SUMMARY
Transcription of tRNA genes by RNA Polymerase III (RNAPIII) is tuned by signaling cascades. The emerging notion of differential tRNA gene regulation implies the existence of additional regulatory mechanisms. However, tRNA gene-specific regulators have not been described. Decoding the local chromatin proteome of a native tRNA gene in yeast revealed reprogramming of the RNAPIII transcription machinery upon nutrient perturbation. Among the dynamic proteins, we identified Fpt1, a protein of unknown function that uniquely occupied RNAPIII regulated genes. Fpt1 binding at tRNA genes correlated with the efficiency of RNAPIII eviction upon nutrient perturbation and required the transcription factors TFIIIB and TFIIIC but not RNAPIII. In the absence of Fpt1, eviction of RNAPIII was reduced and the shutdown of ribosome biogenesis genes was impaired upon nutrient perturbation. Our findings provide support for a chromatin-associated mechanism required for RNAPIII eviction from tRNA genes and for tuning the physiological response to changing metabolic demands.
Keywords: tRNA, tDNA, RNA Polymerase III, Transcription, Chromatin, Chromatin Proteome, Nutrient Signaling
eToc blurb
Van Breugel et al. decoded the chromatin proteome of a single tRNA gene. They observed dynamic behavior of the transcription machinery and Fpt1. Fpt1 preferentially occupied regulated tRNA genes and promoted RNA Polymerase III dynamics. These findings provide molecular insights into the differential regulation of tRNA genes.
Graphical Abstract

INTRODUCTION
Gene transcription is facilitated by RNA polymerase enzyme complexes that collaborate with transcription factors, repressors, chromatin remodelers, and other cellular factors.1–3 RNA polymerases are categorized in three specialized classes.4 RNA Polymerase III (RNAPIII) mainly transcribes short DNA fragments called tDNAs, that code for transfer-RNAs (tRNAs).1,2,5 While tDNAs compromise a small fraction of the genome, their transcription accounts for ~15% of the total RNA pool.1 Transfer-RNA molecules function as amino acid deliverers for mRNA translation but recent studies emphasize that tDNAs and the RNAs they encode are involved in a wide variety of processes such as genome organization, aging, cancer, and other diseases.6–10 In agreement with their cellular abundance and important biological roles, tRNAs are tightly regulated at multiple levels, from transcription, post-transcriptional processing and modification, to transport and degradation.1,5,11–14 Remarkably, chromatin-associated regulatory mechanisms of tDNA transcription are still poorly understood, which sharply contrasts with the wealth of knowledge on regulation of RNA polymerase II (RNAPII).2,3,5,15
From a core transcription perspective, tDNAs are small, self-contained elements.16–18 Transcription of tDNAs is under the control of A- and B-box promoter elements that are located within the gene body coding for the tRNA. In nutrient-rich conditions, assembly factor TFIIIC binds the internal A- and B-box promoter elements and recruits the transcription factor TFIIIB, which promotes binding of RNAPIII to the upstream transcription start site to initiate transcription. Upon transcription initiation, elongation, and termination, RNAPIII dissociates from the tDNA or reinitiates another cycle of transcription in which the same RNAPIII molecule is recycled.5,19 In mammalian cells, the global activator MYC activates tDNA transcription in nutrient-rich conditions.15,20 In repressive conditions, such as nutrient deprivation, tDNA transcription is repressed by Maf1.5 Maf1 is a conserved and well-characterized repressor originally identified in Saccharomyces cerevisiae.21 In nutrient-rich conditions, Maf1 is inactivated by phosphorylation by upstream kinases such as TORC1 and retained in the cytoplasm.5,19 In repressive conditions, Maf1 is dephosphorylated and imported into the nucleus where it represses RNAPIII transcription in trans in a two-step fashion. Maf1 prevents de-novo assembly of TFIIIB onto the DNA by interaction with TFIIIB and Maf1 represses RNAPIII recruitment to the DNA by direct interactions with RNAPIII.22,23 As a consequence, upon nutrient deprivation and other stress signals, RNAPIII recruitment is blocked and tDNA transcription halted.
Interestingly, tRNA genes at different genomic locations but with identical gene bodies, and hence identical A- and B-box promoter elements, can show different expression dynamics. This has been observed in yeast where in repressive conditions (e.g. growth on a non-fermentable carbon source), expression of nearly all tRNA genes was reduced but a small subset of tRNA genes was significantly less repressed and less dependent on Maf1 for reasons still not known.24–26 Non-homogenous transcriptional regulation of tRNA genes has also been observed in human cells.27–31 The relative abundance of individual tRNAs varies considerably across tissues and cell lines, in cells engaged in proliferation or differentiation, and in pathologies such as cancer.32,33 These observations suggest that regulatory mechanisms outside the tDNA elements or embedded in the tDNA chromatin must be at play to tune RNAPIII activity in response to changing cellular demands.34 However, cis-regulatory elements or transcription- and chromatin-factors dedicated to gene-specific tDNA transcription by RNAPIII have not been described.5,15,35,36
Identifying regulators of tRNA genes is hampered by technical challenges. For example, many tRNA genes occur in multiple copies with identical body sequences across the genome, precluding mapping of mature tRNA sequences back to their tRNA gene of origin.37 In addition, tRNA genes are prone to relatively high background in ChIP-assays, presumably due to their high level of transcription and open chromatin structure.38 Moreover, tRNAs are highly structured and harbor many post-transcriptional base modifications, both of which complicate accurate and reproducible detection of tRNA molecules.14,39,40 It is therefore not surprising that little is known about regulation of tRNA genes in their chromatin context. In order to overcome this knowledge gap and to circumvent the technical challenges mentioned above, we employed Epi-Decoder to delineate the local chromatin proteome of a single tRNA gene in budding yeast in a direct and unbiased manner. In Epi-Decoder, the local chromatin abundance of each protein in the cell at a barcoded locus of interest can be measured by chromatin immunoprecipitation followed by DNA-barcode sequencing and counting.41–43 This method is sensitive and quantitative, and overcomes common hurdles associated with other proteomics approaches such as capture of a locus combined with mass spectrometry.44–46
Here we used Epi-Decoder to delineate the chromatin proteome of a single tRNA gene, tP(UGG)M, in Saccharomyces cerevisiae to uncover chromatin-associated regulators of RNAPIII. Comparing the proteome in different metabolic conditions revealed reprogramming of the core RNAPIII transcription machinery and additional known chromatin-binding proteins. In addition, we identified the uncharacterized factor Ykr011c. Ykr011c, which we named Fpt1, specifically occupied RNAPIII-transcribed genes and its abundance varied among tDNAs. In addition, deletion of FPT1 compromised the nutrient-dependent reprogramming of RNAPIII. Together, these results provide evidence for a chromatin-associated regulatory mechanism of RNAPIII assembly at tRNA genes.
RESULTS
Decoding the chromatin proteome of a single native tDNA locus
To delineate the local chromatin proteome of a single tRNA gene and identify potential chromatin-associated RNAPIII regulators, we employed Epi-Decoder (as outlined in Figure 1A).41–43 Many tRNA genes in yeast are flanked by transposons or transposon LTR remnants since retrotransposons in yeast preferentially integrate near tRNA genes.47–49 To capture a representative native chromatin environment, we employed Epi-Decoder on a tRNA gene (tP(UGG)M) flanked by the YMLWTy1–2 retrotransposon. Using CRISPR/Cas9, a library of barcoded yeast strains was generated by inserting a DNA-barcode between the divergent tRNA and Ty1 genes. With high-throughput synthetic genetic array (SGA) methods50, the barcode library was crossed with a genome-wide protein TAP-tag library to create an Epi-Decoder library. In this library, each clone contains a unique barcode/protein TAP-tag combination. To account for individual barcode effects, each TAP-tagged protein was combined with three different DNA barcodes. The three Epi-Decoder libraries were each pooled into single flasks, followed by cross-linking, chromatin shearing, chromatin immunoprecipitation (ChIP), DNA-barcode amplification and sequencing, and barcode counting. Barcode counts (ChIP versus input) served as a readout for occupancy of each tagged protein at the barcoded tDNA-Ty1 locus.
Figure 1. Epi-Decoder reveals rewiring of the proteome of a tDNA locus in response to nutrient availability.
(A) Schematic overview of Epi-Decoder. Using a DNA-barcode repair template library and CRISPR-Cas9 construct, a random DNA-barcode is inserted at the locus of interest. The library of barcoded transformants is arrayed, decoded, and crossed with an arrayed TAP-tag protein library. In the resulting Epi-Decoder library, each clone contains a unique barcode and a different protein tagged. After pooling the library, ChIP is performed and DNA barcodes from ChIP and input are amplified, sequenced, and counted to provide a binding score (ChIP/input) at the locus for all proteins in the library. (B) Epi-Decoder scores for 3707 proteins at the tDNA-Ty1 locus in glucose; arcsinh (fold change ChIP vs input) and FDR (p-value). Subunits that belong to RNAPIII, TFIIIB or TFIIIC are color-coded. Data describes three biological replicates (different DNA-barcode protein-TAG combinations) measured as three technical replicates (same DNA-barcode). Proteins classified as ‘binder’ are indicated with colored dots and significance thresholds with dashed lines (basemean ≥ 400, FDR ≤ 0.01 and log2 fold change ≥ 1). (C) Binding scores for the tDNA-Ty1 locus compared to the HO locus: log2 (fold change ChIPHO vs ChIPtDNA-Ty1) and FDR (p-value) for 154 factors that are ‘binder’ at either of the loci (also described in the Euler diagram). Data describes three biological replicates as described in (B). Colored dots show significant proteins and dashed lines show significance thresholds (FDR ≤ 0.25). (D) As in (C), fold change (log2 ChIPglucose vs ChIPglycerol) between glucose and glycerol 2h (37°C) and FDR (p-value) for 154 ‘binders’ at the tDNA-Ty1 locus in either growth condition (as described in the Euler diagram). (E) Heat maps with average fold change values (ChIP vs input, n = 3) for different protein complexes or families at the tDNA-Ty1 locus, each with a different color scale. Protein names are color-coded based on the Euler diagram in (D).
Decoding the tDNA-Ty1 proteome revealed a wide variety of proteins cross-linking to the locus. Proteins classified as ‘binder’ (base mean ≥ 400, FDR ≤ 0.01 & log2 fold change ≥ 1) included all known RNAPIII subunits and transcription factors present in our library, RNAPII transcription and elongation factors, and metabolic factors (Figure 1B, S1C, Supplemental Table S1 and S2). Histones were also observed but excluded from the analyses for technical reasons (see STAR Methods). Because the barcoded locus contained both an RNAPIII (tDNA) and RNAPII (Ty1 retrotransposon) transcribed gene, and because tDNAs are hotspots for non-specific cross-linking of proteins38, we first wanted to determine the specificity of the binders. To this end, we compared binders at the tDNA-Ty1 locus (n = 87) with binders at the HO locus (n = 135), a previously decoded RNAPII promoter region.42 The majority of proteins (n = 68) occupied both the tDNA-Ty1 and HO locus, including RNAPII transcription and elongation factors, metabolic factors, chromatin remodelers (e.g. FACT, INO80, RSC and PAF1 complexes) and RNA processing factors (Figure 1C, S1A, S1C, Supplemental Table S1 and S2). One major advantage of Epi-Decoder is that binding of each protein is interrogated in a single pool with all other proteins. As a result, potential non-specific cross-linking is expected to be eliminated by internal normalization. Indeed, specific tDNA binders included subunits of the RNAPIII, TFIIIB and TFIIIC complexes suggesting that the tDNA-Ty1 binder set is not affected by potential hotspot artifacts (Figure 1C).
The tDNA proteome is dynamically regulated in response to nutrient availability
In nutrient-rich conditions (Figure 1B), tRNA genes are actively transcribed by RNAPIII, but in repressive conditions, the repressor Maf1 is localized to the nucleus and tDNA transcription is restrained. Maf1 prevents assembly of RNAPIII and TFIIIB onto tDNAs but is itself not a component of the tDNA chromatin and therefore considered a trans-factor.51 To investigate whether specific chromatin-associated regulators of tDNAs exist, we applied Epi-Decoder in a condition of repressed tDNA transcription. After cells were grown to mid-log phase in media containing glucose at 30°C, cells were switched to glycerol at 37°C for 2 hours, a condition associated with increased Maf1 activity and repressed RNAPIII transcription.5,24 Differential analysis showed extensive reprogramming of the general RNAPIII transcription factors upon a switch to repressive conditions (Figure 1D, 1E, S1C, S1D, Supplemental Table S1 and S2). Binding of both RNAPIII and TFIIIB was decreased, corroborating with repressed tDNA transcription. In contrast, binding of TFIIIC was increased. Previous studies on individual subunits of these complexes have suggested that RNAPIII and TFIIIC are inversely correlated due to their competitive binding for the same locus.52,53 Here we confirm and extend this proposed model from the level of individual subunits to their protein complexes. Unexpectedly, the repressed state of the tDNA-Ty1 locus showed an overall increased number of binders (n = 143) compared to the active state (Figure S1B). In repressive conditions, relative binding of the two high-mobility group proteins Nhp6a and Nhp6b, known to aid tDNA transcription54–56, was increased. In addition, we observed an increased relative occupancy of the chromatin remodeling complexes INO80, RSC and the RNAPII elongation complex PAF1, suggesting a broad chromatin adaptation to repressive conditions (Figure 1E, S1D, note that the scale of each heat map is unique due to large differences in cross-linking efficiency between different protein complexes). Since the occupancy of RNAPII subunits was not consistently altered in repressive conditions, the observed changes were most likely not caused by altered transcription of the proximal Ty1 element (Figure S1D). Additionally, we observed increased relative occupancy of heat shock proteins, important chaperones that maintain protein homeostasis, and RNA processing factors. Lastly, we observed increased binding of several transcriptional repressors, suggesting an overall repressed chromatin-state (Figure S1D). To check whether the observed chromatin-proteome perturbations originated from changes in global protein levels, we performed whole-cell proteomics (Supplemental Table S3). The Epi-Decoder and proteomics datasets poorly correlated (Figure S1E), with the exception of a few factors (i.e. Ykr011c, Brf1, and Hsp104). This suggests that generally, changes in protein occupancy at the tRNA gene locus were not caused by changes in global protein levels.
A previously unknown factor enriched at tRNA genes
Having determined that the tDNA chromatin proteome is reprogrammed in changing metabolic conditions, we next asked whether candidate regulatory factors are embedded in the proteome. Focusing on binders that showed dynamic chromatin interactions, the protein Ykr011c drew our attention. Ykr011c was found at the tDNA-Ty1 locus but not the reference HO promoter locus (Figure 1C) and showed increased binding in repressive conditions (Figure 1D). This specific binding and dynamic behavior suggested that Ykr011c could be a bona fide but previously unknown member of the tDNA chromatin proteome. Ykr011c is a protein of unknown function and structure, and phylogenetic analysis showed that this protein is conserved within the Saccharomycetales clade (Figure S2). To validate the Epi-Decoder results, we first determined the genome-wide binding profile of Ykr011c by ChIP-sequencing (TAP-ChIP). An RNAPIII subunit (Rpo31) and RNAPII subunit (Rpb2) were taken along as controls for RNAPIII and RNAPII transcribed regions, respectively. We also included a ribosomal protein (Rpl13a) as a negative control because tRNA genes have been reported to act as sites of non-specific binding.38 Visual inspection of the tDNA-Ty1 Epi-Decoder locus showed that Ykr011c is enriched at the tP(UGG)M tRNA gene and absent from the Ty1 element (Figure 2A). At a genome-wide level, Ykr011c was found to uniquely bind all nuclear tRNA genes and a few other RNAPIII-regulated genes, including RPR1, SNR6, and SCR1 and the tDNA relics ZOD1 and iYGR033c (Figure 2B, S3A). We found no evidence for Ykr011c binding at ETC loci, sites in the genome previously described as regions of extra TFIIIC (ETC) binding57 (Figure S3A). These results confirmed binding of Ykr011c to the tDNA-Ty1 locus and furthermore showed that Ykr011c is a previously unknown specific tDNA-binding protein. From this point onward we will refer to Ykr011c as Fpt1 (Factor in the Proteome of tDNAs number 1).
