Abstract
Mesenchymal stem cells (MSCs) and macrophages collaboratively contribute to bone regeneration after injury. However, detailed mechanisms underlying the interaction between MSCs and inflammatory macrophages (M1) remain unclear. A macrophage-depleted tooth extraction model was generated in 5-wk-old female C57BL/6J mice using clodronate liposome (12.5 mg/kg/mouse, intraperitoneally) or saline injection (control) before maxillary first molar extraction. Mice were sacrificed on days 1, 3, 5, 7, and 10 after tooth extraction (n = 4). Regenerated bone volume evaluation of tooth extraction socket (TES) and histochemical analysis of CD80+M1, CD206+M2 (anti-inflammatory macrophages), PDGFRα+MSC, and TNF-α+ cells were performed. In vitro, isolated MSCs with or without TNF-α stimulation (10 ng/mL, 24 h, n = 3) were bulk RNA-sequenced (RNA-Seq) to identify TNF-α stimulation-specific MSC transcriptomes. Day 7 micro-CT and HE staining revealed significantly lower mean bone volume (clodronate vs control: 0.01 mm3 vs 0.02 mm3, p<.0001) and mean percentage of regenerated bone area per total TES in clodronate group (41.97% vs 54.03%, p<.0001). Clodronate group showed significant reduction in mean number of CD80+, TNF-α+, PDGFRα+, and CD80+TNF-α+ cells on day 5 (306.5 vs 558.8, p<.0001; 280.5 vs 543.8, p<.0001; 365.0 vs 633.0, p<.0001, 29.0 vs 42.5, p<.0001), while these cells recovered significantly on day 7 (493.3 vs 396.0, p=.0004; 479.3 vs 384.5, p=.0008; 593.0 vs 473.0, p=.0010, 41.0 vs 32.5, p=.0003). RNA-Seq analysis showed that 15 genes (|log2FC| > 5.0, log2TPM > 5) after TNF-α stimulation were candidates for regulating MSC’s immunomodulatory capacity. In vivo, Clec4e and Gbp6 are involved in inflammation and bone formation. Clec4e, Gbp6, and Cxcl10 knockdown increased osteogenic differentiation of MSCs in vitro. Temporal reduction followed by apparent recovery of TNF-α-producing M1 macrophages and MSCs after temporal macrophage depletion suggests that TNF-α activated MSCs during TES healing. In vitro mimicking the effect of TNF-α on MSCs indicated that there are 15 candidate MSC genes for regulation of immunomodulatory capacity.
Keywords: cytokines, dental biology, injury healing, osteoimmunology, stem cells
Graphical Abstract
Graphical Abstract.
Introduction
Bone healing after trauma is a complex process comprising 3 stages: inflammation, repair, and remodeling.1 Inflammation initiates the tissue-healing process and protects against damaging stimuli. The sequential healing process relies on the initial inflammatory phase, which is influenced by local and systemic reactions, including immune cells.2-4 Among the immune cells that arrive first at the damaged site, macrophages contribute to inflammation at the injury site by secreting pro- and anti-inflammatory cytokines and by recruiting other immune cells. Macrophages have also been reported to regulate bone regeneration during homeostasis and fracture repair.5,6 Murine studies have also reported that macrophages are closely connected with active osteoblasts, and depletion of macrophages by administration of clodronate liposome results in impaired woven bone formation and healing of the damaged tissue.7,8 Similarly, Sandberg et al. demonstrated that macrophage depletion by clodronate liposomes in tibial metaphyseal screws and drill holes resulted in compromised healing.9 In addition, macrophages secrete cytokines and growth factors because of their functional plasticity to switch from inflammatory (M1) to anti-inflammatory (M2) macrophages.10 M1 macrophages secrete pro-inflammatory mediators such as TNF-α and IL-6, which further recruit inflammatory and immune cells.3,11 The pro-inflammatory cytokine TNF-α functions in a complex manner during bone regeneration, and previous studies have revealed delayed endochondral and intramembranous bone formation in TNF-α receptor-deficient mice and TNF-α knockout mice.12,13
On the other hand, mesenchymal stem cells (MSCs) that live in stem cell niches are multipotent stromal cells and essential for bone regeneration.14,15 MSCs are distinguished by self-renewal capacity and transform into various cell types, such as osteoblasts, chondrocytes, adipocytes, and myoblasts.15 Previous studies have shown that MSCs are possible candidates for both tissue engineering16,17 and immune disease treatment because of their immunomodulatory functions.16,18 MSCs also regulate macrophage chemotaxis and recruitment of macrophages, which are essential for immunomodulatory processes and modulate macrophage phenotype. This could potentially enhance tissue repair, including wound healing.3,19,20 Lu et al. revealed that in a direct co-culture system, the effects of murine bone marrow macrophages, especially of the M1 subtype, had the highest positive impact on osteogenic differentiation of MSCs.21 Macrophages conduct MSC function, but MSCs also conduct macrophage functions as MSCs respond to inflammatory microenvironments via broad immunomodulatory functions.22 Furthermore, another study revealed that TNF-α, which is produced by M1, affects MSCs, which accumulate in tissue healing sites.23 Based on these findings, the interaction between immune cells, such as pro-inflammatory macrophages (M1), and bone marrow-derived MSCs in damaged tissue is vital for tissue regeneration. However, the detailed mechanism of this interaction, particularly in the jawbone area, still needs to be explored, and the manner in which M1-secreted TNF-α initiates and modulates MSC function during bone healing remains unknown.
We used clodronate liposome injection in a maxillary first molar tooth extraction mouse model to verify the interaction between M1 macrophages and MSCs during bone regeneration. Moreover, to clarify the function of TNF-α produced from M1 macrophage-induced bone marrow-derived MSCs, such as osteogenic differentiation and immunomodulation properties during jawbone healing, we performed in vitro studies and analyzed TNF-α-stimulation-specific MSC transcriptomes by RNA-sequencing (RNA-Seq).
Materials and methods
Animal model and materials
Five-week-old C57BL/6J female mice were purchased from CLEA Japan Inc.. We created a tooth extraction model to identify bone regeneration related to macrophages. Mice in the experimental group were intraperitoneally administered clodronate liposomes once before the experiment, whereas the control group was injected with saline and maintained for 24 h. The following day, general anesthesia was administered to the mice by intraperitoneal injection of 35 mg/kg of 2% xylazine (Celactal, Bayer) and 5 mg/kg of ketamine (Ketalar, Daiichi-Sankyo). After the injection, we extracted bilateral maxillary first molars and curetted tooth extraction sockets using a dental excavator from both groups. The control and experimental groups had 4 mice each (n = 4). The mice were sacrificed in chronological order. Maxillae were collected and fixed with 4% paraformaldehyde (PFA).
The Okayama University Research Committee approved the experimental protocols used in this study. All animals were handled in accordance with the guidelines of the Okayama University Animal Research Committee (OKU—2021377).
To prepare the clodronate injection, 12.5 mg/kg of clodronic-acid-encapsulating liposome (Hygieia Bioscience) solution per mouse was kept at room temperature (RT) for 30 min before using.
Micro-CT analysis
After fixing with 4% PFA, the maxillae were analyzed using micro-CT (Skyscan 1174, Bruker). The CT parameters were set to a 6.5 μm pixel size resolution, peak voltage of 50 kV, and 795 μA with a 0.5 mm aluminum filter. Scanned images were reconstructed using NRecon (Bruker) software. The 2D photos were reoriented using DataViewer software and adjusted to the proper position to evaluate the bone volume. The buccal side of the maxillary right first molar was selected, revealing the mesiobuccal root (MBR) and distobuccal root. The images were adjusted by making a straight line crossing the buccal roots of the maxillary first, second, and third molars in the transverse plane.24 This dataset was saved and handled using CtAn (Image Processing Language) for area measurement and segmentation into the binary measure. A total of 22 dataset slices were selected based on defining the top slice (where the whole crown and MBR of the second molar start appearing) and the bottom slice (where the full crown shows with the buccal roots of the second and third molars appearing). The ROIs were measured from binary images at a fixed threshold (lower and upper grey threshold values of 45 and 250, respectively).
