Abstract
Background
The fruit ripening period is an important target trait in fruit tree crop breeding programs. Thus, citrus tree breeders seek to develop extreme early ripening cultivars that allow optimization of citrus maturation periods. In this study, we explored the regulatory network involved in fruit ripening in Citrus sinensis using the ‘Newhall’ navel orange variety and its early-ripening mutant, ‘Gannanzao’. This research will provide a basis for further research on important signaling pathways, gene functions and variety breeding of Citrus sinensis related to fruit ripening period.
Results
Physiological analyses suggested that early fruit ripening in ‘Gannanzao’ is regulated by early accumulation of abscisic acid (ABA), persistently high levels of jasmonic acid (JA), and higher sucrose content in the pericarp. Pericarp samples from ‘Gannanzao’ and ‘Newhall’ navel oranges were sampled for RNA sequencing analysis at 180, 200, and 220 days after flowering; 1430 differentially expressed genes (DEGs) were identified. Functional enrichment analysis indicated that these DEGs were mainly enriched in the plant hormone signal transduction and sugar metabolism pathways, as well as other pathways related to fruit ripening. Important DEGs associated with fruit ripening in ‘Gannanzao’ included genes involved in ABA and JA metabolism and signal transduction, as well as sugar metabolism. Weighted gene co-expression network analysis showed that the deep pink module had the strongest correlations with ABA content, JA content, and early ripening. Based on gene functionality and gene expression analyses of 37 genes in this module, two candidate hub genes and two ethylene response factor 13 (ERF13) genes (Cs_ont_5g000690 and Cs_ont_5g000700) were identified as key genes regulated by ABA and JA signaling. These findings will help to clarify the mechanisms that underlie early citrus fruit ripening and will lead to the development of excellent genetic resources for further breeding of extreme early-ripening varieties.
Conclusions
Through analyses of the ‘Newhall’ navel orange cultivar and its early-ripening mutant ‘Gannanzao’, we identified genes involved in ABA and JA metabolism, signal transduction, and sugar metabolism that were related to fruit ripening. Among these, two ERF13 genes were inferred to be key genes in the regulation of fruit ripening. These findings provide insights into the genetic architecture related to early fruit ripening in C. sinensis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10651-1.
Keywords: Citrus sinensis, Fruit ripening, Transcriptome, WGCNA
Background
The navel orange (Citrus sinensis L. Osbeck) is a non-climacteric fruit for which the ripening mechanism has not been clearly established; therefore, the genes or enzymes involved in its respiration rate at fruit ripening require further exploration [1, 2]. Metabolic and respiratory changes during fruit ripening strongly affect the shelf life and economic value of fruit through effects on color, taste, sugar and acid contents, aroma, texture, and nutritional value [3, 4]. Thus, elucidation of the regulatory pathways and networks associated with fruit ripening is necessary to adjust the ripening period of navel oranges and reveal general regulatory mechanisms involved in the ripening of non-climacteric fruit.
Thus far, few mechanisms and genes regulating fruit ripening in horticultural plants have been discovered. Plant hormones, sugar metabolism, and cell wall metabolism all participate in the regulation of fruit ripening, which is primarily determined by plant hormone biosynthesis, signal transduction, and related pathways. In climacteric fruit, ripening is mainly regulated by ethylene, whereas non-climacteric fruit ripening is mainly regulated by abscisic acid (ABA) [5, 6]. Various components are involved in the ABA metabolism signaling pathways associated with fruit maturation, including 9-cis-epoxycarotenoid dioxygenase 1 (NCED1), PYL9 (PYR/PYL/RCAR), serine/threonine-phosphatase 2 C (PP2C), sucrose non-fermenting1 (SNF1)-regulated protein kinase 2S (SnRK2), and ABA 8’-hydroxylase (CYP707A1) [7–10]. Other phytohormones may also be involved in the regulation of fruit maturation [11, 12]. The involvement of sugar metabolism in fruit development and ripening has been demonstrated in horticultural plants [13]. Among fruit sugars, sucrose functions as a signaling molecule to regulate orange ripening [11]. Fruit softening during the ripening process is associated with the gradual degradation of pectin in the cell wall, leading to modifications of cell wall composition and the softening of parenchyma cells [14, 15].
Various transcription factors (TFs) are also associated with fruit ripening. Based on cloned genes mapped from fruit ripening mutants, three TFs were inferred to be required for fruit ripening in tomato: MADS-box TF ripening inhibitor (RIN), SBP-box TF colorless non-ripening (CNR), and the NAC TF non-ripening (NOR) [16–18]. The functions of orthologs of these three TFs from other species are conserved and similarly regulate fruit ripening [19, 20]. However, in contrast to the many studies of key TFs in fruit ripening, particularly for climacteric fruits, very few studies have examined the mechanisms of ripening in non-climacteric fruits [21, 22]. In one such study, gene expression profile analysis suggested that APETALA2/ethylene-responsive element-binding factor (AP2/ERF) plays roles in color and aroma formation in strawberry fruit [23]. In a recent study, RNA interference-mediated silencing demonstrated that downregulation of the NAC TF FaRIF delays the ripening of strawberry fruit [22].
The molecular mechanisms that underlie fruit ripening have been studied in various citrus mutants. For example, the ‘Fengwan’ sweet orange is a spontaneous late-ripening mutant of the ‘Fengjie 72 − 1’ orange (C. sinensis); the key genes and regulatory pathways involved in ‘Fengwan’ fruit ripening have been identified through small-RNA sequencing, degradome sequencing, and transcriptome and proteome analyses [24, 25]. The regulatory mechanisms of fruit ripening were also analyzed in the spontaneous early-ripening citrus mutants ‘Ganqi 4’ (C. sinensis) and ‘Liuyuezao’ (Citrus maxima) through comparative transcriptome and sRNAome analyses [26, 27]. These studies investigated the roles of TFs, functional genes, and small RNAs related to various metabolic pathways, including those related to sugar metabolism and plant hormones, involved in citrus fruit ripening. However, the molecular regulatory network of citrus fruit ripening requires further in-depth research.
In this study, we investigated the citrus fruit ripening period using the ‘Newhall’ navel orange and its early-ripening mutant cultivar ‘Gannanzao’. We determined the contents of sugars and endogenous hormones, as well as genome-wide gene transcription levels, at various fruit ripening stages to elucidate the molecular mechanisms that underlie early fruit ripening in ‘Gannanzao’. We also conducted transcriptomic analysis and weighted gene co-expression network analysis (WGCNA) to identify hub genes that might regulate fruit ripening. By integrating gene function annotation, expression levels, and correlation with early ripening traits, we identified differentially expressed genes, pathways and core genes that may play a role in regulating citrus early ripening through hormones and sugar metabolism. The core gene identified in this study, along with its homologous genes, have not been previously studied in relation to fruit ripening. Our findings will provide novel insights on the molecular regulation of fruit ripening in non-climacteric fruits and provide valuable insights for the molecular breeding of citrus varieties.