Figure 2. Fpt1 binds uniquely to RNAPIII transcribed genes and is enriched at regulated tDNAs.
(A) Genome tracks with pooled ChIP-seq data (n = 3) for TAP-tagged Fpt1, Rpo31, Rpb2 and Rpl13a at the Epi-Decoder locus: tP(UGG)M tRNA gene and YMLWTy1–2 retrotransposon. (B) As in (A), tRNA genes tL(CAA)G1, tK(UUU)K and tT(CGU)K. (C) ChIP-exo metagene profile of Fpt1 (+/− 500 bp from tDNA midpoint) at 261 tRNA genes from a single representative ChIP-exo experiment. Plots of ChIP-exo tag 5’ ends (exonuclease stop sites) are inverted for tags mapping to the non-transcribed (anti-sense) strand. The y-axis shows linear arbitrary units (AU) which are not equal across plotted datasets. Black box at the top: average position of a representative tRNA gene, start and end are indicated with dashed lines. The insert in the lower right corner shows a schematic tRNA gene, containing the internal A- and B-box promoter elements. (D-H) As in (C), ChIP-exo metagene profiles of Rpo31, Brf1, TBP, Tfc4 and Tfc6 (from Rossi et al.58) overlaid with Fpt1. (I) Fpt1 occupancy (ChIP-seq) at housekeeping (n = 30) and regulated (n = 158) tRNA genes. The used classification of tRNA genes is described in Turowski et al.25 and Star Methods. The average Fpt1 occupancy (13142) across 243 tRNA genes is indicated with a dotted horizontal line. Statistics: Welch-corrected unpaired two-tailed t-test.
To analyze the binding of Fpt1 to tDNAs at higher resolution, we applied ChIP-exo to provide detailed information on the protein-DNA contacts that Fpt1 makes.58,59 ChIP-exo confirmed the binding of Fpt1 to tRNA genes observed by ChIP-seq (Figure 2C). In addition, the tDNA metagene ChIP-exo profile of Fpt1 showed several contact points, both upstream and within the tRNA gene body. Superimposing the ChIP-exo profile of Fpt1 with those of known RNAPIII factors revealed shared tDNA contact points, especially with TFIIIB and TFIIIC (Figure 2D–H). This suggests that Fpt1 makes contact with tDNAs at least in part through the core RNAPIII transcription machinery complex.
Unexpectedly, in contrast to RNAPIII and TFIIIB, which were distributed relatively even across all tRNA genes, ChIP-seq and ChIP-exo analysis displayed varied levels of Fpt1 binding (Figure 2B, S3A–C, Supplemental Table S4). While some tRNA genes were bound by high levels of Fpt1, others were scarcely occupied. In the ChIP-exo data we observed a similar trend for TFIIIC, and the binding patterns of Fpt1 and TFIIIC correlated positively (Figure S3D). To investigate whether this observation for Fpt1 could reflect a functional role, we compared Fpt1 binding at tRNA genes with the previously determined distribution of transcriptionally engaged RNAPIII at tRNA genes in response to changing nutrient conditions.25 Turowksi et al.25 used UV cross-linking and analysis of cDNA (CRAC) to capture nascent RNAs bound by RNAPIII and identified two classes of tRNA genes (Supplemental Table S4 and STAR Methods). One canonical class of ‘regulated tRNA genes’ showed efficient eviction of engaged RNAPIII upon a switch from glucose to glycerol (low RNAPIII retention score) and this repression was dependent on Maf1 (high Maf1 dependency score). Another class of ‘housekeeping tRNA genes’ was less responsive to changing nutrients (high RNAPIII retention score) and less affected by loss of Maf1 (low Maf1 dependency score). Inspection of Fpt1 binding at tRNA genes ranked by their CRAC response score suggested that Fpt1 binding is on average higher at regulated tRNA genes and lower (but still higher than background, Figure 2B, S3A, Supplemental Table S4) at housekeeping tRNA genes (ChIP-seq Figure 2I, ChIP-exo Figure S3G–H). Indeed, Fpt1 occupancy negatively correlated with retention of engaged RNAPIII upon a switch from glucose to glycerol and positively correlated with Maf1-dependent repression (Figure S3E–F) suggesting Fpt1 may be part of a gene-class specific chromatin-associated mechanism of tDNA regulation.
Fpt1 responds to changing nutrient availability
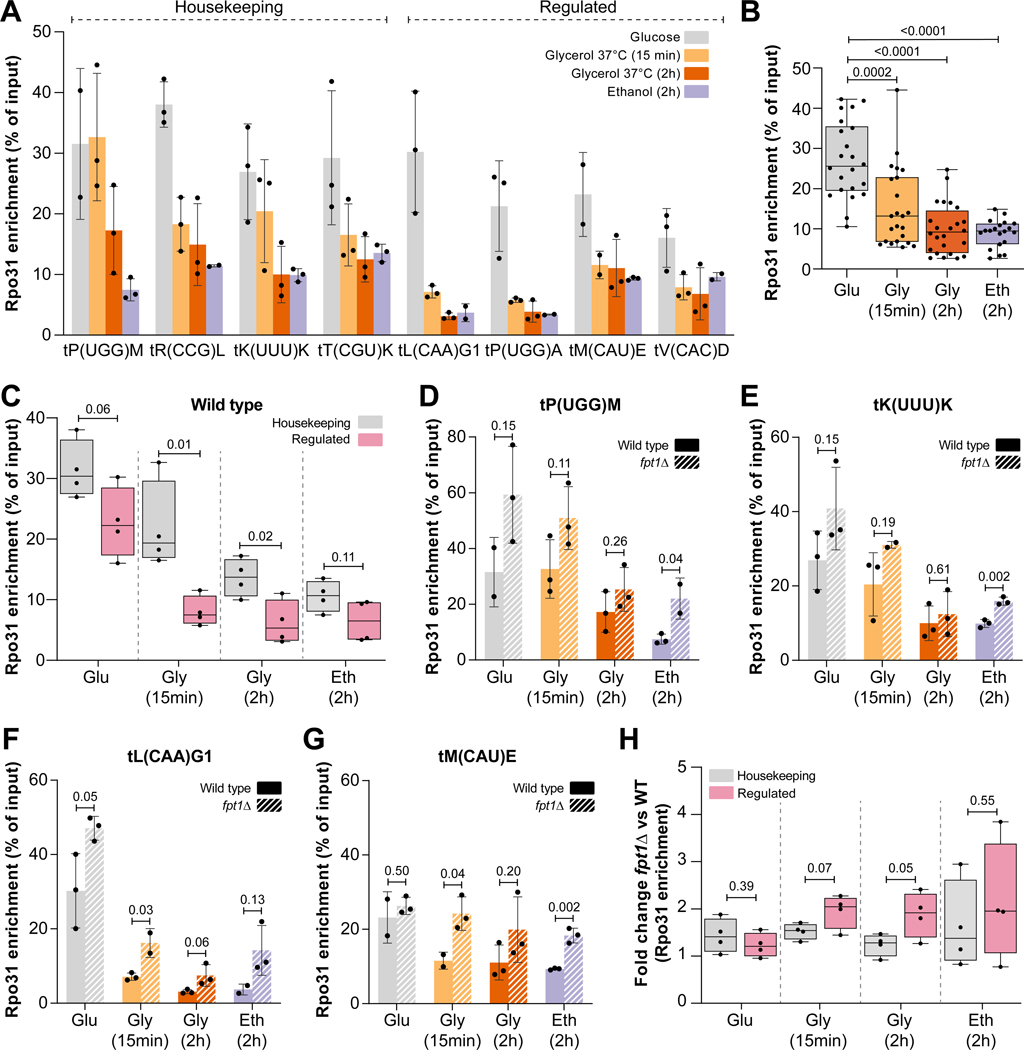
To investigate the role of Fpt1 in tDNA regulation, we first examined Fpt1 binding at several tDNA loci, representing the classes housekeeping (tP(UGG)M, tR(CCG)L, tK(UUU)K, tL(CAA)G1) and regulated (tL(CAA)G1, tP(UGG)A, tM(CAU)E, tV(CAC)D) in conditions of repressed RNAPIII activity. Cells were grown to mid-log phase in glucose and subsequently subjected to two independent repressive conditions by switching to ethanol (30°C) or glycerol (37°C) as a carbon source. Fpt1 occupancy (ChIP/input) increased at all tested tRNA genes in repressive conditions and was on average (n = 8 tRNA genes) highest in the 2h glycerol condition (14.8-fold increase, Figure 3A–B). These results corroborate the nutrient response observed in Epi-Decoder and demonstrate that Fpt1 occupancy generally increased at tRNA genes. Additionally, a similar increase in Fpt1 binding was observed at the tDNA relic ZOD1 and RNAPIII regulated non-tRNA genes SNR6, SCR1 and RPR1 (Figure S4A).
Figure 3. Fpt1 responds to changing nutrient availability.
(A) Fpt1 enrichment (TAP-ChIP) in glucose, glycerol 2h and 15 min (37°C), and ethanol 2h (n = 4 ± SD). (B) Stacked Fpt1 enrichment from (A). Statistics: Welch-corrected unpaired two-tailed t-test. (C) Fpt1-TAP immunoblot (n = 3) in glucose, glycerol 2h and 15 min (37°C), and ethanol 2h. Pgk1 was used as a loading control. BY4741 is a no-tag control. (D) Quantification of (C), n = 3 ± SD. Statistics: unpaired two-tailed t-test. (E) Fpt1-TAP immunoblot (n = 3) in wild type (WT) and Fpt1 overexpression (OE) in glucose. (F) Fpt1 enrichment (TAP-ChIP) in glucose for WT and Fpt1 OE (n = 3 ± SD). (G) Stacked Fpt1 enrichment from (F). Statistics: Welch-corrected unpaired two-tailed t-test. (H) Left, representative images of Fpt1 localization in live yeast cells in glucose. From top to bottom: Fpt1-GFP (yellow), PP7-NLS-mScarlet-I as a nuclear marker (magenta) and the merged channel. Cellular masks to locate cells and nuclei are indicated with white lines. Scale bar: 3 μm. Right, quantification of Fpt1 nuclear enrichment (n = 3) in glucose (n = 1536 cells), glycerol 2h 37°C (n = 327 cells), and ethanol 2h (n = 771 cells). Circles show data for individual cells; box plots show the distribution of the data where the box indicates the quartiles and whiskers extend to show the distribution, except for outliers. Statistics: bootstrapping (MacKinnon et al.67).
To determine whether increased Fpt1 binding to tDNAs during repressive conditions was caused by increased Fpt1 protein expression, we measured global cellular protein levels of Fpt1 in response to changing nutrient conditions. Immunoblotting showed that increased Fpt1 occupancy at tRNA genes was accompanied by an increase in Fpt1 protein levels upon a switch to repressive conditions (1.7-fold in 2h glycerol 37°C, Figure 3C–D). This increase, which was confirmed by whole-cell proteomics analysis (Supplemental Table S3), might at least in part explain the increase in Fpt1 occupancy at tRNA genes. To further explore this, we generated strains in which Fpt1-TAP is overexpressed by the strong TDH3 promoter at the native FPT1 locus. While this led to a 20-fold increase in Fpt1 protein levels (Figure 3E) in glucose, Fpt1 binding was not increased (Figure 3F–G). This suggests that increasing the expression level is not sufficient to increase Fpt1 occupancy at tRNA genes, which indicates additional mechanisms of regulation under repressive conditions. We also determined the cellular localization of Fpt1 in nutrient-rich and repressive conditions. GFP-tagged Fpt1 localized to the nucleus in all tested conditions and showed only a modest increase in nuclear enrichment in repressive conditions (Figure 3H, S4B–D). This suggests that, in contrast to Maf160, Fpt1 is constitutively present in the nucleus and activated by different mechanisms. Altogether, the increased tDNA occupancy and protein levels of Fpt1 in conditions of repressed RNAPIII transcription are in agreement with a potential regulatory role at tDNAs.
Deletion of Fpt1 compromises eviction of RNAPIII upon stress
Studying tDNA transcription poses multiple challenges such as efficient reverse transcription and accurate mapping to the reference genome of mature tRNAs. To circumvent these issues, we focused on RNAPIII occupancy at the chromatin and how it is affected by Fpt1. To this end, we performed ChIP-qPCR of the largest RNAPIII subunit (Rpo31) in both wild type (WT) and fpt1Δ strains. Cells were grown to mid-log phase in glucose and shifted to repressive conditions. A stress response of RNAPIII in WT cells could be observed at all tRNA genes examined and for most tRNA genes, RNAPIII loss could already be observed after 15 minutes in glycerol (37°C) (Figure 4A–B). On the contrary, RNAPIII occupancy at ZOD1, SNR6, SCR1 and RPR1 was less affected by repressive conditions suggesting a different mechanism of regulation (Figure S4E).24 Decreased Rpo31 occupancy in all tested conditions in both WT and fpt1Δ was on average most profound at tRNA genes belonging to the regulated tDNA subset (n = 4 tRNA genes) (Figure 4C, S4F), which is in agreement with previous observations25, although the response was variable among tRNA genes within each class. Looking at the role of Fpt1, we observed that in the absence of Fpt1, more Rpo31 was retained at tRNA genes compared to WT (Figure 4D–G, S4G–J). The gain of Rpo31 occupancy at tRNA genes in fpt1Δ suggests that Fpt1, directly or indirectly, regulates RNAPIII eviction in all tested conditions (Figure S4K). The extent to which Rpo31 occupancy increased in fpt1Δ was variable and distinct across tRNA genes but correlated with the two tRNA gene classes suggesting that regulated tRNA genes are more dependent on Fpt1 (Figure 4H). Immunoblotting and whole-cell proteomics demonstrated that fpt1Δ did not affect global Rpo31 protein levels (Supplemental Tables S3 and S5), suggesting that the increased Rpo31 occupancy at tDNAs in fpt1Δ is not a consequence of increased global Rpo31 protein levels.
Figure 4. Deletion of Fpt1 compromises eviction of RNAPIII upon nutrient perturbation.
(A) Rpo31 enrichment (TAP-ChIP) in glucose, glycerol 2h or 15 min (37°C) and ethanol 2h (n = 3 ± SD). (B) Stacked Rpo31 enrichment from (A). Statistics: Welch-corrected unpaired two-tailed t-test. (C) Rpo31 enrichment (TAP-ChIP) in WT at housekeeping tRNA genes (grey, n = 4) and regulated tRNA genes (pink, n = 4) in glucose, glycerol 2h and 15 min (37°C), and ethanol 2h. For each tRNA gene, the average of three biological replicates is shown. Whiskers indicate the minimum and maximum. Statistics: unpaired two-tailed t-test. (D-G) Rpo31 enrichment (TAP-ChIP) in WT and fpt1Δ (n = 3 ± SD) in glucose, glycerol 2h and 15 min (37°C), and ethanol 2h. Statistics: unpaired two-tailed t-test. (H) Fold change between WT and fpt1Δ for Rpo31 enrichment at housekeeping tRNA genes (grey, n = 4) and regulated tRNA genes (pink, n = 4). For each tRNA gene, the average of three biological replicates is shown. Whiskers indicate the minimum and maximum. Statistics: unpaired two-tailed t-test.
