Abstract
Non-small cell lung cancers (NSCLCs) with activating mutations in the kinase domain of the epidermal growth factor receptor (EGFR) demonstrate dramatic, but transient, responses to the reversible tyrosine kinase inhibitors gefitinib (Iressa) and erlotinib (Tarceva). Some recurrent tumors have a common secondary mutation in the EGFR kinase domain, T790M, conferring drug resistance, but in other cases the mechanism underlying acquired resistance is unknown. In studying multiple sites of recurrent NSCLCs, we detected T790M in only a small percentage of tumor cells. To identify additional mechanisms of acquired resistance to gefitinib, we used NSCLC cells harboring an activating EGFR mutation to generate multiple resistant clones in vitro. These drug-resistant cells demonstrate continued dependence on EGFR and ERBB2 signaling for their viability and have not acquired secondary EGFR mutations. However, they display increased internalization of ligand-activated EGFR, consistent with altered receptor trafficking. Although gefitinib-resistant clones are cross-resistant to related anilinoquinazolines, they demonstrate sensitivity to a class of irreversible inhibitors of EGFR. These inhibitors also show effective inhibition of signaling by T790M-mutant EGFR and killing of NSCLC cells with the T790M mutation. Both mechanisms of gefitinib resistance are therefore circumvented by irreversible tyrosine kinase inhibitors. Our findings suggest that one of these, HKI-272, may prove highly effective in the treatment of EGFR-mutant NSCLCs, including tumors that have become resistant to gefitinib or erlotinib.
Keywords: drug resistance, molecular targeted therapy, non-small cell lung cancer, tyrosine kinase inhibitor
Gefitinib and erlotinib induce dramatic clinical responses in cases of non-small cell lung cancers (NSCLCs) harboring activating mutations in the EGF receptor (EGFR) (1–3), which is targeted by these competitive inhibitors of ATP binding (4, 5). The effectiveness of these tyrosine kinase inhibitors may result both from alterations in the ATP cleft associated with these mutations, which lead to enhanced inhibition of the mutant kinase by these drugs, and from biological dependence of these cancer cells on the increased survival signals transduced by the mutant receptors, a phenomenon described as “oncogene addiction” (6, 7).
Although therapeutic responses to both gefitinib and erlotinib can persist for as long as 2–3 years, the mean duration of response in most cases of NSCLC is only 6–8 months (8–10). The mechanisms underlying acquired drug resistance are not well understood. By analogy with imatinib (Gleevec), which inhibits the BCR-ABL kinase involved in chronic myeloid leukemias (CMLs), the C-KIT kinase implicated in gastrointestinal stromal tumors (GISTs), and the FIP1L1-PDGFR-α kinase in idiopathic hypereosinophilic syndrome (HES), secondary kinase domain mutations can potentially suppress drug binding (11–16). However, recurrent NSCLC is not readily biopsied; hence, only limited clinical specimens are available for analysis. Recently, a single secondary mutation, T790M, within the EGFR kinase domain has been reported in three of six cases with recurrent disease after gefitinib or erlotinib therapy (17, 18). Codon 315 of BCR-ABL, which is analogous to EGFR codon 790, is frequently mutated in imatinib-resistant CML (11, 12), and mutation of the corresponding residue in C-KIT (codon 670) and FIP1L1-PDGFR-α (codon 674) is associated with imatinib-resistant GIST and HES, respectively (15, 16). Early in vitro modeling of resistance to EGFR inhibitors indicated that mutation of codon 790 within the wild-type receptor would similarly suppress inhibition by an EGFR tyrosine kinase inhibitor (19). Recently, transfected EGFR proteins containing activating mutations together with the T790M substitution were shown to exhibit reduced inhibition by gefitinib and erlotinib (17, 18). Although the T790M mutation seems to contribute to acquired resistance in some cases of NSCLC, the mechanisms underlying treatment failure in cases lacking secondary EGFR mutations remain unexplained.
