Abstract
The hemagglutinin, esterase, and fusion (HEF) glycoprotein of influenza C virus possesses receptor binding, receptor destroying, and membrane fusion activities. The HEF cDNAs from influenza C/Ann Arbor/1/50 (HEF-AA) and influenza C/Taylor/1223/47 (HEF-Tay) viruses were cloned and expressed, and transport of HEF to the cell surface was monitored by susceptibility to cleavage by exogenous trypsin, indirect immunofluorescence microscopy, and flow cytometry. Previously it has been found in studies with the C/Johannesburg/1/66 strain of influenza C virus (HEF-JHB) that transport of HEF to the cell surface is severely inhibited, and it is thought that the short cytoplasmic tail, Arg-Thr-Lys, is involved in blocking HEF cell surface expression (F. Oeffner, H.-D. Klenk, and G. Herrler, J. Gen. Virol. 80:363–369, 1999). As the cytoplasmic tail amino acid sequences of HEF-AA and HEF-Tay are identical to that of HEF-JHB, the data indicate that cell surface expression of HEF-AA and HEF-Tay is not inhibited by this amino acid sequence. Furthermore, the abundant cell surface transport of HEF-AA and HEF-Tay indicates that their cell surface expression does not require coexpression of another viral protein. The HEF-AA and HEF-Tay HEF glycoproteins bound human erythrocytes, promoted membrane fusion in a low-pH and trypsin-dependent manner, and displayed esterase activity, indicating that the HEF glycoprotein alone mediates all three known functions at the cell surface.
Influenza A, B, and C viruses are negative-strand, segmented RNA viruses of the Orthomyxoviridae family, differing primarily in the number of integral membrane proteins encoded by the viral genome (reviewed in reference 10). Whereas influenza A and B viruses encode three integral membrane proteins, hemagglutinin (HA), neuraminidase (NA), and M2 (influenza A virus) or NB (influenza B virus), influenza C viruses encode only two integral membrane proteins, HEF and CM2 (9, 17). The lack of a separate protein with receptor-destroying activity is reflected in the influenza C virus genome by the concomitant loss of one RNA segment, as influenza C virus has seven RNA segments (18, 21).
The HEF glycoprotein is a homotrimer which mediates virion binding to the viral receptor, 9-O-N-acetyl neuraminic acid (7), possesses an acetylesterase or receptor-destroying activity (8), and mediates membrane fusion between the influenza C virus and cellular membranes after triggering by low-pH treatment (3, 15). Fusion activity requires that the 90-kDa precursor protein HEF0 be cleaved to the disulfide-linked subunits HEF1 and HEF2 by unidentified cellular protease(s). The recently solved atomic structure of HEF at 3.2 Å resolution shows a marked degree of structural similarity to that of influenza A virus HA, despite very little amino acid homology between the two proteins (22).
Analysis at the cell surface of the three biological activities of HEF has been hindered by an apparent lack of surface transport of the glycoprotein in mammalian cells when the cDNA was derived from influenza C virus strain C/Johannesburg/1/66 (HEF-JHB) (14, 20, 25), although erythrocyte binding has been observed when the C/California/78 HEF cDNA was expressed in cells (26). The lack of surface transport of HEF-JHB has been attributed to a negative regulatory domain (Arg-Thr-Lys) which compromises the predicted cytoplasmic tail of HEF (14). We have studied the expression from cDNA of the HEF glycoproteins from influenza C/Ann Arbor/1/50 virus (HEF-AA) and influenza C/Taylor/1233/47 virus (HEF-Tay) and have determined that the HEF glycoprotein of both strains is transported to the cell surface and exhibits the three known biological activities of HEF when expressed in a variety of cell types and in a variety of expression systems. As HEF-JHB, HEF-AA, and HEF-Tay all possess the same cytoplasmic tail amino acid sequence (2), we interpret our data to indicate that the Arg-Thr-Lys motif is not inhibitory to cell surface transport of HEF-AA and HEF-Tay and that the lack of cell surface expression of the HEF-JHB protein represents either a virus strain phenomenon or a peculiarity of the HEF-JHB cDNA nucleotide sequence (19, 24).
