Abstract
Introduction
Small Cell Lung Cancer (SCLC) can be classified into transcriptional subtypes with distinct degrees of neuroendocrine (NE) differentiation. Recent evidence supports plasticity among subtypes with a bias toward adoption of low-NE states during disease progression or upon acquired chemotherapy resistance. Here, we identify a role for SMARCA4, the catalytic subunit of the SWI/SNF complex, as a regulator of subtype shift in SCLC.
Methods
ATACseq and RNAseq experiments were performed in SCLC cells after pharmacological inhibition of SMARCA4. DNA binding of SMARCA4 was characterized by ChIPseq in high-NE SCLC patient derived xenografts (PDXs). Enrichment analyses were applied to transcriptomic data. Combination of FHD-286 and afatinib was tested in vitro and in a set of chemo-resistant SCLC PDXs in vivo.
Results
SMARCA4 expression positively correlates with that of NE genes in both SCLC cell lines and patient tumors. Pharmacological inhibition of SMARCA4 with FHD-286 induces the loss of NE features and downregulates neuroendocrine and neuronal signaling pathways while activating non-NE factors. SMARCA4 binds to gene loci encoding NE-lineage transcription factors ASCL1 and NEUROD1 and alters chromatin accessibility, enhancing NE programs. Enrichment analysis applied to high-confidence SMARCA4 targets confirmed neuron related pathways as the top GO Biological processes regulated by SMARCA4 in SCLC. In parallel, SMARCA4 also controls REST, a known suppressor of the NE phenotype, by regulating SRRM4-dependent REST transcript splicing. Furthermore, SMARCA4 inhibition drives ERBB pathway activation in SCLC, rendering SCLC tumors sensitive to afatinib.
Conclusions
This study nominates SMARCA4 as a key regulator of the NE state plasticity and defines a novel therapeutic strategy for SCLC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-024-01572-3.
Keywords: Lung Cancer, SCLC, Plasticity, Epigenetics, Targeted therapies
Introduction
Small Cell Lung Cancer (SCLC) is a highly aggressive form of lung cancer accounting for ~ 15% of all lung cancer cases [1]. SCLC can be classified into molecular subtypes based on relative expression of the transcription factors including ASCL1 (SCLC-A), NEUROD1 (SCLC-N) and POU2F3 (SCLC-P) [2]. A more recent and broad classification considering both tumor intrinsic factors and immune-related genes has proposed an inflammatory subtype (SCLC-I) [3]. While generally considered a high-grade neuroendocrine cancer, SCLC tumors demonstrate a spectrum of neuroendocrine (NE) differentiation states [1]. The majority (~ 75%) of SCLC tumors, including those of the SCLC-A and -N subtypes, exhibit a high-NE profile [2, 4].The rarer SCLC-P and SCLC-I typically define low- or non-NE subtypes [2, 3]. Although SCLC tumors can be generally characterized by a dominant subtype, analyses of both human SCLC and murine models of SCLC demonstrate substantial intratumoral cell state heterogeneity, and capacity for transformation between states [4–6]. Murine models of SCLC have demonstrated that Ascl1, a primary driver of the high-NE state, is a required factor in the development of SCLC [7]. Subsequent state plasticity with transition from high- to low-NE or mixed states has been associated with aspects of disease progression, including metastasis and acquired resistance to cytotoxic therapies [1].
State transitions in SCLC appear to be epigenetically determined rather than mutationally defined: no consistent genomic alterations differentiate these subtypes [2]. Some factors have been implicated as regulating high- to low-NE state transition in SCLC. Tumor progression and metastasis in a genetically engineered mouse model with a mutant c-Myc allele, was associated with transition from SCLC-A to SCLC-N subtype and in a subset of tumors to low-NE Yap1+SCLC [6, 8]. The role of YAP1 as a subtype-defining factor in SCLC has been questioned by us and others [3, 4], including a report demonstrating that some long-established YAP1-expressing human SCLC cell lines might in fact be misattributed SMARCA4-deficient undifferentiated tumors [9]. Suppression of NOTCH signaling in a SCLC-A model has been shown to be essential for maintenance of the high-NE state [10], reflecting a similar role for NOTCH in inhibiting NE cell differentiation in fetal lung development [11]. Induction of NOTCH signaling in SCLC-A can promote SCLC state shift through at least two complementary mechanisms: [1] suppression of ASCL1 and ASCL1 target gene expression, and [2] upregulation of REST, a transcription factor that inhibits transcription of a set of NE genes including many non-overlapping from those under ASCL1 control [12]. Inhibition of ASCL1 and activation of REST appear to be required for full transition to low-NE SCLC. Notably, the epigenetic regulators involved in SCLC subtype switching have not been fully defined. Pharmacologically tractable targets to constrain subtype plasticity in SCLC could have substantial clinical utility.
Mammalian SWI/SNF (BAF) ATP-dependent chromatin remodeling complexes are encoded by 29 genes, some of which are commonly mutated in cancer [13]. These complexes are classified into canonical BAF (cBAF), polybromo-associated BAF (PBAF) and noncanonical BAF (ncBAF). All SWI/SNF complexes contain SMARCA2 or SMARCA4 as an ATPase catalytic subunit that drives nucleosome sliding and eviction [14]. SMARCA2 and SMARCA4 demonstrate high homology, and SMARCA2 upregulation can compensate for SMARCA4 loss in some contexts [13]. SWI/SNF complexes modulate promoter and enhancer accessibility and have been shown to control multiple transcription programs including those related to cell and lineage differentiation. BAF complex members have divergent roles depending on the cancer context. As an example, SMARCA4 loss accelerates tumor progression and promotes lung adenocarcinoma (LUAD) dedifferentiation [15], while abrogation of another key SWI/SNF component, Arid1a, suppresses tumor initiation and metastasis in hepatocellular carcinoma [16]. As a transcription factor (TF) implicated in multiple solid and hematologic malignancies, SMARCA4 has recently gained attention as a therapeutic target [17–19]. The observation that SMARCA2 can partially compensate for SMARCA4 loss, and that these two homologous TFs are essential for activity of the SWI/SNF complexes, has led to the development of dual SMARCA2/4 inhibitors [20, 21].
Loss of function mutations in SMARCA4 are uncommon in SCLC (1.5%) but are substantially enriched in the non-NE SCLC tumors [22]. Consistently, SMARCA4 mRNA levels are higher in NE SCLC (SCLC-A and -N) and in POU2F3 cell lines than in either YAP1+SCLC or NSCLC [9]. These observations, together with data pointing to a role for the SWI/SNF complex in lineage differentiation [15, 23], prompted us to study the link between SMARCA4 and subtype plasticity in the high- to low-NE transition in SCLC. Here, we provide evidence for a role of SMARCA4 as a key regulator of the NE phenotype in SCLC, and as a potential target for the treatment of SCLC.
Material and methods
Animal models
Patient derived xenografts (PDXs) were subcutaneously engrafted into female 6-week-old NOD.Cg-Prkdc < scid > Il2rg < tm1Wjl > /SzJ (NSG) mice (5–10 mice per arm, Jackson Labs) whereas RP cells (3.5 million) were injected in one flank of female 8-week-old B6129SF1/J mice (Jackson Labs). Cells were resuspended in a mix of PBS and Matrigel 1:1 prior to injection. When tumors reached 75–100 mm3, mice were randomized and treated with either vehicle, FHD-286 (1.5 mg/kg twice daily dosing (BID) p.o.) or afatinib (15 mg/kg once daily dosing (QD) p.o.). Vehicle for FHD-286 consisted of 20% HP-β-CD in water whereas afatinib was dissolved in 0.5% methylcellulose in water. Tumors were measured twice per week with a caliper. Tumor volume was calculated as ((width)2 × length)/2. All in vivo experiments were performed at Memorial Sloan Kettering Cancer Center (MSKCC) following Animal Care and Use Committee guidelines.
Cell lines
H82, H146, H69, H524, HCC33, SHP77, DMS114, H196, CORL311and H211 were purchased from ATCC except for RP, which was a gift from Sage lab (Stanford), and culture in RPMI 1640 Medium (Gibco) supplemented with 10% tetracycline negative FBS (GeminiBio) and 1% Penicillin–Streptomycin (P/S). 293 T cells were also obtained from ATCC and cultured in DMEM media (Gibco), 10% FBS and 1% P/S. Cells were routinely tested for mycoplasma using the Universal Mycoplasma Detection Kit (ATCC). SMARCA4 and SMARCA2 genetic inhibition was induced by treating the cells with 1 μg/mL at the indicated times for each experiment and renew every 48 h.
Cell proliferation assays and apoptosis
For cell proliferation assays, 2000 cells/well were seeded in 96 well-plates and treated with FHD-286 (5–1000 nM, Foghorn) and/or afatinib (10–150 nM, MedChem Express) for 96 h. Cell viability was determined by using CellTiter-Glo 2.0 Assay (Promega, G9242) following the manufacturer’s instructions. Proliferation was determined by measuring the luminescence (L) at day 0, day 4 without drug and day 4 with drug. Proliferation was calculated by the ratio of L at day 4 with drug minus L at day 0 to the L without drug at day 4 minus L at day 0. In the case of the proliferation assays with genetic knockdowns, proliferation was calculated similarly by normalizing the L of each clone at the end of the experiment minus L at day 0 relative to the L of NTC cells at the end of the experiment minus L at day 0.IC50 was calculated with GraphPad Prism software whereas synergy scores were determined with SynergyFinder web application and using the ZIP method.
For apoptosis experiments, cells were seeded in 6 well plates and treated with FHD-286 (100 nM) and/or afatinib (500 nM) for 5 days. Then, cells were stained with FITC Annexin V and propidium iodide (PI) as indicated by manufacturer (BD Pharmingen™ FITC Annexin V Apoptosis Detection Kit). Cell death was assessed by flow cytometry using a using a LSRFortessa™ Cell Analyzer.
Cell proliferation and apoptosis assays were performed at short intervals following treatment to explore the direct cytotoxic effects of FHD-286 in combination with afatinib.
Plasmid vectors, lentiviral virus production and transductions
To generate SMARCA4 and SMARCA2 knockdown (KD) cell lines, targeting shRNAs were cloned into the vector Tet-on LT3GEPIR (Addgene, #111177) with distinct antibiotic resistance, puromycin for SMARCA4 KD and neomycin for SMARCA2 KD. A non-targeting shRNA vector was used as control (NTC).