In contrast to the cytoplasmic kinase BCR-ABL, signaling by the membrane-bound EGFR involves a complex pathway of ligand binding, receptor homodimerization, and heterodimerization with ERBB2 and other family members, followed by internalization and recycling of the ligand-bound receptor or ubiquitin-mediated receptor degradation (20). Significant EGF-dependent signaling is thought to occur during the process of internalization, which is also associated with the dissociation of EGFR complexes at the low pH of intracellular vesicles. As such, multiple factors modulate the strength and quality of the signal transduced by the receptor, and alterations in EGFR trafficking have been closely linked with the regulation of EGF-dependent cellular responses (20).
Here, we show that even within recurrent gefitinib-resistant NSCLCs containing the secondary T790M EGFR mutation, this acquired mutation is only present in a subset of the resistant tumor cells. In an in vitro model of acquired gefitinib resistance, the T790M mutation is not observed, but increased EGFR internalization is correlated with drug resistance. Irreversible inhibitors, which covalently crosslink the receptor, are effective in cell lines with the T790M mutation and in cells with altered EGFR trafficking, raising the possibility that they may circumvent multiple mechanisms of acquired resistance to gefitinib and erlotinib.
Methods
Analysis of Recurrent NSCLC and Generation of Gefitinib-Resistant NCI-H1650 Cells. Clinical specimens of recurrent NSCLC were obtained at autopsy after appropriate consent. The entire kinase domain of EGFR was sequenced after analysis of uncloned PCR products. Multiple clones of exon 20 were sequenced to examine codon 790. Mutational analysis of EGFR (exons 1–28), ERBB2 (exons 1–24), PTEN (exons 1–9), Kras (codons 12, 13, and 61), and p53 (exons 5–8) in gefitinib-resistant clones as well as the parental NCI-H1650 cell line was performed by automated sequencing of individual exons and flanking intronic sequence (PCR conditions available on request) with bidirectional sequencing by using dye terminator chemistry (bigdye version 1.1, Applied Biosystems). Sequencing reactions were run on an ABI3100 sequencer (Applied Biosystems), and electropherograms were analyzed by using sequence navigator and factura software (Applied Biosystems).
To generate resistant subclones of NCI-H1650 cells, these were treated with ethyl methane sulfonate (EMS; 600 μg/ml), allowed to recover for 72 h, and then seeded at a density of 6 × 104 cells per 10-cm2 dish in 20 μM gefitinib. Relative resistance of these cells to gefitinib, compared with the irreversible inhibitors, was achieved by seeding 5 × 104 cells in six-well plates in 5% FCS and 100 ng/ml EGF (Sigma), in the presence of varying concentrations of drugs, followed after 72 h by fixing cells with 4% formaldehyde, staining with 0.1% crystal violet, and quantifying cell mass by using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). For small interfering RNA (siRNA) knockdown experiments, cells were transfected with double-stranded RNA oligonucleotides targeting EGFR, ERBB2 (both SMARTpool from Dharmacon, Lafayette, CO), or nonspecific control (LRT1B), using X-treme GENE transfection reagent (Roche Applied Science). After 72 h, cells were stained with crystal violet and analyzed on the Odyssey Infrared scanner.
Immunoblotting and Signaling Studies. Inhibition of EGFR signaling by increasing concentrations of gefitinib or the irreversible inhibitors was determined by seeding 9 × 104 cells in 24-well plates, adding the drugs to medium containing 5% FCS for 15 min, followed by a 2-h pulse with 100 ng/ml EGF, and harvesting of lysates. Lysates were prepared in 2× gel loading buffer, sonicated, boiled, and then separated by 10% SDS/PAGE, followed by electrotransfer to polyvinylidene fluoride (PVDF) membranes, and immunoblotting. Antibodies used were phospho-EGFR Y1068 and phospho-mitogen-activated protein kinase (MAPK) (Cell Signaling Technology, Beverly, MA), phospho-AKT (BioSource International, Camarillo, CA), and total EGFR, MAPK, AKT, and tubulin (Santa Cruz Biotechnology).