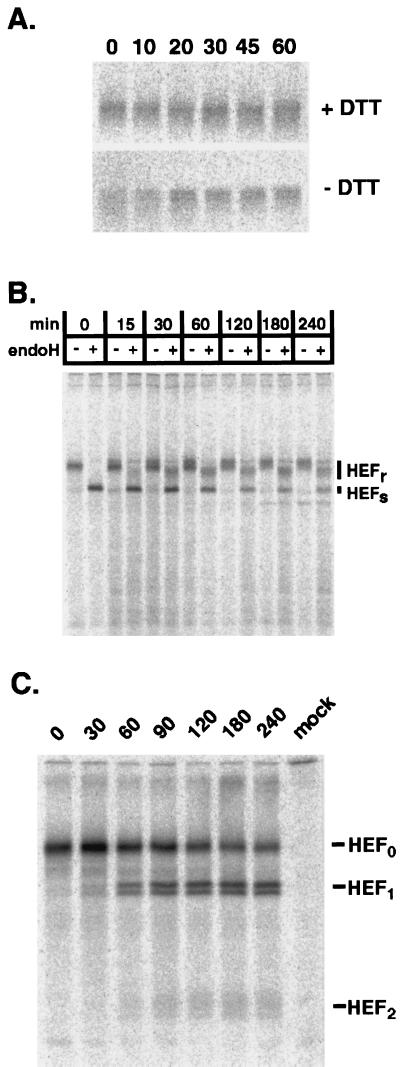
The cDNAs for HEF were obtained by rt-PCR of vRNA from MDCK cell-grown influenza C/AA/1/50 or influenza C/Taylor/1233/47 virus (data not shown; primers were designed based on known influenza C virus RNA segment 4 sequences) (2) and initially cloned into the plasmid pGEM3, under control of the bacteriophage T7 RNA polymerase promoter (16). The pGEM3 HEF-AA plasmid was transfected into HeLa-T4 cells which were infected with a recombinant vaccinia virus (vac-T7) expressing the bacteriophage T7 RNA polymerase (4). The cells were metabolically labeled and incubated in chase medium as previously described (16). At the indicated times, the cells were lysed, and HEF protein was immunoprecipitated with a cocktail of HEF-specific monoclonal antibodies (MAbs) (23). The HEF-AA glycoprotein migrated as a single band of approximately 90 kDa under reducing conditions (Fig. 1A) at all the indicated times. Under nonreducing conditions, the HEF-AA glycoprotein also migrated as a single 90-kDa band, although some heterogeneity was detected after incubation in chase medium for 0, 10, and 20 min, most likely resulting from a lack of proper disulfide bond formation. Conversion of HEF-AA from an 80- to a 100-kDa form under nonreducing conditions, an event associated with but not sufficient to confer cell surface transport of HEF-JHB (14, 25), was not observed.
FIG. 1.
(A) HeLa-T4 cells in 35-mm-diameter tissue culture dishes were infected with vac-T7 at a multiplicity of infection of 5 to 10 PFU per cell for 1 h at 37°C and were then transfected with a plasmid containing the cDNA for HEF-AA under the control of the T7 RNA polymerase promoter (pGEM HEF-AA) as previously described (16). At 5 h posttransfection, the cells were incubated in Dulbecco’s modified Eagle medium deficient in cysteine and methionine, metabolically labelled with [35S]ProMix (100 μCi/ml) for 15 min, and incubated in chase medium for the indicated times (16). The cells were lysed, polypeptides were immunoprecipitated with a cocktail of anti-HEF MAbs, resuspended in sample buffer with or without the reducing agent dithiothreitol (DTT), and boiled for 5 min before analysis of polypeptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% acrylamide gels. (B) Vac-T7-infected, pGEM HEF-AA-transfected HeLa-T4 cells were metabolically labeled, and proteins were immunoprecipitated as described above, the immune complexes were divided into two aliquots and treated with endoglycosidase H (+) or untreated (−) before the analysis of polypeptides by SDS-PAGE on 10% acrylamide gels. (C) Vac-T7 infected, pGEM HEF-AA-transfected HeLa-T4 cell lysates were metabolically labeled as above. Ten minutes prior to the indicated times, TPCK-trypsin (15 μg/ml) was added to the chase media to cleave cell surface HEF0 into HEF1 and HEF2 subunits. At the indicated times, the cells were placed on ice and washed with phosphate-buffered saline containing a cocktail of protease inhibitors, the cells were lysed, proteins were immunoprecipitated, and polypeptides were separated by SDS-PAGE on 15% acrylamide gels as described above. No trypsin was added to the 0-min chase sample.