Fpt1 functions as a regulator of the RNAPIII transcription machinery
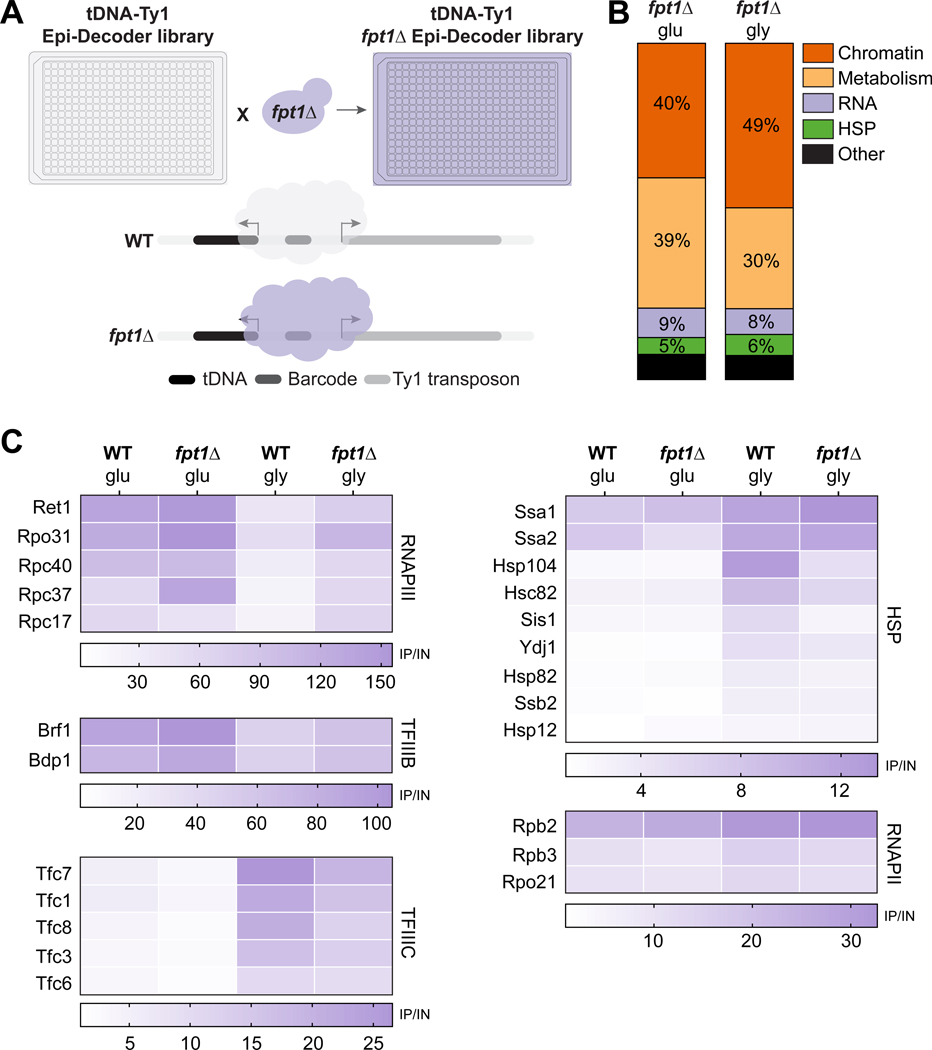
The increased binding of Rpo31 in fpt1Δ raises the question to what extent the full RNAPIII complex and other members of the general tDNA transcription machinery are affected. To address this question, we took advantage of Epi-Decoder to study changes in binding events at the barcoded tDNA locus of all proteins in parallel in WT compared to fpt1Δ. We repeated Epi-Decoder after crossing into the library an fpt1Δ allele as outlined in Figure 5A. Differential analysis was done on binders (base mean ≥ 400, FDR ≤ 0.01 & log2 fold change ≥ 1) in glucose (n = 87 WT, n = 77 fpt1Δ, Figure S5A, Supplemental Table S1 and S2) and 2h glycerol 37°C (n = 143 WT, n = 138 fpt1Δ, Figure S5B, Supplemental Table S1 and S2). Similar to WT, the majority of proteome-members included chromatin remodelers, transcription factors, metabolic enzymes, heat shock proteins and RNA processing factors (Figure 5B). In conditions of active transcription, FPT1 deletion showed mild effects on the relative occupancy of RNAPIII, TFIIIB and TFIIIC. This effect was enhanced when cells were subjected to repressive conditions (Figure 5C). In fpt1Δ, we observed a partial eviction of RNAPIII and TFIIIB subunits compared to WT. On the contrary, binding of TFIIIC subunits was less increased in fpt1Δ. These changes were not caused by altered global protein levels of the subunits of these transcription complexes (Supplemental Table S3). The observed coupling between increased RNAPIII and TFIIIB and decreased TFIIIC occupancy provides further evidence for the previously proposed competitive-binding model.52,53 Additionally, fpt1Δ caused decreased relative binding of several heat shock factors in repressive conditions (Figure 5C). Other factors such as RNAPII subunits (regulating the proximal Ty1 element) and chromatin remodelers remained largely unaffected (Figure 5C, S5C).
Figure 5. Fpt1, embedded in the tDNA proteome, affects the RNAPIII transcription machinery.
(A) Schematic outline: an FPT1 knockout strain is crossed with the WT Epi-Decoder library using SGA, resulting in an fpt1Δ Epi-Decoder library. The tDNA chromatin proteome of both libraries can be compared. (B) Fraction of proteins classified as ‘binder’ in fpt1Δ glucose or glycerol 2h (37°C) for different functional classes indicated (HSP refers to heat shock proteins). (C) Heat maps with average fold change values (ChIP vs input, n = 3) at the tDNA-Ty1 locus for different protein complexes or families for WT or fpt1Δ in glucose and 2h glycerol (37°C).
Fpt1 occupancy at tRNA genes requires TFIIIB and TFIIIC but not RNAPIII
Our data suggest that Fpt1 is part of a previously unknown chromatin-associated mechanism, being in proximity to and regulating the dynamic assembly of the RNAPIII transcription machinery at tRNA genes. To understand how Fpt1 interacts with the core members of the RNAPIII transcription machinery, we first treated cells with 1,10-phenanthroline (PH, Figure 6A) to chemically perturb RNAPIII activity and test if Fpt1 binding requires active transcription. Previously, it has been shown that treatment with PH leads to rapid loss of RNAPII.41 We extended this observation to RNAPIII (Rpo31) and TFIIIB (Brf1), which were evicted from tRNA genes upon incubation for 30 minutes with PH (Figure 6B–C). In agreement with the competitive model between RNAPIII and TFIIIC, TFIIIC (Tfc3) binding increased upon treatment with PH (Figure 6D). Similar to TFIIIC, occupancy of Fpt1 increased in PH treated cells (Figure 6E). Global Fpt1 protein levels also increased (Figure S6A–B), consistent with the higher Fpt1 protein levels in repressive conditions (Figure 3C–D). Rpo31, Brf1, and Tfc3 global protein levels moderately decreased upon treatment with PH (Figure S6A–B). These results suggest that Fpt1 occupancy at tRNA genes does not depend on active RNAPIII, and that Fpt1 behaves similar to TFIIIC. However, PH is a non-specific inhibitor that can have indirect effects, potentially limiting the interpretation of the results.61 To explore the dependency of Fpt1 on the RNAPIII transcription machinery in an independent and more specific way, we used the anchor away system62 to conditionally deplete proteins from the nucleus and check whether Fpt1 binding to tRNA genes is perturbed (Figure 6F). Upon nuclear depletion of Rpo31 (1h rapamycin treatment; Figure 6G, S6C), Fpt1 binding increased at most tRNA genes examined. In sharp contrast, nuclear depletion of TFIIIC (Tfc1 and Tfc3, members of the τA and τB sub complexes of TFIIIC, respectively) and TFIIIB (Brf1 and Bdp1) caused a loss of Fpt1 binding (Figure 6H–K, S6C). Immunoblotting showed that this loss was not caused by altered Fpt1 protein levels (Figure S6D–E). Taken together, our results suggest that Fpt1 occupancy at tRNA genes is not dependent on active RNAPIII, but instead requires both TFIIIB and TFIIIC.
Figure 6. Fpt1 occupancy does not depend on active RNAPIII, but requires TFIIIB and TFIIIC.
(A) Outline of phenanthroline (PH) treatment: cells were grown to mid-log phase and treated with 1,10-phenanthroline (100 μg/mL) or vehicle (0.1% ethanol) for 30 minutes before crosslinking. (B) Rpo31 enrichment (TAP-ChIP) in vehicle and PH-treated cells (n = 3 ± SD). Statistics: unpaired two-tailed t-test. (C-E) As in (B), TAP-ChIP of Brf1, Tfc3, Fpt1 respectively. (F) Schematic of the anchor away (AA) approach. A GFP- and FRB-tagged target protein is depleted from the nucleus after 1 h rapamycin (7.5 μM) treatment. (G) Fpt1 enrichment (TAP-ChIP) upon nuclear depletion of Rpo31 (rapamycin) and DMSO control (n = 3 ± SD). Statistics: unpaired two-tailed t-test. (H-K) As in (G), depletion of Tfc1, Tfc3, Brf1, Bdp1, respectively.
Fpt1 affects tuning of ribosome biogenesis genes and cellular fitness in repressive conditions
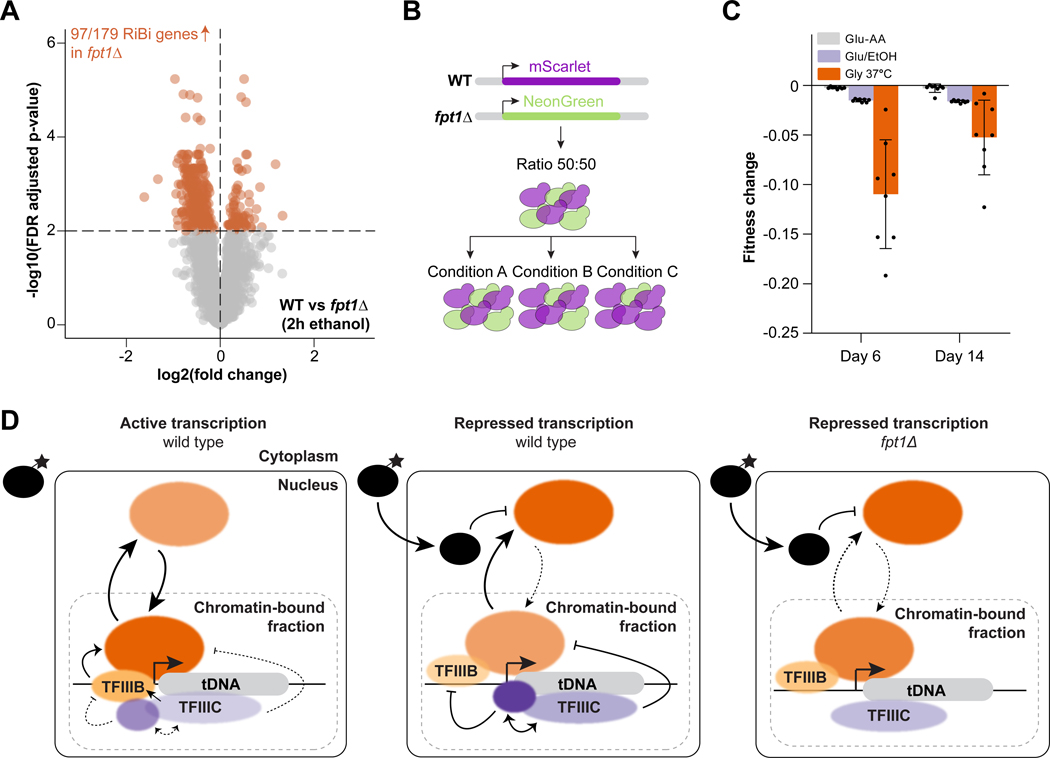
To explore how the altered RNAPIII dynamics in fpt1Δ is linked to general cell physiology, we performed mRNA-sequencing in WT and fpt1Δ. Cells were grown to mid-log phase in glucose containing media and subsequently switched to a non-fermentable carbon source (ethanol) for 2 hours at 30°C to avoid excessive temperature effects on the transcriptome. The switch to a non-fermentable carbon source caused downregulation of genes involved in ribosome biogenesis (RiBi) (Figure S7A–B, Supplemental Table S6). However, in fpt1Δ cells, 97/179 RiBi genes (as defined in Santos et al.63) were relatively upregulated, suggesting an improper shutdown of RiBi gene expression (Figure 7A, S7C, Supplemental Table S6). Fpt1 uniquely binds RNAPIII regulated genes and not RNAPII target genes. Therefore, our results suggest that Fpt1 indirectly affects transcription of ribosome biogenesis genes, most likely through its role in RNAPIII assembly at tDNAs by mechanisms that are currently unknown. This model is supported by the observation that strains expressing a mutant form of the RNAPIII catalytic subunit Rpc128 also show a weak increase in RiBi gene expression.64
Figure 7. Fpt1 affects tuning of ribosome biogenesis genes and cellular fitness in repressive conditions.
(A) Differential gene expression in WT and fpt1Δ in ethanol 2h (n = 4): log2 fold change and FDR (p-value) for 4978 expressed genes. Colored dots represent significant differentially expressed genes (FDR ≤ 0.01). Upregulated genes in fpt1Δ compared to WT are depicted on the left (n = 313 genes), downregulated genes on the right (n = 92 genes). (B) Competitive growth assay: the fluorescent markers mScarlet and NeonGreen were inserted at a neutral intergenic locus in WT and fpt1Δ in both combinations. Red and green cells were mixed in a 50:50 ratio and subjected to different growth conditions to study the effect on the ratio WT and fpt1Δ cells. (C) Relative fitness defect, expressed with the Malthusian coefficient, of fpt1Δ compared to WT in glucose but with alternating levels of non-auxotrophic amino acids (Glu-AA), alternating carbon source (Glu/EtOH), and glycerol 37°C (Gly 37°C). Data of four biological replicates including a color-swap is shown (n = 8). (D) A model for chromatin-associated regulation of the tDNA transcription machinery. For details, see main text.
Finally, we determined the cellular fitness of fpt1Δ cells. Deletion of FPT1 did not lead to observable growth defects in spot test analysis in conditions of active or repressed transcription (Figure S7D). As a more sensitive approach to study cellular fitness of fpt1Δ, we performed a competitive growth assay as outlined in Figure 7B, using different growth regimens (see STAR Methods). Briefly, a different fluorescent reporter (NeonGreen or mScarlet) was inserted at a safe-harbor intergenic locus in WT and fpt1Δ cells. A color-swap was performed to account for reporter effects (Figure S7E–F). At t = 0, WT and fpt1Δ cells were equally mixed and maintained in batch culture using different growth conditions for two weeks. To include previously used repressive conditions, cells were maintained in glycerol at 37°C (Gly 37°C) or in conditions of alternating carbon sources by switching back and forth to ethanol or glucose at 30°C (Glu/EtOH). As a reference, cells were grown in glucose media but with conditions of alternating levels of non-auxotrophic amino acids (Glu-AA). Every few days, cultures were diluted to maintain a low density. Samples were taken at t = 0, t = 6 and t = 14 days to analyze the ratio of green to red cells by flow cytometry. While loss of Fpt1 did not affect fitness in glucose (Glu-AA), in conditions of alternating carbon source (Glu/EtOH) and in glycerol 37°C, fpt1Δ cells showed reduced competitive growth relative to WT cells (Figure 7C). These findings suggest that regulation of tDNA proteome dynamics by Fpt1 is required for optimal growth when yeast cells transiently or constitutively encounter metabolic conditions that require tuning of tDNA repression.
DISCUSSION
Transfer-RNA genes have long been considered as a homogeneous group of genes regulated by general signaling pathways and mechanisms in trans such as Maf1. However, an alternative view has recently emerged. Data from yeast and humans suggest the existence of tDNA regulatory mechanisms acting at the level of chromatin but these mechanisms largely remained elusive.5,15,35,36 Here, by taking advantage of DNA-barcode sequencing and yeast genetics (Epi-Decoder), we observe that the tDNA proteome in yeast is highly dynamic and contains a previously unknown chromatin-associated factor regulating the assembly of the RNAPIII transcription machinery.
By comparing a tDNA proteome in active and repressive conditions, we observed a major reprogramming of the core RNAPIII transcriptional machinery. Previous studies showing an inverse correlation between selected subunits of TFIIIC and RNAPIII led to the model that TFIIIC and RNAPIII directly compete for binding to tDNAs.52,53 Here we extend and thereby confirm this model by demonstrating the same behavior for all TFIIIC and RNAPIII subunits present in our analyses, and by including TFIIIB. Further support comes from the fpt1Δ background, in which reprogramming of the tDNA transcription machinery was compromised, affecting RNAPIII/TFIIIB and TFIIIC in opposite ways. Importantly, our results suggest that this competitive reprogramming extends to other factors, including non-canonical tDNA binding proteins (Figure 1E, S1D, 5C and S5C). However, to determine whether the observed differences of non-canonical tDNA factors in Epi-Decoder are mediated by the tRNA or Ty1 gene and to understand the functional consequences of the observed proteome dynamics, further studies are required. Overall, the dynamic proteome atlas of a barcoded tDNA locus provides a valuable resource for identifying chromatin-associated factors that potentially regulate tDNA biology.