Histological analysis
The maxillae were fixed with 4% PFA and decalcified in 10% EDTA for 3 wk. Samples were embedded in paraffin, and 7 μm thick sections were made and stained with H&E and Masson’s trichrome using standard protocols. Using naphthol AS-MX phosphate (Sigma) in N, N-dimethylformamide as a substrate, tartrate-resistant acid phosphatase (TRAP) staining was performed at pH 5.0, with L(+)-tartaric acid. TRAP-positive cells with more than 2 nuclei on the bone surface were identified as osteoclasts.
Immunofluorescence analysis
PFA-fixed samples were frozen with a super cryoembedding medium (SECTION-LAB Co., Ltd), cut into 5 μm widths, and taken using Kawamoto’s film method.25 Immunofluorescence staining was performed using the antibodies shown in Tables S1 and S2. Tissue sections were blocked with 5% goat serum (Life Technologies) with 0.3% Triton X-100 or 5% chicken serum, New Zealand Origin (Life Technologies) with 0.1% Tween 20 and 1% BSA (Sigma-Aldrich) in PBS with respective antibodies for 1 h at RT. Primary antibodies, such as RUNX2, TNF-α, CD80 for M1 macrophages, CD206 for M2 macrophages, PDGFRα for MSCs, Anti-GBP6 for Gbp6, and Clec-4E (B-7) for Clec4e were diluted at different ratios and kept overnight at 4°C. On the following day, sections were washed and incubated with the secondary antibody: Goat anti-Rabbit for RUNX2 and Gbp6, Goat Anti-Armenian hamster for CD80, Chicken anti-Goat for CD206, and PDGFRα, Goat anti-Mouse for clec4e and single and double staining of TNF-α for 1 h at RT. Sections were mounted with DAPI Fluoromount-G (Southern Biotech). For double fluorescent staining, higher magnification images taken in the same position for all time points were used for quantification analysis. All fluorescent photos were taken using a fluorescence microscope (BZ-710 Keyence).
Isolation of bone marrow MSCs
The femurs and tibias of the mice were flushed with 2% FBS/PBS medium through a 70 μm cell strainer (Greiner Bio-One Inc.) to isolate bone marrow-derived MSCs. The flushed cells were cultured in basal medium with minimum essential medium alpha (α-MEM, Life Technologies) containing 15% FBS (Life Technologies), 100 U/mL penicillin/streptomycin (Sigma-Aldrich), 2 mM glutamate (Life Technologies), and 55 μM 2-mercaptoethanol (2-ME, Life Technologies) in 100 mm dishes (Greiner Bio-One Inc.). After 24 h, the dishes were washed with PBS to remove nonadherent cells, and the cells were maintained at 37°C and 5% CO2. Colony-forming cells were picked and passaged for subculture, and the cells from the second passage were used in the experiments.
Osteogenic differentiation of MSCs
MSCs were seeded in 12-well plates and cultured in basal medium until they reached confluency. For osteogenic differentiation, the basal medium was mixed with 10 mM L-ascorbic acid phosphate (Fujifilm Wako Pure Chemical Corporation), 200 mM β-glycerophosphate (Sigma-Aldrich), and 100 μM dexamethasone. The experimental group was cultured in osteogenic medium, and the control group was cultured in basal medium. After 1 wk, the cells were harvested and used to isolate the total RNA.
siRNA transfection and osteogenic differentiation of MSCs
MSCs were seeded in 6-well plates with basal medium at 60%–80% (0.5 × 106 cells/well) before transfection. Opti-MEM® (Life Technologies) was used to dilute with Lipofectamine® RNAiMAX (Life Technologies) in a 50:3 ratio. For the experimental groups, Silencer® Select Clec4e, Gbp6 (L0C673002), and Cxcl10 (Life Technologies) were used. Silencer® Select Negative Control #1 (Life Technologies) siRNA was used as control. The siRNA was diluted with Opti-MEM® at a ratio of 50:1. Diluted Lipofectamine® RNAiMAX and siRNA were mixed and incubated in one tube for 20 min at RT to form siRNA-Lipofectamine® RNAiMAX complexes. A total of 125 μL siRNA-Lipofectamine® RNAiMAX complexes were added to the wells and mixed gently by rocking the plate back and forth. The medium containing complexes was removed after 4 h and replaced with basal medium without penicillin, and cells were incubated for 24 h at 37°C and 5% CO2. The following day, the control groups without transfection and siRNA-transfected groups were cultured in the osteogenic medium. After 1 wk, the cells were harvested and used to isolate the total RNA.
Real-time reverse transcription-polymerase chain reaction
Real-time reverse transcription-polymerase chain reaction (RT-PCR) was performed to evaluate the gene expression. Following the manufacturer’s instructions, total cellular RNA was extracted from the control and experimental groups of MSCs osteogenic differentiation after one wk using a Purelink RNA Mini Kit (Life Technologies). RNA was reverse-transcribed using an iScript cDNA Synthesis Kit (Bio-Rad. Real-time RT-PCR was performed to quantify target gene expression Using KAPA SYBR FAST qPCR Master Mix (KAPA BIOSYSTEMS) and a CFX96 real-time system (Bio-Rad), as described previously.25 The reference gene ribosomal protein S29 was used to normalize the expression levels of each mRNA. The target genes were Ccl2, Mmp3, Ccl5, Cxcl1, Gbp6, Cxcl10, Cfb, Ifi205, Clec4e, Gbp5, Cxcl5, H2-M2, Ccl20, Cxcl9, Nos2, Runx2, Osterix, and Alp. Primer sequences and siRNA sequences are listed in Tables S3 and S4.
RNA-sequencing
MSCs were seeded in 6-well plates and cultured until they reached confluence. MSCs were stimulated with 10 ng/mL TNF-α (R&D Systems) for 24 h and unstimulated MSCs were used as controls. Total RNA from unstimulated and stimulated MSCs was isolated and purified using the RNeasy Plus Micro Kit (QIAGEN) and treated with DNase (Life Technologies), according to the conventional protocol. RNA concentration and quality were measured using a Tapestation 2200 (Agilent Technologies Inc.) with a High Sensitivity RNA ScreenTape (Agilent Technologies, Inc.). Indexed cDNA libraries were generated using a SMART-Seq Stranded Kit (Takara Bio USA, Inc.). A Tapestation 2200 with a High Sensitivity D5000 ScreenTape (Agilent Technologies Inc.) was used to validate the purified libraries. An Illumina NovaSeq 6000 Sequencer (Illumina) was used for sequencing.
After quality trimming using Trim Galore, STAR aligner26 was used to map the reads according to the reference genome (GRCm39, Ensembl release 107). StringTie27 and SQANTI328 were used to generate and annotate nova transcripts, respectively. Salmon29 was used to re-quantify all samples at the transcript level, and isoformSwitchAnalyzeR,30 based on the mapping results from Salmon, was used to perform additional quality filtering. The median read number of all gene transcripts was used to estimate the gene expression levels. The RUVSeq software31 was used to correct the batch effect at the gene expression level. DESeq232 was used to detect the differentially expressed genes (DEGs). DEGs were defined as False Discovery Rate [or p-value] <.05 and |log2Fold Change| >5. Functional enrichment analysis was performed using publicly accessible DAVID 6.8, and a bubble plot was created using R v4.22. Heatmaps were generated using Microsoft Excel.
