Abstract
Metagenomics involves the study of genetic material obtained directly from communities of microorganisms living in natural environments. The field of metagenomics has provided valuable insights into the structure, diversity and ecology of microbial communities. Once an environmental sample is sequenced and processed, metagenomic binning clusters the sequences into bins representing different taxonomic groups such as species, genera, or higher levels. Several computational tools have been developed to automate the process of metagenomic binning. These tools have enabled the recovery of novel draft genomes of microorganisms allowing us to study their behaviors and functions within microbial communities. This review classifies and analyzes different approaches of metagenomic binning and different refinement, visualization, and evaluation techniques used by these methods. Furthermore, the review highlights the current challenges and areas of improvement present within the field of research.
Keywords: bioinformatics, metagenomics, microorganisms, metagenomic binning
Introduction
Metagenomics is the study of genetic content directly obtained from microbial communities found in various environments [1–4] such as soil, seawater, air, and niches of the human body including the respiratory and gastrointestinal tracts. Samples directly obtained from these environments are processed to isolate the genetic material, which is then sequenced to obtain the nucleotide information. This nucleotide information is analyzed to characterize the different microorganisms present in the microbial community and understand their behaviors and functions. With the advent of high-throughput sequencing approaches, metagenomics has enabled the access and study of genomes from entire microbial communities [2, 5, 6], providing valuable insights into their composition and interactions.
A typical metagenomic analysis pipeline starts by obtaining DNA sequences called reads from a metagenomic sample. Next-Generation Sequencing (NGS) technologies (e.g. Illumina [7] and MGI [8]) generate short reads typically ranging from 50 to 300 base pairs (bp) and are widely used for metagenomic studies. Third Generation Sequencing (TGS) technologies [9] (e.g. Pacific Biosciences and Oxford Nanopore Technologies) that produce long reads typically ranging from 10 kbp to over 1 Mbp recently have gained popularity as well. Then, a process known as assembly is used to connect the sequenced reads to reconstruct the original genome from which the DNA originated [10–12]. This task is computationally challenging because essentially every read must be compared to every other read, and the number of comparisons increases with the square of the number of reads. Sequence assemblers produce longer sequences called contigs which can further be connected to form scaffolds. Metagenomic data is both noisy and redundant; variation in the sequences may arise from sequencing errors or different species in the original samples with different, but highly related sequences. Specialized assemblers known as metagenome assemblers [13] employ heuristics so that they can assemble metagenomic data in a reasonable time (e.g. MEGAHIT [14], metaSPAdes [15] for NGS data, and metaFlye [16] for TGS data). However, they do not always produce complete/near-complete genomes due to the complex composition of metagenomes and the complexity of the calculations. Hence, metagenomic binning and refinement methods are employed to recover draft genomes known as metagenome-assembled genomes (MAGs). In most binning methods, taxonomy-independent, unsupervised techniques are used to place metagenomic sequences into imaginary bins that represent different taxonomic groups such as species, genera, or higher levels [17]. Metagenomic binning solutions have advanced microbial ecology by providing information about community structures, improved human health through pathogen identification and gut microbiome analysis, and progressed biotechnology by enabling the discovery of novel enzymes and metabolic pathways [18–20].
Only a limited number of reviews have been conducted on the computational methods used in taxonomy-independent metagenomic binning [13, 17, 21]. Moreover, new binning methods incorporating new features and techniques have emerged recently. Hence, this review aims to deliver a comprehensive overview of the different types of metagenomic binning methods (Fig. 1), discuss their challenges, study new trends and highlight the areas that require improvement. It must be noted that this review does not focus on benchmarking binning methods, as detailed benchmarking studies on various datasets have already been published [22–26]. This review is a comprehensive starting point for beginners entering the field of computational metagenomics and will pave the way for improvement in the research field.
Figure 1.
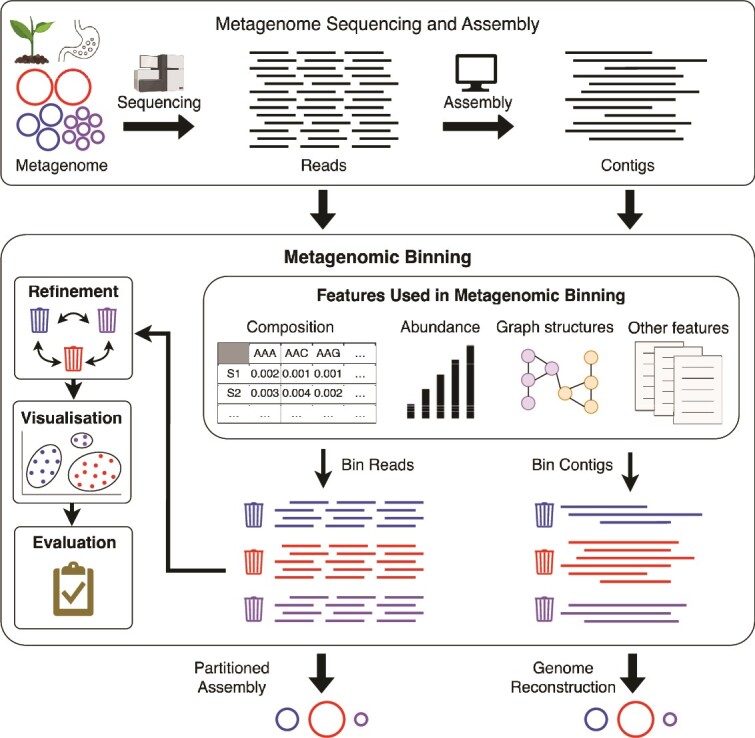
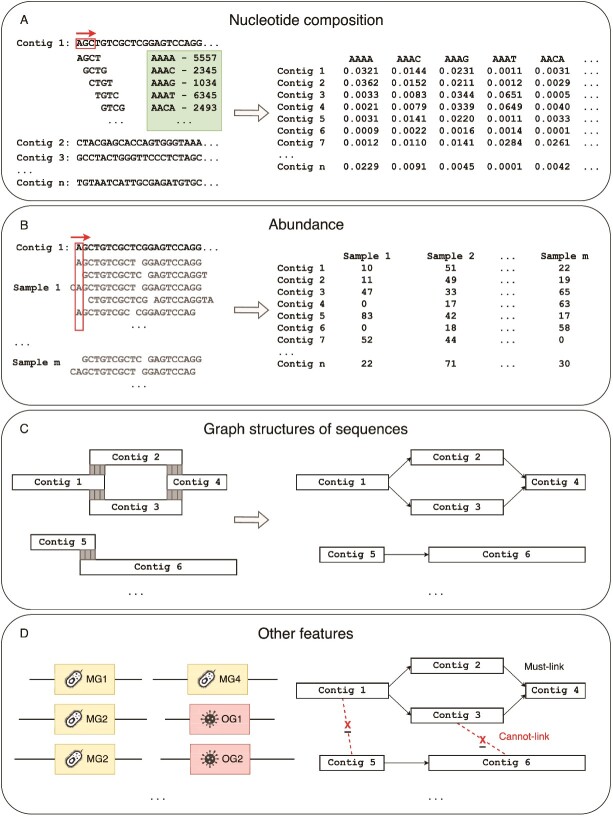
Aspects of metagenomic binning. After obtaining sequences from a metagenome, reads can either be directly binned for partitioned assembly or first assembled into contigs and then binned. Features such as composition, abundance and graph structures of sequences are used during binning. Obtained bins can be refined, visualized, and evaluated prior to obtaining the final set of metagenome-assembled genomes (MAGs).
Features used in metagenomic binning
Metagenomic binning is a clustering problem. To cluster sequences, we must obtain a set of features that characterize the sequences. This section presents the main features used in metagenomic binning; the classic features include (i) nucleotide composition, (ii) abundance, and, more recently, (iii) graph structures of sequences and other biological information (e.g. special genes and constraint information) (Fig. 2). We also present available tools and summarize their details obtained through their relevant publications, published software repositories, and source code.
Figure 2.
Features used in metagenomic binning. The main features include (A) nucleotide composition, (B) abundance, (C) graph structures of sequences, and (D) other features such as special genes and constraint information.
Nucleotide composition
The frequencies of oligonucleotides in genomic sequences (referred to as k-mers) carry taxonomy-specific signals [27, 28]. Composition-based methods have been developed based on the assumption that each taxonomic group has a unique nucleotide composition, and thus their sequences can be separated into bins by comparing the nucleotide content [17], such as the guanine-cytosine (GC) content [29] and normalized frequencies of oligonucleotides [30].
Normalized frequencies of oligonucleotides, especially tetramers or 4-mers [29–36], have become the most popular method to represent the nucleotide composition of sequences in a numerical manner that can be used in computations. Other oligonucleotides such as pentamers [37–40] and hexamers [41] have been used as well. We start by determining all the possible substrings of a given length k (which is why they are known as k-mers) for a given sequence. This is done using a sliding window of size k across the sequence so there are 4k possible k-mers. Next, we count how many times each k-mer appears in the sequence and obtain a k-mer frequency vector with the lexicographic ordering of the k-mers. As DNA is double-stranded and sequencers can sample fragments from both strands, we often combine the counts of each k-mer with its reverse complement, thus reducing the dimension of our k-mer frequency vector to 4k/2 if k is odd or (4k + 4k/2)/2 if k is even [32, 42]. Finally, all the k-mer counts are normalized by the total number of k-mers found in the sequence, resulting in a normalized k-mer frequency vector (Fig. 2A). This normalization assumes that all k-mers are equally likely, but the likelihood of each k-mer depends on the nucleotide composition of the genome. Therefore, other means of representing k-mers include (i) likelihood representations [37, 43] that model the probability of observing a sequence given the k-mer distribution of the genome from which the sequence originates and (ii) Chaos Game Representation (CGR) [44] that provides a visual method for representing one-dimensional sequences as distinctive graphical images enabling pattern identification.
Once the nucleotide composition is obtained, various statistical methods and machine learning-based approaches can be used to cluster the sequences into bins. Table 1 summarizes some of the notable metagenomic binning tools that use composition features.