Results
Fruit ripening in the ‘Gannanzao’ navel orange cultivar
The ‘Gannanzao’ cultivar originated from a bud mutation of the ‘Newhall’ navel orange. ‘Gannanzao’ ripens 1 month earlier and has better fruit quality compared with the original variety [28]. Thus, the onset of color variation and pericarp softening in ‘Gannanzao’ occurs at 180 days after flowering (DAF) (Fig. 1).
Fig. 1.
Performance of fruits of GNZ and Newhall navel orange during fruit ripening
Endogenous hormone contents in ‘Gannanzao’ navel oranges during fruit ripening
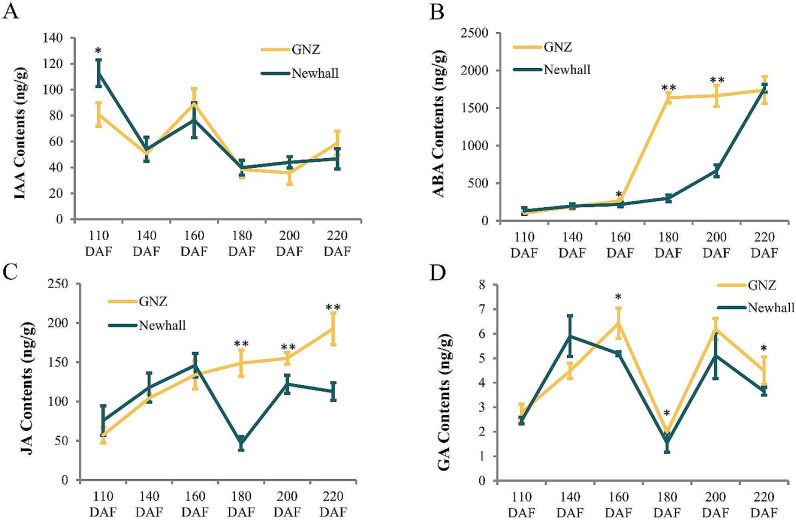
The levels of various endogenous hormones in the pericarp were measured at various time points (110, 140, 160, 180, 200, and 220 DAF) to assess their effects on early ripening (Fig. 2). Auxin (i.e., indole-3-acetic acid [IAA]) levels in the pericarp was significantly lower among ‘Gannanzao’ oranges than among ‘Newhall’ oranges at 110 DAF, and there were no differences between the two cultivars during other stages (Fig. 2A). ABA content in the pericarp of ‘Gannanzao’ oranges sharply increased at 160–180 DAF and remained high thereafter, whereas a corresponding increase in ‘Newhall’ oranges occurred at 200–220 DAF (Fig. 2B). In contrast, a sharp decrease in JA content in the pericarp of ‘Newhall’ oranges occurred at 160–180 DAF; this phenomenon was not observed in ‘Gannanzao’ oranges, leading to significantly higher JA content in the latter cultivar at ≥ 160 DAF (Fig. 2C). Gibberellin (GA) content in the pericarp was significantly higher among ‘Gannanzao’ oranges than among ‘Newhall’ oranges at 160, 180 and 220 DAF, and there were no differences between the two cultivars during other stages (Fig. 2D). Thus, compared with phytohormone levels in the pericarp of ‘Newhall’ oranges, the early accumulation of ABA and persistently high levels of JA in the pericarp of ‘Gannanzao’ oranges suggest that these hormones are involved in early fruit ripening.
Fig. 2.
Endogenous hormone contents during fruit ripening. A-D are represent IAA, ABA, JA and GA contents in pericarp of GNZ and Newhall navel orange during fruit ripening, respectively. ⁎ and ⁎⁎ denote significant difference at the 0.05 and 0.01 probability levels, respectively
Sugar contents in ‘Gannanzao’ navel oranges during fruit ripening
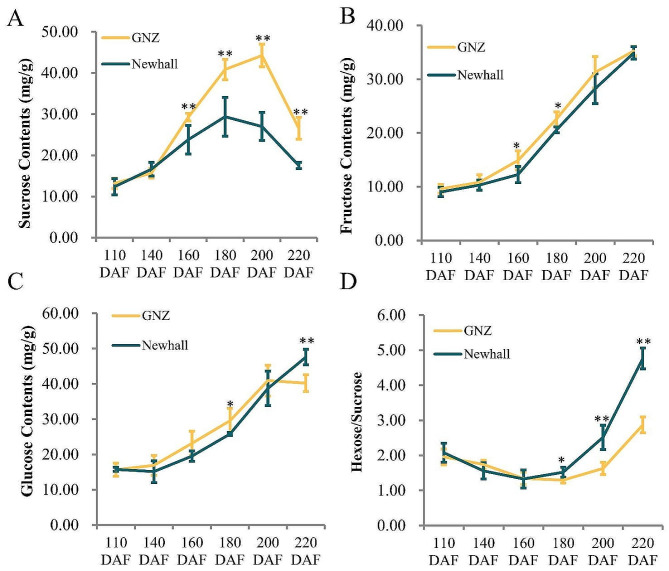
Sugars play various roles in many plant developmental processes, including fruit ripening. The fruit sugar contents of ‘Gannanzao’ and ‘Newhall’ oranges were measured in the pericarp throughout the ripening process, and the ratio of hexose to sucrose was calculated. Sucrose content was significantly higher in ‘Gannanzao’ oranges than in ‘Newhall’ oranges from 160 to 220 DAF (Fig. 3A), whereas fructose contents were higher in ‘Gannanzao’ oranges than in ‘Newhall’ oranges at 160 and 180 DAF; there were no differences between the groups at other stages (Fig. 3B). Glucose content was higher in ‘Gannanzao’ oranges at 180 DAF, but lower at 220 DAF, than in ‘Newhall’ oranges (Fig. 3C). At ≥ 180 DAF, ‘Gannanzao’ oranges showed a larger decrease in the sucrose to hexose ratio compared with ‘Newhall’ oranges (Fig. 3D). These results indicate that sugar content, specifically sucrose content, plays a crucial role in regulating early fruit ripening in ‘Gannanzao’ navel oranges.
Fig. 3.