Quantification and statistical analysis
In the maxillary tooth extraction area, the ROI was defined as the line drawn from the mesial alveolar crest to the interradicular septum and from the distal alveolar crest to the interradicular septum (Figure 1A). We measured the area under these lines until the root tip and used this ROI for all the histological and fluorescence images. In the fluorescence images, 6 segments (Upper, Middle, Lower) from the mesial and distal roots of the tooth extraction socket were selected and measured in 4 samples. The regenerated area and number of positive cells were automatically counted using ImageJ (v1.8.0_345). Statistical analyses were performed using GraphPad Prism (9.5.1). Data described in the figures are expressed as box-and-whisker plots (with median and interquartile range, from maximum to minimum, showing all data points). Comparisons between more than 2 groups were performed using ordinary one-way ANOVA, 2-way ANOVA, and Tukey’s multiple comparison test. A 2-tailed unpaired t-test was used to compare the RT-PCR results. p<.05 was considered to be significantly different.
Figure 1.
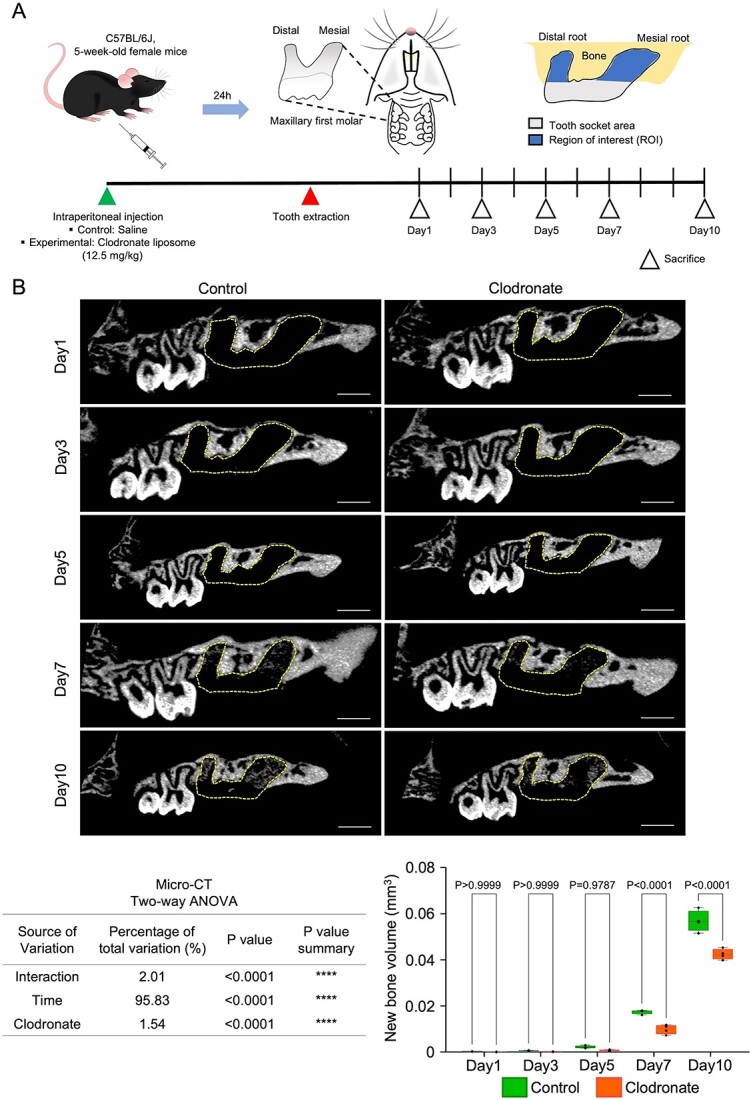
Macrophage depletion related to decreased volume of regenerated bone in the extracted tooth socket. (A) Illustration of tooth extraction mouse model and ROI. (B) Representative micro-CT images of tooth-extracted maxillary bone and quantification of regenerated bone volume in the tooth extraction area. The dotted line-bound area represents the extracted tooth socket area. Scale bar: 900 μm. Statistical analyses were performed using 2-way ANOVA and Tukey’s multiple comparison method. n = 4.
Results
Clodronate injection delayed bone healing in maxillary tooth extraction socket
On day 1 (Figure 1A and B), the CT data showed no significant differences in bone formation between the 2 groups. Similarly, histology of the control and clodronate groups showed only hematoma and necrotic cells in the tooth extraction area (Figure 2A). On day 3, micro-CT showed data similar to those on day 1, and HE data revealed the early stage of inflammation: granulation tissue formation in the healing area of both groups. Granulation tissues comprised newly formed blood vessels, fibroblasts, and infiltrated inflammatory cells. However, the clodronate group showed a smaller granulation tissue area than the control group.
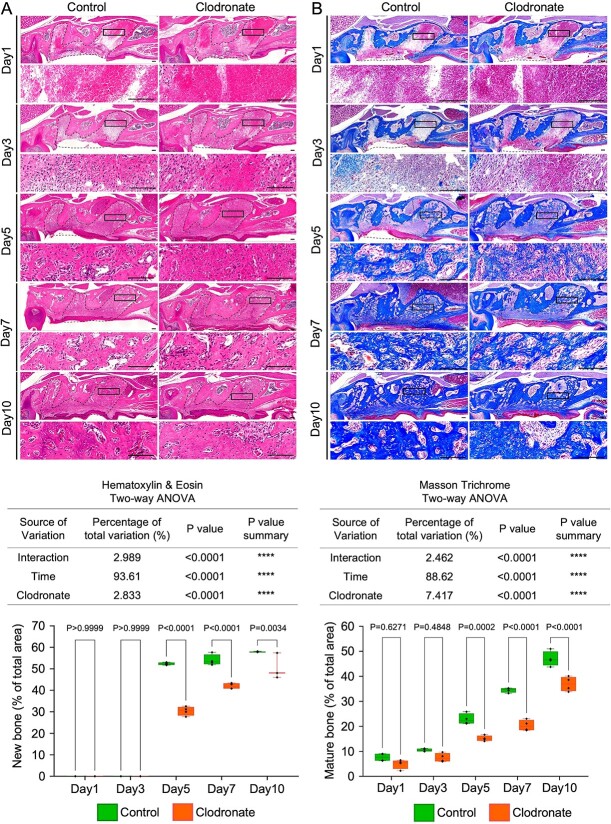
Figure 2.
Rate of new bone formation delayed in the extracted tooth socket. (A) Representative images of HE staining of tooth extraction models and quantification of new bone area per total tooth socket area. (B) Representative images of Masson’s trichrome staining of tooth extraction models and quantification of mature bone area per total tooth socket area. Low- and high-magnification images are depicted. The inset shows the location of the high-magnification image, and the dotted line-bound area shows the extracted tooth socket area. Left panel: control group. Right panel: clodronate group. Scale bar: 100 μm. Statistical analyses were performed using 2-way ANOVA and Tukey’s multiple comparison method. n = 4.
Unlike the observations on days 1 and 3, contrasting information was obtained from the CT and HE data on day 5. Although no changes in bone formation were observed in the CT images of both groups, HE-staining indicated new bone formation in both groups. This difference may result from the newly formed bone density and its relevant radiolucency on CT imaging. Quantification of the regenerated bone area (mean percentage) revealed that the clodronate group showed significantly less new bone area (<40%) than the control group (approximately 55%).
On day 7, CT images showed detectable radiopacity in the tooth socket areas in both groups. However, CT images of the clodronate group showed significantly lower bone formation than those of the control group. In addition, the HE data showed similar results: significantly lower new bone formation was observed in the clodronate group (approximately 40%) than in the control group (>55%).
Moreover, day 10 CT and HE data showed that the clodronate group still had significantly lower new bone formation than the control group. For the HE data, in the control group, approximately 60% of the tooth extraction area was replaced with mature bone with marrow space, whereas the clodronate group showed less mature bone area (approximately 50%).
Masson’s trichrome staining showed a similar trend in the new bone formation with micro-CT and HE data (Figure 2B). In addition, when we observed osteoblast and osteoclast functions in the tooth socket healing process, the cell numbers of TRAP+ osteoclasts and Runx2+ osteoblasts were significantly lower in the clodronate group than in the control group (Figure S1).
These data suggest that clodronate administration in mice before tooth extraction delays the bone healing process in TES.