Table 1.
Comparison of metagenomic binning tools that use composition features
| Binning tool | Year | User interface * | Programming languages used | Sequences binned | Minimum length cut-off (bp) | Main features, techniques, models or algorithms used |
|---|---|---|---|---|---|---|
| TETRA | 2004 | Web GUI | N.D.† | Fragments | 1000 | 4-mers and Maximal-order Markov models |
| CompostBin | 2008 | CLI | C and Matlab | Short reads | 1000 | 6-mers, weighted Principal Component Analysis and Normalized Cut clustering algorithm |
| LikelyBin | 2009 | CLI | Perl and C | Short reads | 400 | 1-mers to 5-mers and Markov chain Monte Carlo (MCMC) methods |
| MetaCluster 2.0 | 2010 | CLI | C | Fragments | 300 | 4-mers and k-means clustering based on Spearman footrule distance |
| SCIMM | 2010 | CLI | Python, C and Matlab | Reads and contigs | 400 | Interpolated Markov models |
| MetaCluster 3.0 | 2011 | CLI | C | Fragments | 500 | 4-mers and k-median clustering based on Spearman distance |
| 2Tbinning | 2012 | N.D. | N.D. | Contigs | N.D. | 4-mers and Gaussian mixture models |
| BiMeta | 2015 | N.D. | N.D. | Short reads | N.D. | 4-mers and k-means clustering |
| VizBin | 2015 | Desktop GUI | Java | Contigs and long reads |
1000 | 5-mers, Barnes-Hut stochastic neighbor embedding and manual clustering |
| MetaProb | 2016 | CLI | C++ | Short reads | N.D. | 4-mers and k-means clustering |
| BusyBee Web | 2017 | Web GUI | N.D. | Contigs and long reads | 500 | 5-mers and bootstrapped supervised binning |
| MetaProb 2 | 2021 | CLI | Python and C++ | Short reads | N.D. | 4-mers and Louvain community detection algorithm |
*CLI, Command line interface; GUI, Graphical user interface.
†N.D., Not defined, no details provided or not available publicly.
Abundance
Methods based on nucleotide composition encounter difficulties when binning sequences from genomes with high genomic similarity [45, 46] and species with low abundance [21]. However, each component of a genome should be present in the sample in the same proportion. Thus, by estimating the abundance of sequences (contigs or reads), we can identify sequences originating from the same chromosome because they should have the same abundance in each sample. These sequences should also belong to the same organism because they should have the same proportion in each sample. Hence, abundance-based binning methods were introduced to overcome the challenges of composition-based methods. These methods have shown improved results for sequences of closely related organisms (e.g. strains of the same species) that have similar composition profiles [21].
Abundance calculations can vary based on the type of sequences binned. The abundance of contigs is generally represented by the coverage which is the average number of reads that align to each base of the contig. This approximates how many copies of that genome are present in the sample. Contig coverage can be calculated by mapping all the reads to the assembled contigs and counting how many reads map to each base of the contig (Fig. 2B). If the contigs are obtained from a cross-assembly of multiple samples (i.e. reads from multiple samples are combined and assembled) or individual assemblies of multiple samples, the reads from individual samples can be mapped to the contigs. Then the coverage for each contig in each sample can be calculated, resulting in a coverage vector for each contig. Recent studies have shown improved binning results of such multi-coverage binning approaches [47].
The abundance of a read in a given dataset involves determining how many overlapping reads can be found within the set of all reads obtained from the given metagenomic dataset. This can be calculated by performing all-vs-all pairwise alignments of reads. We can also use k-mers from reads to calculate the k-mer coverage [48]. The k-mer coverage is defined as the average number of reads covering that k-mer in a genome. The k-mer coverage can be calculated by counting the number of times k-mers are found in the set of reads. The k-mer coverage values for all k-mers in a read can be obtained to form a feature vector or a k-mer coverage histogram [48–51] representing the read.
Abundance-based binning methods can be subdivided into methods that bin (i) a single sample (e.g. AbundanceBin [52] and MBBC [53]) and (ii) multiple samples (e.g. Canopy [54]). Methods using a single sample assume that sequencing follows the Lander-Waterman model [55], meaning that the number of times a base is sequenced follows a Poisson distribution. Methods using multiple samples assume that abundance profiles of sequences vary with changes in the abundance of the underlying organisms across samples or differential abundance [17, 21, 56].
Once the abundance profiles are extracted, various statistical methods and machine learning-based approaches can be used to cluster the sequences into bins. Table 2 summarizes some of the popular metagenomic binning tools that use abundance features.
Table 2.
Comparison of metagenomic binning tools that use abundance features
| Binning tool | Year | User interface * | Programming languages used | Sequences binned | Number of samples | Minimum length cut-off (bp) | Main techniques, models or algorithms used |
|---|---|---|---|---|---|---|---|
| AbundanceBin | 2011 | CLI | C++ | Reads | One | 75 | Expectation- Maximization algorithm |
| Canopy | 2014 | CLI | C++ | Genes | Multiple | N.D.† | Agglomerative clustering based on statistical measures |
| MBBC | 2015 | Desktop GUI and CLI | Java | Reads | One | 75 | Expectation- Maximization algorithm and Markov chains |
*CLI, Command line interface; GUI, Graphical user interface.
†N.D., Not defined, no details provided or not available publicly.
Composition and abundance combined
Composition and abundance-based methods (or hybrid methods) make use of both the variation of oligonucleotide frequencies and coverage information. Once the nucleotide composition and abundance features are calculated for each sequence, they can be either combined (e.g. form a concatenated feature vector for each sequence [19, 57]) to perform clustering or can be used hierarchically (e.g. first cluster using composition features and then using abundance features [48]). These methods have often outperformed composition-based methods and abundance-based methods. Hence, hybrid binning tools have become the preferred choice for binning metagenomic datasets at present.
Once both the composition and abundance features are obtained, techniques such as principal component analysis (PCA), probabilistic models, expectation maximization algorithms and, more recently, machine and deep learning models have been used to develop these hybrid binning tools. Table 3 summarizes some of the popular hybrid metagenomic binning tools.
Table 3.
Comparison of hybrid metagenomic binning tools
| Binning tool | Year | User interface * | Programming languages used | Sequences binned |
Minimum length cut-off (bp) |
Main techniques, models or algorithms used |
|---|---|---|---|---|---|---|
| MetaWatt | 2012 | Desktop GUI and CLI | Java | Contigs | 300 | Interpolated Markov models |
| MetaCluster 4.0 | 2012 | CLI | C | Short reads | 75 | K-median clustering based on Spearman distance |
| MetaCluster 5.0 | 2012 | CLI | C | Short reads | 75 | K-means clustering based on the Spearman distance |
| GroopM | 2014 | CLI | Python | Contigs | 1000 | Heat maps and Gaussian blur filters |
| MaxBin | 2014 | CLI | C++ and Perl | Contigs | 1000 | Expectation maximization algorithm |
| CONCOCT | 2014 | CLI | Python, C and Perl | Contigs | 1000 | Gaussian mixture model fit with a variational Bayesian approximation |
| MetaBAT | 2014 | CLI | C++ | Contigs | 2500 | Modified k-medoid clustering algorithm |
| ABAWACA | 2015 | CLI | C++ | Scaffolds | 5000 | Iterative splitting |
| MaxBin 2.0 | 2016 | CLI | C++ and Perl | Contigs | 1000 | Expectation maximization algorithm |
| MyCC | 2016 | CLI | Python | Contigs | 1000 | Affinity propagation |
| COCACOLA | 2016 | CLI | Matlab | Contigs | 1000 | K-means clustering with L1 distance |
| BinSanity | 2017 | CLI | Python | Contigs | 2500 | Affinity propagation |
| CoMet | 2017 | CLI | R | Contigs | 1000 | DBSCAN clustering |
| BMC3C | 2018 | CLI | N.D.† | Contigs | 1000 | Ensemble k-means and graph partitioning using normalized cut |
| GATTACA | 2018 | CLI | Python and C++ | Contigs | 1000 | Gaussian mixture model with a Dirichlet prior |
| SolidBin | 2019 | CLI | Python | Contigs | 1000 | Constraint-based spectral clustering with normalized cut |
| MetaBAT 2 | 2019 | CLI | C++ | Contigs | 1500 | Graph partitioning using a modified label propagation algorithm |
| MetaBCC-LR | 2020 | CLI | Python and C++ | Long reads | 1000 | Probabilistic sampling and Barnes-Hut t-distributed stochastic neighbor embedding (BH-tSNE) |
| VAMB | 2021 | CLI | Python | Contigs | 100 | Variational autoencoders and iterative medoid clustering |
| LRBinner | 2022 | CLI | Python and C++ | Long reads | 1000 | Variational autoencoders and histogram-based clustering |
| CH-Bin | 2022 | CLI | Python | Contigs | 1000 | Convex hull-based clustering |
| SemiBin | 2022 | CLI | Python | Contigs | 2500 | Siamese neural network and Infomap clustering |
| MetaDecoder | 2022 | CLI | Python | Contigs | 2500 | Dirichlet process Gaussian mixture model |
| binny | 2022 | CLI | Python and Perl | Contigs | 500 | Fast Fourier Transform-accelerated Interpolation-based t-distributed Stochastic Neighbor Embedding |
| CLMB | 2022 | CLI | Python | Contigs | N.D.† | Variational autoencoders and iterative medoid clustering |
| SemiBin2 | 2023 | CLI | Python | Contigs and long reads | 2500 | Siamese neural network, Infomap clustering (contigs), and an ensemble clustering method based on DBSCAN (long reads) |
| AAMB | 2023 | CLI | Python | Contigs | 2000 | Adversarial autoencoders |
| COMEBin | 2024 | CLI | Python | Contigs | 1000 | Feed-forward neural networks and Leiden-based clustering |
*CLI, Command line interface; GUI, Graphical user interface.
†N.D., Not defined, no details provided or not available publicly.