Lentiviral particles were produced by transfecting HEK293T cells (ATCC, no. CRL-1573) with the vector on interest in the presence of pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) packaging vectors (3:2:1 ratio of plasmid of interest: psPAX2:pMD2.G) and with JetPrime transfection reagent (Polyplus) as previously described [24]. Virus was collected after 72 h from transfection and concentrated 1:20 with Lenti-X™ Concentrator following manufacture’s protocol (Takara Bio). Then, isogenic cell lines were spin-transduced (30’ at 800G) with lentiviral particles and selected with the corresponding antibiotic. Doxycycline SMARCA4 and SMARCA2 inducible genetic inhibition was achieved by adding 1 μg/mL of doxycycline every 48 h. All shRNAs and sgRNAs’ sequences are detailed in Sup. Material Table.
Immunohistochemistry
Immunohistochemistry technique was performed as previously described [24]. FFPE slides from NE SCLC PDXs were first deparaffinized and steamed for 45 min in Target Retrieval Solution (Dako). Incubation with primary antibodies anti-NEUROD1 (Abcam, EPR 17084), anti-ASCL1 (BD, 24B72D11.11) and anti-SMARCA4 (Santa Cruz, sc-17796) was carried out following manufacturer instructions. Then, slides were incubated with PV Poly-HRP anti-mouse IgG (Leica Microsystems, #PV6114) followed by a TSA biotin amplification step (Perkin Elmer) with DAB. Finally, slides were counterstained with hematoxylin and scanned on a Ventana DP 200 Slide Scanner (Roche).
Western blotting and PCR
For western blotting, cell pellets were lysed with cold RIPA buffer (Thermo Scientific) and incubated on ice for 30’ followed by a centrifugation at 13,000 rpm at 4 °C for 30’. Protein quantification was performed using Pierce™ BCA Protein Assay Kit (Thermo Scientific). Antibodies used are detailed in Sup. Material Table.
RNA was isolated with the RNeasy Plus Mini Kit (Qiagen) following manufacturer’s instructions and quantified using the NanoDrop ND-2000 spectrophotometer (Thermo Scientific). Then, 250 ng of RNA was retrotranscribed using qScript cDNA SuperMix (Quantabio). PCR reactions were carried out with 50 ng of cDNA using OneTaq Hot Start Quick-Load 2X Master Mix with Standard Buffer (New England Bio Labs), with cycling conditions of 30 s at 94ºC; 40 cycles of 30 s at 94ºC, 30 s at 55ºC and 30 s at 68ºC; and 5 min at 68ºC. The amplified products were analyzed in a 2% agarose gel stained with GelRed Nucleic Acid Stain (MilliporeSigma).
Relative gene expression of REST4 variants was determined by RT-qPCR using SYBR TM Green PCR Master mix (Life Technologies) in a Gene Amp PCR System 9700 (Applied Biosystems). All primers used are detailed in Sup. Material Table.
Publicly available datasets (RNAseq, ChIP-seq and scRNAseq)
RNA levels of SMARCA4, SMARCA2, NE and non-NE markers in SCLC patients’ tumors were assessed using George et al. [25] and Rudin et al. [26] databases. RNA expression levels in cell lines were retrieved from Cancer Cell Line Encyclopedia (CCLE, (https://xenabrowser.net/). Expression levels were downloaded as RPKM (reads per kilobase of transcript per million reads mapped) and represented as RPKM or log (RPKM). SMARCA4 mRNA levels in LUAD and SCLC tumors were obtained from a cohort previously published by Quintanal Villalonga et al. [27] and express as log transformation of Transcripts per million (TPM). ASCL1 and NEURDO1 ChIP-seq datasets were obtained from Borromeo et al. (GSE69394) [7]. The NE score was calculated using Zhang et al. signature [28]. We defined as high NE score when its value was > 0 and low negative score when the value was < 0.
Single cell RNAseq data from SCLC GEMM tumors was previously described and published by Ireland et al. [6].Processed monocle2 cellular trajectory prediction object from Ireland et al. with normalized expression values, pseudotime projections and NE scores based on Zhang et al. [28] signatures were kindly provided by Dr. Trudy Oliver. Using the normalized expression values of RPM1-4 a Seurat object was created, data was scaled using ScaleData function, dimensionality reduction was applied using RunPCA, cellular neighbors were found by FindNeighbors function using the first 20 PCA and clusters were identified by Louvain approach (FindClusters, resolution = 0.5) [29]. 2D embedding was performed using tSNE approach (RunTSNE, dim = 20) and cellular clusters were plotted. Using these embedded coordinates, SMARCA4 log transformed values as well as NE scores were plotted for each RPM cells. Similarly, log expression values for SMARCA4 and SMARCA2 (or any other gene), as well as NE scores for each cell were plotted using previously computed pseudotime profiles by Ireland et al. Code related to this analysis can be found at https://github.com/abcwcm/redin_smarca4.
ATAC-seq
H82 and H146 cells were treated with 100 nM of FHD-286 for 14 days and cryopreserved in cell freezing media (untreated and treated cells) until use. ATAC-seq sample preparation and sequencing was performed at Genewiz. Analysis was performed as previously published [24]. Raw sequencing reads were trimmed with Trim Galore (v0.4.4) (https://github.com/FelixKrueger/TrimGalore) for quality and Illumina adaptor sequences using the pair-end mode. Reads were then aligned to human assembly hg38 using bowtie2 v2.3.4 with the default settings [30]. Picard tool was used to remove reads with same start site and orientation. Enriched open regions for each sample were called using MACS2 and filtered against genomic blacklisted regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/hg38-human/hg38.blacklist.bed.gz) [31]. A union of Peak atlas was later built by merging the filtered peaks within 500 base pairs. Raw read counts were tabulated over this peak atlas using feature Counts v1.6.0 [32]. Differential peaks were called using DESeq2 [33]. For H146, three control samples were sequenced in one batch while one other control and three treated samples were sequenced in the second batch. The batch effect was counted as a co-variant with treatment using the multivariate model in DESeq2 to differentiate open regions in H146. The bigwig format for each sample was created using the BEDTools suite (https://bedtools.readthedocs.io) with the normalization factor from DESeq2 [33]. All bigwig genome tracks on interested gene regions were generated in Integrative Genomics Viewer (IGV) [34]. Replicates were collapsed using bigWigMerge, bedSort and bedGraphToBigWig form UCSC utilities binary tools to merge, sort and convert to bigwig format. The heatmap around significant differential regions with FDR < = 0.01 and FC > = 1.5 for each treatment in the format of collapsed bigwig was visualized using deeptools v3.4.0 [35]. Enriched motifs were identified from differential regions using HOMER v4.7 with mostly default settings [36]. The motifs were scanned in the differential peak regions as size given, controlled against all peaks as background.
Primary targets were identified as those DEG detected at RNAseq with a concordant change in chromatin accessibility nearby (± 10 kb) the TSS. Predicted enhancers shown in Fig. 5F were identified using GeneHancer [37].
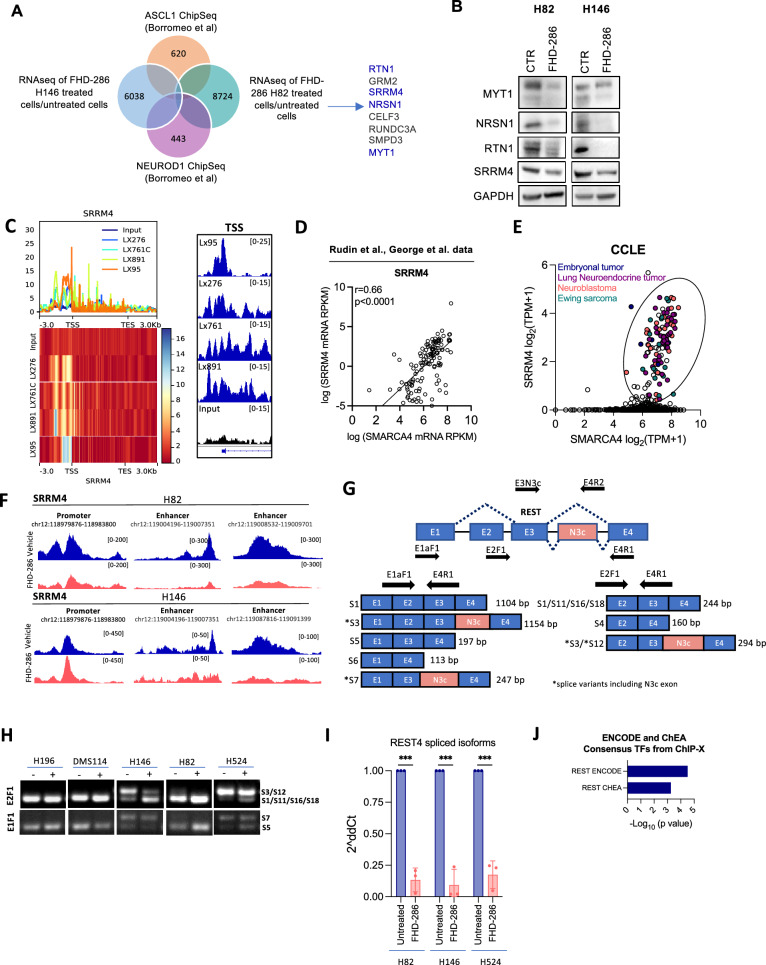
Fig. 5.