Sugar contents during fruit ripening. A-C are represent sucrose, fructose and glucose contents in pericarp of GNZ and Newhall navel orange during fruit ripening, respectively. D is sucrose to hexose ratios in pericarp of GNZ and Newhall navel orange during fruit ripening. ⁎ and ⁎⁎ denote significant difference at the 0.05 and 0.01 probability levels, respectively
Analysis of differentially expressed genes (DEGs) in the pericarp of ‘Gannanzao’ and ‘Newhall’ navel oranges
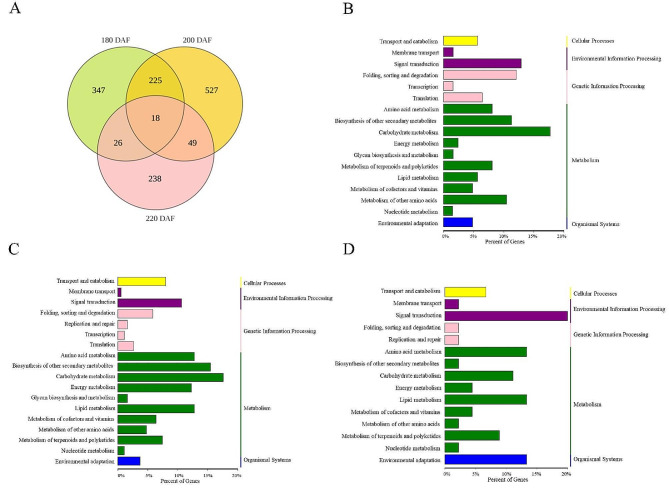
Pericarp samples from ‘Gannanzao’ and ‘Newhall’ navel oranges at 180, 200, and 220 DAF were subjected to RNA sequencing (RNA-seq) analysis to examine differential gene expression between the two cultivars throughout fruit ripening. The boxplot of expression abundance (FPKM) of genes in each sample and the PCA analysis of the transcriptomics was shown in Additional file 1 Fig. S1 A-B. DEGs between the two cultivars were identified according to a false discovery rate (FDR) < 0.05 and |log2(fold change [FC]) | ≥ 1. Based on these criteria, we identified 616 DEGs at 180 DAF (106 upregulated, 510 downregulated), 819 DEGs at 200 DAF (218 upregulated, 601 downregulated), and 331 DEGs at 220 DAF (218 upregulated, 113 downregulated) (Additional file 1 Fig. S1 C-E; Additional file 2–5 Tables S1-S4). Among these DEGs, 225 were identified at both 180 and 200 DAF, 49 were identified at both 200 and 220 DAF, and 26 were identified at both 180 and 220 DAF; in total, 18 DEGs were identified in all three stages (Fig. 4A).
Fig. 4.
Identification and pathway enrichment analysis of DEGs related to fruit ripening. A is venn diagram analysis showing the number of common and unique DEGs identified at three stages. B-D are represent KEGG enrichment pathway (level 1 and 2) annotated classification results of all differentially expressed genes in GNZ and Newhall navel orange at 180-, 200- and 220 DAF respectively
To better understand the functions of these DEGs, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Fig. 4B–D), which identified processes in five main categories: metabolism, genetic information processing, environmental information processing, cell processes, and organismal systems. We identified the top 20 KEGG pathway enrichment terms at all three stages are shown in Additional file 6 Fig. S2. Among these, metabolic-related pathways were most prominent. There were 9 and 4 DEGs significantly enriched in ‘Photosynthesis’ pathway at 200- and 220- DAF, respectively (Additional file 7 Table S5). Among the DEGs, genes encoding chloroplast protein in Photosystem I and II, including Cs_ont_1g005130 (Photosystem I reaction centre subunit N), Cs_ont_2g007730 (Photosystem I psaG/psaK), Cs_ont_2g021750 (Photosystem II Pbs27), Cs_ont_5g010360 (Photosystem I PsaD), Cs_ont_5g011340 (Photosystem II reaction centre W protein) and Cs_ont_7g023170 (Photosystem II protein Y) were relatively low expressed in the transcriptome of ‘Gannanzao’ pericarp at 200- and 220- DAF stages, and possibly related to the color alteration in the pericarp of ‘Gannanzao’ oranges at 200 DAF. There were 24 DEGs significantly enriched in ‘Phenylpropanoid biosynthesis’ pathway at 200 DAF. The ‘Phenylpropanoid biosynthesis’ pathway has been indicated to be involved in the hormone-regulated fruit ripening [29, 30]. We also found 6 and 7 DEGs significantly enriched in ‘Sesquiterpenoid and triterpenoid biosynthesis’ pathway at 180- and 200- DAF, respectively, which is important for ABA biosynthesis [31]. Several pathways enriched in DEGs at all three stages were linked to plant hormone signal transduction and sugar metabolism, although not significantly, reflecting the levels of endogenous hormones and sugars detected in this study.
Hormone-related DEGs regulate early fruit ripening in ‘Gannanzao’ navel oranges
Pathways related to signal transduction, particularly plant hormone signal transduction, were identified as key pathways associated with early fruit ripening in ‘Gannanzao’. Notably, signal transduction pathways had the largest number of upregulated DEGs at 200 DAF; 29 DEGs were enriched in the plant hormone signal transduction pathway, including some homologs to genes that affect ripening in other species (Additional file 7 Table S5). These results suggest that DEGs related to plant hormone signaling are involved in the regulation of fruit ripening.
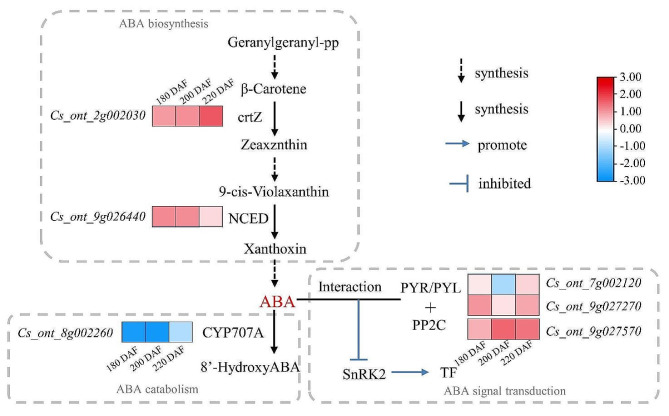
Phytohormones are major regulators of fruit ripening. Navel oranges are non-climacteric fruit, in which ripening is primarily regulated by ABA. Functional annotation of the DEGs identified in this study revealed six DEGs related to ABA (Table 1). Cs_ont_2g002030 (NCED1) and Cs_ont_9g026440 (crtZ) encode key enzymes for ABA biosynthesis and Cs_ont_9g027570 (PP2C) and Cs_ont_9g027270 (PYL8) encode ABA receptors; these genes were highly expressed in ‘Gannanzao’ pericarp compared with ‘Newhall’ pericarp at all three ripening stages. Cs_ont_8g002260 encodes cytochrome P450 707A1, a key enzyme in ABA catabolism, and its expression was downregulated in ‘Gannanzao’ pericarp compared with ‘Newhall’ pericarp at all three ripening stages (Fig. 5A). These results suggest that ABA synthesis and degradation rates are higher and lower, respectively, in ‘Gannanzao’ oranges than in ‘Newhall’ oranges, leading to early accumulation of ABA and enhanced ABA signal transduction in the early-ripening mutant.