Macrophages were depleted at the tooth extraction socket area in clodronate-treated mice, and MSC accumulation at the healing site was positively correlated
Next, to investigate the impact of macrophage depletion on tooth socket healing, the number of M1 and M2 macrophages in the healing area was evaluated with immunofluorescence staining for CD80 and CD206 (Figure 3A,Figure S2).
Figure 3.
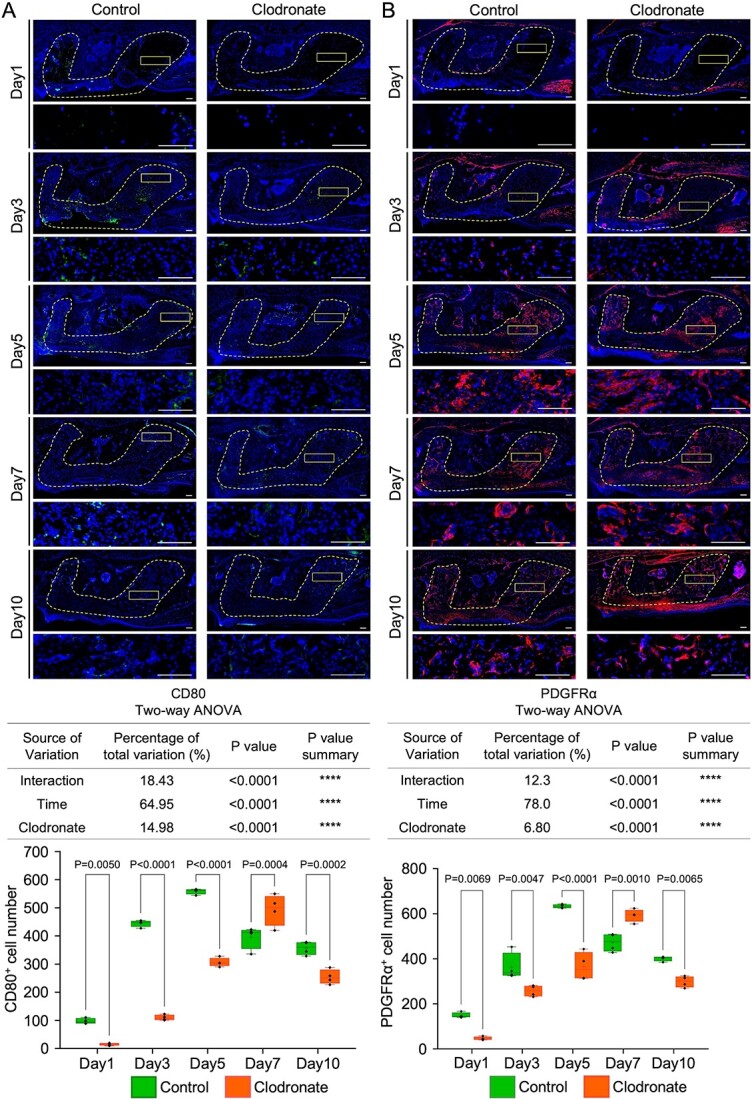
Temporal macrophage depletion by clodronate liposome administration induced a transient reduction and apparent recovery of CD80-positive and PDGFRα-positive cells on day 7 in the extracted tooth socket. (A) Representative images of immunofluorescence CD80 staining in tooth extraction models and quantification of the mean positive cell numbers of CD80 staining per total tooth socket area. (B) Representative images of immunofluorescence PDGFRα staining in tooth extraction models and quantification of the mean number of PDGFRα staining per total tooth socket area. Low- and high-magnification images are depicted. The inset shows the location of the high-magnification image, and the dotted line-bound area shows the extracted tooth socket area. Left panel: control group. Right panel: clodronate group. Scale bar: 100 μm. Statistical analyses were performed using 2-way ANOVA and Tukey’s multiple comparison method. n = 4.
Pro-inflammatory macrophages (M1) are recruited for the initial acute inflammatory response within the injured site.33 However, in the earlier stage, the clodronate liposome injection groups showed a significantly lower mean number of CD80+ M1 infiltrations in the tooth extraction area than the control group: day 1 (16 vs 97, p=.0050), day 3 (110 vs 444.8, p<.0001), and day 5 (306.5 vs 558.8, p<.0001) (Figure 3A). Similarly, the mean number of CD206+ M2 cells was significantly reduced in the clodronate group compared to the control group on days 1 (12.5 vs 26.75, p=.9995), 3 (35.25 vs 95.5, p=.1707), and 5 (180.3 vs 358, p<.0001) (Figure S2). Notably, 7 days after tooth extraction, the clodronate group showed a significant increase in CD80+ M1 infiltration to the healing area compared to the control group (493.3 vs 396, p=.0004). However, the mean number of CD206+ M2 cells was significantly lower in the clodronate group than in the control group on day 7 (277.3 vs 728.5, p<.0001). The mean cell numbers of CD80+ and CD206+ cells were lower in the clodronate group than in the control group on day 10 (254.5 vs 357, p=.0002 and 430 vs 583.3, p<.0001). These results suggest that M1 and M2 macrophages were temporarily depleted during the early stages of healing in the tooth extraction area in the clodronate group. However, the effects of clodronate slowly dissipated, and CD80+ cells were observed on day 7.
MSCs are pluripotent cells that have the function of immunomodulation and osteogenesis properties.15 To identify the distribution of MSCs at the healing site, anti-PDGFRα was used for IHC staining (Figure 3B). Two-way ANOVA revealed that fewer accumulated PDGFRα+ cells were detected in the clodronate group on days 1, 3, and 5 (p=.0069, p=.0047, and p<.0001, respectively). However, the mean number of PDGFRα+ cells was significantly higher in the clodronate group than in the control group on day 7 (593 vs 473, p=.0010). On day 10, the mean number of PDGFRα+ cells was lower in the clodronate group than in the control group (299 vs 402, p=.0065). These results suggest that PDGFRα+ and CD80+ cells accumulated in the clodronate healing area on day 7. The above results show that MSCs and M1 macrophage involvement in tooth socket healing was positively and synergistically related, suggesting that MSCs might be recruited to the healing area on day 7 of the clodronate group by M1 macrophages.
M1 produces TNF-α during the healing process at the extraction socket
TNF-α is an inflammatory cytokine secreted by pro-inflammatory cells, such as M1 and mast cells.34
CD80/TNF-α double-positive cells showed a distribution pattern similar to that of single staining for CD80 and TNF-α. On days 1 and 3, the mean number of CD80+ TNF-α+ cells was lower in the clodronate group than in the control group (p=.0012 and p<.0001, respectively). The decreased number of CD80/TNF-α double-positive cells on day 5 (clodronate vs control: 29.0 vs 42.5, p<.0001) significantly increased in the clodronate group on day 7 (clodronate vs control: 41.0 vs 32.5, p=.0003). On day 10, the mean number of double-positive cells in the clodronate group was lower than that in the control (18.75 vs 27, p=.0005) (Figure 4A and B). These data suggest that CD80+ cells secrete TNF-α in the injured area during bone regeneration. These results showed a positive correlation between M1 macrophages, TNF-α, and MSCs at the extracted tooth socket.
Figure 4.
Restoration of CD80/TNF-α double-positive cell recruitment on day 7 in the macrophage-depleted tooth extraction model. (A) Representative images of CD80 and TNF-α double-positive immunofluorescence staining in tooth extraction models as a reference for high-magnification images for all groups. (B) Representative images of high-magnification double-positive CD80 and TNF-α staining in the control and clodronate groups and quantification of mean positive cell numbers of CD80+ TNF-α+ double staining per selected ROI size in the total tooth socket area. High-magnification images are shown. The inset shows the location of the high-magnification image, and the dotted line-bound area shows the extracted tooth socket area. Upper panel, control group. Lower panel, clodronate group. Scale bar: 100 μm. Statistical analyses were performed using 2-way ANOVA and Tukey’s multiple comparison method. n = 4.