Intracellular transport of HEF-AA to the medial-Golgi apparatus was assessed by determining the rate of resistance to digestion by endoglycosidase H (endo H) of HEF carbohydrate chains in a pulse-chase protocol. HEF-AA began to acquire endo H-resistant carbohydrate modifications after 30 min (Fig. 1B) and reached a maximum (approximately 70%) by 120 min indicating that the majority of the metabolically labeled glycoprotein had been transported to the medial-Golgi apparatus. Transport of HEF-AA glycoprotein to the cell surface was investigated by monitoring the cleavage of HEF0 into HEF1 and HEF2 subunits (6) upon the addition of tosyl phenylalanyl chloromethyl ketone (TPCK)-trypsin to the cell culture media (Fig. 1C). HEF1 and HEF2 were detected 60 min after the pulse-chase, indicating the arrival of HEF0 at the cell surface. As 65% of the HEF-AA glycoprotein was cleaved into HEF1 and HEF2 after 240 min, it is reasonable to conclude that the majority of the HEF-AA glycoprotein is transported to the cell surface when expressed from cDNA by using the vac-T7 expression system.
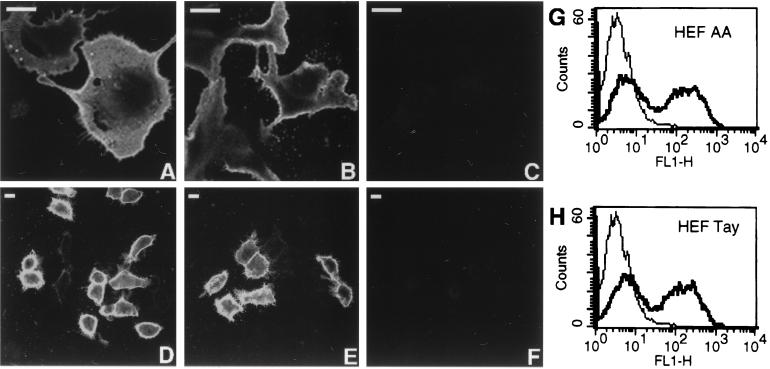
Cell surface expression of HEF-AA and HEF-Tay was also investigated by using two different expression systems, recombinant simian virus 40 (rSV40) (5, 11, 12) and the eukaryotic expression vector pCAGGS (13). The cDNAs for HEF-AA and HEF-Tay were subcloned into the pCAGGS vector and expressed transiently as previously described (17) or subcloned into the plasmid pSV133 and used to generate rSV40s (12). CV-1 cells were infected with rSV40s expressing HEF-AA (Fig. 2A) or HEF-Tay (Fig. 2B) or mock infected (Fig. 2C), whereas HeLa-T4 cells were transfected with pCAGGS HEF-AA (Fig. 2D) or pCAGGS HEF-Tay (Fig. 2E) or mock transfected (Fig. 2F). At 36 h postinfection or 18 h posttransfection, the cells were fixed with 1% formaldehyde, incubated with a cocktail of anti-HEF MAbs, followed by staining with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) and cell surface fluorescence assessed by using a confocal microscope. HEF-AA and HEF-Tay were transported to the cell surface when expressed by using either rSV40 or pCAGGS vectors as indicated by the strong cell surface fluorescence signal compared to mock-infected or transfected cells. Cell surface expression of HEF from the eukaryotic expression vector pCAGGS was quantitated by flow cytometry. At 18 h posttransfection, mock-transfected, pCAGGS HEF-AA-transfected, or pCAGGS HEF-Tay-transfected Vero cells were incubated with a cocktail of anti-HEF MAbs, followed by an FITC-conjugated goat anti-mouse IgG and analyzed by flow cytometry. Vero cells transfected with pCAGGS HEF-AA (Fig. 2G) or pCAGGS HEF-Tay (Fig. 2H) showed a large increase in cell surface fluorescence compared to mock-transfected cells. Quantitation of the histograms in Fig. 2G and H yielded 54.6% positive cells and a mean channel fluorescence of 179.8 for pCAGGS HEF-AA-transfected cells and 56.2% positive cells and a mean channel fluorescence of 169.8 for pCAGGS HEF-Tay-transfected cells (mock-transfected cells were arbitrarily set at a mean channel fluorescence of 3.5). The data shown in Fig. 1 and 2 indicate that the HEF-AA or HEF-Tay glycoproteins are transported to the cell surface when expressed in a variety of expression systems and cell types.