Epi-Decoder revealed the presence of Fpt1, a protein of unknown function. Our studies suggest that Fpt1 is a genuine member of the tDNA chromatin proteome with regulatory functions. Fpt1 binds specifically at RNAPIII-transcribed genes, its abundance is variable among tDNAs across the genome and correlates with the sensitivity to RNAPIII loss at tDNAs, and its abundance increased upon nutrient perturbation. Moreover, reprogramming of the tDNA chromatin proteome in conditions of repressed transcription is compromised in fpt1Δ cells, resulting in retention of RNAPIII and TFIIIB and lower levels of TFIIIC. Together, these findings point to the existence of a chromatin-associated regulatory mechanism of RNAPIII dynamics in which Fpt1 partially explains the differential regulation of tRNA genes. We expect that other chromatin-bound factors and/or cis-regulatory elements are also involved in this differential regulation. Fpt1 seems to directly or indirectly promote eviction of RNAPIII, by mechanisms that are currently unknown. However, the mechanism seems distinct from that of Maf1. While Maf1 is actively transported to the nucleus in conditions of repressed RNAPIII transcription, Fpt1 is persistently present in the nucleus and already bound to tDNAs in conditions of active transcription. Furthermore, while Maf1 does not ChIP well to chromatin in both yeast (Figure 1D and Roberts et al.65) and humans31, Fpt1 is easily detectable and its occupancy at tRNA genes increases in conditions of stress.
Several lines of evidence suggest that Fpt1 contacts tDNAs through interactions with the core RNAPIII transcription machinery. The ChIP-exo pattern of Fpt1 matches individual parts of the TFIIIB (TBP and Brf1) and TFIIIC (Tfc4 and Tfc6) patterns (Figure 2C–H). This correlated pattern indicates that Fpt1 is in very close proximity to both TFIIIB and TFIIIC. This is further supported by the observation that Fpt1 binding to tRNA genes depends on both TFIIIB and TFIIIC (Figure 6G–K), possibly through direct interactions. The Fpt1 ChIP-exo pattern correlates strongest with TFIIIC (Figure S3D) and Fpt1 shows similar dynamics as TFIIIC upon nutrient perturbations (Figures 1D–E and 6). However, it should be noted that ETCs, sites of extra TFIIIC (ETC) without RNAPIII, are not bound by Fpt1. Therefore, TFIIIC alone cannot explain differential binding of Fpt1 at tRNA genes. Fpt1 binding also depends on TFIIIB (Figure 6J–K) but this seems counterintuitive, since the dynamics of Fpt1 and TFIIIB are anti-correlated. Perhaps, TFIIIB is required in a transient manner to recruit or stabilize Fpt1. Further studies are required to fully understand how both TFIIIC and TFIIIB are involved in Fpt1 recruitment, how this interaction affects the dynamics of the core tDNA transcriptional machinery, and why Fpt1 occupancy varies among tRNA genes.
Taken together, our data extend the current knowledge on regulation of tDNAs by RNAPIII and provide evidence for a chromatin-associated regulatory mechanism embedded in the tDNA chromatin proteome (as outlined in Figure 7D). In conditions of active transcription, Maf1 is phosphorylated and located in the cytoplasm. In the nucleus, the chromatin-bound fraction is composed of the RNAPIII transcription machinery that facilitates tDNA transcription. At the tRNA gene, RNAPIII and TFIIIB are strongly enriched while TFIIIC and Fpt1 are moderately enriched. In repressive conditions, Maf1 is dephosphorylated and imported into the nucleus where it directly interacts with RNAPIII and TFIIIB in the unbound fraction. This prevents assembly of TFIIIB and RNAPIII onto the tDNA. In the chromatin-bound fraction, occupancy of TFIIIB and RNAPIII is decreased while TFIIIC and Fpt1 show increased occupancy at the tRNA gene, supporting a model of competition. In repressive conditions, even though Maf1 prevents RNAPIII assembly and is the major repressor, Fpt1 is required to promote RNAPIII eviction and loading of TFIIIC. Fpt1 may do so by acting on RNAPIII via TFIIIB, and/or by stabilizing TFIIIC. The latter would be in line with the ChIP-exo correlations between Fpt1 and TFIIIC and the proposed dual role of TFIIIC, promoting activation as well as repression of tDNA transcription.52,53
Understanding how Fpt1 interplays with the tDNA chromatin proteome and how it affects RNAPIII dynamics and cellular fitness will provide insights into how yeast as a workhorse in biotechnology can be metabolically optimized for production of biomolecules. Phylogenetic analysis indicates that the FPT1 gene evolved in the Saccharomycetales clade closely related to Saccharomyces cerevisiae. Fpt1 likely has a yeast-specific origin and based on the protein sequence, no Fpt1 homologues have been identified in multicellular eukaryotes. However, we expect that chromatin-associated regulatory mechanisms of RNAPIII assembly are present in other eukaryotes as well. Current advances in genome engineering and proteomics may facilitate the development of strategies to decode tDNAs by barcode sequencing or capture approaches in more complex eukaryotes to explore mechanisms of chromatin-associated regulation of RNAPIII. Our study emphasizes the importance of not overlooking uncharacterized proteins in such efforts, as they may possess alternative regulatory roles that could change our views on fundamental cellular processes.66
Limitations of the Study
This study employs Epi-Decoder, a method designed to systematically decode the chromatin proteome of a single genomic locus. We applied Epi-Decoder to study a native tRNA gene, tP(UGG)M. Focusing on one tRNA gene may introduce a bias for certain classes of tRNA genes. Additionally, tP(UGG)M, like many tRNA genes, is flanked by an RNAPII-regulated Ty1-LTR retrotransposon. This could potentially confound the interpretation of the results or mask changes at the proximal tRNA gene. Therefore, in future experiments it will be interesting to apply Epi-Decoder to multiple tRNA genes of different classes and in different genomic contexts. This advocates for further refinement of the Epi-Decoder technology to facilitate screening multiple genomic loci.
In the context of Epi-Decoder analysis, we standardized protein binding scores by normalization against those of all other proteins. This normalization method enables the assessment of relative protein occupancy, providing valuable insights into changes under different conditions. However, this method is potentially sensitive to offsets by global changes in protein occupancy. To acquire information on absolute protein occupancy and to eliminate potential issues with global changes, it will be useful to develop external spike-in strategies.
Through conditional depletion using the anchor away approach, we observed that maintenance of Fpt1 binding to tRNA genes depends on both TFIIIC and TFIIIB, rather than RNAPIII. Prior to rapamycin treatment, RNAPIII could potentially establish a stable and functional Fpt1 on the chromatin in such a way that the presence of RNAPIII is no longer a necessity for sustained Fpt1 binding. This limitation is intrinsic to the acute protein depletion strategy, which was chosen to avoid lethality associated with mutating essential proteins.
STAR METHODS
RESOURCE AVAILABILIY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fred van Leeuwen (fred.v.leeuwen@nki.nl).
Materials availability
All strains and plasmids generated in this study are available upon reasonable request from the lead contact, except for strains derived from the Yeast TAP Tagged ORFs Collection. The Yeast TAP Tagged ORFs Collection (Yeast TAP-Fusion Library; scTAP) is available from Horizon Discovery (#YSC1177).
Data and code availability
Sequencing data for ChIP-seq (including bigwig files), RNA-seq (including FKPM count data) and ChIP-exo (including BedGraphs files) have been deposited at GEO and are publicly available as of the date of publication. De-multiplexed Epi-Decoder sequencing data and TAP-barcode combinations from Epi-Decoder libraries used in this study have been submitted to the NCBI BioProject database and are publicly available as of the date of publication. Proteomics data have been deposited at the PRIDE database and are publicly available as of the date of publication. Previously published ChIP-exo datasets used in this study can be accessed from the GEO database. Previously published Epi-Decoder data from the HO locus can be found at the NCBI BioProject database. All accession numbers are listed in the key resources table. Original western blot images and microscopy data have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgG from rabbit serum | Sigma-Aldrich | Cat# I5006; RRID:AB_1163659 |
| PGK1 monoclonal | Thermo Fisher Scientific | Cat# 459250; RRID:AB_2532235 |
| TAP Tag polyclonal | Thermo Fisher Scientific | Cat# CAB1001; RRID:AB_10709700 |
| IRDye® 800 anti-rabbit | LI-COR Biosciences | Cat# 926-32211; RRID:AB_621843 |
| IRDye® 680 anti-mouse | LI-COR Biosciences | Cat# 926-68070; RRID:AB_10956588 |
| Chemicals, peptides, and recombinant proteins | ||
| Yeast Nitrogen Base w/o AA, Carbohydrate & w/ AS (YNB) (Powder) | US Biological | Cat# Y2025 |
| Drop-out Mix Complete w/o Yeast Nitrogen Base (Powder) | US Biological | Cat# D9515 |
| Bacto™ Agar | Thermo Fisher Scientific | Cat#214030 |
| Bacto™ Peptone | Thermo Fisher Scientific | Cat# 211677 |
| Bacto™ Yeast Extract | Thermo Fisher Scientific | Cat# 212750 |
| D-Glucose | Sigma-Aldrich | Cat# 8270 |
| Glycerol | Honeywell | Cat# 49770 |
| Ethanol | Honeywell | Cat# 32221 |
| Potassium hydrogen phthalate | VWR | Cat# 26948.260 |
| Myo-inositol | Sigma-Aldrich | Cat# I5125 |
| 4-Amino benzoic acid potassium salt | Sigma-Aldrich | Cat# A0254 |
| Geneticin (G418) Sulfate | Santa Cruz Biotech | Cat# sc-29065A |
| Hygromycin B | Gibco™ | Cat# 10687010 |
| Nourseothricin-dihydrogen sulfate (CloNat) | Werner BioAgents | Cat# 5.002.000 |
| 1,10-phenanthroline | Sigma-Aldrich | Cat# 131377 |
| Rapamycin | LC Laboratories | Cat# R-5000 |
| DMSO | Sigma-Aldrich | Cat# D4540 |
| Formaldehyde solution 37% | Sigma-Aldrich | Cat# 252549 |
| Tris(hydroxymethyl)aminomethane | Millipore | Cat# 1083821000 |
| Glycine | Acros Organics | Cat# 220910050 |
| Hydrochloric acid (fuming) | Honeywell | Cat# 30721 |
| Dynabeads™ M-270 Epoxy | Thermo Fisher Scientific | Cat# 14302D |
| PMSF | Roche | Cat# 10837091001 |
| cOmplete™ EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11873580001 |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | Cat# R5000 |
| Proteinase K from Tritirachium album | Sigma-Aldrich | Cat# P2308 |
| Agencourt AMPure XP beads | Beckman Coultier | Cat# A63881 |
| DAPI | Invitrogen™ | Cat# D1306 |
| MyTaq Red Mix 2x | Meridian Bioscience | Cat# BIO-25044 |
| Phusion™ High-Fidelity DNA Polymerase (2 U/μL) | Thermo Fisher Scientific | Cat# F530L |
| Deoxynucleoside Triphosphate Set | Roche | Cat# 11969064001 |
| DNAse I | New England Biolabs | Cat# M0303S |
| Trichloroacetic acid | Sigma-Aldrich | Cat# T6399 |
| Tris/Glycine/SDS running buffer | Bio-Rad | Cat# 161-0772 |
| 40% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad | Cat# 1610149 |
| TEMED | Sigma-Aldrich | Cat# T9281 |
| Ammonium Persulfate | Thermo Fisher Scientific | Cat# 17874 |
| Agarose MP | Sigma-Aldrich | Cat# 11388991001 |
| Trypsin from bovine pancreas | Sigma-Aldrich | Cat# T1426 |
| Guanidinium hydrochloride | Sigma-Aldrich | Cat# G3272 |
| TCEP | Sigma-Aldrich | Cat# C4706 |
| 2-Chloroacetamide | Sigma-Aldrich | Cat# 22790 |
| Critical commercial assays | ||
| QIAquick PCR Purification Kit | Qiagen | Cat# 28106 |
| KAPA Hyper Prep Kit | Roche | Cat# 07962347001 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q33231 |
| High Sensitivity DNA Kit | Agilent | Cat# 5067-4626 |
| SensiFAST SYBR No-ROX Kit | Meridian Bioscience | Cat# BIO-98020 |
| QIAquick Gel Extraction Kit | Qiagen | Cat# 28706 |
| RNeasy Kit | Qiagen | Cat# 74106 |
| TruSeq Stranded mRNA Kit | Illumina | Cat# 20020594 |
| DC™ Protein Assay Kit II | Bio-Rad | Cat# 5000112 |
| Pierce™ Bradford Protein Assay Kit | Thermo Fisher Scientific | Cat# 23200 |
| Deposited data | ||
| ChIP-sequencing data | This study | GEO: GSE227470 |
| RNA-sequencing data | This study | GEO: GSE227470 |
| ChIP-exo data Fpt1 | This study | GEO: GSE227470 |
| ChIP-exo data others | (Rossi et al, 2021) | GEO: GSE147927 |
| Epi-Decoder tDNA-Ty1 locus data | This study | NCBI BioProject: PRJNA945378 |
| Epi-Decoder HO locus data | (Poramba-Liyanage et al, 2020) | NCBI BioProject: PRJNA610036 (SRX7842936) |
| Proteomics data | This study | PXD045463 |
| Immunoblots and microscopy data | This study | Mendeley Data: 10.17632/stwm3b4jsf.1 |
| Experimental models: Organisms/strains | ||
| Please refer to Table S11 | This study | N/A |
| Oligonucleotides | ||
| Please refer to Table S13 | This study | N/A |
| Recombinant DNA | ||
| Please refer to Table S12 | This study | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al, 2012) | RRID:SCR_003070 |
| ChIP-exo data analysis | This study | https://doi.org/10.5281/zenodo.8381412 |
| GraphPad Prism 9 | GraphPad | RRID:SCR_002798 |
| ScriptManager (version v.014) | (Lang et al, 2022) | RRID:SCR_021797 |
| Picard | Picard | RRID:SCR_006525 |
| BWA-MEM (version 0.7.17-r1188 and 0.7.9a) | (Li & Durbin, 2009) | https://github.com/lh3/bwa |
| GenomicRanges | (Lawrence et al, 2013) | RRID:SCR_000025 |
| Rtracklayer | (Lawrence et al, 2009) | RRID:SCR_021325 |
| RStudio | Rstudio | RRID:SCR_000432 |
| Micro-manager | (Edelstein et al, 2014) | RRID:SCR_000415 |
| Tornadoplot R/Github package | This study | https://doi.org/10.5281/zenodo.8348229 |
| DEseq2 | (Love et al, 2014) | RRID:SCR_015687 |
| FlowJo (version 10.6.1) | FlowJo | RRID:SCR_008520 |
| STAR (version 2.7.1a) | (Dobin et al, 2013) | https://github.com/alexdobin/STAR |
| eXpress (Bioconda Package) | (Roberts & Pachter, 2013) | https://anaconda.org/bioconda/express |
| ShinyGO 0.77 | (Ge et al, 2019) | RRID:SCR_019213 |
| EMBOSS (version 6.6.0) | (Rice et al, 2000) | RRID:SCR_008493 |
| BLAST (version 2.2.31+) | (Camacho et al, 2009) | RRID:SCR_004870 |
| OrthoFinder (version 2.2.7) | (Emms & Kelly, 2019) | RRID:SCR_017118 |
| MAFFT (version 7.271) | (Katoh & Standley, 2013) | RRID:SCR_011811 |
| IQ-Tree (version 1.6.8) | (Nguyen et al, 2015) | RRID:SCR_021163 |
| DIA-NN (version 1.8) | (Demichev et al, 2020) | RRID:SCR_022865 |
| Perseus (version 2.0.10.0) | (Tyanova et al, 2016) | RRID:SCR_015753 |
| Other | ||
| Cover Slips round 25mm (#1.5 thickness) | VWR | Cat# 631-0172 |
| Attofluor™ Cell Chamber, for microscopy | Thermo Fisher Scientific | Cat# A7816 |
| Zeiss 4-alpha Plan-Apochromat 1003/1.46NA Oil | Zeiss | Cat# 420792-9800-000 |
| ORCA Flash 4v3 digital sCMOS camera | Hamamatsu | Cat# C13440-20CU |
| UNO Top stage incubator and objective heater | Okolab | N/A |
| Zirconia-Silicate Beads, 0.5 mm | Biospec | Cat# 11079105Z |
| Amersham™ Protran® nitrocellulose membrane | GE Healthcare Life Science | Cat# 10600002 |
| Sep-Pak C18 1cc cartridges | Waters | Cat# WAT054960 |
| Microlab STAR liquid handling system | Hamilton | N/A |
| Rotor HDA | Singer Instruments | N/A |
| Singer Plusplates™ | Singer Instruments | Cat# PLU-003 |
| Singer Repads™ | Singer Instruments | Cat# REP-004 |
| 2100 Bioanalyzer Instrument | Agilent | Cat# G2939BA |
| DynaMag™−2 Magnet | Invitrogen™ | Cat# 12321D |
| Bioruptor® Pico sonication device | Diagenode | Cat# B01060010 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Strain and plasmid construction
Strains, plasmids and oligos used in this study can be found in Supplemental Table S7. Yeast strains were maintained in YEPD at 30°C unless otherwise specified.