SMARCA4 regulates SRRM4 expression to control splicing and activation of REST. A Venn diagram of ASCL1 and NEUROD1 published binding targets from Borromeo et al. [7] overlapping with genes downregulated by FHD-286 in H146 and H82 cells. B Western blots of H82 and H146 cells treated with FHD-286 for 14 days. C Metaplot of SMARCA4 ChIP-seq showing SMARCA4 binding to SRRM4 in 4 NE SCLC PDXs. Range indicates the fold enrichment with respect the input. ChIP-seq genome tracks at SRRM4 TSS. Graphs were obtained from IGV. D Correlation of SMARCA4 and SRRM4 mRNA levels in SCLC patients’ database. Spearman correlation. E Correlation analysis of SRRM4 and SMARCA4 in cancer cell lines retrieved from CCLE. Cell lines with both high SMARCA4 and SRRM4 mRNA levels are highlighted. F Merged ATAC-seq tracks of H82 and H146 parentals cells and FHD-286 treated cells (day 14) at SRRM4 gene locus visualized with IGV. G Graphical representation of REST genomic regions and spliced isoforms with the binding location of the different primers used for PCR. H PCR analysis of REST splicing isoforms using two pairs of primers (E2F1 + E4R1 and E1F1 + E4R1) that span N3c. I RT-qPCR of REST4 isoforms (S3, S7, S12) in H82, H146 and H524 treated with FHD-286 (14 days) versus untreated cells. The pair of primers E3N3c and E4R2 that recognizes all isoforms including exon N3c was used. Student’s two-tailed unpaired t test. ***p < 0.001. The mean ± SD is shown. J Enrich analysis applied to commonly and significantly downregulated genes in both H146 and H82 (n = 904) cell lines identified in the bulk-RNAseq (Fig. 2). See also Fig. S7
ChIP-seq
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed at Active Motif (Carlsbad, CA, USA) using a monoclonal antibody against human SMARCA4 (#ab110641, Abcam). Validation of ChIP was assessed by qPCR before sequencing. A pool of the four PDXs was used an input control. ChIP-Seq libraries were generated from the ChIP-DNA using a custom Illumina library type on an automated system (Apollo 342, Wafergen Biosystems/Takara). ChIP-Seq libraries were sequenced on Illumina NovaSeq 6000 as 75-nt single end reads. Adapter sequences were not trimmed during demultiplexing. Raw reads were processed using the same pipeline described in the ATAC-seq section. Enriched binding regions were called against the input using MACS2 [31] with p value < 0.001. The bigwig format for each sample was created using the BEDTools suite (https://bedtools.readthedocs.io) with the normalization factor 10 million. ChIP density profiles were created with deeptools v3.4.0 [35]. Enrichment pathway analysis of ChIP-seq data was performed using the public web server ChIP-Enrich (http://chip-enrich.med.umich.edu). We used the method Poly-Enrich and the peaks were assigned to the nearest TSS [38, 39]. Motif enrichment analysis on the called peaks was performed using HOMER v4.7 [36]. ChIp-seq data was visualized with the Integrative Genomics Viewer (IGV) [34]. Promoter regions were defined as those within 5 kb from TSS whereas the proximal promoter region was named to the region within 1 kb from TSS.
RNA-seq
RNA isolation and sequencing was performed at Genewiz. RNA integrity and quantity was assessed with Qubit assay. Library preparation and sequencing was conducted with an Illumina sequencer. Fastq files were mapped to the human genome (hg38) and reads counts per gene were quantified using STAR [40] with default parameters and genecode (v28) annotation file. DEGs were identified with DESeq2 [33]. Combination of RNAseq data and public ASCL1 and NEUROD1 ChIP-seq was performed by integrating those genes downregulated at mRNA level upon treatment with FHD-286 (p < 0.1) with previously published ASCL1 and NEUROD1 targets [7]. Integration of RNAseq data with SMARCA4 ChIP-seq data was performed by combining genes downregulated by FHD-286 treatment (p < 0.05) with SMARCA4 binding gene promoters (< 5 kb) detected in at least 2 out of 4 PDXs in the ChIP-seq data.
Pathway enrichment analysis by GSEA, ENRICH and Ingenuity
Gene set enrichment analysis (GSEA, v4.0.2) [41] was conducted using ClusterProfiler R package v3.18 [42]. Analysis was performed on the full set of genes ranked by p value scores computed as -log(p value)*(sign of log2FC) from differential expression analyses between FHD-286 treated cells and parental cells. Gene set annotations were obtained from Molecular Signatures Database (MSigDB v7.0.1 [41, 43]) and the enrichment was calculated by using permutation test with p value adjustment by Benjamin-Hochberg procedure. NE and non-NE gene sets consist of a 25 genes list each from Zhang et al. signature [28]. Normalized enrichment scores (NES) and q or p values are detailed in the figure legends.
ENRICH analysis [44, 45] was applied to all genes significantly (p < 0.05) downregulated between treated and untreated cells in both H82 and H146 cell lines detected at RNAseq as detailed in Figure S7F. ENRICH analysis was performed to those confident targets identified by combining downregulated DEGs (RNAseq) with SMARCA4 targets binding to promoter regions in at least 2 out of 4 PDXs assessed (Fig. S6A). Pathway enrichment analysis with ENRICH was also applied to all genes with a significant (p < 0.05) downregulation in the accessibility detected at any genomic region in both cell lines (Fig. S4E).
To characterize pathways enriched or inhibited after inhibition of SMARCA4 we conducted Ingenuity Pathway Analysis (IPA, Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) on only differential (p < 0.01) upregulated or downregulated genes between FHD-286 treated vs untreated cells detected at RNA-seq data. Data was presented by plotting the Z score, which is calculated based on the data set’s correlation with an activated state and the log transformation of the p value.
Statistical analysis
Statistical comparison between two groups was performed applying unpaired two-tailed Student’s t test (parametric). For multiple comparisons, one- or two-way ANOVA analysis followed by Bonferroni post-hoc test was used. For correlation analysis, Spearman analysis was used. Fisher analysis was performed to explore the association between the NE score (< 0 or > 0) and expression of SMARCA4 (< 0 or > 0). Data was analyzed with GraphPad Prism 9 software and statistical significance was defined as p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****). The analysis used is detailed in the figure legend of each experiment. All functional experiments were replicated a minimum of three times. All western blots were reproduced a minimum of two times with independent protein extracts from biological replicates for a given model, and in a minimum of two different models to support universality of the findings.
Results
SMARCA4 is highly expressed in neuroendocrine SCLC
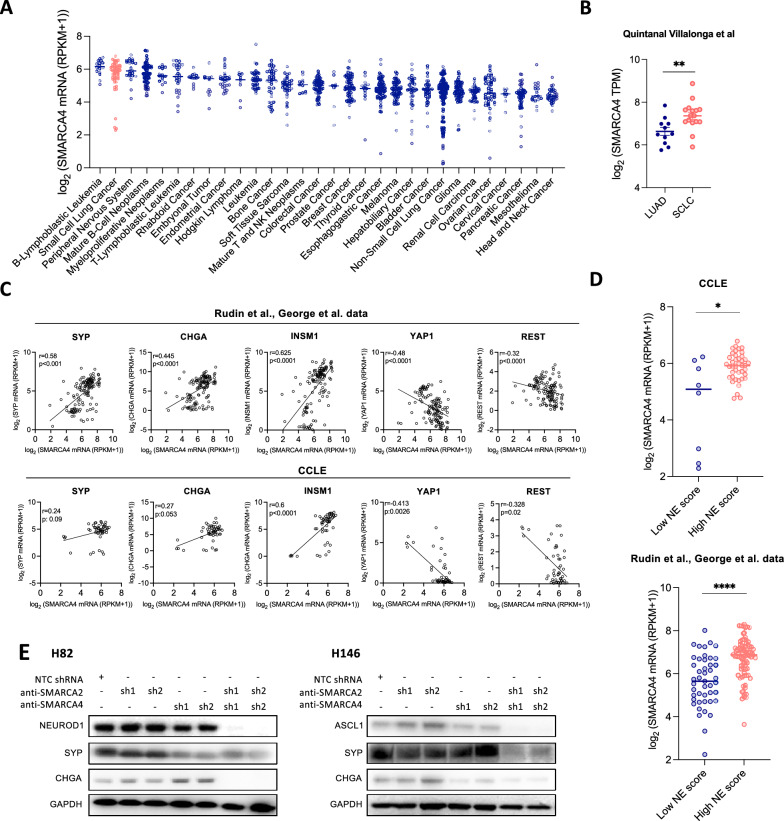
We first sought to evaluate relative expression levels of SMARCA4 across the spectrum of human cancers. SMARCA4 expression was higher in SCLC lines than in any other solid tumor represented in the Cancer Cell Line Encyclopedia (CCLE) (Fig. 1A). Focusing on lung cancer biopsy specimens, SMARCA4 levels were also significantly higher in SCLC than in lung adenocarcinoma (Fig. 1B). Up to 75% of all SCLC tumors are classified as NE-high, based on upregulated expression of ASCL1 and/or NEUROD1 and a variety of NE markers. SMARCA4 expression was positively correlated with multiple NE genes including SYP, CHGA, INSM1, DLL3 and NCAM1 and negatively correlated with non-NE factors REST, NOTCH2, and YAP1 in both SCLC cell lines and patient tumor databases (Figs. 1C and S1A). Stratification of SCLC tumors and cell lines based on the expression of an NE score determined by applying Zhang et al. signature [28] showed significantly lower SMARCA4 expression in low-NE versus high-NE SCLC samples (Fig. 1D).
Fig. 1.
SMARCA4 expression correlates with NE features in SCLC. A SMARCA4 mRNA levels in cell lines derived from 30 tumor types assessed using the Cancer Cell Line Encyclopedia (CCLE). Bars indicate the median expression per tumor type. B SMARCA4 mRNA levels in LUAD and SCLC specimens retrieved from Quintanal Villalonga et al. [27]. Student’s two-tailed unpaired t test. **p < 0.01. C Spearman correlation of SYP, CHGA, INSM1, YAP1 and REST with SMARCA4 mRNA levels in Rudin et al. and George et al. databases and CCLE[25, 26]. D SMARCA4 mRNA expression in low and high NE SCLC tumors in cell lines (CCLE) and clinical specimens (Rudin et al. and George et al.) [25, 26]. One-way ANOVA test followed by Bonferroni post-hoc test. ****p < 0.0001, ***p < 0.001, **p < 0.01. E Western blotting of ASCL1, NEUROD1, SYP and CHGA in isogenic cell lines derived from H82 and H146 expressing different combinations of shRNAs against SMARCA4 and/or SMARCA2. Expression of shRNAs from E was conditional of doxycycline treatment. Protein collection and blotting was performed after 14 days of doxycycline treatment. See also Fig. S1
SMARCA4 expression might be a secondary effect of a NE-high state or might be a factor driving the NE phenotype. To assess whether SWI/SNF activity might promote the expression of NE factors in SCLC, we genetically downregulated the expression of SMARCA4 and/or SMARCA2 using a Tet-On inducible shRNA system. Genetic inhibition of SMARCA4 led to compensatory upregulation of SMARCA2 expression, as previously described [13, 46] (Fig. S1B). Single inhibition of SMARCA2 did not change the protein expression of the master regulators NEUROD1 and ASCL1, or of the NE factors SYP or CHGA, in H82 (SCLC-N) or H146 (SCLC-A). SMARCA4 knockdown slightly reduced some of these markers, and dual inhibition of SMARCA4 and SMARCA2 markedly decreased NE factor expression (Fig. 1E). Single knockdown of each gene did not affect cell proliferation while double knockdown of SMARCA2/4 significantly reduced the proliferative capacity of the cells in vitro (Fig. S1C).
