. In Gene prediction: methods and protocols (ed. Kollmar M), pp. 227–245, vol. 1962. New York, NY: Humana Press. ( 10.1007/978-1-4939-9173-0) [DOI] [PubMed] [Google Scholar]
- 108. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 12, 59–60. ( 10.1038/nmeth.3176) [DOI] [PubMed] [Google Scholar]
- 109. Buchfink B, Reuter K, Drost HG. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 18, 366–368. ( 10.1038/s41592-021-01101-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Galili T. 2015. Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31, 3718–3720. ( 10.1093/bioinformatics/btv428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wickham H. 2022. Ggplot2: elegant graphics for data analysis, 2nd ed. New York, NY: Springer. [Google Scholar]
- 112. Garnier S, et al. 2023. CRAN release V0.6.3. Zenodo. ( 10.5281/zenodo.7890878) [DOI]
- 113. Wickham H. 2007. Reshaping data with the reshape package. J. Stat. Softw. 21, 1–20. ( 10.18637/jss.v021.i12) [DOI] [Google Scholar]
- 114. R Core Team . 2021. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. See https://www.R-project.org. [Google Scholar]
- 115. Dhabalia Ashok A, de Vries J. 2024. Code and data for: Evolutionary assembly of the plant terrestrialization toolkit from protein domains. Zenodo. ( 10.5281/zenodo.11456093) [DOI] [PMC free article] [PubMed]
- 116. de Vries J, Dhabalia Ashok A, Irisarri I, de Vries S. 2024. Data from: Evolutionary assembly of the plant terrestrialization toolkit from protein domains. Dryad Digital Repository. ( 10.5061/dryad.6t1g1jx4q) [DOI] [PMC free article] [PubMed]
- 117. Dhabalia Ashok A, de Vries S, Darienko T, Irisarri I, de Vries J. 2024. Supplementary material from: Evolutionary assembly of the plant terrestrialization toolkit from protein domains. Figshare. ( 10.6084/m9.figshare.c.7370660) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Code is available online [115]. All computational analyses were performed with published tools and are cited in §3. All intermediate files were deposited on Dryad [116].
Supplementary material is available online [117].