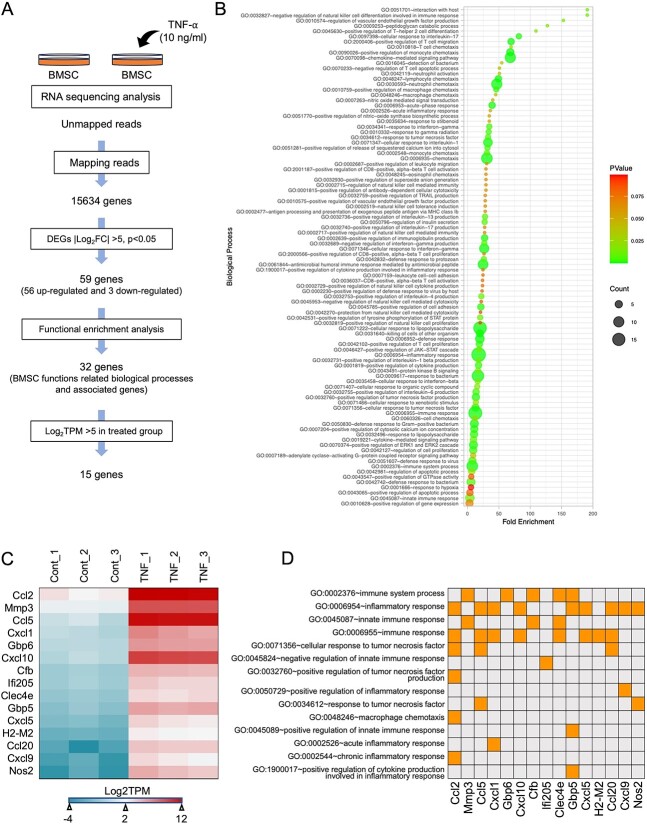
15 upregulated gene candidates for inducing MSC functions during bone repair
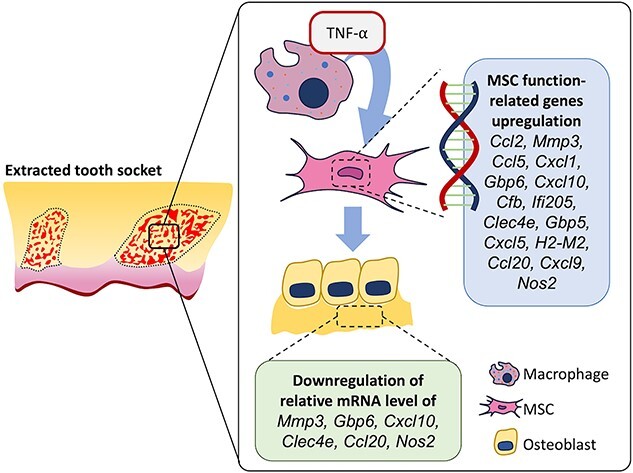
To identify the mechanism underlying the relationship between MSCs and TNF-α, bone marrow-derived MSCs were isolated and cultured until the confluence of the second passage. Seeded MSCs were stimulated with TNF-α (10 ng/mL) for 24 h and unstimulated MSCs were used as controls. Bulk RNA sequencing (RNA-seq) was performed using n = 3 (Figure 5A). After mapping the raw RNA-Seq data and relevant genomes, significant DEGs were detected using DESeq2 software, and a total of 15,534 genes were identified. We filtered the total genes using Log2 fold change (Log2FC), which is greater than 5 or less than −5, and 59 genes were identified, as shown in Table S5. Among them, 56 genes were upregulated, and 3 genes were downregulated after TNF-α stimulation. Furthermore, we identified the biological processes of 59 genes using the database for annotation, visualization, and integrated discovery (DAVID) functional enrichment analysis, and the data are presented as a bubble plot using R programming (Figure 5B). Next, we screened bone marrow MSC functions related to biological processes and sorted the associated genes. We observed that 32 genes were upregulated (Table S6). Among these 32 genes, we isolated 15 that had the highest expression levels (log2TPM > 5) in the TNF-α-induced MSC group (Ccl2, Mmp3, Ccl5, Cxcl1, Gbp6, Cxcl10, Cfb, Ifi205, Clec4e, Gbp5, Cxcl5, H2-M2, Ccl20, Cxcl9, and Nos2) (Figure 5C). Interestingly, these 15 genes were mainly related to biological processes of immune function, as shown in Figure 5D. These data suggest that these 15 genes may be candidate factors for the regulation of the immunomodulatory capacity of MSCs.
Figure 5.
Comprehensive analysis of TNF-α stimulation-specific MSCs transcriptomes revealed that 15 immune-related genes were significantly upregulated in TNF-α-stimulated MSCs. (A) Flow chart of BMSC function-related gene screening. We analyzed DEGs of TNF-α-stimulated and unstimulated MSCs and cut-off values of p<.05 and |log2Fold Change| >5. We performed functional enrichment analysis using DAVID Version 6.8 and selected BMSC function-related biological processes. We observed highly expressed genes in the TNF-α-stimulated MSCs group with absolute log2TPM > 5. (B) Bubble plot for functional enrichment analysis of 59 genes using R v4.22. (C) Heatmap of the expression levels of selected genes with high expression in TNF-α-stimulated MSCs. (D) Table presentation of biological processes related to highly expressed genes in TNF-α-stimulated MSCs.
Clec4e and Gbp6 are involved in the inflammatory stage and decrease when bone formation occurs
The Clec4e and Gbp6 genes were selected to confirm their involvement in the inflammatory stage and bone healing. We investigated the accumulation and distribution of these proteins in the healing area of the tooth extraction model by immunofluorescence staining (Figure 6A). We performed maxillary first molar tooth extraction in mice, euthanized the mice on days 0, 1, 3, and 5, and harvested their maxillae (n = 3–5). The mean number of Clec4e+ cells was not significantly different between days 1 and 0 (259.4 vs 179.3, p=.0761). On day 3, the mean number of Clec4e+ cells was significantly higher than that on day 0 (647.4 vs 179.3, p<.0001). The mean number of Clec4e+ cells on day 5 was significantly lower than that on day 3 (467.7 vs 647.4, p=.0003). Similarly, the mean number of Gbp6+ cells was not significantly different between days 1 and 0 (117 vs 52.67, p=.1669). On day 3, the mean number of Gbp6+ cells was significantly higher than that on day 0 (311.8 vs 52.67, p<.0001). On day 5, the mean number of Gbp6+ cells was significantly lower than that on day 3 (190.3 vs 311.8, p=.0055). These data suggest that Clec4e and Gbp6 are involved in the initial inflammatory stage of bone healing and decrease when bone formation occurs.
Figure 6.