To construct strains, haploid yeast cells (Saccharomyces cerevisiae) were transformed using the LiAc/ssDNA method as described in.68 For standard transformations, BY4741 was used as a parental strain, unless otherwise specified.
Epi-Decoder library manipulations on solid media were performed using synthetic genetic array (SGA) technology50 and a ROTOR instrument (Singer Instruments). The NKI4214 TAP-tag library was made by crossing NKI4212 with the commercially available yeast TAP-tagged ORF library (Horizon Discovery, #YSC1177). Strains NKI5623, NKI4221, NKI5625 and NKI5626 were picked from the NKI4214 library. NKI5601 was made by first integrating a NatMX marker proximal to the locus of interest (between tP(UGG)M and YMLWTy1–2) using plasmid pFvL099 (Supplemental Table S7) and the NatMX Epi-Decoder locus oligos (Supplemental Table S7). The NKI5614 barcode library was made by transforming NKI5601 with the CRISPR-Cas9 plasmid pIla011, containing a gRNA targeting the tDNA-Ty1 locus, and a PCR amplified barcode oligo library (Supplemental Table S7) acting as repair template. The NKI5616 Epi-Decoder library was constructed by crossing the NKI4214 and NKI5614 libraries. Strains NKI5662-NKI5664 were picked from the NKI5616 Epi-Decoder library. NKI5633 was constructed using plasmid pRS400 and fpt1Δ oligos. The NKI5642 fpt1Δ Epi-Decoder library was made by crossing NKI5616 with NKI5633. Strains NKI5665-NKI5667 were picked from the NKI5642 Epi-Decoder library.
NKI5620 was isolated from the YSC1053 knockout library from Open Biosystems. NKI5632 was created using plasmid pFvL029 and Fpt1-TAP oligos. NKI2580 and NKI2581 were constructed using plasmid pFvL099 and maf1Δ oligos. NKI2581 originated from NKI5633. NKI5753 was made using the pKT127 plasmid and Fpt1-GFP oligos. NKI5756 originated from NKI5753, which was transformed with Pac1 digested pTL306 plasmid to insert a fluorescent coat protein and URA3 selection marker at the URA3 locus. NKI5707 and NKI5724 were constructed by amplifying NeonGreen from pET542 with the NeonGreen/Scarlet X-2 locus oligos and using the CRISPR-Cas9 containing pMvB03 plasmid targeting the X-2 locus (a neutral intergenic locus located at ChrX:195625–195645 as described in Mikkelsen et al.69). NKI5712 and NKI5726 were constructed similarly but instead the mScarlet containing pET543 plasmid was used. NKI5636 originated from NKI5623 by amplifying the TDH3 promoter from plasmid pYM-N14 using TDH3-Fpt1 oligos. NKI5771, NKI5772 and NKI5773 were constructed using plasmid pFvL029 and Rpo31-TAP, Brf1-TAP and Tfc3-TAP oligos respectively. NKI5730 originated from BY4741-AA by amplifying the TAP-tag from pFvL029 using the Fpt1-TAP oligos. NKI5744, NKI5747, NKI5748, NKI5749 and NKI5750 originated from NKI5730 by amplifying the FRB-GFP tag from the pTL100 plasmid using the Rpo31-FRB, Bdp1-FRB, Brf1-FRB, Tfc1-FRB and Tfc3-FRB oligos respectively.
Plasmid pMvB03 was constructed by cloning the X-2 gRNA into pML104-NatMX3 using the BclI and SwaI restriction sites. Plasmid pIla011 was constructed by cloning the tDNA-Ty1 gRNA into pML104-URA3 using the BclI and SwaI restriction sites.
Growth conditions and media compositions
For all experiments (except the competitive growth assay and ChIP-exo), liquid yeast cultures were grown to mid-log phase in 2% glucose (YEPD) at 30°C. Optionally, cells were switched to repressive conditions by incubating for 2 hours in 2% ethanol (YEPEtOH) at 30°C or 2% glycerol (YEPgly) at 37°C. For ChIP-exo, cells were grown in 2% glucose (YEPD) at 25°C. In the competitive growth assay, cultures were grown in synthetic complete (SC) media using the following recipes:
Glu-AA
Switching back and forth between SC + 2% glucose (amino acid, AA, complete) and yeast nitrogen base (YNB) + auxotrophic AA + 2% glucose. SC + 2% glucose: 6.7 g YNB w/o amino acids, carbohydrates and with ammonium sulfate + 2 g drop-out mix complete w/o YNB + 100 mL 20% glucose + 900 mL Milli-Q. YNB + auxotrophic AA + 2% glucose: 100 mL 10X YNB (with ammonium sulfate, w/o amino acids) + 200 mL 250 mM potassium hydrogen phthalate + 10 mL methionine (5 mg/mL) + 10 mL histidine (5 mg/mL) + 10 mL leucine (10 mg/mL) + 100 mL uracil (1 mg/mL) + 10 mL adenine (1 mg/mL) + 10 mL myo-inositol (100X solution of 0.2 mg/mL) + 10 mL 4-amino benzoic acid potassium salt (100X solution of 0.02 mg/mL) + 100 mL 20% glucose + 440 mL Milli-Q.
Gly 37°C
Continuous growth in SC + 2% glycerol. SC + 2% glycerol: 6.7 g YNB w/o amino acids, carbohydrates and with ammonium sulfate + 2 g drop-out mix complete w/o YNB + 100 mL 20% glycerol + 900 mL Milli-Q.
Glu/EtOH
Switching back and forth between SC + 2% glucose and SC + 2% ethanol. SC + 2% ethanol: 6.7 g YNB w/o amino acids, carbohydrates and with ammonium sulfate + 2 g drop-out mix complete w/o YNB + 20 mL 100% ethanol + 980 mL Milli-Q.
Media compositions for solid agarose plates used in Epi-Decoder are described in van Breugel et al.43. Solid agarose plates used in spot test analysis (Figure S7D) contained synthetic complete (SC) media including 2% glucose, ethanol or glycerol.
METHOD DETAILS
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as previously described in van Breugel et al.43 with the following modifications. Cell cultures were grown in 75 mL YEPD until mid-log phase (OD660 0.5–0.8). Prior to fixation, 1,10-phenanthroline treated samples were incubated with 100 μg/mL 1,10-phenanthroline for 30 minutes. Control samples were incubated with an equal amount of 100% ethanol (vehicle) for 30 minutes. For anchor away, cells were treated with 7.5 μM rapamycin or DMSO for 60 minutes. Cells were cross-linked for 10 minutes (Rpo31-TAP ChIP) or 15 minutes (all other ChIP experiments) with one-tenth of freshly prepared fix solution (11% formaldehyde, 50 mM Hepes-KOH [pH 7.5], 100 mM NaCl, 1 mM EDTA). Cross-linking was quenched for 5 minutes with glycine (125 mM final concentration) or 1 minute with Tris-HCl pH = 8.0 (750 mM final concentration). Chromatin shearing was performed using the Bioruptor PICO (Diagenode) for 6–10 minutes (depending on the experiment) with 30-second intervals at 4°C. For chromatin immunoprecipitation, one volume of chromatin was mixed with 1/10 volume of Dynabeads M-270 Epoxy (ThermoFisher) coupled to rabbit immunoglobulin G (IgG) and incubated overnight on a turning wheel at 4°C.
ChIP-sequencing
DNA from chromatin immunoprecipitation was prepared for sequencing with the KAPA Hyper Prep Kit (Roche). DNA was amplified with 19 (IP) or 11 cycles (input) and a double sized 0.6X and 1X selection was performed to select for 200–450 bp DNA fragments using AMPure XP beads (Beckman Coultier). An additional 1X size selection was performed on samples that showed high primer-dimer peaks. DNA concentration was measured using a Qubit dsDNA HS Assay Kit (Thermo Fisher) and size-distribution of DNA fragments was visualized with a High Sensitivity DNA Kit (Agilent) and Agilent 2100 Bioanalyzer. Purified DNA was sequenced (single read, 65 bp) on a HiSeq2500 platform (Illumina).
ChIP-exo
Yeast cultures were grown at 25°C in 50 mL YEPD medium to an OD600 of 0.6–0.8. Cells were cross-linked at room temperature with formaldehyde (1% v/v, 15 min) and quenched by glycine (125 mM, 5 min). Cross-linked cells were centrifuged at 4°C (3350 × g, 5 min) and washed with chilled ST buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl). Supernatant was removed and cell pellets were flash frozen and stored at −80°C until further use. Cells were lysed and isolated chromatin was sheared as described previously.70 Briefly, 50 mL culture aliquots were resuspended in 750 μL FA Lysis Buffer (50 mM Hepes-KOH [pH 7.5], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate and complete protease inhibitor (Roche)) and zirconia beads (1 ml, 0.5 mm). Resuspended cells were lysed in a MiniBeadbeater-96 machine (Biospec) by bead beating (4 cycles of 3 min ON and 7 min OFF). Cell lysates were transferred to 1.5 mL Eppendorf tubes and centrifuged at 4°C (16,200 × g, 3 min). Supernatants containing the cytoplasmic fractions were discarded and the obtained chromatin pellets were resuspended in 200 μL FA Lysis Buffer (and 0.1% SDS) along with 75 μL 0.1 mm zirconia beads. Chromatin was sheared in a Bioruptor (Diagenode) by bead beating (4 cycles of 30s ON and 30s OFF). The soluble chromatin fraction was collected by centrifuging the tubes at 4°C (16,200 × g, 10 min). For ChIP-exo, the ChIP-exo 5.0 protocol was performed as described previously.71 Briefly, a single ChIP-exo experiment used chromatin obtained from 50 mL culture and 40 μL rabbit IgG (Sigma) conjugated Dynabeads slurry (~5 μg antibody per ChIP-exo). Chromatin samples mixed with IgG-Dynabeads slurry were incubated overnight at 4°C while rotating to allow for immunoprecipitation. All subsequent steps after immunoprecipitation were essentially performed as described previously.71 ChIP-exo libraries were verified on agarose gels and purified. Purified libraries were sequenced on a NextSeq 500 platform (Illumina, paired-end mode).
ChIP-qPCR
Quantitative PCR was performed on a LightCycler 480 II (Roche) or QuantStudio 5 Real-Time PCR System (Applied Biosystems) and analyzed with software from the manufacturer. In total, 4.2 μL purified DNA from chromatin immunoprecipitation was mixed with 5 μL 2x SensiFAST SYBR No-ROX kit (Bioline) and 0.4 μL forward and reverse primer (10 μM). The quantity of original DNA was determined by interpolating the resulting Cp values from a linear standard curve of values obtained from the dilution-series.
Epi-Decoder
Epi-Decoder was performed as described in van Breugel et al.43. Briefly, a NATMX cassette was inserted in proximity to the tDNA-Ty1 locus. Subsequently, a gRNA-containing CRISPR-Cas9 plasmid (pIla011, Supplemental Table S7) was used to integrate a random 16 bp DNA-barcode oligo library in between the divergent tRNA and Ty1 genes. Colonies were picked and arrayed in a 384-format library to construct a barcoded yeast library. The barcoded yeast library was crossed with a TAP-tag protein library using SGA to create an Epi-Decoder library. Colonies on 384-format Epi-Decoder library plates were pooled together and grown to mid-log phase. Three different barcode-protein combinations represent biological replicates. Just before fixation, the biological replicates were split in three technical replicates (that have the same barcode-protein combinations). Chromatin samples were sonicated for 10 minutes with 30 second intervals. To prepare libraries for sequencing, barcodes in each sample were amplified using unique index primers. In total 23–31 PCR cycles (depending on the experiment and sample) were performed with 25 μL DNA (ChIP undiluted, input 1:50), 8 μL 5x High-Fidelity Phusion buffer (ThermoFisher), 0.4 μL dNTPs (10 mM), 0.4 μL Phusion High-Fidelity DNA Polymerase (ThermoFisher), 0.4 μL primer 1 (p5long-LTR2EF_U5, 10 μM), 0.4 μL primer 2 (sample specific index primer), 0.4 μL P1 Nextera (Illumina) primer (10 μM) and 0.4 μL P2 Nextera (Illumina) primer (10 μM) in a total volume of 40 μL. PCR products were quantified on a 2% agarose gel using ImageJ software.72 Subsequently, PCR products were mixed in equimolar fashion and purified from a 2% agarose gel with a QIAquick gel extraction kit (Qiagen) following the manufacturer’s protocol. Purified DNA was sequenced (single read, 65 bp) on a HiSeq2500 platform (Illumina).
Epi-Decoder validations
Western blot and PCR were used to verify outliers and unexpected hits in the Epi-Decoder screens. We identified a few proteins that lacked a (functional) TAP-tag, were not clonal in the arrayed library, or had a wrong genetic background. Supplemental Table S2 summarizes the excluded proteins. In addition, histone proteins were excluded from the analyses since they are present at very high levels, take up a major part of the read counts, and were spiked-in differently between experiments.
Live cell imaging
Live cell imaging was performed as previously described in detail73 with minor modifications. For Fpt1 imaging in repressive conditions, cells were grown to OD600 0.2–0.4 in SC + 2% glucose and subsequently switched to SC + 2% ethanol (at 30°C) or SC + 2% glycerol (at 37°C) for 2 hours. Cells were imaged on a coverslip with an agarose pad consisting of 2% agarose in SC + 2% glucose, ethanol or glycerol. For validation of anchor away constructs, cells were grown to OD600 0.2–0.4 in SC + 2% glucose and subsequently treated with 7.5 μM rapamycin or DMSO for 60 minutes. Cells were imaged on a coverslip with an agarose pad consisting of 2% agarose in SC + 2% glucose and containing 7.5 μM rapamycin or DMSO. Live cell imaging was performed on a setup consisting of an AxioObserver inverted microscope (Zeiss), an alpha Plan-Apochromat 100× NA 1.46 oil objective, an sCMOS ORCA Flash 4v3 (Hamamatsu) with a 475–570 nm dichroic (Chroma), 570 nm longpass beamsplitter (Chroma), 515/30 nm emission filter (Semrock for GFP) and 600/52 nm emission filter (Semrock for mScarlet), an UNO Top stage incubator (OKOlab) at 30°C, and LED excitation (SpectraX, Lumencor) at 470/24 nm at 100% power for 50 ms exposure (for GFP) and 550/15 nm at 100% power for 150 ms exposure time (for mScarlet-I) resulting in a 3.1 W/cm2 excitation intensity for the GFP signal and 41.3 W/cm2 for the mScarlet-I signal. For each position, a z-stack (9 slices, Δz 0.5 μm) was recorded using Micro-Manager software.74 For each condition, 3–5 images were taken for each of the 3 biological replicates.