Pharmacological inhibition of SMARCA2/4 with FHD-286 downregulates neuroendocrine and neuronal signaling pathways
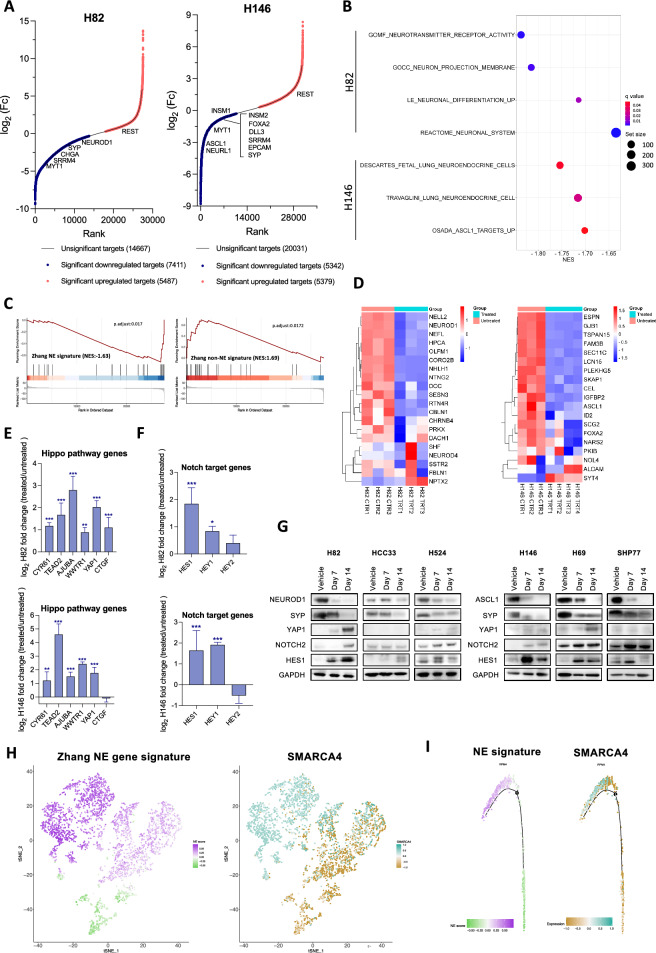
To explore the potential role of SMARCA4 as a regulator of NE cell fate, we used the dual allosteric SMARCA2/SMARCA4 ATPase inhibitor FHD-286 (Foghorn Therapeutics), a small molecule, orally bioavailable, BRG1 and BRM-selective, ATPase inhibitor. Based on its potent pre-clinical activity against cancer cells including leukemia and lung adenocarcinoma cells, FHD286 is currently being evaluated for safety and clinical efficacy in early clinical trials in AML (NCT04891757) [20, 21]. We characterized gene expression changes in H82 (SCLC-N) and H146 (SCLC-A) cells upon treatment with FHD-286 by RNAseq (Figs. 2A and S2A, B). Pharmacological inhibition of SMARCA4 induced downregulation of many key NE factors, and upregulation of factors associated to the low-NE phenotype, including REST (Figs. 2A and S2C, D and Table S1). Gene set enrichment analysis (GSEA) of differentially expressed genes (DEG) revealed downregulation of neuronal and NE pathways in both H82 and H146 cells treated with FHD-286, including decrease in ASCL1 targets in the SCLC-A line H146 (Fig. 2B). Ingenuity pathway enrichment analysis of reduced expressed genes (p < 0.01) confirmed the downregulation of neuronal related pathways (Fig. S2E). GSEA leveraging publicly available high- and low-NE signatures derived from SCLC cell lines [28] supported a shift from a high- to a low-NE phenotypic state (Fig. 2C). Accordingly, upon inhibition of SMARCA4 we observed not only a reduction of ASCL1 and NEUROD1 TFs but also of their most confident targets previously identified by Borromeo et al. [7] (Fig. 2D). Consistent with this high- to low-NE transition, we detected a significant gain in the expression of multiple Hippo signaling targets (YAP1, TEAD2, AJUBA, CYR61, WWTR1) and NOTCH targets (HES1 and HEY1) upon treatment in both models (Fig. 2E, F). GSEA confirmed the activation of Hippo and NOTCH signaling in H82 cell line upon FHD-286 treatment with similar trends observed in H146 (Fig. S2F). We validated the downregulation of the NE markers NEUROD1, ASCL1 and SYP, and increase of NOTCH2 and HES1, both involved in promoting low-NE differentiation in SCLC [10, 12], at the protein level after treatment with FHD-286 (Fig. 2G). Reduction in the expression of NE markers along with increase of NOTCH2, HES1 or YAP1 was also confirmed at the protein level after dual genetic inhibition of SMARCA4/2 (Fig. S2G).
Fig. 2.
SMARCA4 inhibition suppresses the NE phenotype in SCLC. A Hockey-stick plots of DEGs in FHD-286-treated cells after 14 days (100 nM) versus control, untreated cells. (See Table S1). B Dot plots showing negative enrichment in selected neuronal and NE pathways analyzed by GSEA in RNAseq data from H82 and H146 cell lines treated with FHD-286 versus untreated. (See Table S1). C GSEA applying Zhang et al. NE gene signature [28] in H82 cell line treated with FHD-286 versus untreated. D Heatmaps showing the most significant confident targets (top 25 with TPMs > 2) of NEUROD1 (left) and ASCL1 (right) [7], in H82 (left) and H146 (right) bulk RNAseq (FHD-286 treated vs untreated). E Log2 fold change of Hippo pathway genes from data in A. Student’s two-tailed unpaired t test. ***p < 0.001, **p < 0.01. The mean ± SD is shown. F Log2 fold change of NOTCH pathway genes from data in A. Student’s two-tailed unpaired t test. ***p < 0.001, *p < 0.05. The mean ± SD is shown. G Western blotting of H524 (SCLC-N), H82 (SCLC-N), HCC33 (SCLC-N), H69 (SCLC-A), SHP77 (SCLC-A) and H146 (SCLC-A) cells after treatment with 100 nM of FHD-286 for 7 and 14 days. H t-SNE of Zhang NE signature and SMARCA4 levels applied to public scRNAseq data of 4 myc-driven murine (RPM) tumors [6]. I Scoring for Zhang NE signature and SMARCA4 projected in a pseudotime trajectory from early to late time points in a tumor from a Myc-driven murine SCLC model showing subtype plasticity [6]. See also Figs. S2, S3 and Table S1
To study the association of SMARCA4 and NE identity in SCLC at higher resolution, we leveraged a publicly available scRNAseq dataset of 4 tumors derived from the Rb1fl/fl; Trp53fl/fl; MycT58ALSL/LSL (RPM) genetically engineered mouse model (GEMM) of SCLC [6]. In this GEMM, c-Myc promotes tumor transition from a high-NE state into a low-NE state (Fig. S3A). Each cell was assigned with a NE score by applying a previously defined NE gene signature [28] (Fig. 2H). Cells with high SMARCA4 mRNA levels corresponded to those exhibiting a high NE score, whereas low NE cells lacked SMARCA4 expression (Figs. 2H, S3B and Table S1). Fisher analysis applied to these data confirmed the significant association between the presence of SMARCA4 and a high NE score (Fig. S3B). We also analyzed SMARCA4 expression changes in an unsupervised pseudotime trajectory constructed by Ireland et al. [6]. Cells belonging to early pseudotime showed high NE score and high SMARCA4 while in late pseudotime progression, cells had reduced NE score and reduced SMARCA4 (Figs. 2I and S3C, D). No changes in SMARCA2 levels were found along the pseudotime trajectory (Fig. S3D). Taken together, these data suggest that SMARCA4 is required for SCLC cells to maintain high NE identity.