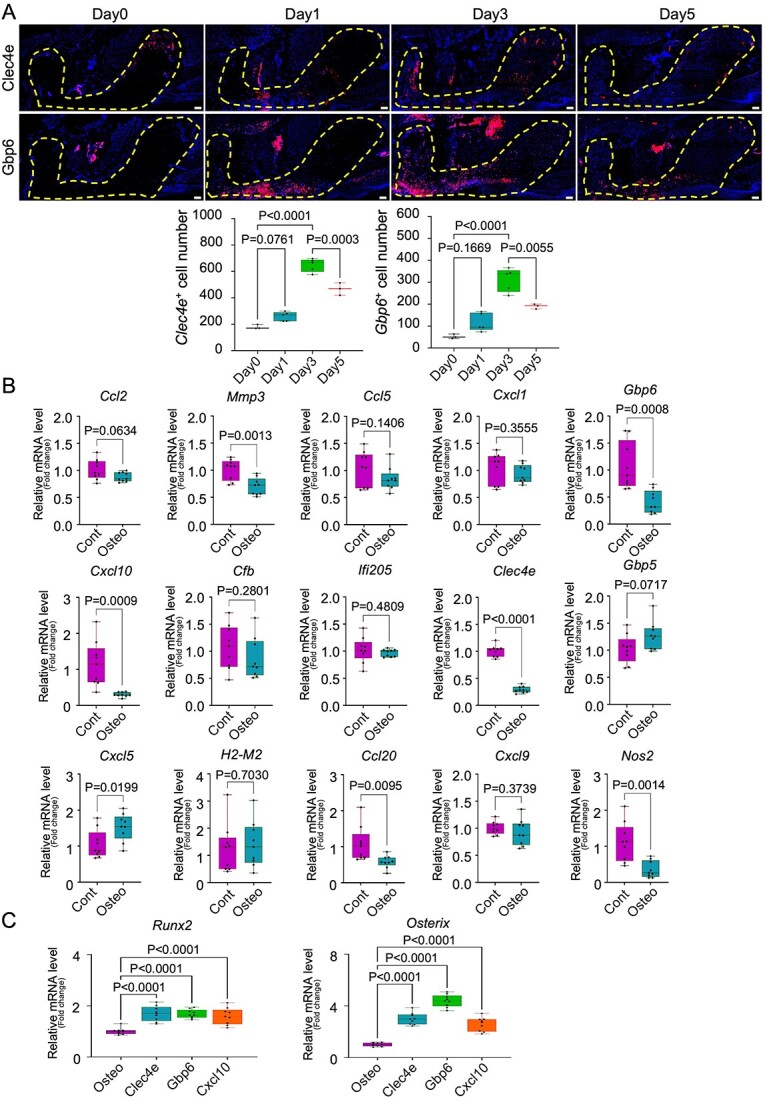
Candidate genes for regulation of the immunomodulatory capacity of MSCs and Clec4e, Gbp6, and Cxcl10 are involved in the inflammatory process and bone healing of the tooth extraction socket. (A) Representative images of immunofluorescence Clec4e and Gbp6 staining in tooth extraction models and quantification of the mean positive cell numbers of Clec4e and Gbp6 staining per total tooth socket area. Low-magnification images are shown, and the dotted line-bound area indicates the extracted tooth socket area. Scale bar: 100 μm. Statistical analyses were performed using one-way ANOVA and Tukey’s multiple comparison method. n = 3–5. (B) Quantification of relative mRNA levels (fold-change) of Ccl2, Mmp3, Ccl5, Cxcl1, Gbp6, Cxcl10, Cfb, Ifi205, Clec4e, Gbp5, Cxcl5, H2-M2, Ccl20, Cxcl9, and Nos2 after induction of osteogenic differentiation in isolated MSCs. MSCs cultured in basal medium were used as controls. Statistical analyses were performed using 2-tailed unpaired t-tests. n = 9. (C) Quantification of the relative mRNA levels (fold-change) of Runx2 and Osterix after siRNA transfection of Clec4e, Gbp6, and Cxcl10. Osteogenic differentiated MSCs without transfection as controls. Statistical analyses were performed using an ordinary one-way ANOVA and Tukey’s multiple comparison method. n = 9.
Mmp3, Gbp6, Cxcl10, Clec4e, Ccl20, and Nos2 may regulate MSCs stemness function for immunomodulation properties
After confirming the involvement of Clec4e and Gbp6 genes in the inflammatory process, we investigated the influence of the 15 candidate genes on MSC function in vitro. First, we isolated bone marrow-derived MSCs from mouse femurs, cultured them until they reached nearly 90% confluence, and induced osteogenic differentiation using osteogenic medium for 1 wk. The cells were harvested, RNA was extracted, and real-time RT-PCR was performed. Primers of osteogenic-related Runx2, Alp, and Osterix (Sp7) genes were used to verify the successful osteogenic differentiation of MSCs shown in Figure S3A. The expression levels of Runx2, Alp, and Osterix (Sp7) were significantly increased (p<.0001, p<.0001, and p<.0001, respectively) in the osteogenic differentiation MSC group. After confirming the osteogenic differentiation of MSCs, primers for identified 15 genes from RNA-Seq data were used to detect relative mRNA levels during osteogenic differentiation of MSCs. As shown in Figure 6 (Figure 6B), the expression level of Cxcl5 was significantly increased (p=.0199), while Gbp5 and H2-M2 expression levels appeared to be higher, with no significant differences (p=.0717 and p=.7030, respectively). Ccl2, Ccl5, Cxcl1, Cfb, ifi205, and Cxcl9 expression levels were not significantly different between the control and experimental groups. Interestingly, the expression of Mmp3, Gbp6, Cxcl10, Clec4e, Ccl20, and Nos2 was significantly downregulated (p=.0013, p=.0008, p=.0009, p<.0001, p=.0095, and p=.0014, respectively). These data suggest that Cxcl5, Gbp5, and H2-M2 are involved in the osteogenic differentiation of MSCs. MSC stemness is important for the immunomodulatory function of MSCs. In this experiment, Mmp3, Gbp6, Cxcl10, Clec4e, Ccl20, and Nos2 were downregulated after MSCs differentiated into bone.
Downregulation of Clec4e, Gbp6, and Cxcl10 genes increase MSCs osteogenic differentiation
To confirm the downregulated genes after osteogenic differentiation of MSCs, we performed siRNA transfection of the Clec4e, Gbp6, and Cxcl10 genes (Figure 6C). Bone marrow-derived MSCs were cultured from mouse femurs until 60%–80% confluent in 6-well plates. Cells were transfected with siRNA and incubated for 24 h. Osteogenic differentiation was induced the following day using osteogenic medium for 1 wk. The cells were harvested for RNA extraction and real-time RT-PCR was performed. Negative siRNA was used as a control to confirm the siRNA transfection. As shown in Figure S4A, the relative mRNA levels of Clec4e, Gbp6, and Cxcl10 were significantly decreased (p<.0001, p=.0002, and p<.0001, respectively) compared to the control and negative siRNA. After siRNA transfection and 1 wk of osteogenic differentiation, the expression levels of Runx2 were significantly increased in the Clec4e, Gbp6, and Cxcl10 transfected groups (p<.0001, p<.0001, p<.0001, respectively). In addition, the expression levels of osterix were significantly higher in Clec4e−, Gbp6, and Cxcl10 transfected groups (p<.0001, p<.0001, and p<.0001, respectively) than in osteogenic differentiated MSCs without transfection. The expression of osterix in the Clec4e and Gbp6 transfected groups was slightly higher than in the Cxcl10 transfected groups. These data suggest that knockdown of Clec4e, Gbp6, and Cxcl10 affects the immunomodulatory function of MSCs and increases osteogenic differentiation.
Discussion
Inflammation initiates the healing process.3 On the other hand, prolonged inflammation is one of the causes of poor healing, suggesting that the healing process is a delicate and fragile process that depends on the homeostasis of the inflammatory process.35 Homeostasis of the inflammatory process largely relies on infiltrated macrophages, which have both pro- and anti-inflammatory properties. M1, a pro-inflammatory macrophage, possesses an early stage of inflammation, and M2, an anti-inflammatory macrophage, acts as a balance of the inflammatory process by neutralizing the effects of M1.36 M1 macrophages secrete pro-inflammatory cytokines that recruit more immune cells and provoke inflammation in the injured area.3,37 In addition, M1 macrophages are involved in bone regeneration during the bone healing process.38 This study evaluated the role of M1 in the bone healing process using an extracted tooth socket healing model created in temporal macrophage-depleted mice. In line with previous reports,39,40 our study showed that temporal macrophage depletion shifted the entire healing process and delayed bone healing by obstructing macrophages and MSCs, which are essential for bone regeneration. Notably, our study indicated that the occurrence of M1 macrophages and MSCs at the injury site was positively correlated.