Analysis of EGFR Internalization. To demonstrate internalization of EGFR by fluorescence microscopy, cells were grown on coverslips and incubated with 1 ng/ml recombinant human (rh) EGF (Molecular Probes, Eugene, OR) for various intervals before fixing in 4% paraformaldehyde for 10 min. Coverslips were washed in PBS and mounted with ProLong Gold antifade reagent (Molecular Probes). To quantify EGFR internalization by cell surface biotinylation, cells were grown to confluency, pretreated with cyclohexamide, incubated on ice for 1 h with 1.5 mg/ml sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin; Pierce), and washed with blocking buffer (50 nM NH4CL/1 mM MgCl2/0.1 mM CaCl2 in PBS) to quench free sulfo-NHS-SS-biotin, followed by several further washes with PBS. The cells were then incubated in culture medium at 37°C for various intervals to allow internalization of the biotinylated molecules, washed twice for 20 min in a glutathione solution (50 mM glutathione/75 mM NaCl/75 mM NaOH/1% BSA) on ice to strip all of the biotinyl groups from the cell surface, and then scraped and lysed in 500 μM radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris·HCl, pH 7.4, with 150 mM NaCL/0.1% SDS/1% Triton X-100) supplemented with NaF, Na-orthovanadate, and protease inhibitors. Cell extracts were centrifuged, and the supernatants were incubated with streptavidin beads (Sigma) to collect the biotinylated proteins, which were then analyzed by SDS/PAGE and immunoblotting with anti-EGFR antibody (SC-03, Santa Cruz Biotechnology) or antibody against transferrin receptor (Santa Cruz Biotechnology).
Results and Discussion
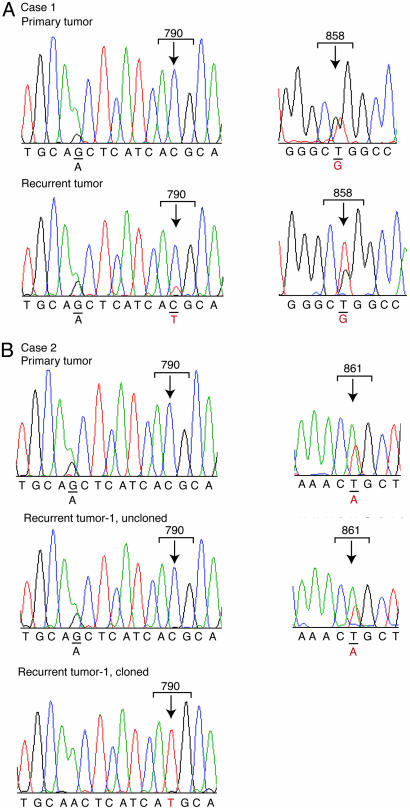
Analysis of Recurrent Lung Cancers with Acquired Resistance to Gefitinib. Recurrent gefitinib-resistant NSCLC developed in two patients whose tumors had harbored an activating mutation of the EGFR kinase at the time of diagnosis and who had shown a dramatic initial clinical response to the drug (1). In both cases, progressive metastatic disease in the liver led to the patients' demises, 1–2 years after initiation of treatment. In case 1, analysis of the major liver metastasis obtained at the time of autopsy indicated persistence of the sensitizing EGFR mutation (L858R), as well as the presence of a newly acquired T790M mutation (Fig. 1A). Interestingly, analysis of uncloned PCR products showed the initial L858R mutation to be present at an abundance consistent with a heterozygous mutation that is present in all tumor cells, whereas the secondary T790M mutation was seen at approximately one-fifth the abundance of the corresponding wild-type allele. Thus, this resistance-associated mutation seems to be present in only a fraction of cells within the recurrent tumor.
Fig. 1.
EGFR sequence analysis in recurrent metastatic lesions from two NSCLC patients with acquired gefitinib resistance. (A) Case 1. The T790M mutation in EGFR is present in a recurrent liver lesion after the development of clinical gefitinib resistance. (Left) The mutation was not detected in the primary lung lesion at the time of diagnosis. (Right) Both the primary lung tumor and the recurrent liver lesion harbor the L858R gefitinib-sensitizing mutation. Of note, the L858R mutation is present in the expected ratio for a heterozygous mutation in both primary and recurrent lesions, whereas T790M is detectable at low levels compared with the wild-type allele. A polymorphism (G/A) is shown in the same tracing to demonstrate equivalent representation of the two alleles in the uncloned PCR product. (B) Case 2. The T790M mutation is present within a small minority of gefitinib-resistant cells. (Left) The T790M mutation was undetectable either in the lung primary tumor or in eight recurrent liver lesions from this case by sequencing uncloned PCR products. Heterozygosity at an adjacent polymorphism (G/A) confirms amplification of both EGFR alleles from these specimens. The heterozygous gefitinib-sensitizing mutation, L861Q, was detected at the expected ratio within the primary lung tumor as well as each of the eight recurrent liver lesions.