FIG. 2.
CV-1 cells grown on glass coverslips in 35-mm-diameter culture dishes were infected with rSV40 HEF-AA (A), rSV40 HEF-Tay (B), or mock infected (C) and incubated at 37°C for 36 h. HeLa-T4 cells were transfected with pCAGGS HEF-AA (D) pCAGGS HEF-Tay (E), or mock transfected (F) and incubated at 37°C for 18 h. Cell monolayers were fixed with 1% formaldehyde in phosphate-buffered saline, and surface HEF glycoprotein was detected by indirect immunofluorescence (16) by using a 1:500 dilution of an anti-HEF MAb cocktail followed by FITC-conjugated goat anti-mouse IgG and visualized with a Zeiss LSM 410 confocal microscope. Bar, 25 μM. Quantitation of HEF cell surface expression in Vero cells transfected with pCAGGS HEF-AA (G) or pCAGGS HEF-Tay (H). At 18 h posttransfection, the cells were incubated with a 1:500 dilution of a cocktail of anti-HEF MAbs, followed by FITC-conjugated goat anti-mouse IgG, and analyzed by flow cytometry. The relative fluorescence intensity of 10,000 cells is depicted in the histograms. For comparative purposes, each histogram displays the relative fluorescence of mock-transfected Vero cells (thin line, same population in each histogram) as well as cells transfected with the indicated plasmids (thick line).
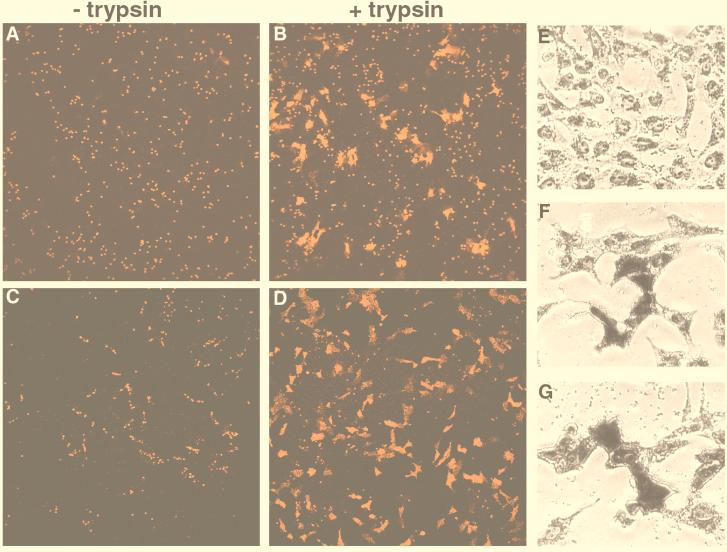
To determine whether expression of HEF from cDNA resulted in cell surface accumulation of a functional glycoprotein, HEF-AA and HEF-Tay glycoproteins were tested for receptor-binding and fusion activities by using a modified lipid mixing assay involving fusion of glycoprotein-expressing cells to octadecyl rhodamine B (R-18)-labeled human erythrocytes (1). Vero cells transfected with plasmids encoding pCAGGS HEF-AA (Fig. 3A and B) or pCAGGS HEF-Tay (Fig. 3C and D) were assayed at 18 h posttransfection for binding to and fusion with R-18-labeled human erythrocytes as previously described (1). Both HEF-AA- and HEF-Tay-expressing cells were able to bind R-18-labeled human erythrocytes (Fig. 3A and C) and displayed trypsin-dependent cleavage and low pH-induced fusion, as assayed by the transfer of R-18 dye from the bound erythrocytes to the Vero cell plasma membranes (Fig. 3B and D). HEF esterase activity was analyzed by using a commercially available α-naphthyl esterase/Fast Blue BB detection kit, which results in the formation of a dark precipitate on the surface of cells possessing esterase activity. As shown in Fig. 3, cells transfected with pCAGGS HEF-AA (Fig. 3F) and pCAGGS HEF-Tay (Fig. 3G) showed a precipitate indicating expressed esterase activity, whereas cells transfected with pCAGGS vector alone (Fig. 3E) showed no detectable precipitate. Thus, the data indicate that HEF glycoprotein expressed at the cell surface from the HEF cDNA is biologically functional.
FIG. 3.