SMARCA4 inactivation alters chromatin accessibility in neuroendocrine SCLC
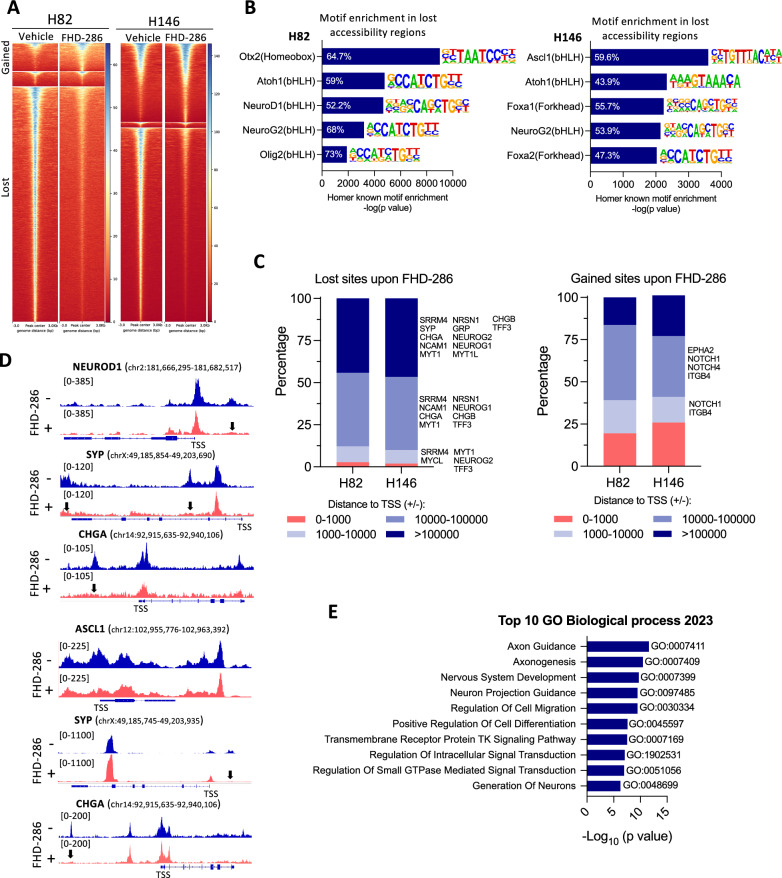
We next explored whether SMARCA4 could control the chromatin accessibility of NE and non-NE genes as mechanism of regulating their mRNA expression. Suppression of SMARCA4 activity by FHD-286 induced global changes in the accessibility with a predominance in the number of lost regions: > 35,000 sites lost in both H82 and H146 cells (Figs. 3A and S4A–C). Notably, reduced accessible genomic regions upon FHD-286 had a striking enrichment for the DNA-binding proneuronal and NE genes motifs ASCL1, NEUROD1, OLIG2, ATOH1, NEUROG2, FOXA2, FOXA1 and OTX2 (Figs. 3B and S4D). OTX2 is selectively expressed in NEUROD1high SCLC cells, and its DNA motif is also enriched at NEUROD1-bound sequences [7]. Changes in gene loci accessibility were mainly located at TSS distal regions (< 10 kb from TSS), as observed in other tumors such as lung adenocarcinoma [20] (Fig. 3C and Table S2). Among the genes with reduced distal accessibility changes we identified relevant NE genes, but we did not find evidence of increased accessibility in non-NE genes previously found upregulated at the mRNA level after FHD-286 treatment or reduced accessibility around the TSS of ASCL1, NEUROD1, SYP or CHGA among others (Fig. 3C, D). Pathway enrichment analysis of genes with sites of lost accessibility (p < 0.05) in both cell lines (n = 6666), revealed a strong enrichment in neuronal pathways, supporting a role for SMARCA4 in regulating chromatin accessibility of NE genes (Figs. 3E and S4E). Lastly, we integrated the differential ATAC-seq peaks within 10 kb up or downstream of gene TSS with our RNAseq data to identify SMARCA4 primary targets. In line with our previous findings, only 12.5% (H82) and 21.4% (H146) of genes downregulated, and 20.6% (H82) and 26.6% (H146) or genes upregulated, showed a concordant change in accessibility around the promoter region (± 10 kb) (Table S2).
Fig. 3.
SMARCA4 inactivation alters chromatin accessibility in NE-high SCLC. A Heatmap showing ATACseq chromatin accessibility changes (FDR:0.01, FC > 1.5) in H82 and H146 cells after treatment with FHD-286 (100 nM, 14 days). B Enrichment of neuronal and NE HOMER transcription factor-binding DNA motifs in ATAC-seq peaks lost after treatment with FHD-286 (100 nM, 14 days). The percentage indicates the amount of target sequences with motif. C Genomic localization of lost and gained accessible sites upon FHD-286 treatment in H82 and H146 cells. D ATACseq genome tracks of NEUROD1, SYP and CHGA in H82 and H146 cells after treatment with FHD-286. Peaks with a significant reduction in chromatin accessibility are indicated with arrows. E Enrich analysis applied to all genes with lost sites (across all gene body) following FHD-286 treatment. Top 10 GO Biological processes enriched are shown. See also Fig. S4
SMARCA4 binds to neuronal and NE lineage TF genes in SCLC
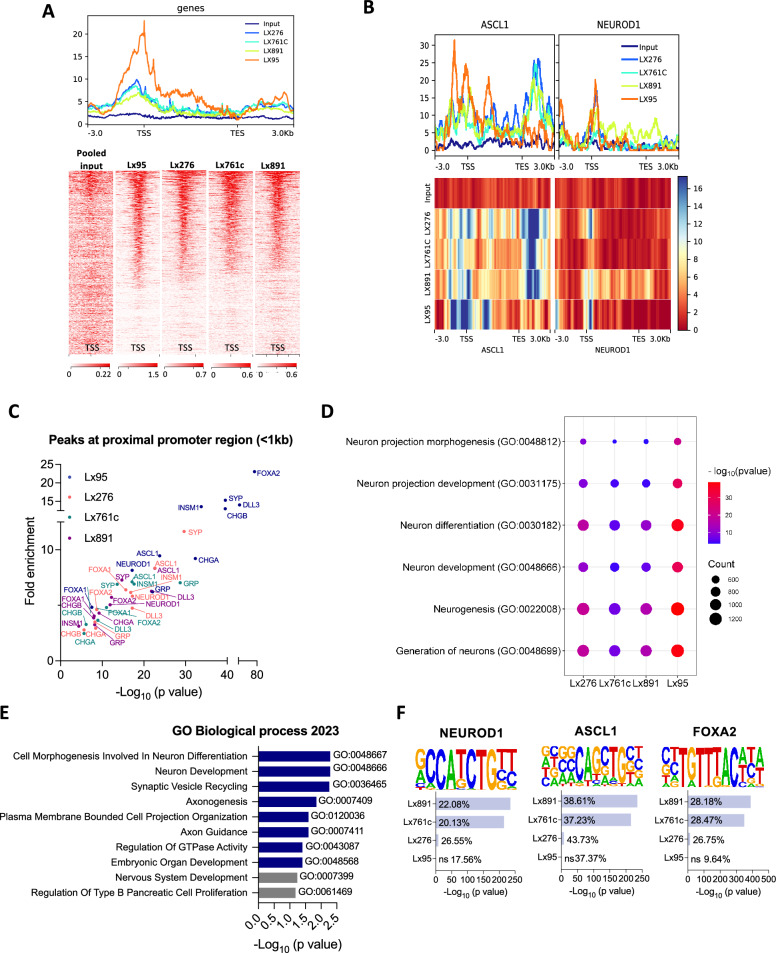
To better understand how the SMARCA4-containing SWI/SNF complex controls NE cell fate in SCLC, we performed ChIP-seq of SMARCA4 in four NE SCLC patient-derived xenografts (PDXs) with high levels of SMARCA4 (Fig. S5A) and no mutations in any of the SWI/SNF complex subunits (Fig. S5B). SMARCA4 binding peaks were detected in promoter regions (within 5 kb upstream of TSS), 5’UTR, exons, introns and 3’UTR (Figs. 4A and S5C). Peak annotation identified 20,754 (Lx95), 17,994 (Lx276), 16,556 (Lx761c) and 15,655 (Lx891) SMARCA4 candidate gene targets. SMARCA4-bound promoters included those of the lineage-specifying TFs ASCL1 and NEUROD1 and many other NE genes (SYP, CHGA, INSM1, FOXA2, DLL3, GRP, FOXA1) (Figs. 4B, C and S5D). Interestingly, SMARCA4 binding to ASCL1 was not only detected at the TSS (as is also the case for NEUROD1) but at sites along the entire ASCL1 gene body (Fig. 4B). We next explored whether the downregulation of ASCL1 and NEUROD1 top confident targets, observed following SMARCA4 inhibition (Fig. 2D), could be a consequence of SMARCA4 direct binding [7]. Consistent with this hypothesis, 96% of ASCL1 targets and 80% of NEUROD1 targets were detected as SMARCA4-bound genes in at least 3 out of 4 PDXs analyzed, suggesting that SMARCA4 binding might be required to fully activate their transcription (Fig. S5E). Poly-Enrich analysis of SMARCA4 ChIP-seq binding profile predicted strong enrichment in neuron development and differentiation biological processes based on SMARCA4 targets (Fig. 4D). We also found enrichment in regulators of NOTCH signaling, including negative regulators of this pathway, and chromatin remodeling and organization processes (Fig. S5F).
Fig. 4.
SMARCA4 binds to neuronal and NE lineage TF genes in SCLC. A Heatmap and metaplot showingSMARCA4 binding profile determined by ChIP-seq in 4 NE SCLC PDXs and a pooled input. The range under the map indicates the ChIP-seq signal intensity. B Metaplots of ASCL1 and NEUROD1 in all PDXs and input. Heatmaps showing the binding of SMARCA4 to ASCL1 and NEUROD1 gene bodies. The range indicates the normalized enrichment along the respective gene regions. C NE lineage TFs and gene promoter proximal regions (within 1 kb of TSS) bound by SMARCA4 in NE SCLC PDXs. D Dot plot of Poly-Enrich analysis applied to SMARCA4 ChIP-seq peaks. Fold enrichment refers to the fold increase in the signal for a particular gene relative to the background signal. The counts refer to the number of genes detected in the ChIP-seq data that are part of the indicated pathways. E Enrich analysis of 617 consensus genes selected by combining RNAseq from Fig. 2 and ChIP-seq data. See also Fig. S5E. F Enrichment analysis of TF-binding motifs in the SMARCA4 ChIP-seq data identified with HOMER. See also Figs. S5, S6 and Table S3
We next cross-referenced SMARCA4 binding promoter regions (< 5 kb) identified by ChIP-seq with genes downregulated by FHD-286 treatment as determined by RNAseq to identify high-confidence SMARCA4 targets. This analysis nominated 617 common SMARCA4 targets in both ASCL1 and NEUROD1 SCLC subtypes (Fig. S6A and Table S3). Pathway enrichment analysis of these confident targets again showed neuron related processes among the top GO Biological processes regulated by SMARCA4 (Fig. 4E). With the aim of identifying which DNA-binding motifs are the most enriched within SMARCA4 ChIP-seq peaks we performed HOMER analysis. Remarkably, 52.7% of all motifs detected overlapped in at least 3 out of the 4 PDXs analyzed (Fig. S6B). We found a significant enrichment in known motifs of the neuronal and NE lineage TFs NEUROD1, ASCL1, FOXA2, ATOH1 and NEUROG2 (Figs. 4F and S6C). Remarkably, most of these motifs matched those with reduced accessibility after FHD-286 treatment and identified by ATACseq (Fig. 3B). A complete list of the gene motifs detected in the ChIP-seq data is found in Table S3.