Case 2 involved eight distinct recurrent metastases in the liver after the failure of gefitinib therapy. In all of these independent lesions, the sensitizing L861Q EGFR mutation was present at the expected ratio for a heterozygous mutation. No secondary EGFR mutation was detectable by analysis of uncloned PCR products from any of these metastases. However, after subcloning of the PCR products, the T790M mutation was found to be present at very low frequency in two of the four metastatic tumors analyzed (T790M, 2 of 50 clones sequenced from lesion 1 and 1 of 56 from lesion 2), but not from two other recurrent metastases (0 of 55 clones from lesion 3 and 0 of 59 from lesion 4), or the primary tumor (0 of 75 clones) (Fig. 1B and Table 1). Taken together, these results are consistent with previous reports that the T790M mutation is present in some, but not all, cases of acquired gefitinib resistance (three of seven tumors; see refs. 17, 18, and 21). Furthermore, as previously noted (18), even in some cases with this resistance-associated mutation, it seems to be present in only a small fraction of tumor cells within a recurrent lesion. These observations suggest that additional mechanisms of resistance are involved in cases without a secondary EGFR mutation and that such mechanisms coexist with the T790M mutation in other cases.
Table 1. Presence of EGFR T790M mutation at very low frequency in recurrent tumors from case 2.
| No. of clones
|
||
|---|---|---|
| Tumor | T790M mutant | Wild type |
| Primary | 0 | 75 |
| Recurrent 1 | 2 | 48 |
| Recurrent 2 | 1 | 55 |
| Recurrent 3 | 0 | 55 |
| Recurrent 4 | 0 | 59 |
Sequencing of large numbers of cloned PCR products revealed that a minority of alleles within two of four liver lesions contain the T790M mutation.
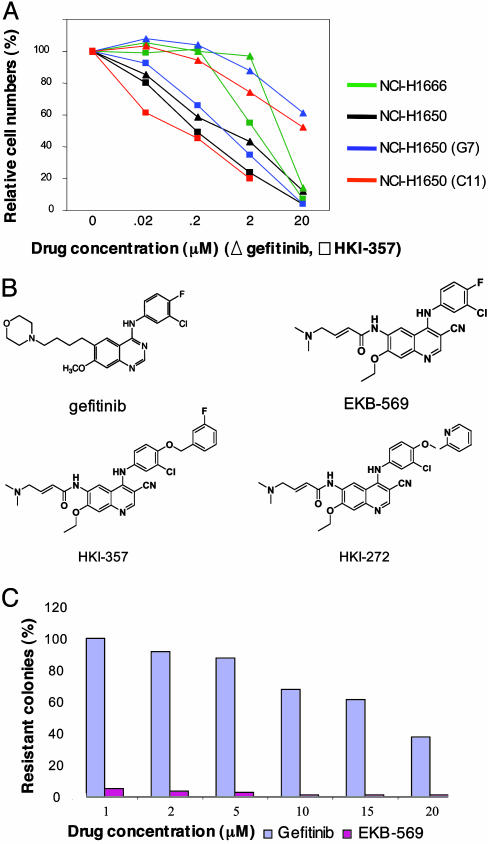
Generation of Gefitinib-Resistant Cell Lines with Susceptibility to Irreversible Inhibitors. Given the excellent correlation between the clinical responsiveness of EGFR-mutant NSCLC and the enhanced gefitinib-sensitivity of NSCLC cell lines with these mutations (2, 6, 22, 23), and the limited availability of clinical specimens from relapsing patients, we modeled gefitinib resistance in vitro. We cultured the bronchoalveolar cancer cell line NCI-H1650, which has an in-frame deletion of the EGFR kinase (delE746-A750), in 20 μM gefitinib, either with or without prior exposure to the mutagen ethyl methane sulfonate. This cell line exhibits 100-fold increased sensitivity to gefitinib, compared with some NSCLC lines expressing wild-type EGFR (6). Whereas the vast majority of these cells are efficiently killed by 20 μM gefitinib, drug-resistant colonies were readily observed at a frequency of ≈10–5, irrespective of mutagen treatment. Forty-nine independent drug-resistant clones were isolated, showing an average 50-fold decrease in gefitinib sensitivity (Fig. 2A). All of these showed persistence of the sensitizing mutation without altered expression of EGFR, and none had acquired a secondary EGFR mutation or new mutations in ERBB2, p53, Kras, or PTEN (data not shown). Gefitinib-resistant clones demonstrated comparable resistance to related inhibitors of the anilinoquinazoline class (data not shown). Remarkably, however, they displayed persistent sensitivity to three inhibitors of the ERBB family (Fig. 2A and data not shown): HKI-272 (24) and HKI-357 (compound 7f in ref. 25), which are dual inhibitors of EGFR and ERBB2 (IC50 values of 92 and 34 nM, respectively, for EGFR and 59 and 33 nM, respectively, for ERBB2), and EKB-569 (26), a selective