Growth assays and flow cytometry
Growth on solid media was measured using spot test analysis. Serial ten-fold dilutions were spotted on plates containing SC media including 2% glucose, glycerol or ethanol. Growth was assessed after 2 days at 30°C or 37°C. For competitive growth assays in liquid media, cells were cultured 48h in pre-competition media (SC + 2% glucose). Wild type and fpt1Δ cells (0.5*106), labelled with mScarlet and NeonGreen fluorescent markers, were mixed in 1 mL SC + 2% glucose (t = 0). A color swap was done to account for reporter effects on cell fitness. Additionally, NeonGreen labelled wild type and mScarlet labelled wild type were mixed to measure fitness defects caused by the fluorescent reporters. Equally mixed NeonGreen and mScarlet labelled cells were inoculated in 5 mL YNB + auxotrophic amino acids + 2% glucose (Glu-AA), SC + 2% ethanol (Glu/EtOH) and SC + 2% glycerol (Gly 37°C) and incubated at 30°C, except for glycerol which was incubated at 37°C. Glu-AA samples were switched to SC + 2% glucose every other day. Glu/EtOH samples were alternated between 24h in SC + 2% glucose and 48h in SC + 2% ethanol. Glycerol samples were diluted every 3–4 days to prevent saturation of the culture. At t = 0, 6 and 14 days, aliquots were collected to make glycerol stocks for storage at −80°C. To analyze samples with flow cytometry, samples were thawed and spun down for 3 minutes at 3824 × g. Glycerol was removed and cells were resuspended in 1 mL SC + 2% glucose and briefly vortexed. Cells were sonicated for 10 seconds at level 3 (Diagenode, Bioruptor). DAPI was used as a dead-cell marker (1:50) and 50.000 events were measured for each sample on a BD LSRFortessa cell analyzer (BD Biosciences) with the following laser-settings: BL(D) 530/30 (NeonGreen), YG(D) 610/20 (mScarlet) and V(F) 450/50 (DAPI).
RNA-sequencing
Cells were grown in 10 mL YEPD to mid-log phase. Half of the culture was spun down (840 × g, 5 minutes) and media was changed to YEP + 2% ethanol. Cells were grown for another 2h. Pellets were harvested by a 2-minute spin at 1881 × g 4°C, washed once with water and stored at −80°C. RNA was isolated using the RNeasy Kit (Qiagen) following the manufacturer’s protocol. DNaseI treatment was performed on the column (NEB) and RNA was stored at −80°C. The Illumina TruSeq Stranded mRNA kit was used to make cDNA library by following the manufacturer’s protocol. Sequencing (single read, 65 bp) of cDNA was done on a HiSeq2500 platform (Illumina).
Immunoblotting
Cells were grown in 15 mL YEPD to mid-log phase. Optionally, cells were switched to repressive conditions or treated with either 1,10-phenanthroline or rapamycin. To collect samples, cell pellets were harvested by spinning cells at 840 × g for 5 minutes. The pellet was washed with 1 mL TE + PMSF (1:500 100 mM). Pellets were stored at −80°C until further processing. Cell pellets were resuspended in 400 μL SUME buffer (1% SDS, 8M Urea, 10mM MOPS, pH 6.8, 10mM EDTA, 0.01% bromophenol blue) containing proteinase inhibitors (proteinase inhibitor cocktail, EDTA-free, Roche). Cells were lysed for 3 minutes by bead beating using zirconia silica beads (BioSpec, 0.5 mm). Samples were incubated for 10 minutes at 65°C and centrifuged at 16,200 × g for 10 minutes to pellet beads and insoluble proteins. Protein concentration of the supernatant was measured using a DC protein assay (Bio-Rad) following the manufacturer’s protocol. Approximately 20–30 μg protein was separated on a polyacrylamide gel in 1x TGS (Tris-Glycine-SDS) running buffer. Proteins were transferred overnight to 0.45 μM nitrocellulose blotting membrane using 0.1 A at 4°C.
Fpt1-TAP immunoblot from Figure 3C
The protocol is similar as described above but with the following modifications. Cell pellets were washed with 1 mL 20% trichloroacetic acid (TCA) and resuspended in 200 μL 20% TCA. Cells were lysed for 3 minutes by bead beating using zirconia silica beads (Biospec, 0.5 mm). Beads were washed 3 times with 200 μL 5% TCA and supernatant was pooled. Cells were centrifuged for 10 minutes at 900 × g at 4°C. Pellets were resuspended in 100 μL Laemmli Buffer and 50 μL 2M Tris pH 8.5 was added to neutralize the solution. Samples were incubated for 5 minutes at 95°C and centrifuged for 10 minutes at 900 × g. Protein extract (6 μL) was separated on a 10% polyacrylamide gel in 1x TGS (Tris-Glycine-SDS) running buffer. Proteins were transferred to 0.45 μM nitrocellulose blotting membrane for 2h using 1 A at 4°C.
Membranes were blocked with 5% milk powder (Nutrilon) in PBS for 30 minutes at room temperature. Primary antibody staining for anti-TAP (Invitrogen CAB1001 rabbit anti-TAP, 1:2500) was done in 2% milk in TBS-T for 2h. After washing three times with TBS-T, membranes were incubated with the secondary antibody (goat anti-rabbit antibody IRDye 800, Licor, 1:10000) in 2% milk in TBS-T for 45 minutes. Pgk1 was stained with Pgk1 primary mouse antibody (Invitrogen 22C508, 1:2000) and secondary antibody (donkey anti-mouse antibody IRDye 680, Licor, 1:10000) for 2h and 45 minutes respectively in 2% milk in TBS-T. Membranes were washed once in PBS before imaging on a LI-COR Odyssey DLx Imager (LI-COR Biosciences).
Mass spectrometry
Cells were grown in 100 mL YEPD until mid-log phase and split in two before harvesting. Half of the culture (50 mL) was spun down by a 5-minute spin at 840 × g and subsequently switched to 2% glycerol at 37°C for 2 hours. Samples were harvested by a 5-minute spin at 840 × g at 4°C, and washed once with 10 mL PBS. Pellets were stored at −80°C until further processing. Cell pellets were resuspended in 400 μL lysis buffer (6 M guanidinium hydrochloride [GuHCl], 5 mM tris(2-carboxyethyl)phosphine, 10 mM chloroacetamide, 100 mM Tris-HCl [pH = 8.5]) and lysed using zirconia silica beads (Biospec, 0.5 mm) in 3× 1.5-minute intervals. Protein lysates were boiled for 10 minutes at 95°C. To pellet debris, lysates were spun 5 minutes at maximum speed. Protein concentration was determined with a Pierce Coomassie (Bradford) Protein Assay Kit (Thermo Scientific), according to the manufacturer’s instructions. After dilution to 2M GuHCl, aliquots corresponding to 200 μg of protein were digested twice (overnight and 4h) with trypsin at 37°C, enzyme/substrate ratio 1:75. Digestion was quenched by the addition of formic acid (final concentration 5%), after which the peptides were desalted on a Sep-Pak C18 cartridge (Waters, Massachusetts, USA). Samples were dried in a vacuum centrifuge. Prior to mass spectrometry analysis, the peptides were reconstituted in 2% formic acid. Peptide mixtures were analyzed by nanoLC-MS/MS on an Orbitrap Exploris 480 Mass Spectrometer equipped with an EASY-NLC 1200 system (Thermo Scientific). Samples were directly loaded onto the analytical column (ReproSil-Pur 120 C18-AQ, 2.4 μm, 75 μm × 500 mm, packed in-house). Solvent A was 0.1% formic acid/water and solvent B was 0.1% formic acid/80% acetonitrile. Samples were eluted from the analytical column at a constant flow of 250 nL/min in a 90-minute gradient, containing a 78-minute linear increase from 6% to 30% solvent B, followed by a 12-minute wash at 90% solvent B. The Exploris 480 was run in data-independent acquisition (DIA) mode, with full MS resolution set to 120,000 at m/z 200, MS1 mass range was set from 350–1400, normalized AGC target was 300% and maximum IT was 45ms. DIA was performed on precursors from 400–1000 in 48 windows of 12.5 m/z with an overlap of 1 m/z. Resolution was set to 30,000 and normalized CE was 27.
QUANTIFICTION AND STATISTICAL ANALYSIS
Statistical details of experiments can be found in the figure legends. In the figure legends, ‘n’ represents the number of biological replicates unless otherwise specified. Statistical analyses were performed using Prism 9 V9.4.1.
ChIP-sequencing data analysis
ChIP-sequencing reads were mapped to the sacCer3 reference genome downloaded from UCSC using the BWA-MEM program (version 0.7.17-r1188)75 using the option ‘-M’. No filtering for mapping quality was performed. Coverage tracks were created using the GenomicRanges76 and rtracklayer77 R/Bioconductor packages. Specifically, unique reads were extended 200 bp from their 5’ end and received a penalty for the number of alternative mapping positions. Coverage was down sampled to integrate to 1x genome size (reads per genomic content, RPGC). For counting reads in gene bodies, we took gene bodies from Ensembl’s R64–1-1.108 GTF annotation78 and extended these by 150 bp in both directions, specifically to more generously count reads at tDNA positions. Counting was then performed using the ‘summarizeOverlaps’ function from the GenomicAlignments R/Bioconductor package76, wherein parameters were set such that the minimum mapping quality was greater than 10 and no duplicated reads were counted. To make the tornado heat map of ChIP-seq and ChIP-exo signal at tDNAs, excluding mitochondrial tDNAs, we used the tornadoplot R/GitHub package (10.5281/zenodo.8348229) with arguments set to ‘width = 2000, binwidth = 1’ and sorted on Fpt1 signal. R version 4.2.1 and Bioconductor release 3.15 were used.
ChIP-exo data analysis
Obtained reads were aligned to the S. cerevisiae genome (sacCer3) using BWA-MEM (version 0.7.9a).75 PCR duplicates and non-unique alignments were subsequently removed. All data files were deduplicated and analyses were performed on ScriptManager (version v.014).79 ChIP-exo 5’ tags of TAP-tagged strains were NCIS normalized relative to the no tag negative control (PMID: 33692541), and mapped to the TSSs of tRNA genes. The data analysis parameters used in ScriptManager can be accessed via https://github.com/CEGRcode/2023-Breugel_JournalXXXX/releases.
ChIP-sequencing and ChIP-exo data processing
Count data at individual tRNA genes from ChIP-seq and ChIP-exo and an overview of tDNAs included in the ChIP-seq heat maps can be found in Supplemental Table S4.
ChIP-sequencing
Genome tracks as in Figure 2A–B and S3A (bigwig files) were displayed in the UCSC genome browser. For Fpt1 counts at tRNA genes (as in Figure 2I) in the ChIP-seq dataset, tRNA genes with <5 mapped read counts for Rpo31 and Fpt1 (n = 20) were excluded.
ChIP-exo
Hypervariable tRNA genes were excluded from the analysis leaving 261 tRNA genes. Selection criteria and coordinate reference files were obtained from a previously published study to analyze Fpt1 (and related) datasets.58 ChIP-exo plots were smoothed using 10 neighbors and 4th order (Prism 9 V9.4.1). Heat maps were based on NCIS normalized CDT files that contain strand-combined data.
Classification of housekeeping and regulated tRNA genes
Based on UV cross-linking and analysis of cDNA (CRAC) to capture nascent RNAs bound by RNAPIII, two classes of tRNA genes have been identified by Turowski et al.25 (Supplemental Table S4). The CRAC dataset contains data for 275 tRNA genes. One canonical class of ‘regulated tRNA genes’ shows loss of engaged RNAPIII upon a switch from glucose to glycerol (RNAPIII CRAC gly/glu ratio < 1, here described as low RNAPIII retention score) and repression that is dependent on Maf1 (RNAPIII CRAC maf1Δ/wt ratio > 1, here described as high Maf1 dependency score). The other class of ‘housekeeping tRNA genes’ is defined as RNAPIII CRAC gly/glu ratio > 1 (high RNAPIII retention score), and RNAPIII CRAC maf1Δ/wt ratio < 1 (low Maf1 dependency score). For Figures 2I and S3E (ChIP-seq), a subset of tRNA genes was used. After filtering for mapped reads, tRNA genes that had gly/glu or maf1Δ/wt ratios classified as outliers (determined with a box plot) were not included (n = 12), resulting in 243 tRNA genes. The same tRNA genes were excluded for Figures S3F and S3G (ChIP-exo), resulting in 249 tRNA genes.
Epi-Decoder data analysis
Samples were de-multiplexed using the 12 bp sample index resulting in 1.5–6M reads per sample. Barcode counting and data preprocessing and filtering were performed as described previously42 with minor modifications. To optimize the normalization step and since most analyses only required a specific subset of the data, two subsets were created: 1) only tDNA samples (Epi-Decoder tDNA-Ty1 locus) named ‘tDNA’ and 2) only HO samples (Epi-Decoder HO locus) named ‘ho’. In the ‘tDNA’ subset, counts of technical replicates were summed to a biological replicate, in the ‘ho’ subset the mean was taken. For normalization, the relative library size factor was calculated. For the ‘tDNA’ subset, this factor was based on the total library size and for the ‘ho’ subset on the sample median because the ‘ho’ subset is screened in sub pools. Additionally, a combined set of the two subsets was created: ‘tDNA_ho’. The counts of the two underlying subsets were normalized using a relative size factor based on total library size. For differential analysis of enrichment (ChIP) in ‘tDNA’ and ‘tDNA_ho’, extra columns were created with ChIP (IP) values corrected for differences in input (IN). For this, a relative size factor was calculated per row (protein) for each of the IN conditions to correct the corresponding IP. Differential analysis was based on a paired analysis with DEseq280 using the ‘tDNA’ subset with the following two exceptions. Differential analysis of IP and input at the HO locus in glucose was based on the ‘ho’-subset and was done paired. Differential analysis of IP between tDNA and HO was done based on the ‘tDNA_ho’ subset using the corrected IP values and was done unpaired.
Live cell imaging data analysis
For localization of Fpt1, the microscopy data was analyzed using custom Python software (10.5281/zenodo.7650172). First, a maximum intensity projection was made. Next, the mScarlet-I channel was used to segment cells using Otsu thresh-holding and water shedding. For each cell, the total nuclear mScarlet-I intensity was determined in each of the 9 slices in the z-stack, and the z-slice with the maximum signal was taken to be the slice where the nucleus is in focus. In this z-slice, the nuclear and cytoplasmic intensity were determined and background correction was performed on nuclear and cytoplasmic intensities by subtracting the median value of all intensities measured in the same z-slice outside the cells. For each cell, the nuclear enrichment was taken to be the ratio between the median nuclear and median cytoplasmic signal.
Competitive growth assay and flow cytometry analysis
Flow cytometry data was processed and analyzed using FlowJo software (version 10.6.1). The Malthusian coefficient m was calculated as described previously81 using in which MUTend and WTend are the fraction of fpt1Δ and wild type cells at t = 14, MUTstart and WTstart the fraction at t = 0 and t is the number of generations. The number of generations was determined based on known growth curves. The Malthusian coefficient was corrected for reporter effects determined by the wild type/wild type mixtures.
RNA-sequencing data analysis
Reads were mapped to SacCer3 using STAR (version 2.7.1a)82 and count tables were generated using the bioconda package eXpress.83 Differential expression analysis was performed with DEseq2.80 Gene ontology analysis on differentially expressed genes was performed using ShinyGO (version 0.77).84
Immunoblotting analysis
Quantification of gels was done using ImageJ software72 by applying the area under the curve method. Pgk1 protein levels were used to normalize target protein levels.