SMARCA4 regulates common ASCL1 and NEUROD1 targets and induces REST splicing by SRRM4
We next sought to identify convergent downstream targets of ASCL1 and NEUROD1 under the control of SMARCA4, with a potential role in NE differentiation. Combining our RNAseq data (genes inhibited by FHD-286) and publicly available ChIP-seq data of ASCL1 and NEUROD1 we identified 8 common targets (Fig. 5A) [7]. Among these candidates, we selected Reticulon 1 (RTN1), Neurensin 1 (NRSN1), Myelin transcription factor (MYT1) and Serine/Arginine Repetitive Matrix (SSRM4), as promising targets of the SMARCA4/ASCL1/NEUROD1 axis because of their suggested roles in sustaining the NE phenotype and neuronal development [47–50]. Western blotting revealed a strong inhibition of all four targets upon treatment with FHD-286 and after genetic inhibition of SMARCA4/2 (Figs. 5B and S7A). SMARCA4 ChIP-seq showed binding of SMARCA4 to the TSS of all four genes, defining them as high confidence targets of SMARCA4 (Figs. 5C and S7B). Analysis of RTN1, NRSN1, MYT1 and SRRM4 levels in scRNAseq pseudotime trajectory of the GEMM SCLC model demonstrating subtype plasticity (Fig. 2H, I) [6] also confirmed loss of expression of these four genes in the transition from high- to low-NE state in SCLC (Fig. S7C). Consistently, we observed a positive correlation of SMARCA4 expression with that of all 4 genes in patients’ SCLC samples (Figs. 5D and S7D). Additional correlation analysis between SMARCA4 and SRRM4 across the pan-cancer CCLE dataset revealed two well-defined groups: one including cell lines expressing SMARCA4 and lacking SRRM4, and another group with a strong positive correlation between these genes. The cell lines belonging to the latter group were almost entirely comprised of tumor types with NE/neuronal features, suggesting that SRRM4 expression may be restricted to NE tumors (Fig. 5E).
Given the activity of SRRM4 in regulating RNA splicing of REST, a known transcriptional driver of low-NE cell fate in SCLC [12, 51], we decided to delve deeper into its role. Alternative splicing of REST by SRRM4 induces the incorporation of the exon N3c into the transcript, leading to the expression of the truncated and non-functional derivative, REST4. Reduction of active REST by SRRM4-driven splicing has been shown to promote a NE phenotype in prostate tumors [50, 52, 53]. In addition to SMARCA4 binding to the SRRM4 promoter, we found that pharmacological targeting of SMARCA2/4 significantly reduced DNA accessibility of SRRM4 regulatory elements, including promoter (for H82) and enhancers regions (Figs. 5F, 3C). To investigate the role of SMARCA4 in REST splicing through SRRM4, we first analyzed the different splicing isoforms of REST harboring N3c exon (Fig. 5G). Inactive REST4 variants (S3, S7 and S12) were consistently present in high-NE and undetectable in low-NE SCLC cell lines (Fig. S7E). Pharmacological inhibition of SMARCA4 with FHD-286 increased the abundance of non-N3c active isoforms (S11/S16/S18) including the canonical one (S1) while reducing inactive REST4 isoforms S3 and S12. The variant S7 did not change after treatment (Fig. 5H). To quantify the relative amount of REST4 isoforms after treatment with FHD-286, we performed RT-qPCR using a pair of primers (E3N3c/E4R2) spanning N3c of all three REST4 variants (Fig. 5I). Pharmacological inhibition of SMARCA4 strikingly reduced the relative levels of inactive REST4 (S3, S7 and S12) in all NE cell lines tested (Fig. 5I). Consistent with these results, Enrichment analysis performed on those genes commonly and significantly downregulated at mRNA level (n = 904; Fig. S7F) nominated REST as the top and only significant TF involved in the loss of NE markers after SMARCA4 inhibition (Fig. 5J). Taken together, these findings demonstrate that SMARCA4 controls REST splicing by sustaining the expression of SRRM4.
SMARCA4 suppression by FHD-286 activates ERBB pathways and sensitizes to afatinib
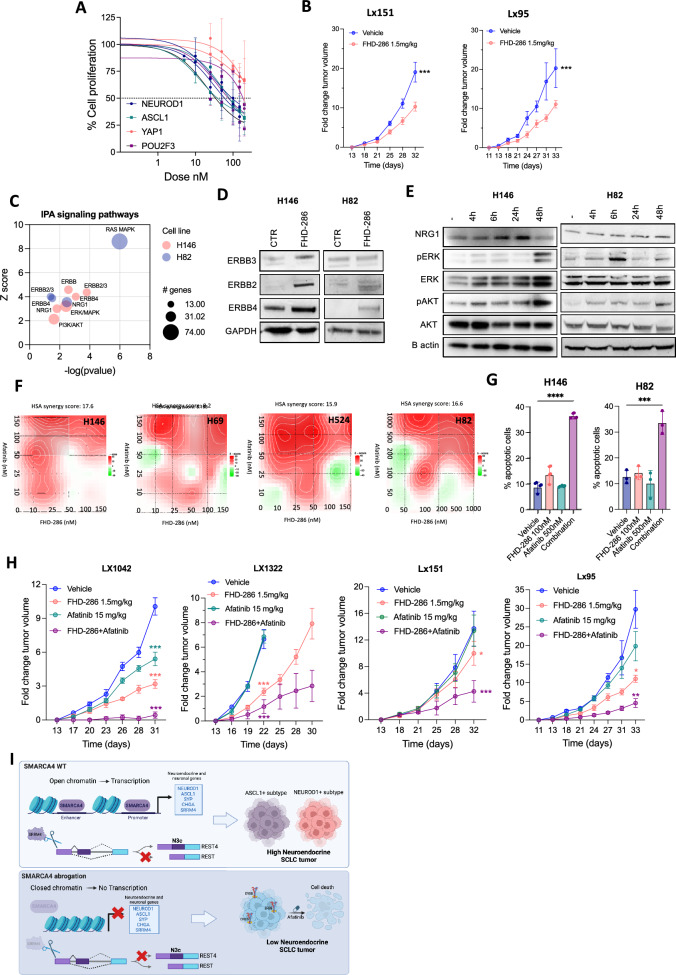
Finally, we evaluated the potential of SMARCA4 pharmacological inhibition as a therapeutic approach for SCLC tumors. Cell proliferation assays in vitro showed response to FHD-286 across a panel of SCLC lines, in the nanomolar range (median IC50 of 90 ± 45.9 nM), except for YAP1+SCLC low-NE lines, which showed IC50 values above 200 nM (Figs. 6A and S8A). In vivo treatment of two high-NE SCLC PDX models with single agent FHD-286 at a dose of 1.5 mg/kg twice daily demonstrated limited growth inhibition (Fig. 6B).
Fig. 6.
SMARCA4/2 inhibition by FHD-286 induces ERBB signaling and sensitivity to afatinib in SCLC. A Proliferation curves of SCLC-A, -N, -P and -Y SCLC cell lines treated with FHD-286 for 96 h. The mean ± SD is shown. B Tumor growth of Lx151 and Lx95 SCLC PDXs implanted in NSG mice and treated with 1.5 mg/kg BID p.o. of FHD-286. Student’s two-tailed unpaired t test. ***p < 0.001. C IPA analysis on significantly upregulated genes in FHD-286-treated cells versus control untreated cells. D Immunoblot of ERBB family proteins in H146 and H82 cells after treatment with 100 nM of FHD-286 for 14 days. E Western blots of FHD-286 (100 nM) treated cells at the indicated times. F Synergy plots of FHD-286 and afatinib in NE SCLC cell lines. G Cell death quantification by flow cytometry at day 5 of H146 and H82 cells after treatment with FHD-286, afatinib or both. One way ANOVA followed by Bonferroni comparison test. ***p < 0.001, ****p < 0.0001. H Normalized tumor growth of Lx1042 (SCLC-N), Lx1322 (SCLC-P), Lx151 (SCLC-A) and Lx95 (SCLC-A) relative to day 1 of treatment. Two-way ANOVA followed by Bonferroni comparison test. *p < 0.05, **p < 0.01, ***p < 0.001. I Schematic representation of the role of SMARCA4 in sustaining the NE phenotype in SCLC
We sought to identify vulnerabilities induced by SMARCA4 inactivation. Ingenuity pathway analysis (IPA) of differential upregulated genes (p < 0.01) detected by RNAseq in treated vs untreated cells suggested activation of ERBB and Neuroregulin-1 (NRG1) pathways upon FHD-286 treatment (Fig. 6C). Consistently, FHD-286 treatment in two NE SCLC cell lines induced protein upregulation of ERBB family receptors ERBB2, ERBB3 and ERBB4 (Fig. 6D), and of NRG1 (Figs. 6E and S8B), a direct ligand and activator of ERBB proteins. In line with ERBB pathway activation, we observed increased phosphorylation of the downstream targets ERK and AKT (Figs. 6E and S8B). Addition of recombinant NRG1 to the NE SCLC cell lines H82 and H146 supported a role of NRG1 as ligand and activator of ERBB pathway in SCLC, inducing phosphorylation of ERK and AKT (Fig. S8C). These results suggest that SMARCA4 inhibition might drive the activation of the NRG1-ERBB pathway in SCLC.
We therefore investigated whether the pharmacological blockade of ERBB pathway with the irreversible inhibitor afatinib could synergize with FHD-286. Drug combination assays demonstrated a strong synergy between these drugs in all 4 SCLC subtypes cell lines tested (hsa synergy score: 8–17.5) (Figs. 6F and S8D), accompanied by increased cell death relative to either single agent treatment (Fig. 6G). Accordingly, ectopic silencing of either SMARCA4 or SMARCA4/SMARCA2 increased the effectiveness of afatinib in vitro (Fig. S8E).
In the light of these results, we explored the combination of FHD-286 and afatinib in vivo in a set of chemo-resistant SCLC PDXs and in an immunocompetent mouse model. Afatinib monotherapy did not reduce tumor growth in any of the models tested except for LX1042, a PDX derived from an EGFR-mutant adenocarcinoma that transformed to SCLC on targeted therapy (Fig. 6H). SMARCA4/2 inhibition with FHD-286 monotherapy slightly decreased tumor growth in all models tested; in contrast, the combination of FHD-286 with afatinib induced strong growth-suppressive responses in all models assessed (Figs. 6H and S8F).
Discussion
SCLC is the most lethal form of lung cancer, with limited therapeutic options. Transcriptional profiling has been used to classify SCLC into high-NE (ASCL1 and/or NEUROD1 +) and low-NE (POU2F3 and/or Inflamed) states [2, 3]. ASCL1 and NEUROD1 are well established transcription activators of NE genes, but it is unclear which factors enforce the maintenance of the NE-high state, or regulate cell state transitions between high- and low-NE phenotypes [7, 54–57]. Here, we report SMARCA4 as a critical regulator of the NE phenotype and as a therapeutic vulnerability in SCLC (Fig. 6I).