As shown in Table 3, most of the tools are designed for contigs and developed as command line tools. Moreover, most tools have a contig cut-off length of 1000 bp that discards short sequences. Furthermore, these tools use various traditional clustering methods (e.g. k-means, k-medoids and DBSCAN) [57–65], traditional machine learning techniques (label propagation and Gaussian mixture models) [19, 66–68] and deep learning techniques (variational autoencoders [69, 70], Siamese neural networks [71, 72]. adversarial autoencoders [73] and feed-forward neural networks [74]) to bin sequences.
The number of unique k-mers increases exponentially with the increasing size of k. Hence, most binning tools only consider one k value to represent the nucleotide information. The most popular k-mer size is k = 4 (tetranucleotides), as it results in a reasonably long feature vector with 136 dimensions after combining reverse complements. Some tools use trinucleotides (e.g. MetaBCC-LR [48] and LRBinner [50, 51]) that will further reduce the vector space to 32 dimensions. However, some tools use the nucleotide composition from a combination of small k-mer sizes (e.g. ABAWACA [75] uses k = 1,2,3 and binny uses k = 2,3,4). Furthermore, tools such as CH-Bin [76] implement high-dimensional clustering techniques that use combinations of slightly larger k-mer sizes (k = 4,5,6 by default) for binning, thus increasing the resolution of composition patterns.
Early binning tools used nucleotide composition and coverage to perform stepwise binning. For example, GroopM [77] mainly uses differential coverage patterns across multiple samples to bin contigs. The bins are refined by splitting and merging operations based on the nucleotide composition and marker gene information. However, recent tools have combined both the coverage and nucleotide composition features either into one single concatenated vector for each sequence (e.g. CONCOCT [19] and SolidBin [63]) or into one probabilistic or distance metric (e.g. MaxBin [78], MaxBin 2.0 [79], MetaBAT [60], and MetaBAT 2 [80]). Normalization techniques such as normalizing over sequence lengths, normalizing over samples and z-scaling are used when combining the coverage and nucleotide composition features. Also, the concatenated vectors can have a large number of dimensions and can affect the efficiency of the downstream clustering steps. Hence, dimension reduction techniques such as PCA and Barnes-Hut t-distributed stochastic neighbor embedding (BH-tSNE) are employed to obtain a low-dimensional representation of the feature space [48, 81]. These techniques have markedly enhanced the speed and performance of metagenomic binning tools, allowing them to handle large-scale metagenomic datasets.
Graph structures of sequences
Until the late 2010s, metagenomic binning methods mainly relied on the nucleotide composition and abundance features to bin sequences. In most binning tools, sequences are represented as feature vectors and binning is done based on distance or probability calculations. These tools treat sequences as individual data points rather than considering that some sequences may originate from consecutive genomic regions.
A graph is a data structure consisting of a set of vertices or nodes and a set of connections between vertices known as edges [82]. In some graphs, the edges can have weights that represent the strength of the connection. Graphs can represent complex relationships and neighborhood information among their nodes that may not be captured in the Euclidean or probabilistic space. Hence, metagenomic binning has shifted towards using graph structures to represent sequences in binning.
Tools such as MetaBAT 2 [80] build a graph using feature similarity between contigs. However, these graphs lack the location, orientation and connectivity information of contigs within the constituent genomes. Graph structures containing contigs and their orientation information were already available from metagenomic assemblers in the form of assembly graphs and metagenomic studies have been using assembly graphs for the manual curation of contigs [83]. Subsequently, assembly graphs [84, 85] and other graph structures, such as Hi-C contact maps (e.g. bin3C [86] and HiCBin [87]) and read overlap graphs (e.g. OBLR [88]) were introduced in automated binning tools (Fig. 2C). Table 4 presents a comparison of metagenomic binning tools that use special graph structures.
Table 4.
Comparison of metagenomic binning tools that use special graph structures
| Binning tool | Year | User interface * | Programming languages used | Graph structure used | Main techniques, models or algorithms used |
|---|---|---|---|---|---|
| bin3C | 2019 | CLI | Python | Hi-C contact maps | Infomap clustering algorithm |
| MetaCoAG | 2022 | CLI | Python | Assembly graph | Graph matching and label propagation |
| RepBin | 2022 | CLI | Python | Assembly graph | Graph convolutional networks |
| OBLR | 2022 | CLI | Python and C++ | Read overlap graph | Graph Sample and Aggregate (GraphSAGE) |
| GraphMB | 2022 | CLI | Python | Assembly graph | Variational autoencoders and Graph Neural Networks |
| HiCBin | 2022 | CLI | Python | Hi-C contact maps | Leiden clustering algorithm |
| CCVAE | 2022 | CLI | Python | Assembly graph | Connectivity-constrained variational autoencoders |
| UnitigBIN | 2024 | CLI | Python | Assembly graph | Variational autoencoders and graph convolutional networks |
| hmBin | 2024 | CLI | Python | Assembly graph | t-distributed stochastic neighbor embedding (tSNE) and a distance-based clustering algorithm |
*CLI, Command line interface.
Recently, assembly graphs have become the most common graph structure used to represent contigs in metagenomic binning, as they are readily available from the assembler output. Assemblers start by identifying overlaps between sequences (it can be reads or k-mers depending on the assembly paradigm [12]) and form a graph structure where sequences are vertices and overlaps are edges [89, 90]. After performing several simplification steps we get the final assembly graph where vertices represent contigs and edges represent overlaps between the contigs [15]. Normally these overlaps represent prefix-suffix overlaps (i.e. the suffix of the first contig overlaps the prefix of the second contig) denoting that the two contigs are placed one after the other along their genome with relevant orientation information (Fig. 2C). With the introduction of the assembly graph as a feature in automated metagenomic bin refinement [84], many stand-alone metagenomic binning tools that incorporate the connectivity information of assembly graphs (e.g. MetaCoAG [45, 46], RepBin [91], GraphMB [92], CCVAE [93], UnitigBIN [94], and hmBin [95]) have been developed (Table 4). Moreover, read binning tools such as OBLR [88] employ read overlap graphs that hold neighborhood information of overlapping reads. The use of the read overlap graph has greatly increased the accuracy of binning yet requires efficient means to handle large overlap graphs with millions of reads.
Other features
In addition to the primary binning categories mentioned above, some tools have used other features to perform metagenomic binning and improve the binning results (Fig. 2D). BMC3C [62] utilizes codon usage in addition to the composition and coverage information. COCACOLA [57] considers linkage information from paired-end reads to improve the binning process, although that information is also included in some of the assembly graphs. mBin [96] and nanodisco [97] use bacterial DNA methylation profiles to accurately map mobile genetic elements to their corresponding host bacterial bins.
Another common feature used to assist binning is single-copy marker genes (Fig. 2D). Single-copy marker genes are special genes found in the majority of bacterial genomes and they appear only once in each genome [78, 98, 99]. Hence, some binning tools have utilized single-copy marker genes to estimate the number of bins during binning and to refine the binning results. The BV-BRC metagenomic binning algorithm [100], MaxBin [78], MaxBin 2.0 [79], MetaCoAG [45], and SingleM [101] use single-copy marker genes to estimate the number of bins for the initialization of the binning process. Tools such as GroopM [77] and MyCC [64] use single-copy marker genes to refine the final binning results.
Two common constraints, must-link and cannot-link are frequently used to determine whether a pair of contigs should be placed in the same bin or different bins (Fig. 2D). Some tools employ taxonomic annotations to determine must-link and cannot-link constraints. For example, SolidBin [63] aligns contigs to reference genomes and generates must-link constraints if contigs align to the same species and cannot-link constraints if contigs align to different genera. Taxonomic annotations can be obtained using public databases such as the National Center for Biotechnology Information (NCBI) databases [102] or the Genome Taxonomy Database (GTDB) [103].
As viruses do not encode single-copy marker genes, binning tools specifically designed for viruses incorporate virus-specific information for binning; VRhyme [104] uses protein redundancy scores, CoCoNet [105] is trained on the NCBI RefSeq viral database [106], PHAMB [107] uses viral orthologous groups and ViralCC [108] uses virus-host proximity structure. The use of virus-specific information has enabled viral binning tools to recover viral metagenome-assembled genomes (vMAGs) from metagenomic data.
Ensemble binners
Ensemble binners combine the results from multiple metagenomic binning approaches to optimize and improve the accuracy of genome binning results (e.g. DAS Tool [109], MetaWRAP [110], MetaBinner [111], and BASALT [112]). These tools use different metrics and additional information such as single-copy marker genes to determine a set of non-redundant bins from multiple binning results. Table 5 summarizes some of the popular metagenomic ensemble binning tools.
Table 5.
Comparison of metagenomic ensemble binning tools
| Binning tool | Year | User interface * | Programming languages used | Features used | Main techniques, models or algorithms used |
|---|---|---|---|---|---|
| DAS Tool | 2018 | CLI | R | Single-copy genes | Iterative selection |
| MetaWRAP | 2018 | CLI | Python | Single-copy genes from CheckM | Read mapping and re-assembly |
| MetaBinner | 2023 | CLI | Python and Perl | Composition, coverage and Single-copy genes | k-means++ clustering and Binning_refiner |
| BASALT | 2024 | CLI | Python | Tetranucleotide frequency and coverage correlation coefficient | Feedforward neural network |
*CLI, Command line interface.
Bin refinement
Bin refinement tools accept a binning result from an existing tool and attempt to improve the quality and accuracy of the resulting genomic bins. Table 6 presents a summary of the available metagenomic bin refinement tools.
Table 6.
Comparison of metagenomic bin refinement tools
| Bin refinement tool | Year | User interface * | Programming languages used | Features used | Main techniques, models or algorithms used |
|---|---|---|---|---|---|
| Binning_refiner | 2017 | CLI | Python and R | Sequence similarity | Pairwise nucleotide BLAST |
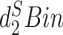
|
2017 | CLI | Python and C | Dissimilarity between contigs |
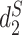 measure, Markov models and k-means clustering
measure, Markov models and k-means clustering |
| GraphBin | 2020 | CLI | Python | Assembly graph | Label propagation |
| GraphBin2 | 2020 | CLI | Python | Assembly graph and contig coverage | Iterative label propagation |
| METAMVGL | 2020 | CLI | Python | Assembly graph and paired-end graph | Multi-view label propagation |
| UGMAGrefiner | 2023 | N.D.† | N.D. | Assembly graph | Graph algorithms and Gaussian Mixture models |
*CLI, Command line interface.