Phylogenetic analysis
Orthologous sequences of YKR011C were retrieved from 77 publicly available yeast genomes. For 41 genomes, gene model annotations and the corresponding amino acid sequences were publicly available (“annotated proteins” column in Supplemental Table S7). For the remaining 36 genomes, the protein annotations were not available, and putative orthologues of YKR011C were predicted with a sequence similarity search (“predicted protein” column in Supplemental Table S7). Putative orthologous were retrieved by stringent sequence similarity search (e-value < 10e-40) using EMBOSS (version 6.6.0)85 and BLAST (version 2.2.31+)86 and using the 24 previously identified YKR011C orthologues as query sequences. Yeast proteomes were clustered in gene families using OrthoFinder (version 2.2.7).87 Protein sequences were aligned with MAFFT (version 7.271)88 and a Maximum Likelihood tree was inferred with IQ-Tree (version 1.6.8).89 Hanseniaspora sequences were highly divergent from other YKR011C orthologues and therefore dropped from the alignment.
Mass spectrometry data analysis
Raw data were analyzed by DIA-NN (version 1.8)90 without a spectral library and with “Deep learning” option enabled. The Swissprot Yeast database (6,727 entries, release 2023_07) was added for the library-free search. The Quantification strategy was set to Robust LC (high accuracy) and MBR option was enabled. The other settings were kept at the default values. The protein groups report from DIA-NN was used for downstream analysis in Perseus (version 2.0.10.0).91 Values were Log2-transformed, after which proteins were filtered for at least 75% valid values in at least one sample group. Missing values were replaced by imputation based a normal distribution using a width of 0.3 and a minimal downshift of 2.4. Differentially expressed proteins were determined using a Student’s t-test (minimal threshold: –log(p-value) ≥ 1.3 and [x-y] ≥ 0.5 | [x-y] ≤ −0.5).
ADDITIONAL RESOURCES
YKR011C is referred to as FPT1 (Factor in the Proteome of tDNAs number 1). This name has been reserved in the Yeast Genome Database: www.yeastgenome.org/locus/S000001719.
Supplementary Material
Supplemental Table S1. Barcode counts Epi-Decoder HO locus glucose (wild type) and tDNA-Ty1 locus glucose and glycerol (2h) 37°C in both wild type and fpt1Δ, related to Figures 1, S1, 5 and S5.
Supplemental Table S2. Epi-Decoder DEseq2 analysis to determine binding scores, differential binders between two conditions or loci and a list of proteins excluded from Epi-Decoder analysis, related to Figures 1, S1, 5 and S5.
Supplemental Table S3. Proteomics data for wild type and fpt1Δ in glucose and glycerol (2h) 37°C, related to Figure S1.
Supplemental Table S4. Information on tRNA genes, including CRAC data, housekeeping/regulated classification and ChIP-seq and ChIP-exo counts at tRNA genes, related to Figures 2 and S3.
Supplemental Table S7. List of strains, plasmids and oligos used in this study and genome versions of species described in the ML tree, related to STAR Methods and Figure S2.
Highlights.
The chromatin proteome of a tRNA gene locus is dynamically regulated
Fpt1 differentially and dynamically occupies RNA Polymerase III transcribed genes
Occupancy of Fpt1 depends on TFIIIB and TFIIIC, not RNA Polymerase III
Fpt1 promotes eviction of RNA Polymerase III
ACKNOWLEDGEMENTS
This research was supported by an institutional grant of the Dutch Cancer Society and of the Dutch Ministry of Health, Welfare and Sport, by the Dutch Research Council (grant NWO-NCI-LIFT-731.015.405 to F.v.L. and grant 016.Veni.192.071 to I.B.), by the US National Institutes of Health (grant GM145217 to B.F.P.), by the Dutch NWO X-omics Initiative (to L.H.) and by support for T.L.L. by the Oncode Institute (which is partly financed by the Dutch Cancer Society) and the European Research Council (ERC Starting Grant 755695 BURSTREG). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank David Tollervey and Tomasz Turowksi for sharing the CRAC dataset on RNAPIII transcripts and discussing the project. We thank Evelina Tuttuci for sharing the pET542 and pET543 plasmids and Johan van Heerden and Frank Bruggeman for discussing the project. We thank Ben Morris for help with re-arraying yeast libraries by robotics and Pascale Daran-Lapujade for advice regarding the neutral X-2 locus. We thank João Caetano and Mireia Novell Cardona for their input in the 1,10-phenanthroline and anchor away studies. We thank Folkert van Werven and Fabian Moretto for analysis of Ty1 expression. We thank the Genomics Core Facility, Robotics Facility, High Throughput Screening Facility, Proteomics Facility, High Performance Computing Facility and Flow Cytometry Facility of the NKI for assistance. We thank members of the F.v.L. lab for helpful discussions. We acknowledge the following resources: Yeast Genome Database (SGD Project), BioRender.com, Platform for Epigenomic and Genomic Research, RRID:SCR_021861; Cornell University Biotechnology Resource Center Epigenomics Core Facility, RRID:SCR_021287; Cornell University BRC Genomics Core Facility, RRID:SCR_021727; Pennsylvania State University’s Institute for Computational and Data Sciences Advanced Cyberinfrastructure.
B.F.P. is an owner of and has a financial interest in Peconic, which uses the ChIP-exo technology (U.S. Patent 20100323361A1) implemented in this study and could potentially benefit from the outcomes of this research.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Turowski TW, and Tollervey D. (2016). Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem. Soc. Trans 44, 1367–1375. 10.1042/BST20160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girbig M, Misiaszek AD, and Müller CW (2022). Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat. Rev. Mol. Cell Biol 23, 603–622. 10.1038/s41580-022-00476-9. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P. (2019). Organization and regulation of gene transcription. Nature 573, 45–54. 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 4.Vannini A, and Cramer P. (2012). Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446. 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Willis IM, and Moir RD (2018). Signaling to and from the RNA Polymerase III Transcription and Processing Machinery. Annu. Rev. Biochem 87, 75–100. 10.1146/annurev-biochem-062917-012624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orellana EA, Siegal E, and Gregory RI (2022). tRNA dysregulation and disease. Nat. Rev. Genet 23, 651–664. 10.1038/s41576-022-00501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta T, Malkin MG, and Huang S. (2022). tRNA Function and Dysregulation in Cancer. Front. cell Dev. Biol 10, 886642. 10.3389/fcell.2022.886642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyczewska A, Rzepczak A, Sobańska D, and Grzywacz K. (2023). The emerging roles of tRNAs and tRNA-derived fragments during aging: Lessons from studies on model organisms. Ageing Res. Rev 85, 101863. 10.1016/j.arr.2023.101863. [DOI] [PubMed] [Google Scholar]
- 9.Guimarães AR, Correia I, Sousa I, Oliveira C, Moura G, Bezerra AR, and Santos MAS (2021). tRNAs as a Driving Force of Genome Evolution in Yeast. Front. Microbiol 12, 634004. 10.3389/fmicb.2021.634004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheepbouwer C, Aparicio-Puerta E, Gomez-Martin C, Verschueren H, van Eijndhoven M, Wedekind LE, Giannoukakos S, Hijmering N, Gasparotto L, van der Galien HT, et al. (2023). ALL-tRNAseq enables robust tRNA profiling in tissue samples. Genes Dev. 37, 243–257. 10.1101/gad.350233.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leśniewska E, and Boguta M. (2017). Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes. Open Biol. 7, 170001. 10.1098/rsob.170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, and Gartenberg MR (2014). Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 28, 959–970. 10.1101/GAD.236729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee K, Nostramo RT, Wan Y, and Hopper AK (2018). tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim. Biophys. Acta 1861, 373. 10.1016/J.BBAGRM.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phizicky EM, and Hopper AK (2023). The Life and Times of a tRNA. RNA. 10.1261/rna.079620.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber A, Ito K, Chu C-S, and Roeder RG (2020). Gene-Specific Control of tRNA Expression by RNA Polymerase II. Mol. Cell 78, 765–778.e7. 10.1016/j.molcel.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J, Aiello U, Clement Y, Haidara N, Girbig M, Schmitzova J, Pena V, Müller CW, Libri D, and Porrua O. (2022). An integrated model for termination of RNA polymerase III transcription. Sci. Adv 8, eabm9875. 10.1126/sciadv.abm9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J-L, Lee Y-S, Kunkeaw N, Kim S-Y, Kim I-H, and Lee YS (2017). Epigenetic regulation of noncoding RNA transcription by mammalian RNA polymerase III. Epigenomics 9, 171–187. 10.2217/epi-2016-0108. [DOI] [PubMed] [Google Scholar]
- 18.Acker J, Conesa C, and Lefebvre O. (2013). Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta 1829, 283–295. 10.1016/j.bbagrm.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Moir RD, and Willis IM (2013). Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta 1829, 361–375. 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Roman N, Grandori C, Eisenman RN, and White RJ (2003). Direct activation of RNA polymerase III transcription by c-Myc. Nature 421, 290–294. 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 21.Boguta M, Czerska K, and Żołądek T. (1997). Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185, 291–296. 10.1016/S0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- 22.Vorländer MK, Baudin F, Moir RD, Wetzel R, Hagen WJH, Willis IM, and Müller CW (2020). Structural basis for RNA polymerase III transcription repression by Maf1. Nat. Struct. Mol. Biol 27, 229–232. 10.1038/s41594-020-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai N, Lee JH, Upadhya R, Chu Y, Moir RD, and Willis IM (2005). Two Steps in Maf1-dependent Repression of Transcription by RNA Polymerase III. J. Biol. Chem 280, 6455–6462. 10.1074/JBC.M412375200. [DOI] [PubMed] [Google Scholar]
- 24.Ciesla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, Balicki K, Conesa C, Lefebvre O, and Boguta M. (2007). Maf1 Is Involved in Coupling Carbon Metabolism to RNA Polymerase III Transcription. Mol. Cell. Biol 27, 7693–7702. 10.1128/mcb.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turowski TW, Leśniewska E, Delan-Forino C, Sayou C, Boguta M, and Tollervey D. (2016). Global analysis of transcriptionally engaged yeast RNA polymerase III reveals extended tRNA transcripts. Genome Res. 26, 933–944. 10.1101/gr.205492.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloom-Ackermann Z, Navon S, Gingold H, Towers R, Pilpel Y, and Dahan O. (2014). A Comprehensive tRNA Deletion Library Unravels the Genetic Architecture of the tRNA Pool. PLOS Genet. 10, e1004084. 10.1371/journal.pgen.1004084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aharon-Hefetz N, Frumkin I, Mayshar Y, Dahan O, Pilpel Y, and Rak R. (2020). Manipulation of the human tRNA pool reveals distinct tRNA sets that act in cellular proliferation or cell cycle arrest. Elife 9. 10.7554/eLife.58461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dittmar KA, Goodenbour JM, and Pan T. (2006). Tissue-Specific Differences in Human Transfer RNA Expression. PLOS Genet. 2, e221. 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Ye Y, Gong J, Ruan H, Liu C-J, Xiang Y, Cai C, Guo A-Y, Ling J, Diao L, et al. (2018). Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun. Biol 1, 234. 10.1038/s42003-018-0239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. (2014). A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292. 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Orioli A, Praz V, Lhôte P, and Hernandez N. (2016). Human MAF1 targets and represses active RNA polymerase III genes by preventing recruitment rather than inducing long-term transcriptional arrest. Genome Res. 26, 624–634. 10.1101/GR.201400.115/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thandapani P, Kloetgen A, Witkowski MT, Glytsou C, Lee AK, Wang E, Wang J, LeBoeuf SE, Avrampou K, Papagiannakopoulos T, et al. (2022). Valine tRNA levels and availability regulate complex I assembly in leukaemia. Nature 601, 428–433. 10.1038/s41586-021-04244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earnest-Noble LB, Hsu D, Chen S, Asgharian H, Nandan M, Passarelli MC, Goodarzi H, and Tavazoie SF (2022). Two isoleucyl tRNAs that decode synonymous codons divergently regulate breast cancer metastatic growth by controlling translation of proliferation-regulating genes. Nat. Cancer 3, 1484–1497. 10.1038/s43018-022-00469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rak R, Dahan O, and Pilpel Y. (2018). Repertoires of tRNAs: The Couplers of Genomics and Proteomics. Annu. Rev. Cell Dev. Biol 34, 239–264. 10.1146/annurev-cellbio-100617-062754. [DOI] [PubMed] [Google Scholar]
- 35.Arimbasseri AG, and Maraia RJ (2016). RNA Polymerase III Advances: Structural and tRNA Functional Views. Trends Biochem. Sci 41, 546–559. 10.1016/j.tibs.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt BM, Rudolph KLM, Karagianni P, Fonseca NA, White RJ, Talianidis I, Odom DT, Marioni JC, and Kutter C. (2014). High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA–tRNA interface. Genome Res. 24, 1797–1807. 10.1101/gr.176784.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189. 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teytelman L, Thurtle DM, Rine J, and Van Oudenaarden A. (2013). Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U. S. A 110, 18602–18607. 10.1073/PNAS.1316064110/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behrens A, Rodschinka G, and Nedialkova DD (2021). High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 10.1016/j.molcel.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837. 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poramba-Liyanage DW, Korthout T, Cucinotta CE, van Kruijsbergen I, van Welsem T, El Atmioui D, Ovaa H, Tsukiyama T, and van Leeuwen F. (2020). Inhibition of transcription leads to rewiring of locus-specific chromatin proteomes. Genome Res. 10.1101/gr.256255.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korthout T, Poramba-Liyanage DW, van Kruijsbergen I, Verzijlbergen KF, van Gemert FPA, van Welsem T, and van Leeuwen F. (2018). Decoding the chromatin proteome of a single genomic locus by DNA sequencing. PLOS Biol. 16, e2005542. 10.1371/journal.pbio.2005542 S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Breugel ME, and van Leeuwen F. (2022). Epi-Decoder: Decoding the Local Proteome of a Genomic Locus by Massive Parallel Chromatin Immunoprecipitation Combined with DNA-Barcode Sequencing. Methods Mol. Biol 2458, 123–150. 10.1007/978-1-0716-2140-0_8. [DOI] [PubMed] [Google Scholar]
- 44.Gauchier M, van Mierlo G, Vermeulen M, and Déjardin J. (2020). Purification and enrichment of specific chromatin loci. Nat. Methods 17, 380–389. 10.1038/s41592-020-0765-4. [DOI] [PubMed] [Google Scholar]
- 45.Wierer M, and Mann M. (2016). Proteomics to study DNA-bound and chromatin-associated gene regulatory complexes. Hum. Mol. Genet 25, R106–R114. 10.1093/hmg/ddw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigismondo G, Papageorgiou DN, and Krijgsveld J. (2022). Cracking chromatin with proteomics: From chromatome to histone modifications. Proteomics 22, e2100206. 10.1002/pmic.202100206. [DOI] [PubMed] [Google Scholar]
- 47.Cheung S, Manhas S, and Measday V. (2018). Retrotransposon targeting to RNA polymerase III-transcribed genes. Mob. DNA 9, 14. 10.1186/s13100-018-0119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Saïb A, Werner M, Voytas DF, and Lesage P. (2015). An RNA polymerase III subunit determines sites of retrotransposon integration. Science (80-. ) 348. 10.1126/science.1259114. [DOI] [PubMed] [Google Scholar]
- 49.Devine SE, and Boeke JD (1996). Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10, 620–633. 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 50.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368. 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 51.Hummel G, Warren J, and Drouard L. (2019). The multi-faceted regulation of nuclear tRNA gene transcription. IUBMB Life 71, 1099–1108. 10.1002/iub.2097. [DOI] [PubMed] [Google Scholar]
- 52.Cieśla M, Skowronek E, and Boguta M. (2018). Function of TFIIIC, RNA polymerase III initiation factor, in activation and repression of tRNA gene transcription. Nucleic Acids Res. 46, 9444. 10.1093/NAR/GKY656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts DN, Stewart AJ, Huff JT, and Cairns BR (2003). The RNA polymerase III transcriptome revealed by genome-wide localization and activity–occupancy relationships. Proc. Natl. Acad. Sci 100, 14695–14700. 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez S, Livingstone-Zatchej M, Jourdain S, Thoma F, Sentenac A, and Marsolier M-C (2001). High-Mobility-Group Proteins NHP6A and NHP6B Participate in Activation of the RNA Polymerase III SNR6 Gene. Mol. Cell. Biol 21, 3096. 10.1128/MCB.21.9.3096-3104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kassavetis GA, and Steiner DF (2006). Nhp6 Is a Transcriptional Initiation Fidelity Factor for RNA Polymerase III Transcription in Vitro and in Vivo. J. Biol. Chem 281, 7445–7451. 10.1074/JBC.M512810200. [DOI] [PubMed] [Google Scholar]
- 56.Braglia P, Dugas SL, Donze D, and Dieci G. (2007). Requirement of Nhp6 Proteins for Transcription of a Subset of tRNA Genes and Heterochromatin Barrier Function in Saccharomyces cerevisiae. Mol. Cell. Biol 27, 1545–1557. 10.1128/MCB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moqtaderi Z, and Struhl K. (2004). Genome-Wide Occupancy Profile of the RNA Polymerase III Machinery in Saccharomyces cerevisiae Reveals Loci with Incomplete Transcription Complexes. Mol. Cell. Biol 24, 4118:4127. 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi MJ, Kuntala PK, Lai WKM, Yamada N, Badjatia N, Mittal C, Kuzu G, Bocklund K, Farrell NP, Blanda TR, et al. (2021). A high-resolution protein architecture of the budding yeast genome. Nature 592, 309–314. 10.1038/s41586-021-03314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee HS, and Pugh BF (2012). Genome-wide structure and organization of eukaryotic preinitiation complexes. Nature 483, 295–301. 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oler AJ, and Cairns BR (2012). PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J. 31, 1440. 10.1038/EMBOJ.2011.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zencir S, Dilg D, Shore D, and Albert B. (2022). Pitfalls in using phenanthroline to study the causal relationship between promoter nucleosome acetylation and transcription. Nat. Commun 2022 131 13, 1–4. 10.1038/s41467-022-30350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haruki H, Nishikawa J, and Laemmli UK (2008). The Anchor-Away Technique: Rapid, Conditional Establishment of Yeast Mutant Phenotypes. Mol. Cell 31, 925–932. 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 63.Santos DA, Shi L, Tu BP, and Weissman JS (2019). Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 47, 4974–4985. 10.1093/NAR/GKZ205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudzińska I, Cieśla M, Turowski TW, Armatowska A, Leśniewska E, and Boguta M. (2021). Reprogramming mRNA Expression in Response to Defect in RNA Polymerase III Assembly in the Yeast Saccharomyces cerevisiae. Int. J. Mol. Sci 22, 7298. 10.3390/ijms22147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts DN, Wilson B, Huff JT, Stewart AJ, and Cairns BR (2006). Dephosphorylation and Genome-Wide Association of Maf1 with Pol III-Transcribed Genes during Repression. Mol. Cell 22, 633. 10.1016/J.MOLCEL.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kustatscher G, Collins T, Gingras AC, Guo T, Hermjakob H, Ideker T, Lilley KS, Lundberg E, Marcotte EM, Ralser M, et al. (2022). Understudied proteins: opportunities and challenges for functional proteomics. Nat. Methods 2022 197 19, 774–779. 10.1038/s41592-022-01454-x. [DOI] [PubMed] [Google Scholar]
- 67.MacKinnon JG (2007). Bootstrap Hypothesis Testing. https://ideas.repec.org/p/qed/wpaper/1127.html.