inhibitor of EGFR (IC50 values of 39 nM for EGFR and 1.3 μM for ERBB2) (Wyeth) (Fig. 2B). All three drugs are irreversible inhibitors, most likely via a covalent bond with the cys773 residue within the EGFR catalytic domain or the cys805 of ERBB2. Like gefitinib, these compounds demonstrate increased killing of NSCLC cells harboring an EGFR mutation, compared with cells expressing wild-type receptor (Fig. 2A). However, in contrast to gefitinib, against which resistant clones are readily generated, even at high drug concentrations, we were unable to establish clones of cells that were resistant to the irreversible inhibitors at concentrations above 10 μM, even after ethyl methane sulfonate mutagenesis (Fig. 2C).
Fig. 2.
Acquired resistance to gefitinib in bronchoalveolar cancer cell lines and persistent sensitivity to irreversible ERBB family inhibitors. (A) Inhibition by tyrosine kinase inhibitors of proliferation of bronchoalveolar cancer cell lines with wild-type EGFR (NCI-H1666), the activating delE746-A750 mutation in EGFR (NCI-H1650), or two representative gefitinib-resistant subclones of NCI-H1650 (G7 and C11). The effect of the reversible inhibitor gefitinib is compared with that of the irreversible inhibitor HKI-357. Comparable results were observed with the other irreversible inhibitors. Cell numbers were measured by crystal violet staining, after culture in 5% FCS, with 100 ng/ml EGFR, at 72 h after exposure to indicated drug concentrations. Each data point represents the mean of four samples. (B) Chemical structure of gefitinib, a reversible inhibitor of EGFR; EKB-569, an irreversible inhibitor of EGFR; and HKI-272 and HKI-357, two irreversible dual inhibitors of EGFR and ERBB2. (C) Generation of drug-resistant NCI-H1650 cells after treatment with varying concentrations of gefitinib or the irreversible ERBB inhibitor EKB-569. Colonies were stained after 12 days in culture in the presence of inhibitors.
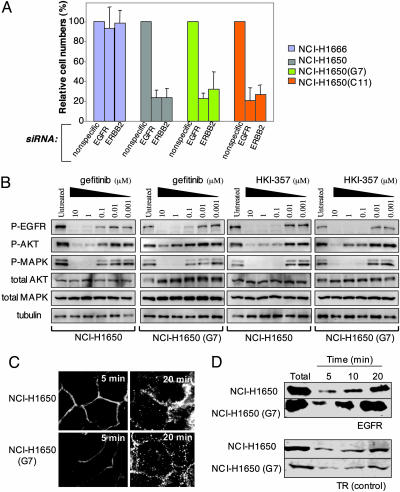
Dependence of Gefitinib-Resistant Cells on EGFR and ERBB2 Expression. To gain insight into the mechanisms underlying the acquisition of gefitinib resistance and the persistent sensitivity to the irreversible inhibitors, we first determined whether resistant cell lines remain dependent upon EGFR for their viability. We have previously shown that siRNA-mediated knockdown of EGFR triggers apoptosis in cells harboring mutant EGFRs, but not in those with wild-type alleles (6). Significantly, parental NCI-H1650 cells as well as their gefitinib-resistant derivatives showed comparable reduction in cell viability after transfection with siRNA targeting EGFR (Fig. 3A). Thus, acquisition of gefitinib-resistance does not involve EGFR-independent activation of downstream effectors. Because HKI-272 and HKI-357 target both EGFR and ERBB2, we also tested suppression of this related receptor. Knockdown of ERBB2 in NCI-H1650 and its gefitinib-resistant derivatives also caused loss of viability (Fig. 3A), suggesting a role for EGFR–ERBB2 heterodimers in transducing essential survival signals in tumor cells harboring EGFR mutations. Inhibition of EGFR alone by an irreversible inhibitor seems to be sufficient to induce apoptosis in gefitinib-resistant cells, as demonstrated by the effectiveness of EKB-569, which primarily targets EGFR (26). However, given the potentially complementary effects of targeting both EGFR and ERBB2 by using siRNA and the availability of irreversible inhibitors that target both of these family members, the potential benefit of dual inhibition warrants consideration.