†N.D., Not defined, no details provided or not available publicly.
During bin refinement, several processing steps are performed to improve bins including:
Merging bins—bins can be incomplete and fragmented and should be combined to form one bin.
Splitting bins—bins can be erroneously merged due to high similarity and should be split into the correct number of bins.
Correcting mis-binned sequences—contigs may be erroneously placed into incorrect bins and should be reassigned to their correct bins.
Recovering sequences discarded by the initial binning tool—binning tools can discard short sequences (shorter than a given minimum length cut-off) that can contain short repeat sequences. Such short sequences should be placed in their relevant bins.
Early bin-refinement tools make use of sequence similarity within bins for refinement [113, 114]. However, recent tools use the connectivity information of the assembly graph for refinement (e.g. GraphBin [84], GraphBin2 [115, 116], METAMVGL [117] and UGMAGrefiner [118]). The results produced from bin refinement tools can depend on the quality of the initial binning result. In some cases, the errors in the initial binning result can be propagated, leading to even worse results. Furthermore, most of these tools adjust contigs among bins and do not adjust the number of bins during refinement.
Bin visualization
Binning sequences can be challenging to understand with all the complex algorithms and models used. Bin visualization tools can help biologists understand how the sequences were grouped together and even identify potentially incorrect results. For example, visualizing bins can show how similar sequences were clustered, detect anomalies within bins that may indicate mis-binned sequences and determine sequences with irregular coverage patterns. Table 7 summarizes some of the tools that can visualize metagenomic binning results. Some tools are stand-alone binning tools that provide the functionality to visualize bins.
Table 7.
Comparison of metagenomic bin visualization tools
| Bin visualization tool | Year | User interface * | Programming languages used | Features used | Main techniques, models or algorithms used |
|---|---|---|---|---|---|
| MetaWatt | 2012 | Desktop GUI and CLI | Java | GC content, coverage and contig length | Contour plots |
| Blobology (now known as BlobTools) | 2013 | CLI | Perl and R | GC content and coverage | Scatter plots |
| GroopM | 2014 | CLI | Python | Differential coverage | Heat maps and Gaussian blur filters |
| VizBin | 2015 | Desktop GUI | Java | 5-mer frequency vectors | Barnes-Hut Stochastic Neighbor Embedding (BH-SNE) and scatter plots |
| gbtools | 2015 | CLI | R and Perl | GC content and coverage | Scatter plots |
| BusyBee Web | 2017 | Web GUI | N.D.† | 5-mer frequency vectors | BH-SNE and scatter plots |
*CLI, Command line interface; GUI, Graphical user interface.
†N.D., Not defined, no details provided or not available publicly.
Most of the visualization tools employ coverage and GC content to visualize bins. They generate coverage versus GC content plots of sequences using different types of plots such as scatter plots [38, 39, 119, 120], heatmaps [77] and contour plots [68]. Tools such as Blobology [119] and gbtools [120] use scatter plots where each point represents a sequence. In contrast, MetaWatt [68] uses contour plots to visualize the boundaries of sequences within bins. Moreover, only a single sample can be visualized in the coverage versus GC content plots. However, gbtools allows the user to visualize differential coverage plots. Furthermore, tools such as Blobology and gbtools can accept taxonomic annotations results or taxonomic markers and represent sequences in corresponding colors.
Bin evaluation
With the availability of metagenomic binning tools and ample computing power, draft microbial genomes are rapidly generated from various environmental samples. To draw reliable conclusions about environmental dynamics from the growing availability of draft microbial genomes, it is crucial to determine the quality of genomes [121, 122]. Moreover, as new metagenomic binning tools are being developed, it is essential to evaluate the accuracy of their results and ensure they operate as desired. Collective efforts have led to the organization of worldwide challenges such as the Critical Assessment of Metagenome Interpretation (CAMI) [23, 26] that have provided gold standard truth datasets to facilitate standard benchmarking of these methods. Several metrics have been proposed to evaluate metagenomic binning results including, precision, recall, F1-score, purity, completeness, and contamination [123, 124]. Table 8 summarizes some of the automated tools that evaluate metagenomic binning results and calculate these quality metrics.
Table 8.
Comparison of metagenomic bin evaluation tools
| Bin evaluation tool | Year | User interface * | Programming languages used | Information used | Main techniques, models or algorithms used |
|---|---|---|---|---|---|
| CheckM | 2015 | CLI | Python | Single-copy marker genes | Hidden Markov Model (HMM) profiles |
| BUSCO | 2015 | CLI | Python | Benchmarking sets of universal single-copy orthologs (BUSCO) | HMM profiles |
| AMBER | 2018 | CLI | Python | Ground truth annotations from CAMISIM or alignment to reference genomes | Calculation of performance metrics used in the CAMI challenges |
| CheckM2 | 2023 | CLI | Python | Simulated genomes with known levels of completeness and contamination | Artificial neural networks and gradient-boosted decision trees |
*CLI, Command line interface.
CheckM [123] and BUSCO [125] are two popular bin evaluation tools widely used in metagenomic studies. They can determine the quality of MAGs from real metagenomic data as well as benchmark binning results from simulated or mock datasets. To determine bin quality, CheckM uses single-copy marker genes whereas BUSCO [125] uses the benchmarking sets of universal single-copy orthologs (BUSCO) identified using OrthoDB [126]. However, these methods can fail to accurately evaluate genomes from novel lineages due to the lack of high-quality genomes and the absence of certain marker genes. CheckM2 [127] was introduced to overcome these challenges and evaluate genomes/MAGs without directly considering the taxonomic information of marker genes. Moreover, AMBER [124] relies on metrics such as precision, recall, F1-score and Adjusted Rand Index (ARI) based on a known ground truth (where you know the microbial composition of your metagenome and which sequence belongs to which reference genome) [128]. Hence, AMBER can be applied for simulated or mock metagenomes. AMBER is often used during development to evaluate and benchmark tools using datasets with known ground truth. Additionally, assembly evaluation tools such as QUAST [129] and metaQUAST [130] have been used to measure genome completeness after assembling long-read bins [48] and duplication of bin sizes [131].
Discussion
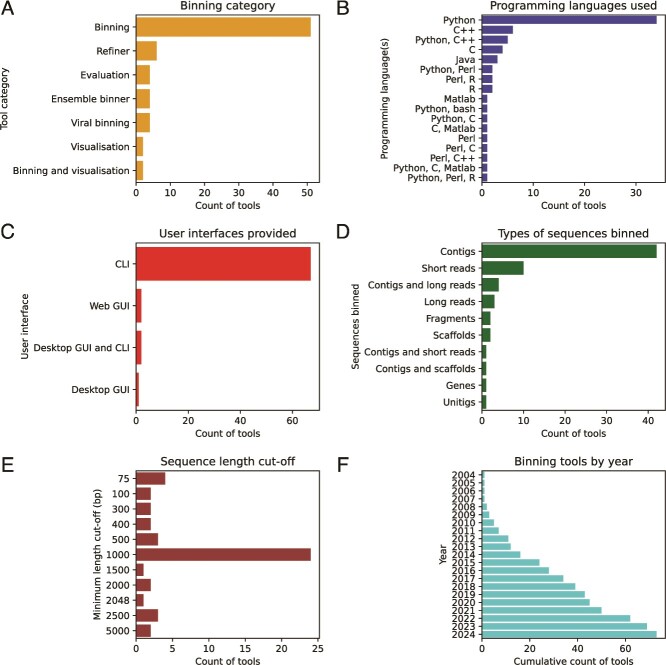
Metagenomic binning tools have encountered rapid growth over the past two decades. Each year, new tools are introduced claiming to be better than existing tools. Figure 3 summarizes 73 tools related to metagenomic binning which have been discussed in this paper based on different aspects (refer to the section ‘Data and code availability’ for links to the details of the tools). Even though new tools are published each year, the field still encounters various challenges and care should be taken to address them wherever possible. In this section, we explore trends, issues, and insights that researchers and developers should consider when creating new metagenomic binning tools.
Figure 3.
Summary plots of binning tools. Summary count plots of tools by (A) binning category, (B) programming languages used, (C) user interfaces provided, (D) types of sequences binned, (E) sequence length cut-off (in base pairs), and (F) cumulative count of tools by year.
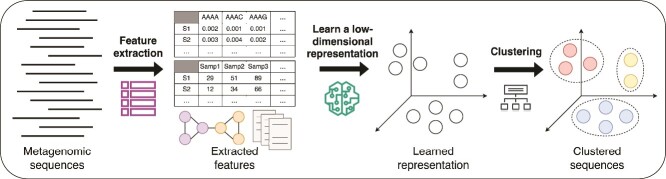
Rising use of deep learning techniques
Many recent metagenomic binning tools (those developed since 2021, Table 4) have shown a trend towards using deep learning techniques. The main idea is to learn a low-dimensional representation or embedding of the sequence features and obtain a clustering of these embeddings to produce bins (Fig. 4). VAMB was among the early tools to use deep learning techniques in metagenomic binning. Subsequently, many deep learning-based binning tools emerged, including those using variational autoencoders (CLMB and LRBinner), Siamese neural networks (SemiBin and SemiBin2), feed-forward neural networks (COMEBin) and graph neural networks (RepBin and UnitigBIN). The use of such deep learning techniques has enabled metagenomic binning tools to bin large-scale complex datasets accurately and efficiently.
Figure 4.
Workflow of a typical deep learning-based approach for metagenomic binning. Firstly, features (including nucleotide composition, abundance, graph structures, and other features) are extracted from the metagenomic sequences. Then a low-dimensional representation or embedding of the sequence features is learned using a deep learning model. Finally, the embeddings of the sequences are clustered to obtain bins.