Table 1.
Important differentially expressed genes of different pathways related to fruit ripening in pericarps of navel orange
| Gene ID | Annotation | 180 DAF | 200 DAF | 220 DAF | |||
|---|---|---|---|---|---|---|---|
| GNZ | Newhall | GNZ | Newhall | GNZ | Newhall | ||
| ABA metabolism and signal transduction | |||||||
| Cs_ont_2g002030 | NCED1 | 37.38 | 19.049 | 45.136 | 21.197 | 102.88 | 32.177 |
| Cs_ont_9g026440 | Beta-carotene hydroxylase, crtZ | 911.065 | 411.183 | 990.257 | 457.486 | 929.496 | 749.646 |
| Cs_ont_8g002260 | Cytochrome P450 707A1 | 1.293 | 8.211 | 0.677 | 4.573 | 0.663 | 1.291 |
| Cs_ont_7g002120 | Abscisic acid receptor PYL4 | 11.805 | 10.564 | 7.12 | 14.623 | 4.334 | 3.369 |
| Cs_ont_9g027270 | Abscisic acid receptor PYL8 | 41.217 | 20.198 | 17.461 | 14.966 | 23.917 | 13.318 |
| Cs_ont_9g027570 | Probable protein phosphatase 2 C | 87.056 | 52.02 | 156.415 | 54.096 | 371.114 | 142.466 |
| JA signal transduction | |||||||
| Cs_ont_7g027580 | Protein TIFY 9, JAZ | 15.938 | 54.707 | 36.086 | 66.021 | 8.466 | 6.404 |
| Cs_ont_1g018990 | Protein TIFY 10 A, JAZ | 14.175 | 47.427 | 48.574 | 56.051 | 9.667 | 7.423 |
| Cs_ont_1g019000 | Protein TIFY 10 A, JAZ | 32.516 | 75.723 | 77.133 | 106.642 | 22.309 | 24.43 |
| Starch and sucrose metabolism | |||||||
| Cs_ont_1g020030 | Beta-fructofuranosidase, insoluble isoenzyme CWINV1 | 1.628 | 4.21 | 1.058 | 0.892 | 6.283 | 0.548 |
| Cs_ont_1g020150 | Beta-fructofuranosidase, insoluble isoenzyme CWINV1 | 1.04 | 2.749 | 0.66 | 0.598 | 3.989 | 0.372 |
| Cs_ont_2g010790 | Endoglucanase 12 | 0.721 | 1.752 | 0.822 | 1.336 | 0.729 | 0.378 |
| Cs_ont_3g010740 | Trehalose-phosphate phosphatase, TPP | 2.905 | 7.045 | 1.843 | 3.982 | 0.307 | 0.448 |
| Cs_ont_4g026880 | Alpha, alpha-trehalose-phosphate synthase | 6.281 | 12.854 | 9.624 | 5.704 | 8.741 | 6.306 |
| Cs_ont_6g008700 | Beta-fructofuranosidase, insoluble isoenzyme, CWINV1 | 0.346 | 2.591 | 0.043 | 1.999 | 0.596 | 0.378 |
| Cs_ont_9g014140 | Sucrose-phosphate Synthase 3 | 15.67 | 7.168 | 18.183 | 8.392 | 14.508 | 11.876 |
| Cs_ont_2g006970 | Beta-glucosidase 46 | 2.478 | 2.574 | 1.558 | 3.525 | 1.304 | 4.103 |
| Cs_ont_8g027180 | Beta-glucosidase 11 | 94.099 | 89.626 | 47.447 | 96.714 | 21.144 | 22.638 |
| Cs_ont_2g006950 | Glucose-6-phosphate/phosphate translocator 2 | 43.219 | 79.713 | 100.127 | 95.966 | 42.614 | 18.839 |
| Cs_ont_3g005060 | Alpha-amylase | 33.794 | 23.534 | 18.465 | 12.502 | 13.643 | 5.929 |
| Cs_ont_8g021290 | Glucose-1-phosphate adenylyltransferase | 150.729 | 96.512 | 106.336 | 79.453 | 134.214 | 38.921 |
Fig. 5.
Expression analysis of key DEGs involved in ABA metabolism and signal transduction. Each column represents an experimental condition, and each row represents a gene. Red means the up-regulated expression of a DEG and blue means the down-regulated expression
In addition, three DEGs (Cs_ont_7g027580, Cs_ont_1g018990, and Cs_ont_1g019000) encode repressors of jasmonate zinc-finger inflorescence meristem (JAZ) domain proteins, which are JA-associated TFs. These genes were downregulated in ‘Gannanzao’ pericarp compared with ‘Newhall’ pericarp at 180 DAF (Table 1), possibly related to the sharp reduction in JA content detected in the pericarp of ‘Newhall’ oranges at 160–180 DAF.
Responses of sugar-related DEGs to early fruit ripening in ‘Gannanzao’ navel oranges
Sugar, the main component of citrus fruits, regulates pericarp softening and ripening. KEGG pathway analysis revealed that DEGs between the two navel orange cultivars were mainly enriched in metabolic pathways at all three stages. Specifically, carbohydrate metabolism pathways showed the greatest gene enrichment (58 DEGs), suggesting that these genes are related to fruit softening and sugar metabolism (Additional file 7 Table S5).
Sucrose is involved in orange fruit ripening processes and may interact with plant hormones [11]. We detected 12 DEGs involved in starch and sucrose metabolism (Table 1). Among these, Cs_ont_9g014140, encoding a sucrose phosphate synthase with an important role in sucrose metabolism, showed 2.19-, 2.17-, and 1.22-fold increases in ‘Gannanzao’ at 180, 200, and 220 DAF, respectively. Three DEGs identified in this study (Cs_ont_1g020030, Cs_ont_1g020150, and Cs_ont_6g008700) encode beta-fructofuranosidase, an insoluble isoenzyme that limits the rate of sucrose metabolism; these three genes were downregulated in ‘Gannanzao’ compared with ‘Newhall’ at 180 DAF. Several DEGs involved in monosaccharide metabolism, such as fructose 1,6-bisphosphate (Cs_ont_6g015890) and fructose-bisphosphate aldolase 2 (Cs_ont_8g020190), were significantly downregulated at 200 and 220 DAF; fructose-2,6-bisphosphatase (Cs_ont_8g000380) was significantly downregulated at all three stages; and glucose-1-phosphate adenylyltransferase (Cs_ont_8g021290), glucose-6-phosphate translocator (Cs_ont_2g006950), and glyceraldehyde-3-phosphate dehydrogenase (Cs_ont_9g028230) were significantly upregulated at 220 DAF (Additional file 7 Table S5). These results suggest that sucrose and monosaccharide metabolic processes are involved in the formation of early-ripening traits in ‘Gannanzao’.