There is evidence that MSCs and M1 cells modulate each other and promote bone regeneration. Omar et al. reported that the expression of osteogenic differentiation-related genes such as BMP-2, RUNX2, and ALP in MSCs increased in LPS-stimulated monocyte-conditioned media.41 In a follow-up study, the same group showed that exosomes are secreted by LPS-activated monocytes, which partially explains the induction of BMP-2 and RUNX2 in MSCs.42 Similarly, Lu et al. reported that M1 macrophages showed the highest effect compared to other subtypes of macrophages in enhancing bone formation during direct co-culture with murine bone marrow MSCs.21 Moreover, pro-inflammatory cytokines (TNF-α, IL-1α, or IL-1β) combined with IFN-γ activate MSCs in inflamed or damaged tissues to produce several chemokines and mediate the immunomodulation of infiltrated macrophages,43 potentially enhancing wound healing19 and tissue regeneration.20 In this study, we identified the downstream effects of TNF-α stimulation on MSCs using RNA-seq analysis. We observed that TNF-α secreted by M1 macrophages is critical for recruiting MSCs in bone regeneration. RNA-Seq analysis of the TNF-α-stimulated specific MSC transcriptomes revealed that (Ccl2, Mmp3, Ccl5, Cxcl1, Gbp6, Cxcl10, Cfb, Ifi205, Clec4e, Gbp5, Cxcl5, H2-M2, Ccl20, Cxcl9, and Nos2) genes were highly differentially expressed.
It has been reported that in the presence of insufficient levels of TNF-α and IFN-γ, MSCs enhance immune responses by producing chemokines such as Cxcl9, Cxcl10, and Cxcl11.43,44 In addition, macrophage-inducible C-type lectin (Mincle) encoding the CLEC4E gene in macrophages showed increased expression upon stimulation with inflammatory agents and pro-inflammatory cytokines such as LPS, IFN-γ, TNF-α, and IL-6.45 Mincle is also a crucial regulatory element in inflammation, decreasing pro-inflammatory responses and promoting anti-inflammatory responses.46 Conversely, in the nuclei of adipose tissue-derived stem cells and osteosarcoma, Ifi205 localizes, interacts with several transcription factors, and induces adipogenic differentiation.47,48 Inflammatory stimuli induce MSCs to secrete molecules associated with the regulation of tissue homeostasis including NO, PGE2, CCL2, IL-10, and galectins.49 These previous findings are similar to those of our RNA-Seq analysis, which showed that 15 candidate genes might be involved in the different functions of bone marrow-derived MSCs.
In vivo studies have also revealed that Clec4e and Gbp6 genes are involved in the inflammatory stage and slowly disappear when bone formation occurs. Interestingly, on confirmation of the involvement of these genes in the osteogenic differentiation of MSCs, 6 out of these 15 candidate genes (Mmp3, Gbp6, Cxcl10, Clec4e, Ccl20, and Nos2) were significantly decreased in their mRNA levels, indicating that these 6 genes might be candidates for regulating MSC stemness, which is to maintain immature stem cells. Our results revealed that the knockdown of Clec4e, Gbp6, and Cxcl10 in vitro increased the osteogenic differentiation of MSCs. Future in vivo and in vitro studies involving detailed investigations into the 15 candidate genes involved in the relationship between bone marrow-derived MSCs and TNF-α from M1 macrophages will be of great interest for clinical applications in bone regeneration, immune-related diseases, and stem cell therapies. It is well-known that, besides TNF-α, various pro-inflammatory cytokines are present in the local environment of wound healing. In this study, we highlighted the potential involvement of TNF-α, a pro-inflammatory cytokine, in the functional expression of MSCs, emphasizing a previously unexplored relationship between macrophages and MSCs.
Conclusion
Temporal macrophage depletion by clodronate administration in mice immediately before tooth extraction delayed bone wound healing of the tooth extraction socket, induced a temporary reduction of TNF-α-producing M1 macrophages, and subsequently recovered MSC recruitment in the tooth extraction socket. From the RNA-seq results, 15 candidate genes that activate MSC function upon TNF-α stimulation were identified. Among them, Clec4e, Gbp6, and Cxcl10 were suggested to negatively regulate osteoblast differentiation.
Supplementary Material
Contributor Information
Aung Ye Mun, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Kentaro Akiyama, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Ziyi Wang, Department of Molecular Biology and Biochemistry, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8558, Japan.
Jiewen Zhang, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Wakana Kitagawa, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan; Department of Molecular Biology and Biochemistry, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8558, Japan.
Teisaku Kohno, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Ryuji Tagashira, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Kei Ishibashi, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan; Department of Molecular Biology and Biochemistry, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8558, Japan.
Naoya Matsunaga, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Tingling Zou, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Mitsuaki Ono, Department of Molecular Biology and Biochemistry, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8558, Japan.
Takuo Kuboki, Department of Oral Rehabilitation and Regenerative Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 2-5-1 Shikata-cho, Kita-ku, Okayama 700-8525, Japan.
Author contributions
Kentaro Akiyama conceptualized, designed, and managed the study; performed animal experiments; and wrote the original draft of the manuscript. Aung Ye Mun performed animal experiments, histological analysis, RNA-sequencing analysis, immunohistochemistry, cell culture, gene expression analysis, and data analysis, and wrote the original draft of the manuscript. Ziyi Wang performed RNA-sequencing analysis. Jiewen Zhang performed cell culture, gene expression analysis, and immunohistochemistry. Wakana Kitagawa performed the RNA sequencing analysis. Teisaku Kohno and Ryuji Tagashira performed histological analyses and animal experiments. Kei Ishibashi and Naoya Matsunaga performed the micro-CT and data analyses. Tingling Zou performed the animal experiments and immunohistochemistry. Mitsuaki On performed the RNA sequencing analysis. Takuo Kuboki supervised Kentaro Akiyama and Aung Ye Mun and edited the manuscript.
Aung Ye Mun (Data curation, Formal analysis, Investigation, Methodology, Writing—original draft), Kentaro Akiyama (Conceptualization, Funding acquisition, Project administration, Writing—review & editing), Ziyi Wang (Data curation, Investigation), Jiewen Zhang (Data curation, Investigation), Wakana Kitagawa (Data curation), Teisaku Kohno (Data curation), Ryuji Tagashira (Data curation), Kei Ishibashi (Data curation), Naoya Matsunaga (Data curation), Tingling Zou (Data curation), Mitsuaki Ono (Investigation, Methodology), and Takuo Kuboki (Conceptualization, Supervision, Writing—review & editing)
Funding
This research was funded by Japanese Society for the Promotion of Science, JSPS KAKENHI (Grant Numbers JP20K23080, JP21H03131, and JP22K17138).
Conflicts of interest
The authors declare no conflicts of interest.
Data availability
The RNA-Seq datasets generated in this study were available in the Gene Expression Omnibus (GEO) (GSE271144) at the National Center for Biotechnology Information (NCBI).