Recently, binning tools have incorporated unsupervised representation learning techniques such as contrastive learning [132] into the process of learning low-dimensional embeddings of sequence features. The goal of contrastive learning is to learn a representation of the data such that similar points are positioned closely within the representation space and dissimilar points are placed at a greater distance from one another. Binning tools such as RepBin have incorporated contrastive learning using must-link and cannot-link constraints based on the presence of single-copy marker genes. If two contigs are connected together in the assembly graph, they are modelled as must-link constraints. If two contigs have the same single-copy marker gene, it means that the two contigs come from two different genomes. Hence, such pairs of contigs are modelled as cannot-link constraints. During the learning process, contigs with must-link constraints are positioned together, and contigs with cannot-link constraints are placed further apart in the learned representation. Such constraints have improved the learning process and produced accurate results.
Machine/deep learning-based metagenomic binning tools can effectively utilize the efficient and enhanced numerical computation capabilities of General-Purpose Graphics Processing Units (GPGPUs/GPUs), making them ideal for binning large-scale metagenomic datasets with millions of sequences. However, most available tools must load the entire dataset into the memory for feature vector calculations, which can be challenging if computational memory is scarce. For very large datasets and specific use cases, batch-wise processing techniques can be beneficial for handling and processing data in machine/deep learning applications.
Incorporation of taxonomic information
There has been a growing interest in using taxonomic information for metagenomic binning. For example, SolidBin and SemiBin2 model must-link and cannot-link constraints using the taxonomic annotations of contigs from databases such as NCBI or GTDB. The viral binning tool CoCoNet is trained on the NCBI RefSeq viral database [106].
The databases used by binning tools are from well-studied organisms and often generalize the strain-level information into more common signatures within species. While such information is sufficient in contrasting species, it may not be adequate to resolve hidden strains that might be driving the differences in an environment. Furthermore, the results are entirely dependent on the chosen database and are susceptible to missing novel species or discarding organisms that are not from a kingdom represented in the chosen databases.
Challenges in biological delineation of metagenomic data
Determining the exact number of underlying genomes in a metagenomic sample is a computationally challenging task. Tools such as MaxBin and SolidBin use counts of contigs containing single-copy marker genes to initialize the number of bins. This initialization method relies on marker genes being properly assembled within contigs and can be affected when marker genes are fragmented across contigs or when the open reading frames cannot be predicted correctly. To overcome this issue, tools such as MetaCoAG have adapted strategies to dynamically adjust the number of bins during the binning process. Moreover, certain tools use alternate approaches such as agglomerative clustering (Canopy), histogram-based clustering (LRBinner), and iterative clustering (VAMB) that do not rely on an initial estimate of the number of bins.
Short sequences may not provide accurate genome-specific signals due to the low resolution in a single sample. The composition profiles of such short sequences can be sparse and highly deviated from that of their constituent genomes. Binning such short contigs solely based on composition and coverage information can be challenging. Hence, the majority of the available metagenomic binning tools discard these short contigs during the binning process (as denoted by the minimum length cut-off in Table 4). Bin refinement tools such as GraphBin and GraphBin2 have utilized the connections between short contigs and large components in the assembly graph to accurately place them in their corresponding bins. It is worth exploring such methods to recover these short contigs as they can contain useful biological information including short repetitive sequences in viruses [133].
As microbial communities can be composed of different taxonomic groups, the sequencing data obtained from these samples can be very complex and heterogeneous. Even for the same genus, there can be several different species with very similar genomic signatures, and they may co-exist in similar abundance. Moreover, analyzing strain-level variations of species can be a computationally challenging task. Most of the binning methods are unable to generate MAGs at strain-level resolution due to their extremely high similarity and poor assembly quality. They often bin contigs from similar strains together and result in highly contaminated bins [23, 95]. Tools such as VAMB [69] have attempted to address this issue by individually assembling samples, binning all the resulting contigs, and separating MAGs from different samples. However, it is still challenging to recover strain-level MAGs within a single sample due to their highly similar genomic composition and the inherent problems in assembly. Hence, further investigation is required to develop methods capable of recovering MAGs of closely related microorganisms. While this problem may not be entirely solvable computationally due to the high similarity of contigs from different strains, combining long-read and short-read sequencing techniques, along with tools designed to handle both types of data, could offer a viable solution.
Different bacterial genomes in a metagenomic sample may share similar genes and genomic regions [1], which is a major challenge in assembling metagenomic reads into contigs [15]. Therefore, some assembled contigs may be shared by multiple genomes in the sample. However, most of the binning tools will assign these contigs to a single bin based on feature similarity to simplify the computational process. This can affect the completeness of the MAGs constructed afterwards. Very few binning tools support overlapped binning where shared contigs are assigned to their corresponding bins. Among such tools, GraphBin2 has attempted to address this challenge using the assembly graph and coverage information of contigs (e.g. if a contig is shared between two genomes, then its coverage will be elevated and should be close to the sum of the coverages of the two genomes). As shared contigs correspond to shared vertices between different genomic paths on the assembly graph [15], it is worth exploring methods to infer such shared contigs from the assembly graph without additional sequencing requirements.
Most metagenomic binning tools focus only on bacteria and archaea (especially those that rely on bacterial and archaeal single-copy marker genes). Such tools can incorrectly bin or even discard viral sequences as viruses lack universal marker genes [134]. Moreover, micro-eukaryotes such as fungi and protists remain under-characterized in metagenomic studies even though it is possible to adapt existing methods based on their single-copy marker genes [135, 136]. However, identifying fungal genomes is challenging as some DNA sequences can be mixed with closely related genera and require multiple molecular markers and precise sequence selection methods from databases [137, 138]. For example, the nuclear ribosomal RNA (rRNA) gene internal transcribed spacers (ITS) region, widely used for fungal identification, includes ITS1, 5.8S gene, and ITS2, but lacks sufficient resolution power for differentiating closely related fungi. Prahl et al. [137] demonstrated that combining ITS2 sequences and their secondary structures can effectively differentiate true Ampelomyces ITS sequences from incorrectly identified ones derived from environmental DNA. Additionally, the complexity of eukaryotic gene architecture, particularly exon-intron structures, complicates binning efforts. Moreover, the current lack of comprehensive eukaryotic databases impedes marker gene-based analyzes. To address these challenges, there is a clear need for metagenomic tools that focus on a more holistic approach to binning while maximizing the use of kingdom-specific information.
Issues with binning methods
Previous studies have shown that the nucleotide content of certain bacterial genomes can vary locally along chromosomes, especially in terms of GC content [139, 140]. For example, protein-coding regions tend to have higher GC content than non-coding regions [141]. The nucleotide composition of microbial genomes can vary based on factors such as genome size, oxygen requirement and nitrogen abundance [142–144]. Moreover, repeat regions, low complexity regions, homopolymer stretches, library preparation steps, and sequencing biases can result in genomic sequences with uneven sequencing coverage. Hence, there can be high variance in nucleotide composition and abundance features even among the sequences that originate from the same genome. Such genomic sequences that do not match either the average nucleotide composition of the genome or the average abundance of the genome are often incorrectly binned [45]. Approximately one-third of the genes do not match the modal codon usage of their genomes [145] and thus are likely to be misplaced in bins. These genes are often acquired through horizontal gene transfer (e.g. plasmids and mobile genetic elements like prophages). Moreover, prophages have different GC composition compared to the bacterial chromosome backbone [146] precluding them from binning.
During the assembly process, very similar reads are collapsed into single contigs, and then when reads are mapped back to estimate coverage, those contigs appear over-represented compared to their cognate genome. Highly conserved regions including rRNA gene repeats and genes encoding transfer RNA (tRNA) are frequently mis-binned due to their high similarity and repetitive nature, and many bins do not contain any rRNA encoding regions. Similarly, repetitive sequences like transposable elements (including transposons and insertion sequences) and bacteriophages are also rarely binned correctly. Additional steps may be required at the end of binning to accurately add these sequences back to their appropriate genomes, such as marker gene analysis. Marker gene analysis, using databases such as Rfam [147, 148] or SILVA [149], can help identify and potentially correct the placement of rRNA and tRNA genes in bins.
Software best practices
Many metagenomic binning tools are available as open-source software through public software repositories such as GitHub and Bitbucket. Even though most of them are CLI-based software (Fig. 3C), authors should ensure that their code is easy to install, executable, and well-maintained, especially around dependency updates. Proper testing should be carried out before publishing software to ensure that the core algorithms work as desired and produce the correct outputs. Simple test data sets should be included in the installation process so that users can quickly ascertain they are achieving expected outcomes. Software repositories such as GitHub and Bitbucket support continuous integration where workflows for automated building and testing can be set up before merging the changes to the main repository. Furthermore, software should be well-documented with detailed instructions on how to install and execute.
Some binning tools require certain additional files as input for calculations, e.g., Binary Alignment Map files to calculate the average coverage of contigs. Instead of executing these commands separately, one seamless workflow providing these pre-processing steps and the binning tool in just one command would be ideal. For this purpose, we can use workflow managers such as Snakemake [150] or a workflow template such as Snaketool [151]. Moreover, containerized binning tools, especially those utilizing software like Docker to bundle code and all necessary dependencies, are becoming popular as they allow the tools to run quickly and reliably on different computers. However, licensing issues, including potential incompatibilities between open-source software licenses, can sometimes hinder their widespread adoption and publication. Binning tools can also be published on package managers such as Bioconda and PyPI for Python, CRAN for R, or CPAN for Perl. Authors should take advantage of these platforms and services to ensure that the tools are correctly distributed, installed, and run.
Moving beyond binning to obtain contiguous genomes
Most of the available metagenomic binning tools produce MAGs with contigs in a random order. The contigs are not ordered as they are found in the genome and binning tools do not resolve genomic paths from contigs (i.e. determine the ordering of contigs in the genome). This can also be challenging, especially for large genomes such as those from bacteria or micro-eukaryotes as they can result in tangled and complex assemblies from NGS data. One solution is to extract just those reads that appear in each bin, and separately assemble the reads, without the confounding issues associated with the rest of the metagenome sequences [152]. Another alternative is to bin contigs and resolve the contigs in MAGs to construct complete and contiguous genomes. This has been done for bacterial MAGs to reconstruct strains in STRONG [153] and vMAGs to obtain contiguous bacteriophage genomes in Phables [133]. The assembly graph is the key feature used to determine the order of contigs. The connectivity information of contigs from the assembly graph is used to determine genomic paths and resolve representative genomes.