Co-expression network analysis
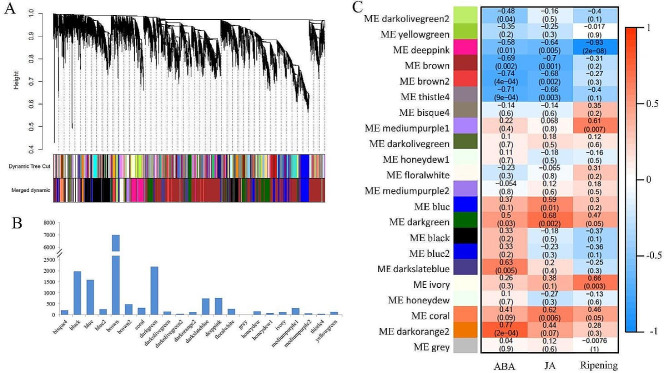
To further explore transcriptional changes in the two navel orange cultivars during fruit ripening, we performed co-expression network analysis using the WGCNA package in R software (R Core Team, Vienna, Austria). Genes with similar expression patterns were grouped into 22 modules, which were assigned different colors (Fig. 6A). Of these, the brown and gray modules contained the highest and lowest numbers of genes: 3157 and 13, respectively (Fig. 6B). Analyses of module–trait relationships showed that the 11 identified modules were significantly correlated with ABA content, JA content, and early ripening (P < 0.05; Fig. 6C). The deep pink module was strongly correlated with all three traits; therefore, we inferred that it is a crucial module related to early ripening.
Fig. 6.
identification modules related to fruit ripening by gene co-expression network and module–trait relationships analysis. (A) Weighted gene co-expression network analysis identified 22 co-expression modules. (B) Number of genes in each module. (C) Module–trait relationships plot. The module sample correlation and corresponding p values are shown in parentheses. The panel on the left shows 22 modules. The color code on the right shows the module feature correlation − 1 (blue) to 1 (red)
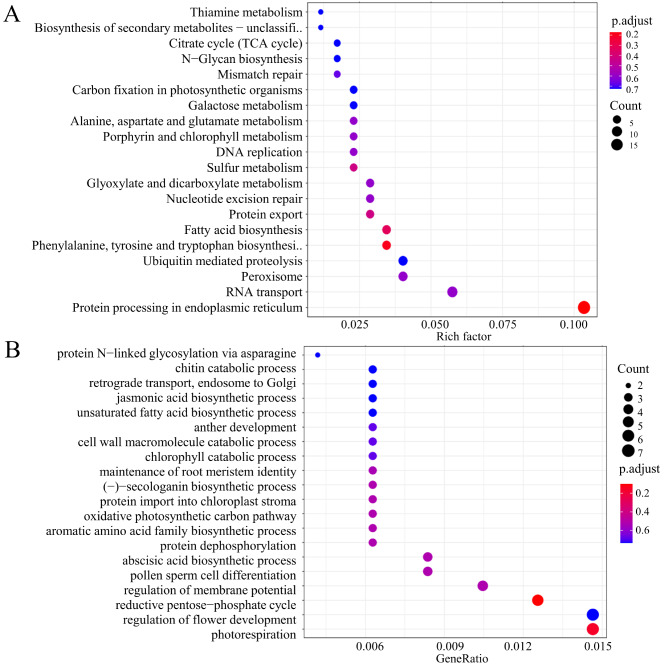
To explore the functions of genes within the deep pink module during fruit ripening, we performed KEGG and Gene Ontology (GO) pathway analyses of these genes. Top 20 KEGG pathway enrichments with DEGs were showed (Fig. 7A). Consistent with the DEG enrichment analysis results, the deep pink module was enriched in pathways related to genetic information processing (protein processing in the endoplasmic reticulum, RNA transport, protein export, and DNA replication) and sugar metabolism (galactose metabolism, N-glycan biosynthesis, and the citrate acid cycle). In GO terms of biological processes, genes in the deep pink module were mainly enriched in JA and ABA biosynthesis and cell wall metabolism (chitin and cell wall macromolecule catabolism) (Fig. 7B). Some genes in the deep pink module were enriched in chlorophyll catabolism and protein import into the chloroplast stroma, implying that they are involved in fruit coloring. Taken together, these results suggest that genes in the deep pink module play roles in the formation of early-ripening traits in the ‘Gannanzao’ cultivar. Therefore, the deep pink module was identified as a key module in early citrus fruit ripening.
Fig. 7.
Enrichment analyses of genes in the deeppink module. (A) KEGG enrichment analysis of genes in deeppink module. (B) Biological_process (GO) enrichment analyses for genes in deeppink module
The main gene nodes were selected according to CytoHubba maximal clique centrality (MCC) scores. The 37 genes with the highest connectivity scores (i.e., the top 5%) in the deep pink module were selected as candidate hub genes; the homologous functions of these genes were annotated via BLAST searches of the Swiss-Prot and Pfam databases (Additional file 8 Table S6); the correlation (Gene Significance, GS) between these gene expression levels and trait was shown in Additional file 9 Table S7. Among these candidate hub genes, only two genes were identified: Cs_ont_5g000690 and Cs_ont_5g000700, which encode the TF ethylene-responsive factor 13 (ERF13). AP2/ERF TFs mediate ethylene, JA, and ABA signaling; they are involved in JA and ABA stimulus and signaling responses [32–34]. Both genes expression level in the pericarp of ‘Gannanzao’ oranges consistently reduced during fruit ripening, whereas a corresponding reduce in ‘Newhall’ oranges occurred at 200–220 DAF. Cs_ont_5g000690 was downregulated in ‘Gannanzao’ pericarp compared with ‘Newhall’ pericarp at 200, and 220 DAF and Cs_ont_5g000700 was downregulated at 180, 200, and 220 DAF (Additional file 10 Fig. S3). The correlation coefficients between Cs_ont_5g000690 expression and ABA content, JA content, and early ripening trait were − 0.81(p < 0.01), -0.79(p < 0.01) and 0.72(p < 0.01), respectively; the correlation coefficients between Cs_ont_5g000700 expression and ABA content, JA content, and early ripening trait were − 0.73(p < 0.01), -0.51(p < 0.05) and 0.60(p < 0.01), respectively (Additional file 9 Table S7). This result indicating that they are key genes involved in early citrus fruit ripening and may play key roles in downstream gene regulation.