Fig. 3.
Persistent dependence on EGFR and ERBB2 signaling in gefitinib-resistant cells, and altered receptor trafficking. (A) Cell viability after siRNA-mediated knockdown of EGFR and ERBB2 in bronchoalveolar cell lines with wild-type EGFR (NCI-H1666), compared with cells with the activating delE746-A750 mutation in EGFR (NCI-H1650) and two gefitinib-resistant derivatives (G7 and C11). Viable cells were counted 72 h after treatment with double-stranded RNA and are shown as a fraction relative to cells treated with nonspecific siRNA, with standard deviations based on triplicate samples. (B) Inhibition of EGFR autophosphorylation (Y1068) and phosphorylation of downstream effectors AKT and MAPK (ERK) in cells treated with increasing concentrations of gefitinib or the irreversible inhibitor HKI-357, followed by a 2-h pulse with EGF. The parental cell line NCI-H1650 is compared with a representative gefitinib-resistant line, G7. Total AKT and MAPK are shown as controls; tubulin is used as loading control for total EGFR levels, which are at the lower limit of detection in these cells. (C) Altered EGFR internalization in gefitinib-resistant NCI-H1650 (G7) cells, compared with the sensitive NCI-H1650 parental cell line. Rhodamine-tagged EGF is used to label EGFR at 5 and 20 min, after addition of ligand. The increased internalization of EGFR in NCI-H1650 (G7) cells is most evident at 20 min. (Zeiss microscope, ×63 magnification). (D) Immunoblotting of internalized EGFR from NCI-H1650 parental cells and the resistant derivative G7 after pulse labeling of cell surface proteins by biotinylation and chase over 20 min. The increased intracellular EGFR in NCI-H1650 (G7) cells is compared with the unaltered transferrin receptor (TR) internalization.
We compared the ability of gefitinib and irreversible ERBB family inhibitors to suppress signaling via downstream effectors of EGFR that mediate its proliferative and survival pathways. HKI-357 was 10-fold more effective than gefitinib in suppressing EGFR autophosphorylation (measured at residue Y1068), and AKT and MAPK phosphorylation in parental NCI-H1650 cells harboring the delE746-A750 EGFR mutation (Fig. 3B). In a gefitinib-resistant derivative, NCI-H1650(G7), gefitinib exhibited considerably reduced efficacy in suppressing AKT phosphorylation, a key EGFR signaling effector linked to gefitinib responsiveness (6), whereas HKI-357 demonstrated persistent activity (Fig. 3B).
Altered EGFR Internalization in Gefitinib-Resistant Clones. Given the absence of secondary mutations in EGFR and the persistent susceptibility of gefitinib-resistant cells to siRNA-mediated suppression of EGFR, we tested whether the mechanism underlying the differential inhibition of EGFR signaling in gefitinib-resistant cells by reversible and irreversible inhibitors might be correlated with alterations in receptor trafficking, a well documented modulator of EGFR-dependent signaling (20). Indeed, analysis of EGFR trafficking in NCI-H1650-derived resistant cells demonstrated a consistent increase in EGFR internalization, compared with the parental drug-sensitive cells, as measured both by internalization of fluorescein-labeled EGF (Fig. 3C) and quantitation of cytoplasmic biotinylated EGFR (Fig. 3D). No such effect was observed with the transferrin receptor, suggesting that this did not result from a generalized alteration in all receptor processing. Although further work is required to define the precise mechanism for this alteration in EGFR trafficking, a complex process in which numerous regulatory proteins have been implicated, these results suggest that gefitinib's ability to inhibit EGFR activation is compromised in these cells, whereas the action of the irreversible inhibitors are not detectably affected.