With the rising developments in TGS technologies, particularly longer read lengths that can span repetitive regions and provide increased overlap between reads [154], along with reduced error rates, it is now possible to obtain near-finished bacterial genomes, even from metagenomes [155]. Hence, it is worth exploring methods to move beyond binning and produce not just a set of contigs that constitute a genome, but to connect these contigs and produce the contiguous genome.
Conclusion
The history of metagenomic binning tools dates back to the early 2000s when they were created to automate the process of binning short DNA fragments obtained from environmental samples [156, 157]. Since then, various approaches have been introduced to bin different types of sequences such as short reads, assembled contigs and even error-prone long reads. This article presented a review of metagenomic binning tools and the various aspects including refinement, visualization, and evaluation. Recently, binning tools have incorporated new features such as graph data structures (e.g. assembly graph and read overlap graph) to capture accurate neighborhood information of sequences. The combination of all these features has advanced the recovery of microbial genomes from environmental samples.
Finally, we have discussed new trends in metagenomic binning and existing challenges, despite the significant advances in binning algorithms. Addressing these issues requires a collective effort from the scientific community and well-established community standards. As we move forward, more deep learning-based solutions will emerge, allowing the field of metagenomics to study large-scale metagenomes while harnessing the power of machine learning techniques and high-performance computing platforms. Moreover, attention should be paid to software best practices and making tools more user-friendly, which will widen the user base of binning tools. Overcoming these challenges will allow us to fully harness the capabilities of metagenomic binning approaches, thereby further facilitating pioneering discoveries in the fields of microbial ecology, human health, and biotechnology.
Key Points
Metagenomic binning is a crucial step in metagenomic analyzes that allows the study of uncultured microorganisms. It involves clustering DNA sequences obtained from environmental samples into bins representing different taxonomic groups.
Classic features such as nucleotide composition and abundance, and novel features including special graph structures of sequences used in existing metagenomic binning tools are compared in this study.
New trends including the rising use of deep learning techniques and challenges such as the biological delineation of closely related taxonomic groups are discussed, and further avenues of improvement are presented.
Researchers should be aware of the challenges associated with metagenomic binning and carefully evaluate their binning results to fully leverage metagenomic binning approaches.
Acknowledgements
This work is dedicated to the memory of the late Dr Yu Lin (The Australian National University) who was the PhD advisor of VM, AW, and HX, and whose guidance and support were instrumental in shaping this work. His wisdom and mentorship will be deeply missed.
Conflict of interest: None declared.
Contributor Information
Vijini Mallawaarachchi, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Adelaide, SA 5042, Australia.
Anuradha Wickramarachchi, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Westmead, NSW 2145, Australia.
Hansheng Xue, School of Computing, National University of Singapore, Singapore 119077, Singapore.
Bhavya Papudeshi, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Adelaide, SA 5042, Australia.
Susanna R Grigson, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Adelaide, SA 5042, Australia.
George Bouras, Adelaide Medical School, Faculty of Health and Medical Sciences, The University of Adelaide, Adelaide, SA 5005, Australia; The Department of Surgery—Otolaryngology Head and Neck Surgery, University of Adelaide and the Basil Hetzel Institute for Translational Health Research, Central Adelaide Local Health Network, Adelaide, SA 5011, Australia.
Rosa E Prahl, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Westmead, NSW 2145, Australia.
Anubhav Kaphle, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Westmead, NSW 2145, Australia.
Andrey Verich, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Westmead, NSW 2145, Australia; The Kirby Institute, The University of New South Wales, Randwick, Sydney, NSW 2052, Australia.
Berenice Talamantes-Becerra, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Westmead, NSW 2145, Australia.
Elizabeth A Dinsdale, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Adelaide, SA 5042, Australia.
Robert A Edwards, Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University, Adelaide, SA 5042, Australia.
Funding
This work is supported by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases [RC2DK116713] and the Australian Research Council [DP220102915].
Data and code availability
The data summarizing the features of the different tools and the code used to draw plots in Fig. 3 can be found at https://github.com/metagentools/metagenomic_binning_review.
Author contributions
All authors participated in the writing of the original draft, as well as its review and editing.
References
- 1. Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet 2004;38:525–52. [DOI] [PubMed] [Google Scholar]
- 2. Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. Microb Inform Exp 2012;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edwards RA, Haggerty JM, Cassman N. et al. Microbes, metagenomes and marine mammals: enabling the next generation of scientist to enter the genomic era. BMC Genomics 2013;14:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pargin E, Roach MJ, Skye A. et al. The human gut virome: composition, colonization, interactions, and impacts on human health. Front Microbiol 2023;14:963173. 10.3389/fmicb.2023.963173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quince C, Walker AW, Simpson JT. et al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35:833–44. [DOI] [PubMed] [Google Scholar]
- 6. Dinsdale EA, Edwards RA, Hall D. et al. Functional metagenomic profiling of nine biomes. Nature 2008;452:629–32. [DOI] [PubMed] [Google Scholar]
- 7. Canard B, Sarfati RS. DNA polymerase fluorescent substrates with reversible 3′-tags. Gene 1994;148:1–6. [DOI] [PubMed] [Google Scholar]
- 8. Drmanac R, Sparks AB, Callow MJ. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010;327:78–81. [DOI] [PubMed] [Google Scholar]
- 9. Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet 2010;19:R227–40. [DOI] [PubMed] [Google Scholar]
- 10. Baker M. De novo genome assembly: what every biologist should know. Nat Methods 2012;9:333–37. [Google Scholar]
- 11. Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform 2009;10:354–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z, Chen Y, Mu D. et al. Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-bruijn-graph. Brief Funct Genomics 2012;11:25–37. [DOI] [PubMed] [Google Scholar]
- 13. Yang C, Chowdhury D, Zhang Z. et al. A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data. Comput Struct Biotechnol J 2021;19:6301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li D, Liu C-M, Luo R. et al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015;31:1674–76. [DOI] [PubMed] [Google Scholar]
- 15. Nurk S, Meleshko D, Korobeynikov A. et al. metaSPAdes: a new versatile metagenomic assembler. Genome Res 2017;27:824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolmogorov M, Bickhart DM, Behsaz B. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods 2020;17:1103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sedlar K, Kupkova K, Provaznik I. Bioinformatics strategies for taxonomy independent binning and visualization of sequences in shotgun metagenomics. Comput Struct Biotechnol J 2017;15:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 2004;68:669–85. 10.1128/mmbr.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alneberg J, Bjarnason BS, de Bruijn I. et al. Binning metagenomic contigs by coverage and composition. Nat Methods 2014;11:1144–46. [DOI] [PubMed] [Google Scholar]
- 20. Qin J, Li R, Raes J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sangwan N, Xia F, Gilbert JA. Recovering complete and draft population genomes from metagenome datasets. Microbiome 2016;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papudeshi B, Haggerty JM, Doane M. et al. Optimizing and evaluating the reconstruction of metagenome-assembled microbial genomes. BMC Genomics 2017;18:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sczyrba A, Hofmann P, Belmann P. et al. Critical assessment of metagenome interpretation-a benchmark of metagenomics software. Nat Methods 2017;14:1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yue Y, Huang H, Qi Z. et al. Evaluating metagenomics tools for genome binning with real metagenomic datasets and CAMI datasets. BMC Bioinformatics 2020;21:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Borderes M, Gasc C, Prestat E. et al. A comprehensive evaluation of binning methods to recover human gut microbial species from a non-redundant reference gene catalog. NAR Genom Bioinform 2021;3:lqab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meyer F, Fritz A, Deng Z-L. et al. Critical assessment of metagenome interpretation: the second round of challenges. Nat Methods 2022;19:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlin S, Ladunga I. Comparisons of eukaryotic genomic sequences. Proc Natl Acad Sci 1994;91:12832–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet 1995;11:283–90. [DOI] [PubMed] [Google Scholar]
- 29. Saeed I, Tang S-L, Halgamuge SK. Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acids Res 2012;40:e34. 10.1093/nar/gkr1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dick GJ, Andersson AF, Baker BJ. et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol 2009;10:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teeling H, Waldmann J, Lombardot T. et al. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 2004;5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang B, Peng Y, Leung HCM. et al. MetaCluster: unsupervised binning of environmental genomic fragments and taxonomic annotation. Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology, New York, NY, USA: ACM; 2010. 10.1145/1854776.1854803. [DOI] [Google Scholar]
- 33. Leung HCM, Yiu SM, Yang B. et al. A robust and accurate binning algorithm for metagenomic sequences with arbitrary species abundance ratio. Bioinformatics 2011;27:1489–95. [DOI] [PubMed] [Google Scholar]
- 34. Van Vinh L, Van Lang T, Binh LT. et al. A two-phase binning algorithm using l-mer frequency on groups of non-overlapping reads. Algorithms Mol Biol 2015;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Girotto S, Pizzi C, Comin M. MetaProb: accurate metagenomic reads binning based on probabilistic sequence signatures. Bioinformatics 2016;32:i567–75. [DOI] [PubMed] [Google Scholar]