Coexistence of NE and non-NE cells in GEMM models was one of the first observations pointing to cell state plasticity in SCLC [58]. Activation of c-Myc can facilitate transition of SCLC-A tumors to SCLC-N and Yap1+SCLC in a GEMM [6]. Abrogation of epigenetic regulators including EZH2, LSD1 and KMD6A have been also associated to phenotypic switching between subtypes in SCLC [59–61]. Simultaneous detection of molecular subtypes, and shifts associated with disease progression, have been observed in human SCLC [6, 62]. Here we show that the chromatin remodeler SMARCA4 sustains the NE phenotype in both ASCL1 and NEUROD1 SCLC subtypes, and that its inactivation promotes a shift toward a low-NE state. In silico analysis showed a strong correlation between levels of SMARCA4 and NE markers in both SCLC patient tumors and cell lines. ChIP-seq of SMARCA4 in NE-high SCLC PDXs revealed binding to regulatory elements of lineage TFs including ASCL1, NEUROD1, FOXA2 and INSM1 as well as to relevant genes implicated in axonogenesis, synapse formation, and neuropeptide signaling pathways. Several genes identified as high-confidence SMARCA4 binding targets overlapped with ASCL1 and NEUROD1 targets, suggesting a role for SMARCA4 in regulating ASCL1 and NEUROD1 downstream transcriptional programs [63]. Accordingly, we found reduction in chromatin accessibility at distal regions across a spectrum of NE genes when SMARCA4 was pharmacologically inhibited. Phenotypic changes driven by SMARCA4 inactivation have been previously described in other tumors, including lung adenocarcinoma, where SMARCA4 has a cell-type specificity role in lineage transformation and exhibits divergent functions depending on the cell of origin [15]. mSWI/SNF complex has been recently reported as a dependency in POU2F3 SCLC tumors [64, 65]. Intriguingly, SMARCA4/2 inhibition affects distinct programs in POU2F3 SCLC cells than those we have observed in SCLC-A and SCLC-N, suggesting a different function for SMARCA4 in high vs low-NE SCLC subtypes.
Notably, SMARCA4 binds to several known regulators of NOTCH signaling. Activation of NOTCH has been shown to promote non-NE fate by increasing REST and HES1 in SCLC [12, 66]. Whether SMARCA4 functions as a transcriptional repressor of some of these NOTCH regulators in SCLC is still unknown and requires further investigation. REST is a key regulator of non-NE differentiation, and its activation appears necessary to achieve transition to a non-NE state in SCLC [12, 67, 68]. REST has been shown to be spliced to encode the inactive REST isoform REST4 by SRRM4 in NE prostate and SCLC tumors [50, 52, 53, 69, 70]. However, upstream molecular mechanisms underlying SRRM4 activation in NE tumors had not been defined. We confirmed REST splicing into inactive REST4 variants in NE-high SCLC and demonstrated that SMARCA4 inhibition with FHD-286 reduced the levels of inactive REST4 through downregulation of SRRM4. SMARCA4 binds to the SRRM4 promoter and its inhibition reduces the chromatin accessibility of SRRM4. Enrichment analysis of DEG identified REST as the top TF associated with SMARCA4 driven non-NE SCLC transition. Interestingly, a recent study has shown that REST and ASCL1 regulate distinct cell fate targets in SCLC and suggested that inhibition of ASCL1 and activation of REST are both required to promote a NE to non-NE transition [12]. Our work proposes a unified upstream regulatory mechanism in which SMARCA4 sustains the NE phenotype through regulation of ASCL1 and NEUROD1 transcriptional programs and concurrently controls REST expression by SRRM4-driven splicing.
SCLC is considered a recalcitrant malignancy, with patients in critical need of novel therapeutic options. A surprising finding of this study is the activation of ERBB/MAPK mitogenic signaling, suppressed in NE-high SCLC, following pharmacological inhibition of SMARCA4/2. Activation of the MAPK pathway selectively induces cell death of ASCL1+SCLC and reduces the expression of NE markers [71]. ERK activity appears to be limited to low-NE cells in SCLC, in line with the phenotypic changes we observe when SMARCA4 is inhibited [58]. Inactivation of SMARCA4 induced the expression of ERBB family receptors and the cognate ligand NRG1. Consistent with this observation, a previous report showed that SMARCA4 directly regulates NRG1 levels in candida albicans through induction of an antisense NRG1 transcript, suggesting that NRG1 dysregulation by SMARCA4 could be a conserved mechanism [72]. Remarkably, combined SMARCA4/ERBB inhibition showed efficacy in delaying tumor growth, even in PDXs derived from tumors after several lines of treatment, supporting the potential of this combinatorial therapy as a therapeutic strategy for the treatment of SCLC. Our results provide insight into how intrinsic SCLC plasticity is controlled and can be exploited to induce clinically favorable states associated with a therapeutic vulnerability.
Conclusions
In conclusion, our data uncover a critical role for SMARCA4 in sustaining high-NE states in SCLC and define a resulting potential therapeutic vulnerability.
Supplementary Information
Acknowledgements
We acknowledge the help from the Pathology Core, Gene Editing & Screening Core Facility and Integrated Genomics Operation Core at MSKCC. We thank Foghorn Therapeutics for providing us with FHD-286 for use in this study, and Murphy Hentemann for productive discussions. We thank Abbie Ireland and Dr. Trudy Oliver for providing the processed scRNAseq data. We also acknowledge all members of the Rudin laboratory for their scientific input.
Abbreviations
- NE
Neuroendocrine
- SCLC
Small cell lung cancer
- SCLC-A
Small cell lung cancer-ASCL1
- SCLC-N
Small cell lung cancer-NEUROD1
- SCLC-P
Small cell lung cancer-POU2f3
- SCLC-I
Small cell lung cancer-inflamed
- SMARCA4
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
- SMARCA2
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
- NOTCH
Notch receptor
- SWI/SNF
SWItch/sucrose non-fermentable
- BAF
BRG1/BRM-associated factor
- cBAF
Canonical BAF
- PBAF
Polybromo-associated BAF
- ncBAF
Non-canonical BAF
- LUAD
Lung adenocarcinoma
- ARID1A
AT-rich interaction domain 1A
- mRNA
Messenger RNA
- ATPase
Adenosine triphosphatase
- ASCL1
Achaete-scute family bHLH transcription factor 1
- NEUROD1
Neuronal differentiation 1
- SYP
Synaptophysin
- FOXA1
Forkhead box A1
- FOXA2
Forkhead box A2
- INSM1
INSM transcriptional repressor 1
- CHGA
Chromogranin A
- NCAM1
Neural cell adhesion molecule 1
- DLL3
Delta like canonical Notch ligand 3
- REST
RE1 silencing transcription factor
- REST4
RE1 silencing transcription factor isoform4
- NOTCH2
Notch receptor 2
- YAP1
Yes1 associated transcriptional regulator
- POU2F3
POU class 2 homeobox 3
- SRRM4
Serine/arginine repetitive matrix
- NRG1
Neuroregulin-1
- ERBB
Erythroblastic leukemia viral oncogene homologue
- HES1
Hes family bHLH transcription factor 1
- HEY1
Hes related family bHLH transcription factor with YRPW motif 1
- TEAD2
TEA domain transcription factor 2
- AJUBA
Ajuba LIM protein
- CYR61
Cysteine rich angiogenic inducer 61
- WWTR1
WW domain containing transcription regulator 1
- Rb1
RB transcriptional corepressor 1
- TP53
Tumor protein p53
- MYC
MYC proto-oncogene
- OTX2
Orthodenticle homeobox 2
- GRP
Gastrin releasing peptide
- ERK
Extracellular signal-regulated kinase
- AKT
AKT serine/threonine kinase 1
- ERBB3
Erb-b2 receptor tyrosine kinase 3
- EZH2
Enhancer of zeste 2 polycomb repressive complex 2 subunit
- LSD1
Lysine-specific histone demethylase 1A
- KMD6A
Lysine demethylase 6A
- MAPK
Mitogen-activated protein kinases
- RTN1
Reticulon1
- NRSN1
Neurensin1
- MYT1
Myelin transcription factor1
- GEMM
Genetically engineered mouse model
- GSEA
Gene enrichment analysis
- DEG
Differentially expressed genes
- CCLE
Cancer cell line encyclopedia
- PDX
Patient-derived xenograft
- DNA
Deoxyribonucleic acid
- 5’UTR
Eukaryotic 5’ untranslated region
- 3’UTR
3’ untranslated regions
- TF
Transcription factor
- HOMER
Hypergeometric Optimization of Motif EnRichment
- ChIP-seq
Chromatin immunoprecipitation followed by sequencing
- ATAC-seq
Assay for transposase-accessible chromatin with sequencing
- RNA-seq
RNA sequencing
Author contributions
Conceptualization: ER, CMR, AQV; Methodology: ER, HS, YAZ, BPM, HZ, MH, VD, AS, PM, IL, JQ, ES, MA, DB; Data Analysis: ER, YAZ, MA. Writing–Original Draft: ER, AQV, YAZ, CMR; Review & Editing: All authors; Supervision: AQV and CMR; Funding acquisition: CMR and AQV. All authors read and approved the final version of the manuscript.
Funding
This study was supported by Ramon Areces Foundation (to ER), Druckenmiller Center for Lung Cancer Research (to ER, AQV and CMR), NIH T32 CA1600001 (to AQV), American Lung Association (to AQV) NCI R35 CA263816, U24 CA213274, P30 CA008748, the Robert J. and Helen C. Kleberg Foundation, and the Van Andel Research Institute – Stand Up to Cancer Epigenetics Dream Team (to CMR).
Availability of data and materials
All raw and processed data generated in this study can be found at GEO repository: GSE256345, GSE256346, GSE256347.
Declarations
Ethics approval and consent to participate
All in vivo experiments were performed at Memorial Sloan Kettering Cancer Center (MSKCC) following Animal Care and Use Committee guidelines.
Consent for publication
Not applicable.