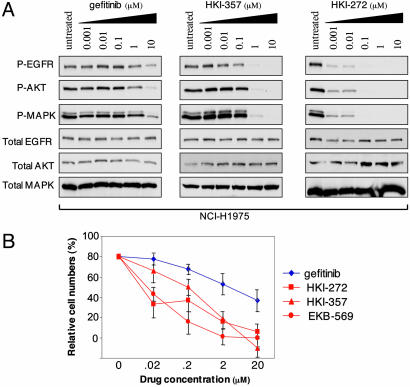
Inhibition of T790M EGFR Signaling and Enhanced Cell Killing by Irreversible Inhibitors. The enhanced suppression of EGFR signaling by irreversible ERBB inhibitors raised the possibility that these drugs may also exhibit persistent activity in the context of cells harboring the T790M secondary mutation in EGFR. We therefore tested the effect of these inhibitors on the NCI-H1975 bronchoalveolar cancer cell line, which harbors both L858R and T790M mutations in EGFR (18). Significantly, this cell line was derived from a patient that had not been treated with an EGFR inhibitor, indicating that this mutation is not uniquely associated with acquired drug resistance. Both HKI-357 and HKI-272 were considerably more effective than gefitinib in suppressing ligand-induced EGFR autophosphorylation and its downstream signaling, as determined by AKT and MAPK phosphorylation (Fig. 4A). Similarly, all three irreversible inhibitors suppressed proliferation in this cell line under conditions where it is resistant to gefitinib (Fig. 4B). Thus, irreversible ERBB inhibitors seem to be effective in cells harboring the T790M EGFR as well as in cells with altered trafficking of the wild-type receptor.
Fig. 4.
Effectiveness of irreversible ERBB inhibitors in suppressing the T790M EGFR mutant. (A) Comparison of gefitinib and two irreversible inhibitors, HKI-357 and HKI-272, in their ability to suppress EGFR autophosphorylation (Y1068) and phosphorylation of downstream effectors AKT and MAPK (ERK) in the NCI-H1975 bronchoalveolar cell line, harboring both a sensitizing mutation (L858R) and the resistance-associated mutation (T790M). Total EGFR, AKT, and MAPK are shown as loading controls. (B) Suppression of proliferation in NCI-H1975 cells harboring the L858R and T790M mutations by the three irreversible ERBB family inhibitors, compared with gefitinib.
Conclusion
Our results confirm the report of T790M mutations in EGFR as secondary mutations that arise in previously sensitive NSCLCs harboring an activating mutation, associated with the emergence of acquired resistance (17, 18). However, this mutation is present only in a subset of cases, and even tumors that harbor the T790M mutation may contain only a small fraction of cells with this mutation. These observations imply that multiple resistance mechanisms can coexist in recurrent tumors after an initial response to gefitinib or similar reversible EGFR inhibitors. Moreover, these findings suggest that T790M-independent resistance mechanisms may be equally, if not more, effective than the T790M substitution itself in conferring drug resistance and may explain why recurrent tumors rarely exhibit clonality for T790M (17, 18). In vitro mechanisms of acquired gefitinib resistance do not involve secondary EGFR mutations at a significant frequency, but instead are correlated with altered receptor trafficking. However, it should be noted that we have not examined EGFR trafficking in all of the resistant clones that we established in vitro, and it remains possible that additional mechanisms may contribute to gefitinib resistance in some of the clones. Nonetheless, virtually all gefitinib-resistant clones exhibited comparable sensitivity to the irreversible ERBB inhibitors.