- 36. Andreace F, Pizzi C, Comin M. MetaProb 2: metagenomic reads binning based on assembly using minimizers and K-Mers statistics. J Comput Biol 2021;28:1052–62. [DOI] [PubMed] [Google Scholar]
- 37. Kislyuk A, Bhatnagar S, Dushoff J. et al. Unsupervised statistical clustering of environmental shotgun sequences. BMC Bioinformatics 2009;10:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laczny CC, Sternal T, Plugaru V. et al. VizBin - an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 2015;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laczny CC, Kiefer C, Galata V. et al. BusyBee web: metagenomic data analysis by bootstrapped supervised binning and annotation. Nucleic Acids Res 2017;45:W171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmartz GP, Hirsch P, Amand J. et al. BusyBee web: towards comprehensive and differential composition-based metagenomic binning. Nucleic Acids Res 2022;50:W132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chatterji S, Yamazaki I, Bai Z. et al. CompostBin: A DNA composition-based algorithm for binning environmental shotgun reads. In: Vingron M, Wong L, (eds.), Research in Computational Molecular Biology. RECOMB 2008. Lecture Notes in Computer Science; 4955. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008, p. 17–28. 10.1007/978-3-540-78839-3_3. [DOI] [Google Scholar]
- 42. Gori F, Mavroedis D, Jetten MSM. et al. Genomic signatures for metagenomic data analysis: Exploiting the reverse complementarity of tetranucleotides. 2011 IEEE International Conference on Systems Biology (ISB). Zhuhai, China: IEEE; 2011, p. 149–54. 10.1109/isb.2011.6033147. [DOI] [Google Scholar]
- 43. Kelley DR, Salzberg SL. Clustering metagenomic sequences with interpolated Markov models. BMC Bioinformatics 2010;11:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeffrey HJ. Chaos game representation of gene structure. Nucleic Acids Res 1990;18:2163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mallawaarachchi V, Lin Y. MetaCoAG: Binning metagenomic contigs via composition, coverage and assembly graphs. In: Pe'er I, (ed), Research in Computational Molecular Biology. RECOMB 2022. Lecture Notes in Computer Science; 13278. Cham: Springer International Publishing; 2022, p. 70–85. 10.1007/978-3-031-04749-7_5. [DOI] [Google Scholar]
- 46. Mallawaarachchi V, Lin Y. Accurate binning of metagenomic contigs using composition, coverage, and assembly graphs. J Comput Biol 2022;29:1357–76. 10.1089/cmb.2022.0262. [DOI] [PubMed] [Google Scholar]
- 47. Mattock J, Watson M. A comparison of single-coverage and multi-coverage metagenomic binning reveals extensive hidden contamination. Nat Methods 2023;20:1170–73. [DOI] [PubMed] [Google Scholar]
- 48. Wickramarachchi A, Mallawaarachchi V, Rajan V. et al. MetaBCC-LR: metagenomics binning by coverage and composition for long reads. Bioinformatics 2020;36:i3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wickramarachchi A. Models and algorithms for metagenomics analysis and Plasmid classification. Canberra, Australia: The Australian National University Open Research Repository, 2022. 10.25911/V200-9J82. [DOI] [Google Scholar]
- 50. Wickramarachchi A, Lin Y. Binning long reads in metagenomics datasets using composition and coverage information. Algorithms Mol Biol 2022;17:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wickramarachchi A, Lin Y. LRBinner: Binning Long Reads in Metagenomics Datasets. 21st International Workshop on Algorithms in Bioinformatics (WABI 2021). Leibniz International Proceedings in Informatics (LIPIcs); 201, Schloss Dagstuhl – Leibniz-Zentrum für Informatik, Wadern, Germany; 2021, p. 11:1–11:18. 10.4230/LIPIcs.WABI.2021.11. [DOI] [Google Scholar]
- 52. Wu Y-W, Ye Y. A novel abundance-based algorithm for binning metagenomic sequences using l-tuples. J Comput Biol 2011;18:523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y, Hu H, Li X. MBBC: an efficient approach for metagenomic binning based on clustering. BMC Bioinformatics 2015;16:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nielsen HB, Almeida M, Juncker AS. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 2014;32:822–28. [DOI] [PubMed] [Google Scholar]
- 55. Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 1988;2:231–39. [DOI] [PubMed] [Google Scholar]
- 56. Dutilh BE, Schmieder R, Nulton J. et al. Reference-independent comparative metagenomics using cross-assembly: crAss. Bioinformatics 2012;28:3225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu YY, Chen T, Fuhrman JA. et al. COCACOLA: binning metagenomic contigs using sequence COmposition, read CoverAge. CO-alignment and paired-end read LinkAge. Bioinformatics 2017;33:791–98. [DOI] [PubMed] [Google Scholar]
- 58. Wang Y, Leung HCM, Yiu SM. et al. MetaCluster 4.0: a novel binning algorithm for NGS reads and huge number of species. J Comput Biol 2012;19:241–49. [DOI] [PubMed] [Google Scholar]
- 59. Wang Y, Leung HCM, Yiu SM. et al. MetaCluster 5.0: a two-round binning approach for metagenomic data for low-abundance species in a noisy sample. Bioinformatics 2012;28:i356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kang DD, Froula J, Egan R. et al. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015;3:e1165. 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herath D, Tang S-L, Tandon K. et al. CoMet: a workflow using contig coverage and composition for binning a metagenomic sample with high precision. BMC Bioinformatics 2017;18:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu G, Jiang Y, Wang J. et al. BMC3C: binning metagenomic contigs using codon usage, sequence composition and read coverage. Bioinformatics 2018;34:4172–79. [DOI] [PubMed] [Google Scholar]
- 63. Wang Z, Wang Z, Lu YY. et al. SolidBin: improving metagenome binning with semi-supervised normalized cut. Bioinformatics 2019;35:4229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin H-H, Liao Y-C. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep 2016;6:24175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Graham ED, Heidelberg JF, Tully BJ. BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 2017;5:e3035. 10.7717/peerj.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Popic V, Kuleshov V, Snyder M. et al. Fast metagenomic binning via hashing and Bayesian clustering. J Comput Biol 2018;25:677–88. [DOI] [PubMed] [Google Scholar]
- 67. Liu C-C, Dong S-S, Chen J-B. et al. MetaDecoder: a novel method for clustering metagenomic contigs. Microbiome 2022;10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Strous M, Kraft B, Bisdorf R. et al. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol 2012;3:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nissen JN, Johansen J, Allesøe RL. et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat Biotechnol 2021;39:555–60. [DOI] [PubMed] [Google Scholar]
- 70. Zhang P, Jiang Z, Wang Y. et al. CLMB: Deep Contrastive Learning for Robust Metagenomic Binning. In: Pe'er I, (ed.), Research in Computational Molecular Biology. RECOMB 2022. Lecture Notes in Computer Science; 13278. Cham: Springer International Publishing; 2022, p. 326–48. 10.1007/978-3-031-04749-7_23. [DOI] [Google Scholar]
- 71. Pan S, Zhu C, Zhao X-M. et al. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments. Nat Commun 2022;13:2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pan S, Zhao X-M, Coelho LP. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing. Bioinformatics 2023;39:i21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Líndez PP, Johansen J, Kutuzova S. et al. Adversarial and variational autoencoders improve metagenomic binning. Commun Biol 2023;6:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Z, You R, Han H. et al. Effective binning of metagenomic contigs using contrastive multi-view representation learning. Nat Commun 2024;15:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brown CT, Hug LA, Thomas BC. et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 2015;523:208–11. [DOI] [PubMed] [Google Scholar]
- 76. Chandrasiri S, Perera T, Dilhara A. et al. CH-bin: a convex hull based approach for binning metagenomic contigs. Comput Biol Chem 2022;100:107734. 10.1016/j.compbiolchem.2022.107734. [DOI] [PubMed] [Google Scholar]
- 77. Imelfort M, Parks D, Woodcroft BJ. et al. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2014;2:e603. 10.7717/peerj.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu Y-W, Tang Y-H, Tringe SG. et al. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2014;2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016;32:605–07. [DOI] [PubMed] [Google Scholar]
- 80. Kang DD, Li F, Kirton E. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019;7:e7359. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hickl O, Queirós P, Wilmes P. et al. Binny: an automated binning algorithm to recover high-quality genomes from complex metagenomic datasets. Brief Bioinform 2022;23:bbac431. 10.1093/bib/bbac431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. West DB. Introduction to Graph Theory. New Jersey, USA: Prentice Hall, 2001. [Google Scholar]
- 83. Barnum TP, Figueroa IA, Carlström CI. et al. Genome-resolved metagenomics identifies genetic mobility, metabolic interactions, and unexpected diversity in perchlorate-reducing communities. ISME J 2018;12:1568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mallawaarachchi V, Wickramarachchi A, Lin Y. GraphBin: refined binning of metagenomic contigs using assembly graphs. Bioinformatics 2020;36:3307–13. [DOI] [PubMed] [Google Scholar]
- 85. Mallawaarachchi V. Metagenomics binning using assembly graphs. Canberra, Australia: The Australian National University Open Research Repository, 2022. 10.25911/STAT-HC66. [DOI] [Google Scholar]
- 86. DeMaere MZ, Darling AE. bin3C: exploiting hi-C sequencing data to accurately resolve metagenome-assembled genomes. Genome Biol 2019;20:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Du Y, Sun F. HiCBin: binning metagenomic contigs and recovering metagenome-assembled genomes using hi-C contact maps. Genome Biol 2022;23:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wickramarachchi A, Lin Y. Metagenomics binning of long reads using read-overlap graphs. In: Jin L, Durand D, (eds.), Comparative Genomics, RECOMB-CG 2022. Lecture Notes in Computer Science; 1323. Cham: Springer International Publishing; 2022, p. 260–78. 10.1007/978-3-031-06220-9_15. [DOI] [Google Scholar]
- 89. Kececioglu JD, Myers EW. Combinatorial algorithms for DNA sequence assembly. Algorithmica 1995;13:7–51. [Google Scholar]
- 90. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xue H, Mallawaarachchi V, Zhang Y. et al. RepBin: constraint-based graph representation learning for metagenomic binning. Proc Conf AAAI Artif Intell 2022;36:4637–45. [Google Scholar]
- 92. Lamurias A, Sereika M, Albertsen M. et al. Metagenomic binning with assembly graph embeddings. Bioinformatics 2022;38:4481–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lamurias A, Tibo A, Hose K. et al. Metagenomic Binning using Connectivity-constrained Variational Autoencoders. In: Krause A, Brunskill E, Cho K, Engelhardt B, Sabato S, Scarlett J, editors. Proceedings of the 40th International Conference on Machine Learning, vol. 202, PMLR; JMLR.org, 23--29 Jul 2023, p. 18471–81.