Discussion
Fruit ripening traits are important targets of crop and horticultural research because short fruit ripening periods have long hindered the development of citrus cultivars in China. In recent years, the appearance of early- and late-ripening citrus cultivars has led to new investigations of citrus ripening periods [35]. The ‘Gannanzao’ navel orange examined in this study recently originated from a bud mutation of the ‘Newhall’ navel orange in China [28]. This new variety ripens 1 month earlier than its parental cultivar and can adapt to any environment where navel oranges are grown; thus, it is suitable for widespread production.
Citrus is a non-climacteric fruit, in which ripening is mainly regulated by ABA [6, 11]. In the present study, ABA synthesis and degradation rates were higher and lower, respectively, among ‘Gannanzao’ navel oranges than among ‘Newhall’ navel oranges, leading to early accumulation of ABA at 180 DAF in the early-ripening cultivar (Fig. 2B). Subsequently, these high ABA levels inhibit the downstream gene expression by enhancing the expression levels of genes related to ABA signal transduction (Fig. 4). These findings explain the much higher number of downregulated genes compared with upregulated genes at 180 and 200 DAF. In addition, JA plays a crucial role in regulating fruit ripening [11, 36]. We observed changes in JA content and identified genes involved in the JA signaling pathway that were significantly downregulated in ‘Gannanzao’ at 180 DAF. The signal transduction pathways of various hormones form a complex and sophisticated network in plants; many JA-responsive genes are also induced by ABA [37, 38]. Further research is required to investigate whether JA is transcriptionally regulated by ABA signaling in ‘Gannanzao’ navel oranges. Pericarp IAA content did not differ between ‘Gannanzao’ and ‘Newhall’ navel oranges; however, multiple DEGs enriched in plant hormone signal transduction pathways were involved in auxin signaling (Additional file 7 Table S5). GO enrichment analysis of co-expression network modules significantly correlated with ABA content indicated that the expression of DEGs in these modules may be regulated by ABA. For example, the dark orange 2 module was enriched in negative regulation of the auxin-mediated signaling pathway (Additional file 11 Fig. S4). Our sugar content analysis revealed that sucrose content showed the greatest difference between the two navel orange cultivars. Previous studies have demonstrated the involvement of ABA in the regulation of sucrose-induced functional gene transcription in plants [39]. In summary, our results indicate that ABA metabolism and signal transduction are critical factors involved in the formation of early-ripening traits in the ‘Gannanzao’ navel orange cultivar.
ERF, a member of the APETALA2/ERF family, is a plant-specific TF with a variety of functions, including biotic and abiotic stress responses, as well as the regulation of hormone biosynthesis and fruit development, ripening, and senescence [40–42]. Recent studies have confirmed the involvement of ERF proteins in fruit ripening [40, 43], and our WGCNA identified two ERF13 genes (Cs_ont_5g000690 and Cs_ont_5g000700) as potential key regulatory genes of early fruit ripening. ERF13 is regulated by both ABA and JA [33, 34]. The expression patterns of coupling element binding factors were analyzed in Arabidopsis overexpression lines to investigate their functions in vivo, revealing that AtERF13 imparts ABA hypersensitivity [33]. AtERF13 is also responsive to JA signaling via calcium-dependent protein kinase-related protein kinase 2 [34]. ERF13 proteins are involved in biotic and abiotic stress responses [34, 44]. However, this is the first study to investigate the involvement of ERF13 in fruit ripening regulation. Through a combination of WGCNA and gene expression analyses, we revealed that two ERF13 genes (Cs_ont_5g000690 and Cs_ont_5g000700) are key TFs regulated by ABA and JA signaling; they subsequently regulate downstream genes involved in sugar and cell wall metabolism, thereby influencing the fruit ripening period.
Methods
Plant materials
The early-ripening ‘Gannanzao’ navel orange, which exhibits a bud mutation, was originally isolated from the ‘Newhall’ navel orange in Ganzhou, China. All plant materials were grown in the fields of the experimental nursery at Gannan Normal University. The cultivation conditions for early-ripening plants were identical to the conditions for ‘Newhall’ navel orange plants. Pericarp samples were collected from ‘Gannanzao’ and ‘Newhall’ plants at 110, 140, 160, 180, 200, and 220 DAF; immediately frozen in liquid nitrogen; and stored at − 80 °C until use.
Quantification of endogenous hormones and sugar
The contents of endogenous hormones including IAA, GA, JA, and ABA were determined using high-performance liquid chromatography, as previously described [45]. The extraction and quantitative analysis of sugars including sucrose, fructose, and glucose were performed using reverse HPLC, as previously described [46]. All analyses were performed with three biological replicates; mean values were compared between the ‘Gannanzao’ and ‘Newhall’ cultivars using Student’s t-test.
RNA preparation
Total RNA was extracted from tissues using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). RNA quality and concentration were determined by electrophoresis and spectrophotometry (Nanodrop ND-2000, Thermo Fisher Scientific, Waltham, MA, USA). First-strand complementary DNA was synthesized using a reverse-transcription kit (TaKaRa Bio, Shiga, Japan).
RNA-seq analysis
Transcriptome analyses of ‘Gannanzao’ pericarp tissue were performed via RNA sequencing, using ‘Newhall’ pericarp tissue as the control. Pericarp tissue was sampled at 180, 200, and 220 DAF, then subjected to extraction of total RNA. Library construction and sequencing were conducted using the Illumina HiSeq platform (Illumina, San Diego, CA, USA). Clean reads were mapped to the C. sinensis v3.0 reference genome (http://citrus.hzau.edu.cn) using HISAT2 [47]. Differential expression (|log2(FC)| ≥ 1, FDR < 0.05) was analyzed using DESeq2 [48]. All experiments were performed with three biological replicates.
WGCNA
Transcriptome data for 18 samples were normalized to fragments per kilobase of transcript per million mapped reads (FPKM) to select genes for gene co-expression network analysis of C. sinensis. Co-expression-network analysis was conducted with the WGCNA package in R software, as previously described [49, 50]. After the network had been generated, key modules that might be related to fruit ripening in C. sinensis were identified according to correlations between modules and traits. Functional annotation of genes within the module of interest was conducted by searching against the GO and KEGG databases [51, 52], and the network was mapped with Cytoscape v3.8.0 (http://apps.cytoscape.org/apps/cytohubba) [53]. Based on the intersecting network, hub genes were identified using the CytoHubba plug-in with MCC algorithms [54].