Competing interests
AQV has received honoraria from Astra Zeneca. CMR has consulted regarding oncology drug development with AbbVie, Amgen, Astra Zeneca, D2G, Daiichi Sankyo, Epizyme, Genentech/Roche, Ipsen, Jazz, Kowa, Lilly, Merck, and Syros. He serves on the scientific advisory boards of Auron, Bridge Medicines, DISCO, Earli, and Harpoon Therapeutics. All other authors declare no conflicts of interest.
Footnotes
Lead Contact: Charles M. Rudin
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/29/2024
A Correction to this paper has been published: 10.1186/s13045-024-01609-7
Contributor Information
Álvaro Quintanal-Villalonga, Email: quintaa@mskcc.org.
Charles M. Rudin, Email: rudinc@mskcc.org
References
- 1.Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39(3):346–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baine MK, Hsieh MS, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol. 2020;15(12):1823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai L, De Berardinis RJ, Xie Y, Minna JD, Xiao G. A comparative study of neuroendocrine heterogeneity in small cell lung cancer and neuroblastoma. Mol Cancer Res. 2023;21(8):795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38(1):60–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16(5):1259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2017;31(2):270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng J, Cai L, Girard L, Prall OWJ, Rajan N, Khoo C, et al. Molecular and pathologic characterization of YAP1-expressing small cell lung cancer cell lines leads to reclassification as SMARCA4-deficient malignancies. [cited 2024 Jan 31]; Available from: https://git.biohpc.swmed.edu/BICF/Astrocyte/r [DOI] [PMC free article] [PubMed]
- 10.Oser MG, Sabet AH, Gao W, Chakraborty AA, Schinzel AC, Jennings RB, et al. The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev. 2019;33(23–24):1718–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiyokawa H, Morimoto M. Notch signaling in the mammalian respiratory system, specifically the trachea and lungs, in development, homeostasis, regeneration, and disease. Dev Growth Differ. 2020;62:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shue YT, Drainas AP, Li NY, Pearsall SM, Morgan D, Sinnott-Armstrong N, et al. A conserved YAP/Notch/REST network controls the neuroendocrine cell fate in the lungs. Nat Commun. 2022;13(1):2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centore RC, Sandoval GJ, Soares LMM, Kadoch C, Chan HM. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 2020;36(12):936–50. 10.1016/j.tig.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Mittal P, Roberts CWM. The SWI/SNF complex in cancer—biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17(7):435–48. 10.1038/s41571-020-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Concepcion CP, Ma S, Lafave LM, Bhutkar A, Liu M, Deangelo LP, et al. Smarca4 inactivation promotes LineageSpecific transformation and early metastatic features in the lung. Cancer Discov. 2022;12(2):562–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32(5):574–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun. 2020;11(1):5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navickas SM, Giles KA, Brettingham-Moore KH, Taberlay PC. The role of chromatin remodeler SMARCA4/BRG1 in brain cancers: a potential therapeutic target. Oncogene. 2023;42:2363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mardinian K, Adashek JJ, Botta GP, Kato S, Kurzrock R. SMARCA4: Implications of an altered chromatin-remodeling gene for cancer development and therapy. Mol Cancer Ther. 2021;20:2341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Miguel FJ, Gentile C, Feng WW, Silva SJ, Sankar A, Exposito F, et al. Mammalian SWI/SNF chromatin remodeling complexes promote tyrosine kinase inhibitor resistance in EGFR-mutant lung cancer. Cancer Cell. 2023;41(8):1516–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiskus W, Piel J, Collins M, Hentemann M, Cuglievan B, Mill CP, et al. BRG1/BRM inhibitor targets AML stem cells and exerts superior preclinical efficacy combined with BET or Menin inhibitor. bioRxiv. 2023; 2023.09.28.560054. http://biorxiv.org/content/early/2023/10/01/2023.09.28.560054.abstract. [DOI] [PubMed]
- 22.Sivakumar S, Moore JA, Montesion M, Sharaf R, Lin DI, Colón CI, et al. Integrative analysis of a large real-world cohort of small cell lung cancer identifies distinct genetic subtypes and insights into histologic transformation. Cancer Discov. 2023;13(7):1572–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017 [cited 2024 Jan 31];8. https://pubmed.ncbi.nlm.nih.gov/28262751/. [DOI] [PMC free article] [PubMed]
- 24.Quintanal-Villalonga A, Durani V, Sabet A, Redin E, Kawasaki K, Shafer M, et al. Exportin 1 inhibition prevents neuroendocrine transformation through SOX2 down-regulation in lung and prostate cancers. Sci Transl Med. 2023;15(707):eadf7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George J, Lim JS, Jang SJ, Cun Y, Ozretia L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44(10):1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, et al. Multiomic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov. 2021;11(12):3028–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res. 2018;7(1):32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Stein TI, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017. 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch RP, Lee C, Imbriano PM, Patil S, Weymouth TE, Smith RA, et al. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res. 2014;42(13):e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CT, Cavalcante RG, Lee C, Qin T, Patil S, Wang S, et al. Poly-Enrich: count-based methods for gene set enrichment testing with genomic regions. NAR Genom Bioinform. 2020;2(1):lqaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Januario T, Ye X, Bainer R, Alicke B, Smith T, Haley B, et al. PRC2-mediated repression of SMARCA2 predicts EZH2 inhibitor activity in SWI/SNF mutant tumors. Proc Natl Acad Sci USA. 2017;114(46):12249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez AM, Traunmüller L, Scheiffele P. Neurexins: molecular codes for shaping neuronal synapses. Nat Rev Neurosci. 2021;22:137–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasconcelos FF, Sessa A, Laranjeira C, Raposo AASF, Teixeira V, Hagey DW, et al. MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep. 2016;17(2):469–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steiner P, Kulangara K, Sarria JCF, Glauser L, Regazzi R, Hirling H. Reticulon 1-C/neuroendocrine-specific protein-C interacts with SNARE proteins. J Neurochem. 2004;89(3):569–80. [DOI] [PubMed] [Google Scholar]
- 50.Lee AR, Gan Y, Tang Y, Dong X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine. 2018;35:167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545(7654):360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Coleman IM, Brown LG, True LD, Kollath L, Lucas JM, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(20):4698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Labrecque MP, Brown LG, Coleman IM, Lakely B, Brady NJ, Lee JK, et al. RNA splicing factors SRRM3 and SRRM4 distinguish molecular phenotypes of castration-resistant neuroendocrine prostate cancer. Cancer Res. 2021;81(18):4736–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127(18):3913–21. [DOI] [PubMed] [Google Scholar]
- 55.Neptune ER, Podowski M, Calvi C, Cho JH, Garcia JGN, Tuder R, et al. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283(30):21160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osada H, Tomida S, Yatabe Y, Tatematsu Y, Takeuchi T, Murakami H, et al. Roles of achaete-scute homologue 1 in DKK1 and E-cadherin repression and neuroendocrine differentiation in lung cancer. Cancer Res. 2008;68(6):1647–55. [DOI] [PubMed] [Google Scholar]
- 57.Bohuslavova R, Fabriciova V, Smolik O, Lebrón-Mora L, Abaffy P, Benesova S, et al. NEUROD1 reinforces endocrine cell fate acquisition in pancreatic development. Nat Commun. 2023;14(1):55546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19(2):244–56. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen EM, Taniguchi H, Chan JM, Zhan YA, Chen X, Qiu J, et al. Targeting lysine-specific demethylase 1 rescues major histocompatibility complex class I antigen presentation and overcomes programmed death-ligand 1 blockade resistance in SCLC. J Thorac Oncol. 2022;17(8):1014–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahadevan NR, Knelson EH, Wolff JO, Vajdi A, Saigí M, Campisi M, et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov. 2021;11(8):1952–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duplaquet L, Li Y, Booker MA, Xie Y, Olsen SN, Patel RA, et al. KDM6A epigenetically regulates subtype plasticity in small cell lung cancer. Nat Cell Biol. 2023;25(9):1346–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39(11):1479–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pozo K, Kollipara RK, Kelenis DP, Rodarte KE, Ullrich MS, Zhang X, et al. ASCL1, NKX2-1, and PROX1 co-regulate subtype-specific genes in small-cell lung cancer. iScience. 2021;24(9):102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He T, Xiao L, Qiao Y, Klingbeil O, Young E, Wu XS, et al. Targeting the mSWI/SNF complex in POU2F-POU2AF transcription factor-driven malignancies. bioRxiv : the preprint server for biology. 2024. [DOI] [PubMed]
- 65.Leslie Duplaquet KS, Alexander W Ying XLYLXQRL. Mammalian SWI/SNF complex activity regulates POU2F3 and constitutes a targetable dependency in small cell lung cancer. bioRxiv. 2024. [DOI] [PMC free article] [PubMed]
- 66.Olsen RR, Ireland AS, Kastner DW, Groves SM, Spainhower KB, Pozo K, et al. ASCL1 represses a SOX9+ neural crest stem-like state in small cell lung cancer. Genes Dev. 2021;38(11–12):847–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flores-Morales A, Bergmann TB, Lavallee C, Batth TS, Lin D, Lerdrup M, et al. Proteogenomic characterization of patient-derived xenografts highlights the role of REST in neuroendocrine differentiation of castration-resistant prostate cancer. Clin Cancer Res. 2019;25(2):595–608. [DOI] [PubMed] [Google Scholar]
- 68.Puca L, Vlachostergios PJ, Beltran H. Neuroendocrine differentiation in prostate cancer: emerging biology, models, and therapies. Cold Spring Harb Perspect Med. 2019;9(2):a030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Donmez N, Sahinalp C, Xie N, Wang Y, Xue H, et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2017;71(1):68–78. [DOI] [PubMed] [Google Scholar]
- 70.Shimojo M, Shudo Y, Ikeda M, Kobashi T, Ito S. The small cell lung cancer-specific isoform of RE1-silencing transcription factor (REST) is regulated by neural-specific Ser/Arg repeat-related protein of 100 kDa (nSR100). Mol Cancer Res. 2013;11(10):1258–68. [DOI] [PubMed] [Google Scholar]
- 71.Caeser R, Hulton C, Costa E, Durani V, Little M, Chen X, et al. MAPK pathway activation selectively inhibits ASCL1-driven small cell lung cancer. iScience. 2021;24(11):103224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol. 2012;85(3):557–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed data generated in this study can be found at GEO repository: GSE256345, GSE256346, GSE256347.