References
- 1. Claes L, Recknagel S, Ignatius A. Fracture healing under healthy and inflammatory conditions. Nat Rev Rheumatol. 2012;8(3):133–143. 10.1038/nrrheum.2012.1 [DOI] [PubMed] [Google Scholar]
- 2. Pahwa R, Goyal A, Jialal I. Chronic inflammation. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023: http://www.ncbi.nlm.nih.gov/books/NBK493173/. [Google Scholar]
- 3. Pajarinen J, Lin T, Gibon E, et al. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials. 2019;196:80–89. 10.1016/j.biomaterials.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maruyama M, Rhee C, Utsunomiya T, et al. Modulation of the inflammatory response and bone healing. Front Endocrin. 2020;11:386. 10.3389/fendo.2020.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Odgren PR, Witwicka H, Reyes-Gutierrez P. The cast of clasts: catabolism and vascular invasion during bone growth, repair, and disease by osteoclasts, chondroclasts, and septoclasts. Connect Tissue Res. 2016;57(3):161–174. 10.3109/03008207.2016.1140752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davison NL, Gamblin A-L, Layrolle P, Yuan H, Bruijn JDD, Groot FB. Liposomal clodronate inhibition of osteoclastogenesis and osteoinduction by submicrostructured beta-tricalcium phosphate. Biomaterials. 2014;35(19):5088–5097. 10.1016/j.biomaterials.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 8. Alexander KA, Chang MK, Maylin ER, et al. Osteal macrophages promote in vivo intramembranous bone healing in a mouse tibial injury model. J Bone Miner Res. 2011;26(7):1517–1532. 10.1002/jbmr.354 [DOI] [PubMed] [Google Scholar]
- 9. Sandberg OH, Tätting L, Bernhardsson ME, Aspenberg P. Temporal role of macrophages in cancellous bone healing. Bone. 2017;101:129–133. 10.1016/j.bone.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 10. Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med. 2011;13:e23. 10.1017/S1462399411001943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodman SB, Pajarinen J, Yao Z, Lin T. Inflammation and bone repair: from particle disease to tissue regeneration. Front Bioeng Biotechnol. 2019;7:230. 10.3389/fbioe.2019.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karnes JM, Daffner SD, Watkins CM. Multiple roles of tumor necrosis factor-alpha in fracture healing. Bone. 2015;78:87–93. 10.1016/j.bone.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 13. Gerstenfeld LC, Cho T-J, Kon T, et al. Impaired fracture healing in the absence of TNF-α signaling: the role of TNF-α in endochondral cartilage resorption. J Bone Miner Res. 2003;18(9):1584–1592. 10.1359/jbmr.2003.18.9.1584 [DOI] [PubMed] [Google Scholar]
- 14. Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of Guinea-pig bone marrow and spleen cells. Cell Prolif. 1970;3(4):393–403. 10.1111/j.1365-2184.1970.tb00347.x [DOI] [PubMed] [Google Scholar]
- 15. Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- 16. Kikuiri T, Kim I, Yamaza T, et al. Cell-based immunotherapy with mesenchymal stem cells cures bisphosphonate-related osteonecrosis of the jaw-like disease in mice. J Bone Miner Res. 2010;25(7):1668–1679. 10.1002/jbmr.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagata S, Hirano K, Kanemori M, Sun L-T, Tada T. Self-renewal and pluripotency acquired through somatic reprogramming to human cancer stem cells. PLoS One. 2012;7(11):e48699. 10.1371/journal.pone.0048699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akiyama K, Chen C, Wang D, et al. Mesenchymal-stem-cell-induced immunoregulation involves FAS-ligand-/FAS-mediated T cell apoptosis. Cell Stem Cell. 2012;10(5):544–555. 10.1016/j.stem.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L, Tredget EE, Wu PYG, Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. 2008;3(4):e1886. 10.1371/journal.pone.0001886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernardo ME, Fibbe WE. Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell. 2013;13(4):392–402. 10.1016/j.stem.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 21. Lu LY, Loi F, Nathan K, et al. Pro-inflammatory M1 macrophages promote osteogenesis by mesenchymal stem cells via the COX-2-prostaglandin E2 pathway. J Orthop Res. 2017;35(11):2378–2385. 10.1002/jor.23553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8(9):726–736. 10.1038/nri2395 [DOI] [PubMed] [Google Scholar]
- 23. Ueda M, Fujisawa T, Ono M, et al. A short-term treatment with tumor necrosis factor-alpha enhances stem cell phenotype of human dental pulp cells. Stem Cell Res Ther. 2014;5(1):31. 10.1186/scrt420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chavez MB, Chu EY, Kram V, de Castro LF, Somerman MJ, Foster BL. Guidelines for micro–computed tomography analysis of rodent dentoalveolar tissues. JBMR Plus. 2021;5(3):e10474. 10.1002/jbm4.10474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komori T, Ono M, Hara ES, et al. Type IV collagen α6 chain is a regulator of keratin 10 in keratinization of oral mucosal epithelium. Sci Rep. 2018;8(1):2612. 10.1038/s41598-018-21000-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tardaguila M, de la Fuente L, Marti C, et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 2018;28(3):396–411. 10.1101/gr.222976.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vitting-Seerup K, Sandelin A. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics. 2019;35(21):4469–4471. 10.1093/bioinformatics/btz247 [DOI] [PubMed] [Google Scholar]
- 31. Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32(9):896–902. 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCauley J, Bitsaktsis C, Cottrell J. Macrophage subtype and cytokine expression characterization during the acute inflammatory phase of mouse bone fracture repair. J Orthop Res. 2020;38(8):1693–1702. 10.1002/jor.24603 [DOI] [PubMed] [Google Scholar]
- 34. Horiuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology. 2010;49(7):1215–1228. 10.1093/rheumatology/keq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin T, Tamaki Y, Pajarinen J, et al. Chronic inflammation in biomaterial-induced periprosthetic osteolysis: NF-κB as a therapeutic target. Acta Biomater. 2014;10(1):1–10. 10.1016/j.actbio.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fullerton JN, Gilroy DW. Resolution of inflammation: a new therapeutic frontier. Nat Rev Drug Discov. 2016;15(8):551–567. 10.1038/nrd.2016.39 [DOI] [PubMed] [Google Scholar]
- 37. Loi F, Córdova LA, Pajarinen J, Lin T, Yao Z, Goodman SB. Inflammation, fracture and bone repair. Bone. 2016;86:119–130. 10.1016/j.bone.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang Y, He B, Wang L, et al. Bone marrow mesenchymal stem cell-derived exosomes promote rotator cuff tendon-bone healing by promoting angiogenesis and regulating M1 macrophages in rats. Stem Cell Res Ther. 2020;11(1):496. 10.1186/s13287-020-02005-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vi L, Baht GS, Whetstone H, et al. Macrophages promote osteoblastic differentiation In vivo: implications in fracture repair and bone homeostasis. J Bone Miner Res. 2015;30(6):1090–1102. 10.1002/jbmr.2422 [DOI] [PubMed] [Google Scholar]
- 40. Schlundt C, El Khassawna T, Serra A, et al. Macrophages in bone fracture healing: their essential role in endochondral ossification. Bone. 2018;106:78–89. 10.1016/j.bone.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 41. Omar OM, Granéli C, Ekström K, et al. The stimulation of an osteogenic response by classical monocyte activation. Biomaterials. 2011;32(32):8190–8204. 10.1016/j.biomaterials.2011.07.055 [DOI] [PubMed] [Google Scholar]
- 42. Ekström K, Omar O, Granéli C, Wang X, Vazirisani F, Thomsen P. Monocyte exosomes stimulate the osteogenic gene expression of mesenchymal stem cells. PLoS One. 2013;8(9):e75227. 10.1371/journal.pone.0075227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren G, Zhang L, Zhao X, et al. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell. 2008;2(2):141–150. 10.1016/j.stem.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 44. Li W, Ren G, Huang Y, et al. Mesenchymal stem cells: a double-edged sword in regulating immune responses. Cell Death Differ. 2012;19(9):1505–1513. 10.1038/cdd.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsumoto M, Tanaka T, Kaisho T, et al. A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J Immunol. 1999;163(9):5039–5048. 10.4049/jimmunol.163.9.5039 [DOI] [PubMed] [Google Scholar]
- 46. Patin EC, Orr SJ, Schaible UE. Macrophage inducible C-type lectin as a multifunctional player in immunity. Front Immunol. 2017;8:8. 10.3389/fimmu.2017.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu F, Jiao Y, Zhu Z, Sun C, Li H. Interferon-inducible protein 205 (p205) plays a role in adipogenic differentiation of mouse adipose-derived stem cells. Mol Cell Endocrinol. 2014;392(1–2):80–89. 10.1016/j.mce.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 48. Asefa B, Dermott JM, Kaldis P, Stefanisko K, Garfinkel DJ, Keller JR. p205, a potential tumor suppressor, inhibits cell proliferation via multiple pathways of cell cycle regulation. FEBS Lett. 2006;580(5):1205–1214. 10.1016/j.febslet.2006.01.032 [DOI] [PubMed] [Google Scholar]
- 49. Shi Y, Su J, Roberts AI, Shou P, Rabson AB, Ren G. How mesenchymal stem cells interact with tissue immune responses. Trends Immunol. 2012;33(3):136–143. 10.1016/j.it.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq datasets generated in this study were available in the Gene Expression Omnibus (GEO) (GSE271144) at the National Center for Biotechnology Information (NCBI).