Conclusion
We conducted physiological analyses of the ‘Newhall’ navel orange and its early-ripening mutant cultivar, ‘Gannanzao’; we found that ABA, JA, and sucrose levels in ‘Gannanzao’ orange pericarp may regulate early fruit ripening in this cultivar. Through RNA-seq analysis, we identified 1430 DEGs between the two navel orange cultivars. Functional enrichment analysis revealed that these DEGs were mainly enriched in the plant hormone signal transduction and sugar metabolism pathways, as well as other pathways related to fruit ripening. A combination of WGCNA, pathway enrichment analysis, and gene functionality and expression analyses showed that the deep pink module was a key module; 37 genes were identified as candidate hub genes. Among these, two ERF13 genes (Cs_ont_5g000690 and Cs_ont_5g000700) were inferred to be key genes that are regulated by ABA and JA signaling; they regulate downstream genes. Overall, our findings provide insights into citrus fruit ripening and establish a foundation for the breeding of citrus cultivars with extreme early ripening periods.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- WGCNA
Weighted gene co-expression network analyses
- ABA
Abscisic acid
- JA
Jasmonic acid
- DEGs
Differentially expressed genes
- ERF13
Ethylene response factor 13
- NCED1
9-cis-epoxycarotenoid dioxygenase 1
- PYL9
PYR/PYL/RCAR
- PP2C
Serine/threonine-phosphatase 2 C
- CYP707A1
ABA 8’-hydroxylase
- TFs
Transcription factors
- RIN
Ripening inhibitor
- CNR
Colorless non-ripening
- NOR
Non-ripening
- AP2/ERF
APETALA2/ethylene-responsive element-binding facto
- DAF
Days after flowering
- IAA
indole-3-acetic acid
- GA
Gibberellin
- FC
Fold change
- FPKM
Fragments per kilobase of transcript per million mapped reads
- FDR
False discovery rate
Author contributions
J.C. and S.W. conceived and designed the study. J.C. and L.X., performed many of the experiments and wrote the manuscript. S.W. and B.Z. advised on the experiments and modified the manuscript. Y.L.took part in the RNA extraction and assay of endogenous hormones. All authors read and approved the final manuscript.
Funding
This research was supported financially by the National Natural Science Foundation of China (32060667 and 32302493), the Key Research and Development Program of Jiangxi Province (20223BBF61004), and the Scientific Research Project of Education Department of Jiangxi Province (GJJ2201258). The funders provided the financial support to the research, but had no role in the design of the study, analysis, interpretations of data and in writing the manuscript.
Data availability
Sequence data that support the findings of this study have been deposited in the uploaded to National Center for Biotechnology Information with the primary accession code SRP500014.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Paul V, Pandey R, Srivastava GC. The fading distinctions between classical patterns of ripening in climacteric and non-climacteric fruit and the ubiquity of ethylene—an overview. J Food Sci Technol. 2012;49(1):1–21. 10.1007/s13197-011-0293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kou X, Wu M. Characterization of climacteric and non-climacteric fruit ripening. In: Guo Y, editor. Plant Senescence: methods and protocols. New York, NY, USA: Springer; 2018. pp. 89–102. [DOI] [PubMed] [Google Scholar]
- 3.Castellarin SD, Gambetta GA, Wada H, Krasnow MN, Cramer GR, Peterlunger E, Shackel KA, Matthews MA. Characterization of major ripening events during softening in grape: Turgor, sugar accumulation, abscisic acid metabo-lism, colour development, and their relationship with growth. J Exp Bot. 2016;67:709–22. 10.1093/jxb/erv483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pott DM, Vallarino JG, Osorio S. Metabolite changes during postharvest storage: effects on fruit quality traits. Mtabolites. 2020;10:187. 10.3390/metabo10050187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenn MA, Giovannoni JJ. Phytohormones in fruit development and maturation. Plant J. 2021;105:446–58. 10.1111/tpj.15112 [DOI] [PubMed] [Google Scholar]
- 6.Li BJ, Grierson D, Shi Y, Chen KS. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic Res. 2022;9:uhac089. 10.1093/hr/uhac089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, Yuan B, Leng P. Cloning of 9-cis-epoxycarotenoid dioxygenase (NCED) gene and the role of ABA on fruit ripening. Plant Signal Behav. 2009;4:460–3. 10.4161/psb.4.5.8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirayama T, Umezawa T. The PP2C-SnRK2 complex: the central regulator of an abscisic acid signaling pathway. Plant Signal Behav. 2010;5:160–3. 10.4161/psb.5.2.10460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Peng F, Xiao Y, Wang G, Luo J. Overexpression of PpSnRK1α in Tomato promotes Fruit Ripening by En-hancing RIPENING INHIBITOR Regulation Pathway. Front Plant Sci. 2018;9:1856. 10.3389/fpls.2018.01856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia M, Feng J, Zhang L, Zhang S, Xi W. PaPYL9 is involved in the regulation of Apricot Fruit ripening through ABA signaling pathway. Hortic Plant J. 2021;8:461–73. 10.1016/j.hpj.2021.11.012 [DOI] [Google Scholar]
- 11.Zhang YJ, Wang XJ, Wu JX, Chen SY, Chen H, Chai LJ, Yi H-L. Comparative transcriptome analyses between a spontaneous late-ripening sweet orange mutant and its wild type suggest the functions of ABA, sucrose and JA during citrus fruit ripening. PLoS ONE 2014; 9, e116056. [DOI] [PMC free article] [PubMed]
- 12.Niu H, Wang H, Zhao B, He J, Yang L, Ma X, Cao J, Li Z, Shen J. Exogenous auxin-induced enhancer of shoot re-generation 2 (ESR2) enhances femaleness of cucumber by activating the csacs2 gene. Hortic Res. 2022;9:uhab085. 10.1093/hr/uhab085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desnoues E, Baldazzi V, Genard M, Mauroux JB, Lambert P, Confolent C, Quilot-Turion B. Dynamic QTLs for sugars and enzyme activities provide an overview of genetic control of sugar metabolism during peach fruit development. J Exp Bot. 2016;67:3419–31. 10.1093/jxb/erw169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu M, Zhou WQ, Geng WJ, Zhao SR, Pan Y, Fan GQ, Zhang SK, Wang YT, Liao K. Transcriptome analysis insight into ethylene metabolism and pectinase activity of apricot (Prunus armeniaca L.) development and ripening. Sci Rep. 2021;11(1):13569. 10.1038/s41598-021-92832-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jara K, Castro RI, Ramos P, Parra-Palma C, Valenzuela-Riffo F, Morales-Quintana L. Molecular insights into FaEG1, a strawberry endoglucanase enzyme expressed during strawberry fruit ripening. Plants. 2019;8:140. 10.3390/plants8060140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–6. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni JJ. Genetic regulation of Fruit Development and Ripening. Plant Cell. 2004;16:S170–80. 10.1105/tpc.019158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–52. 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- 19.Ríos P, Argyris J, Vegas J, Leida C, Kenigswald M, Tzuri G, Troadec C, Bendahmane A, Katzir N, Picó B, et al. ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J Cell Mol Biol. 2017;91:671–83. 10.1111/tpj.13596 [DOI] [PubMed] [Google Scholar]