Vero cells were grown on glass coverslips in 35-mm-diameter culture dishes, transfected with pCAGGS HEF-AA (A and B) or pCAGGS HEF-Tay (C and D) as described previously (17), and treated as indicated with TPCK-trypsin (15 μg/ml) for 15 min at 18 h posttransfection. Human erythrocytes labeled with octadecyl rhodamine B (R18) were then added to the cells and incubated at 4°C for 30 min (1). The cells were washed extensively with phosphate-buffered saline, pH 7.4, incubated in phosphate-buffered saline, pH 5.4 for 10 min at 37°C, and analyzed immediately for lipid mixing by using a confocal microscope. Vero cells were grown on glass coverslips in 35-mm culture dishes, transfected with pCAGGS vector alone (E), pCAGGS HEF-AA (F), or pCAGGS HEF-Tay (E) as previously described (17), and analyzed at 18 h posttransfection with an α-naphthyl esterase detection kit (Sigma Diagnostics, St. Louis, Mo.) as per manufacturer’s instructions. The cells were photographed by using a Nikon Diaphot inverted, phase-contrast microscope (Nikon Corp., Tokyo, Japan).
Our data contrast with previously published reports on the expression of HEF-JHB from cDNA (14, 20, 25). When expressed from cDNA, the HEF-JHB glycoprotein accumulates in an 80-kDa, presumably misfolded, form and is retained in the endoplasmic reticulum (25). Amino acid substitutions in or deletion of the HEF-JHB cytoplasmic tail restored the 100-kDa form of the protein but failed to restore cell surface transport, indicating conversion of the 80-kDa form of HEF to the 100-kDa form alone was insufficient to promote cell surface transport (25). Subsequently, Oeffner and colleagues (14) showed that replacing the HEF cytoplasmic tail with that of the influenza A virus HA or the cellular protein gp40 restored both conversion to the 100-kDa form of HEF and cell surface transport, implicating the HEF cytoplasmic tail as being inhibitory to cell surface expression (14). It must be noted that the Arg-Thr-Lys motif was not shown to be sufficient in and of itself to prevent cell surface transport of other glycoproteins. Our data indicate that the inhibitory effect of the HEF cytoplasmic tail on HEF cell surface transport is an influenza C virus strain-specific phenomenon, since HEF-AA, HEF-Tay, and HEF-JHB contain the exact same cytoplasmic tail amino acid sequence (2). It remains to be determined whether coexpression of some other viral protein is required for HEF-JHB cell surface transport or whether any of the amino acid differences between the original HEF-JHB sequence (19) and that used by Szepanski and colleagues (24) perturb the folding and/or intracellular transport of the HEF-JHB protein.
Acknowledgments
We thank K. Nakamura, Yamagata University School of Medicine, Japan, for the generous gift of anti-HEF monoclonal antibodies and the members of the Lamb laboratory for helpful discussions.
This research was supported by research grant R37 AI-20201 from the National Institute of Allergy and Infectious Diseases. A.P. is an associate and R.A.L. is an investigator at the Howard Hughes Medical Institute.
REFERENCES
- 1.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buonagurio D A, Nakada S, Desselberger U, Krystal M, Palese P. Noncumulative sequence changes in the hemagglutinin genes of influenza C virus isolates. Virology. 1985;146:221–232. doi: 10.1016/0042-6822(85)90006-6. [DOI] [PubMed] [Google Scholar]
- 3.Formanowski F, Wharton S A, Calder L J, Hofbauer C, Meier-Ewert H. Fusion characteristics of influenza C viruses. J Gen Virol. 1990;71:1181–1188. doi: 10.1099/0022-1317-71-5-1181. [DOI] [PubMed] [Google Scholar]
- 4.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gething M J, Sambrook J. Cell-surface expression of influenza haemagglutinin from a cloned DNA copy of the RNA gene. Nature. 1981;293:620–625. doi: 10.1038/293620a0. [DOI] [PubMed] [Google Scholar]
- 6.Herrler G, Compans R W, Meier-Ewert H. A precursor glycoprotein in influenza C virus. Virology. 1979;99:49–56. doi: 10.1016/0042-6822(79)90035-7. [DOI] [PubMed] [Google Scholar]