- 68.Gietz RD, and Woods RA (2006). Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol 313, 107–120. 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 69.Mikkelsen MD, Buron LD, Salomonsen B, Olsen CE, Hansen BG, Mortensen UH, and Halkier BA (2012). Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng 14, 104–111. 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Mittal C, Lang O, Lai WKM, and Pugh BF (2022). An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters. Genes Dev. 36, 985–1001. 10.1101/gad.350026.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rossi MJ, Lai WKM, and Pugh BF (2018). Simplified ChIP-exo assays. Nat. Commun 9, 2842. 10.1038/s41467-018-05265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brouwer I, Patel HP, Meeussen JVW, Pomp W, and Lenstra TL (2020). Single-Molecule Fluorescence Imaging in Living Saccharomyces cerevisiae Cells. STAR Protoc. 1, 100142. 10.1016/j.xpro.2020.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N. (2014). Advanced methods of microscope control using μManager software. J. Biol. methods 1 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H, and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for Computing and Annotating Genomic Ranges. PLOS Comput. Biol 9, e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence M, Gentleman R, and Carey V. (2009). rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842. 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. (2022). Ensembl 2022. Nucleic Acids Res. 50, D988–D995. 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lang O, Pugh BF, and Lai WKM (2022). ScriptManager: An Interactive Platform for Reducing Barriers to Genomics Analysis. In Practice and Experience in Advanced Research Computing PEARC ‘22. (Association for Computing Machinery). 10.1145/3491418.3535161. [DOI] [Google Scholar]
- 80.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hittinger CT, and Carroll SB (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681. 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 82.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts A, and Pachter L. (2013). Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10, 71–73. 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge SX, Jung D, and Yao R. (2019). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rice P, Longden I, and Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 86.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emms DM, and Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Katoh K, and Standley DM (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol 30, 772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen L-T, Schmidt HA, von Haeseler A, and Minh BQ (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32, 268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Demichev V, Messner CB, Vernardis SI, Lilley KS, and Ralser M. (2020). DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44. 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Barcode counts Epi-Decoder HO locus glucose (wild type) and tDNA-Ty1 locus glucose and glycerol (2h) 37°C in both wild type and fpt1Δ, related to Figures 1, S1, 5 and S5.
Supplemental Table S2. Epi-Decoder DEseq2 analysis to determine binding scores, differential binders between two conditions or loci and a list of proteins excluded from Epi-Decoder analysis, related to Figures 1, S1, 5 and S5.
Supplemental Table S3. Proteomics data for wild type and fpt1Δ in glucose and glycerol (2h) 37°C, related to Figure S1.
Supplemental Table S4. Information on tRNA genes, including CRAC data, housekeeping/regulated classification and ChIP-seq and ChIP-exo counts at tRNA genes, related to Figures 2 and S3.
Supplemental Table S7. List of strains, plasmids and oligos used in this study and genome versions of species described in the ML tree, related to STAR Methods and Figure S2.
Data Availability Statement
Sequencing data for ChIP-seq (including bigwig files), RNA-seq (including FKPM count data) and ChIP-exo (including BedGraphs files) have been deposited at GEO and are publicly available as of the date of publication. De-multiplexed Epi-Decoder sequencing data and TAP-barcode combinations from Epi-Decoder libraries used in this study have been submitted to the NCBI BioProject database and are publicly available as of the date of publication. Proteomics data have been deposited at the PRIDE database and are publicly available as of the date of publication. Previously published ChIP-exo datasets used in this study can be accessed from the GEO database. Previously published Epi-Decoder data from the HO locus can be found at the NCBI BioProject database. All accession numbers are listed in the key resources table. Original western blot images and microscopy data have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgG from rabbit serum | Sigma-Aldrich | Cat# I5006; RRID:AB_1163659 |
| PGK1 monoclonal | Thermo Fisher Scientific | Cat# 459250; RRID:AB_2532235 |
| TAP Tag polyclonal | Thermo Fisher Scientific | Cat# CAB1001; RRID:AB_10709700 |
| IRDye® 800 anti-rabbit | LI-COR Biosciences | Cat# 926-32211; RRID:AB_621843 |
| IRDye® 680 anti-mouse | LI-COR Biosciences | Cat# 926-68070; RRID:AB_10956588 |
| Chemicals, peptides, and recombinant proteins | ||
| Yeast Nitrogen Base w/o AA, Carbohydrate & w/ AS (YNB) (Powder) | US Biological | Cat# Y2025 |
| Drop-out Mix Complete w/o Yeast Nitrogen Base (Powder) | US Biological | Cat# D9515 |
| Bacto™ Agar | Thermo Fisher Scientific | Cat#214030 |
| Bacto™ Peptone | Thermo Fisher Scientific | Cat# 211677 |
| Bacto™ Yeast Extract | Thermo Fisher Scientific | Cat# 212750 |
| D-Glucose | Sigma-Aldrich | Cat# 8270 |
| Glycerol | Honeywell | Cat# 49770 |
| Ethanol | Honeywell | Cat# 32221 |
| Potassium hydrogen phthalate | VWR | Cat# 26948.260 |
| Myo-inositol | Sigma-Aldrich | Cat# I5125 |
| 4-Amino benzoic acid potassium salt | Sigma-Aldrich | Cat# A0254 |
| Geneticin (G418) Sulfate | Santa Cruz Biotech | Cat# sc-29065A |
| Hygromycin B | Gibco™ | Cat# 10687010 |
| Nourseothricin-dihydrogen sulfate (CloNat) | Werner BioAgents | Cat# 5.002.000 |
| 1,10-phenanthroline | Sigma-Aldrich | Cat# 131377 |
| Rapamycin | LC Laboratories | Cat# R-5000 |
| DMSO | Sigma-Aldrich | Cat# D4540 |
| Formaldehyde solution 37% | Sigma-Aldrich | Cat# 252549 |
| Tris(hydroxymethyl)aminomethane | Millipore | Cat# 1083821000 |
| Glycine | Acros Organics | Cat# 220910050 |
| Hydrochloric acid (fuming) | Honeywell | Cat# 30721 |
| Dynabeads™ M-270 Epoxy | Thermo Fisher Scientific | Cat# 14302D |
| PMSF | Roche | Cat# 10837091001 |
| cOmplete™ EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11873580001 |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | Cat# R5000 |
| Proteinase K from Tritirachium album | Sigma-Aldrich | Cat# P2308 |
| Agencourt AMPure XP beads | Beckman Coultier | Cat# A63881 |
| DAPI | Invitrogen™ | Cat# D1306 |
| MyTaq Red Mix 2x | Meridian Bioscience | Cat# BIO-25044 |
| Phusion™ High-Fidelity DNA Polymerase (2 U/μL) | Thermo Fisher Scientific | Cat# F530L |
| Deoxynucleoside Triphosphate Set | Roche | Cat# 11969064001 |
| DNAse I | New England Biolabs | Cat# M0303S |
| Trichloroacetic acid | Sigma-Aldrich | Cat# T6399 |
| Tris/Glycine/SDS running buffer | Bio-Rad | Cat# 161-0772 |
| 40% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad | Cat# 1610149 |
| TEMED | Sigma-Aldrich | Cat# T9281 |
| Ammonium Persulfate | Thermo Fisher Scientific | Cat# 17874 |
| Agarose MP | Sigma-Aldrich | Cat# 11388991001 |
| Trypsin from bovine pancreas | Sigma-Aldrich | Cat# T1426 |
| Guanidinium hydrochloride | Sigma-Aldrich | Cat# G3272 |
| TCEP | Sigma-Aldrich | Cat# C4706 |
| 2-Chloroacetamide | Sigma-Aldrich | Cat# 22790 |
| Critical commercial assays | ||
| QIAquick PCR Purification Kit | Qiagen | Cat# 28106 |
| KAPA Hyper Prep Kit | Roche | Cat# 07962347001 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q33231 |
| High Sensitivity DNA Kit | Agilent | Cat# 5067-4626 |
| SensiFAST SYBR No-ROX Kit | Meridian Bioscience | Cat# BIO-98020 |
| QIAquick Gel Extraction Kit | Qiagen | Cat# 28706 |
| RNeasy Kit | Qiagen | Cat# 74106 |
| TruSeq Stranded mRNA Kit | Illumina | Cat# 20020594 |
| DC™ Protein Assay Kit II | Bio-Rad | Cat# 5000112 |
| Pierce™ Bradford Protein Assay Kit | Thermo Fisher Scientific | Cat# 23200 |
| Deposited data | ||
| ChIP-sequencing data | This study | GEO: GSE227470 |
| RNA-sequencing data | This study | GEO: GSE227470 |
| ChIP-exo data Fpt1 | This study | GEO: GSE227470 |
| ChIP-exo data others | (Rossi et al, 2021) | GEO: GSE147927 |
| Epi-Decoder tDNA-Ty1 locus data | This study | NCBI BioProject: PRJNA945378 |
| Epi-Decoder HO locus data | (Poramba-Liyanage et al, 2020) | NCBI BioProject: PRJNA610036 (SRX7842936) |
| Proteomics data | This study | PXD045463 |
| Immunoblots and microscopy data | This study | Mendeley Data: 10.17632/stwm3b4jsf.1 |
| Experimental models: Organisms/strains | ||
| Please refer to Table S11 | This study | N/A |
| Oligonucleotides | ||
| Please refer to Table S13 | This study | N/A |
| Recombinant DNA | ||
| Please refer to Table S12 | This study | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al, 2012) | RRID:SCR_003070 |
| ChIP-exo data analysis | This study | https://doi.org/10.5281/zenodo.8381412 |
| GraphPad Prism 9 | GraphPad | RRID:SCR_002798 |
| ScriptManager (version v.014) | (Lang et al, 2022) | RRID:SCR_021797 |
| Picard | Picard | RRID:SCR_006525 |
| BWA-MEM (version 0.7.17-r1188 and 0.7.9a) | (Li & Durbin, 2009) | https://github.com/lh3/bwa |
| GenomicRanges | (Lawrence et al, 2013) | RRID:SCR_000025 |
| Rtracklayer | (Lawrence et al, 2009) | RRID:SCR_021325 |
| RStudio | Rstudio | RRID:SCR_000432 |
| Micro-manager | (Edelstein et al, 2014) | RRID:SCR_000415 |
| Tornadoplot R/Github package | This study | https://doi.org/10.5281/zenodo.8348229 |
| DEseq2 | (Love et al, 2014) | RRID:SCR_015687 |
| FlowJo (version 10.6.1) | FlowJo | RRID:SCR_008520 |
| STAR (version 2.7.1a) | (Dobin et al, 2013) | https://github.com/alexdobin/STAR |
| eXpress (Bioconda Package) | (Roberts & Pachter, 2013) | https://anaconda.org/bioconda/express |
| ShinyGO 0.77 | (Ge et al, 2019) | RRID:SCR_019213 |
| EMBOSS (version 6.6.0) | (Rice et al, 2000) | RRID:SCR_008493 |
| BLAST (version 2.2.31+) | (Camacho et al, 2009) | RRID:SCR_004870 |
| OrthoFinder (version 2.2.7) | (Emms & Kelly, 2019) | RRID:SCR_017118 |
| MAFFT (version 7.271) | (Katoh & Standley, 2013) | RRID:SCR_011811 |
| IQ-Tree (version 1.6.8) | (Nguyen et al, 2015) | RRID:SCR_021163 |
| DIA-NN (version 1.8) | (Demichev et al, 2020) | RRID:SCR_022865 |
| Perseus (version 2.0.10.0) | (Tyanova et al, 2016) | RRID:SCR_015753 |
| Other | ||
| Cover Slips round 25mm (#1.5 thickness) | VWR | Cat# 631-0172 |
| Attofluor™ Cell Chamber, for microscopy | Thermo Fisher Scientific | Cat# A7816 |
| Zeiss 4-alpha Plan-Apochromat 1003/1.46NA Oil | Zeiss | Cat# 420792-9800-000 |
| ORCA Flash 4v3 digital sCMOS camera | Hamamatsu | Cat# C13440-20CU |
| UNO Top stage incubator and objective heater | Okolab | N/A |
| Zirconia-Silicate Beads, 0.5 mm | Biospec | Cat# 11079105Z |
| Amersham™ Protran® nitrocellulose membrane | GE Healthcare Life Science | Cat# 10600002 |
| Sep-Pak C18 1cc cartridges | Waters | Cat# WAT054960 |
| Microlab STAR liquid handling system | Hamilton | N/A |
| Rotor HDA | Singer Instruments | N/A |
| Singer Plusplates™ | Singer Instruments | Cat# PLU-003 |
| Singer Repads™ | Singer Instruments | Cat# REP-004 |
| 2100 Bioanalyzer Instrument | Agilent | Cat# G2939BA |
| DynaMag™−2 Magnet | Invitrogen™ | Cat# 12321D |
| Bioruptor® Pico sonication device | Diagenode | Cat# B01060010 |