- 20.Migicovsky Z, Yeats TH, Watts S, Song J, Forney CF, Burgher-MacLellan K, Somers DJ, Gong Y, Zhang Z, Vrebalov J, et al. Apple Ripening is controlled by a NAC Transcription Factor. Front Genet. 2021;12:908. 10.3389/fgene.2021.671300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyano E, Martínez-Rivas FJ, Blanco-Portales R, Molina-Hidalgo FJ, Ric-Varas P, Matas-Arroyo AJ, Caballero JL, Muñoz-Blanco J, Rodríguez-Franco A. Genome-wide analysis of the NAC transcription factor family and their ex-pression during the development and ripening of the Fragaria × ananassa fruits. PLoS ONE. 2018;13:e0196953. 10.1371/journal.pone.0196953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Pizarro C, Vallarino JG, Osorio S, Meco V, Urrutia M, Pillet J, Casanal A, Merchante C, Amaya I, Willmitzer L, et al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 2021;33:1574–93. 10.1093/plcell/koab070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng L, Ma C, Chen Y, Gao H, Wang J. Genome-wide screening of AP2 transcription factors involving in Fruit Color and Aroma Regulation of Cultivated Strawberry. Genes. 2021;12:530. 10.3390/genes12040530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Xu Z, Zhang Y, Chai L, Yi H, Deng X. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J Exp Bot. 2014;65:1651–71. 10.1093/jxb/eru044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu JX, Zheng SS, Feng GZ, Yi HL. Comparative analysis of miRNAs and their target transcripts between a spontaneous late-ripening Sweet Orange Mutant and its wild-type using small RNA and degradome sequencing. Front Plant Sci. 2016;7:1416. 10.3389/fpls.2016.01416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi LF, Ma D, Lv SP, Xu SB, Zhong BL, Peng T, Liu DC, Liu Y. Comparative transcriptome and sRNAome analyses reveal the regulatory mechanisms of fruit ripening in a spontaneous early-ripening navel orange mutant and its wild type. Genes. 2022;13(10):1706. 10.3390/genes13101706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan H, Lyu S, Chen Y, Xu S, Ye J, Chen G, Wu S, Li X, Chen J, Pan D. MicroRNAs and transcripts Associated with an early ripening mutant of Pomelo (Citrus grandis Osbeck). Int J Mol Sci. 2021;22:9348. 10.3390/ijms22179348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu LH, Chen JM, Xie LH, Zhong BL, Mi LF. Changes of main quality indexes of ‘Gannanzao’ navel orange during fruit ripening. South China Fruits. 2016;45(2):65–8. [Google Scholar]
- 29.Breitel DA, Chappell-Maor L, Meir S, Panizel I, Aharoni. A. AUXIN RESPONSE FACTOR 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLoS Genet. 2016;12(3):e1005903. 10.1371/journal.pgen.1005903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattus-Araya E, Guajardo J, Herrera R, Moya-León MA. ABA speeds up the progress of color in developing F.chiloensis fruit through the activation of PAL, CHS and ANS, key genes of the phenylpropanoid/flavonoid and anthocyanin pathways. Int J Mol Sci. 2022;23(7):3854. 10.3390/ijms23073854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–85. 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Huang Z, Xie B, Chen Q, Tian X, Zhang X, Zhang H, Lu X, Huang D, Huang R. The ethylene-, jasmonate-, abscisic acid-and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta. 2004;220:262–70. 10.1007/s00425-004-1347-x [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Park JH, Lee MH, Yu JH, Kim SY. Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol. 2010;10:277. 10.1186/1471-2229-10-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto T, Uemura T, Nemoto K, Daito M, Nozawa A, Sawasaki T, Arimura G. Tyrosine kinase-dependent defense responses against herbivory in Arabidopsis. Front Plant Sci. 2019;10:776. 10.3389/fpls.2019.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Deng XX. Advances in research of citrus cultivars selected by bud mutation and the mechanism of formation of mutated characteristics. J Fruit Sci. 2006;23:871–6. [Google Scholar]
- 36.Gao HN, Jiang H, Cui JY, You CX, Li YY. The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021;312:111024. 10.1016/j.plantsci.2021.111024 [DOI] [PubMed] [Google Scholar]
- 37.Steffens B, Wang J, Sauter M. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta. 2005;223:604–12. 10.1007/s00425-005-0111-1 [DOI] [PubMed] [Google Scholar]
- 38.Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot. 2008;59:2991–3007. 10.1093/jxb/ern155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in arabidopsis. New Phytol. 2008;179(4):1004–16. 10.1111/j.1469-8137.2008.02511.x [DOI] [PubMed] [Google Scholar]
- 40.Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng Y-Q, Luo X, Yang J. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011;157:216–28. 10.1104/pp.111.179945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Li XA, Yang X, Wassie M, Shi HY. Genome-wide identification and molecular characterization of the AP2/ERF superfamily members in sand pear (Pyrus pyrifolia). BMC Genomics. 2023;24:32. 10.1186/s12864-022-09104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahid A, Ghazanfar A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. Plant Physiol. 2006;163:723–30. 10.1016/j.jplph.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 43.Freiman ZE, Rosianskey Y, Dasmohapatra R, Kamara I, Flaishman MA. The ambiguous ripening nature of the fig (Ficus carica L.) fruit: a gene-expression study of potential ripening regulators and ethylene-related genes. J Exp Bot. 2015;66:3309–24. 10.1093/jxb/erv140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Yang JC, Gao Q, He S, Xu YM, Luo ZP, Liu PP, Wu MZ, Xu X, Ma LX. The transcription factor NtERF13a enhances abiotic stress tolerance and phenylpropanoid compounds biosynthesis in tobacco. Plant Sci. 2023;334:111772. 10.1016/j.plantsci.2023.111772 [DOI] [PubMed] [Google Scholar]
- 45.Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat Protoc. 2010;5:986–92. 10.1038/nprot.2010.37 [DOI] [PubMed] [Google Scholar]
- 46.Albertini MV, Carcouet E, Pailly O, Gambotti C, Luro F, Berti L. Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. J Agricultural Food Chem. 2006;54(21):8335–9. 10.1021/jf061648j [DOI] [PubMed] [Google Scholar]
- 47.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langfelder P, Horvath SWGCNA. An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y, Wang Z, Ge H, Liu Y, Chen H. Weighted gene co-expression network analysis identifies genes related to an-thocyanin biosynthesis and functional verification of hub gene SmWRKY44. Plant Sci. 2021;309:110935. 10.1016/j.plantsci.2021.110935 [DOI] [PubMed] [Google Scholar]
- 51.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto SKEGG. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape. A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;Suppl 4:S11. 10.1186/1752-0509-8-S4-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the uploaded to National Center for Biotechnology Information with the primary accession code SRP500014.