- 7.Herrler G, Klenk H-D. The surface receptor is a major determinant of the cell tropism of influenza C virus. Virology. 1987;159:102–108. doi: 10.1016/0042-6822(87)90352-7. [DOI] [PubMed] [Google Scholar]
- 8.Herrler G, Rott R, Klenk H-D, Muller H-P, Shukla A K, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetyl esterase. EMBO J. 1985;4:2711–2720. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hongo S, Sugawara K, Nishimura H, Muraki Y, Kitame F, Nakamura K. Identification of a second protein encoded by influenza C virus RNA segment 6. J Gen Virol. 1994;75:3503–3510. doi: 10.1099/0022-1317-75-12-3503. [DOI] [PubMed] [Google Scholar]
- 10.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology (3rd ed.). New York, N.Y: Lippincott-Raven Press; 1996. pp. 1353–1394. [Google Scholar]
- 11.Lamb R A, Lai C-J. Spliced and unspliced messenger RNAs synthesized from cloned influenza virus M DNA in an SV40 vector: expression of the influenza virus membrane protein (M1) Virology. 1982;123:237–256. doi: 10.1016/0042-6822(82)90258-6. [DOI] [PubMed] [Google Scholar]
- 12.Naim H Y, Roth M G. SV40 virus expression vectors. Methods Cell Biol. 1994;43:113–136. doi: 10.1016/s0091-679x(08)60601-9. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 14.Oeffner F, Klenk H D, Herrler G. The cytoplasmic tail of the influenza C virus glycoprotein HEF negatively affects transport to the cell surface. J Gen Virol. 1999;80:363–369. doi: 10.1099/0022-1317-80-2-363. [DOI] [PubMed] [Google Scholar]
- 15.Ohuchi M, Ohuchi R, Mifune K. Demonstration of hemolytic and fusion activities of influenza C virus. J Virol. 1982;42:1076–1079. doi: 10.1128/jvi.42.3.1076-1079.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pekosz A, Lamb R A. The CM2 protein of influenza C virus is an oligomeric integral membrane glycoprotein structurally analogous to influenza A virus M2 and influenza B virus NB proteins. Virology. 1997;237:439–451. doi: 10.1006/viro.1997.8788. [DOI] [PubMed] [Google Scholar]
- 17.Pekosz A, Lamb R A. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc Natl Acad Sci USA. 1998;95:13233–13238. doi: 10.1073/pnas.95.22.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petri T, Meier-Ewert H, Crumpton W M, Dimmock N J. RNA’S of influenza C virus strains. Arch Virol. 1979;61:239–243. doi: 10.1007/BF01318058. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer J B, Compans R W. Structure of the influenza C glycoprotein gene as determined from cloned DNA. Virus Res. 1984;1:281–296. doi: 10.1016/0168-1702(84)90017-0. [DOI] [PubMed] [Google Scholar]
- 20.Pleschka S, Klenk H D, Herrler G. The catalytic triad of the influenza C virus glycoprotein HEF esterase: characterization by site-directed mutagenesis and functional analysis. J Gen Virol. 1995;76:2529–2537. doi: 10.1099/0022-1317-76-10-2529. [DOI] [PubMed] [Google Scholar]
- 21.Racaniello V R, Palese P. Isolation of influenza C virus recombinants. J Virol. 1979;32:1006–1014. doi: 10.1128/jvi.32.3.1006-1014.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenthal P B, Zhang X, Formanowski F, Fitz W, Wong C-H, Meier-Ewert H, Skehel J J, Wiley D C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugawara K, Nishimura H, Kitame F, Nakamura K. Antigenic variation among human strains of influenza C virus detected with monoclonal antibodies to gp88 glycoprotein. Virus Res. 1986;6:27–32. doi: 10.1016/0168-1702(86)90054-7. [DOI] [PubMed] [Google Scholar]
- 24.Szepanski S, Gross H J, Brossmer R, Klenk H D, Herrler G. A single point mutation of the influenza C virus glycoprotein (HEF) changes the viral receptor-binding activity. Virology. 1992;188:85–92. doi: 10.1016/0042-6822(92)90737-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szepanski S, Veit M, Pleschka S, Klenk H D, Schmidt M F, Herrler G. Post-translational folding of the influenza C virus glycoprotein HEF: defective processing in cells expressing the cloned gene. J Gen Virol. 1994;75:1023–1030. doi: 10.1099/0022-1317-75-5-1023. [DOI] [PubMed] [Google Scholar]
- 26.Vlasak R, Krystal M, Nacht M, Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]