- 94. Xue H, Mallawaarachchi V, Xie L. et al. Encoding Unitig-level Assembly Graphs with Heterophilous Constraints for Metagenomic Contigs Binning. The Twelfth International Conference on Learning Representations, 2024.
- 95. Feng X, Li H. Evaluating and improving the representation of bacterial contents in long-read metagenome assemblies. Genome Biol 2024;25:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Beaulaurier J, Zhu S, Deikus G. et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol 2018;36:61––69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tourancheau A, Mead EA, Zhang X-S. et al. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat Methods 2021;18:491–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dupont CL, Rusch DB, Yooseph S. et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 2012;6:1186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Albertsen M, Hugenholtz P, Skarshewski A. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 2013;31:533–38. [DOI] [PubMed] [Google Scholar]
- 100. Olson RD, Assaf R, Brettin T. et al. Introducing the bacterial and viral bioinformatics resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 2023;51:D678–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Woodcroft BJ, Aroney STN, Zhao R. et al. SingleM and Sandpiper: Robust microbial taxonomic profiles from metagenomic data. bioRxiv: 2024. 10.1101/2024.01.30.578060. [DOI] [Google Scholar]
- 102. National Center for Biotechnology Information. National Center for Biotechnology Information; 2024. https://www.ncbi.nlm.nih.gov/ (accessed February 26, 2024). [Google Scholar]
- 103. Parks DH, Chuvochina M, Chaumeil P-A. et al. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol 2020;38:1079–86. [DOI] [PubMed] [Google Scholar]
- 104. Kieft K, Adams A, Salamzade R. et al. vRhyme enables binning of viral genomes from metagenomes. Nucleic Acids Res 2022;50:e83. 10.1093/nar/gkac341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Arisdakessian CG, Nigro OD, Steward GF. et al. CoCoNet: an efficient deep learning tool for viral metagenome binning. Bioinformatics 2021;37:2803–10. [DOI] [PubMed] [Google Scholar]
- 106. O’Leary NA, Wright MW, Brister JR. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Johansen J, Plichta DR, Nissen JN. et al. Genome binning of viral entities from bulk metagenomics data. Nat Commun 2022;13:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Du Y, Fuhrman JA, Sun F. ViralCC retrieves complete viral genomes and virus-host pairs from metagenomic hi-C data. Nat Commun 2023;14:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sieber CMK, Probst AJ, Sharrar A. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 2018;3:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018;6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang Z, Huang P, You R. et al. MetaBinner: a high-performance and stand-alone ensemble binning method to recover individual genomes from complex microbial communities. Genome Biol 2023;24:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Qiu Z, Yuan L, Lian C-A. et al. BASALT refines binning from metagenomic data and increases resolution of genome-resolved metagenomic analysis. Nat Commun 2024;15:2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Song W-Z, Thomas T. Binning_refiner: improving genome bins through the combination of different binning programs. Bioinformatics 2017;33:1873–75. [DOI] [PubMed] [Google Scholar]
- 114. Wang Y, Wang K, Lu YY. et al. Improving contig binning of metagenomic data using d2S oligonucleotide frequency dissimilarity. BMC Bioinformatics 2017;18:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mallawaarachchi VG, Wickramarachchi AS, Lin Y. GraphBin2: Refined and overlapped binning of metagenomic contigs using assembly graphs. 20th International Workshop on Algorithms in Bioinformatics (WABI 2020). Leibniz International Proceedings in Informatics (LIPIcs); 172, Wadern, Germany: Schloss Dagstuhl – Leibniz-Zentrum für Informatik; 2020, p. 8:1–8:21. 10.4230/LIPICS.WABI.2020.8. [DOI] [Google Scholar]
- 116. Mallawaarachchi VG, Wickramarachchi AS, Lin Y. Improving metagenomic binning results with overlapped bins using assembly graphs. Algorithms Mol Biol 2021;16:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang Z, Zhang L. METAMVGL: a multi-view graph-based metagenomic contig binning algorithm by integrating assembly and paired-end graphs. BMC Bioinformatics 2021;22:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xiang B, Zhao L, Zhang M. Unitig level assembly graph based metagenome-assembled genome refiner (UGMAGrefiner): a tool to increase completeness and resolution of metagenome-assembled genomes. Comput Struct Biotechnol J 2023;21:2394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kumar S, Jones M, Koutsovoulos G. et al. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet 2013;4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Seah BKB, Gruber-Vodicka HR. gbtools: interactive visualization of metagenome bins in R. Front Microbiol 2015;6:1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mardis E, McPherson J, Martienssen R. et al. What is finished, and why does it matter. Genome Res 2002;12:669–71. [DOI] [PubMed] [Google Scholar]
- 122. Chain PSG, Grafham DV, Fulton RS. et al. Genomics. Genome project standards in a new era of sequencing. Science 2009;326:236–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Parks DH, Imelfort M, Skennerton CT. et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Meyer F, Hofmann P, Belmann P. et al. AMBER: assessment of metagenome BinnERs. Gigascience 2018;7:giy069. 10.1093/gigascience/giy069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Simão FA, Waterhouse RM, Ioannidis P. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31:3210–12. [DOI] [PubMed] [Google Scholar]
- 126. Waterhouse RM, Tegenfeldt F, Li J. et al. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res 2013;41:D358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chklovski A, Parks DH, Woodcroft BJ. et al. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods 2023;20:1203–12. [DOI] [PubMed] [Google Scholar]
- 128. Fritz A, Hofmann P, Majda S. et al. CAMISIM: simulating metagenomes and microbial communities. Microbiome 2019;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gurevich A, Saveliev V, Vyahhi N. et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mikheenko A, Saveliev V, Gurevich A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics 2016;32:1088–90. [DOI] [PubMed] [Google Scholar]
- 131. Harris PNA, Bauer MJ, Lüftinger L. et al. Rapid nanopore sequencing and predictive susceptibility testing of positive blood cultures from intensive care patients with sepsis. Microbiol Spectr 2024;12:e03065-23. 10.1128/spectrum.03065-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen T, Kornblith S, Norouzi M. et al. A Simple Framework for Contrastive Learning of Visual Representations. In: Iii HD, Singh A, editors. Proceedings of the 37th International Conference on Machine Learning, vol. 119, PMLR; JMLR.org, 13--18 Jul 2020, p. 1597–607.
- 133. Mallawaarachchi V, Roach MJ, Decewicz P. et al. Phables: from fragmented assemblies to high-quality bacteriophage genomes. Bioinformatics 2023;39:btad586. 10.1093/bioinformatics/btad586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Caetano-Anollés G, Claverie J-M, Nasir A. A critical analysis of the current state of virus taxonomy. Front Microbiol 2023;14:1240993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Cissé OH, Stajich JE. FGMP: assessing fungal genome completeness. BMC Bioinformatics 2019;20:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Eren AM, Esen ÖC, Quince C. et al. Anvi’o: an advanced analysis and visualization platform for 'omics data. PeerJ 2015;3:e1319. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Prahl RE, Khan S, Deo RC. The role of internal transcribed spacer 2 secondary structures in classifying mycoparasitic Ampelomyces. PloS One 2021;16:e0253772. 10.1371/journal.pone.0253772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Prahl RE, Khan S, Deo RC. Ampelomyces mycoparasites of powdery mildews – a review. Can J Plant Pathol 2023;45:391–404. [Google Scholar]
- 139. Bohlin J, Eldholm V, Pettersson JHO. et al. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genomics 2017;18:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bohlin J, Snipen L, Hardy SP. et al. Analysis of intra-genomic GC content homogeneity within prokaryotes. BMC Genomics 2010;11:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bohlin J, Skjerve E, Ussery DW. Investigations of oligonucleotide usage variance within and between prokaryotes. PLoS Comput Biol 2008;4:e1000057. 10.1371/journal.pcbi.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. McEwan CE, Gatherer D, McEwan NR. Nitrogen-fixing aerobic bacteria have higher genomic GC content than non-fixing species within the same genus. Hereditas 1998;128:173–78. [DOI] [PubMed] [Google Scholar]
- 143. Mitchell D. GC content and genome length in Chargaff compliant genomes. Biochem Biophys Res Commun 2007;353:207–10. [DOI] [PubMed] [Google Scholar]
- 144. Naya H, Romero H, Zavala A. et al. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J Mol Evol 2002;55:260–64. [DOI] [PubMed] [Google Scholar]
- 145. Davis JJ, Olsen GJ. Modal codon usage: assessing the typical codon usage of a genome. Mol Biol Evol 2010;27:800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kang HS, McNair K, Cuevas DA, Bailey BA, Segall AM, Edwards RA. Prophage genomics reveals patterns in phage genome organization and replication. bioRxiv: 2017. 10.1101/114819. [DOI] [Google Scholar]
- 147. Kalvari I, Nawrocki EP, Argasinska J. et al. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 2018;62:e51. 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kalvari I, Nawrocki EP, Ontiveros-Palacios N. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res 2021;49:D192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Quast C, Pruesse E, Yilmaz P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 2012;28:2520–22. [DOI] [PubMed] [Google Scholar]
- 151. Roach MJ, Pierce-Ward NT, Suchecki R. et al. Ten simple rules and a template for creating workflows-as-applications. PLoS Comput Biol 2022;18:e1010705. 10.1371/journal.pcbi.1010705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dutilh BE, Cassman N, McNair K. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 2014;5:4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Quince C, Nurk S, Raguideau S. et al. STRONG: metagenomics strain resolution on assembly graphs. Genome Biol 2021;22:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Jain M, Olsen HE, Paten B. et al. The Oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 2016;17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sereika M, Kirkegaard RH, Karst SM. et al. Oxford nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat Methods 2022;19:823–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Venter JC, Remington K, Heidelberg JF. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004;304:66–74. [DOI] [PubMed] [Google Scholar]
- 157. Tyson GW, Chapman J, Hugenholtz P. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004;428:37–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data summarizing the features of the different tools and the code used to draw plots in Fig. 3 can be found at https://github.com/metagentools/metagenomic_binning_review.