Our results indicate striking differences between competitive EGFR inhibitors such as gefitinib, whose effectiveness is limited by the rapid development of drug resistance in vitro, and irreversible inhibitors, to which acquired resistance appears to be rare (Fig. 2C). We speculate that increased internalization of ligand-bound EGFR in resistant cells may be linked to dissociation of the gefitinib–EGFR complex at the low pH of intracellular vesicles. In contrast, irreversible cross-linking of the receptor would be unaffected by such alterations in receptor trafficking. Acquired resistance to gefitinib is stably maintained after passage of cells for up to 20 generations in the absence of drug, suggesting that genetic or epigenetic alterations in genes that modulate EGFR turnover may underlie this phenomenon. Because receptor trafficking cannot be readily studied by using available clinical specimens, identification of such genomic alterations may be required before clinical correlations are possible. Nonetheless, such a mechanism may contribute to in vivo acquired gefitinib-resistance in patients with recurrent disease who do not have secondary mutations in EGFR.
Of the three irreversible ERBB inhibitors described here, both HKI-272 and EKB-569 have been subjected to phase I clinical testing. Clinical studies in EGFR mutant NSCLC are required to determine whether these drugs have persistent activity in NSCLC cases that have become refractory to gefitinib or erlotinib and whether they potentially induce longer lasting responses in untreated patients. If validated in such clinical trials, the design of additional irreversible tyrosine kinase inhibitors targeting cancer-associated receptor tyrosine kinases may warrant consideration.
Acknowledgments
We thank Drs. Lee Greenberger, Bruce Chabner, and David Louis for helpful comments. This work was supported by National Institutes of Health Grant PO1 95281 (to D.A.H. and D.W.B.), the Doris Duke Charitable Foundation (to D.A.H.), the Sandler Family Foundation (to D.A.H. and D.W.B.), the V Foundation Award (to J.S.), the Samuel Waxman Cancer Research Foundation (to J.S.), the Saltonstall Scholarship (to J.S.), the Cole-Angelus Fund, the Romaine Fund, and Sue's Fund for lung cancer research (to T.J.L.).
Author contributions: E.L.K., R.S., D.W.B., N.G.-H., K.J.I., J.S., and D.A.H. designed research; E.L.K., R.S., D.W.B., N.G.-H., R.A.O., B.W.B., P.L.H., D.R.D., and S.V.S. performed research; E.L.K., R.S., D.W.B., N.G.-H., R.A.O., B.W.B., P.L.H., D.R.D., P.F., T.J.L., S.K.R., J.P.M., A.W., and S.V.S. contributed new reagents/analytic tools; E.L.K., R.S., D.W.B., N.G.-H., R.A.O., B.W.B., P.L.H., D.R.D., P.F., T.J.L., S.K.R., J.P.M., A.W., S.V.S., K.J.I., J.S., and D.A.H. analyzed data; and E.L.K., R.S., D.W.B., N.G.-H., J.S., and D.A.H. wrote the paper.
Abbreviations: NSCLC, non-small cell lung cancer; EGFR, EGF receptor; siRNA, small interfering RNA; MAPK, mitogen-activated protein kinase.
Note. Irreversible ERBB inhibitors also seem to be effective in overcoming gefitinib resistance mediated by the T790M mutation, an effect that presumably results from the preservation of inhibitor binding despite alteration of this critical residue. While this work was in progress, another irreversible inhibitor of EGFR [CL-387,785, Calbiochem (27)] was shown to inhibit the kinase activity of the T790M EGFR mutant (17). The effectiveness of CL-387,785 in the context of T790M was proposed to result from the absence of a chloride at position 3 of the aniline group, which is present in gefitinib and was postulated to interfere sterically with binding to the mutant methionine at codon 790. However, EKB-569, HKI-272, and HKI-357 all have chloride moieties at that position in the aniline ring, suggesting that their shared ability to bind irreversibly to EGFR is likely to explain their effectiveness, rather than the absence of a specific steric interaction with T790M (24–26). Thus, these irreversible inhibitors may prove to be broadly effective in circumventing a variety of resistance mechanisms, in addition to the T790M mutation.
References
- 1.Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., et al. (2004) N. Engl. J. Med. 350, 2129–2139. [DOI] [PubMed] [Google Scholar]
- 2.Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman N., Boggon, T. J., et al. (2004) Science 304, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3.Pao, W., Miller, V., Zakowski, M., Doherty, J., Politi, K., Sarkaria, I., Singh, B., Rusch, V., Fulton, L., Mardis, E., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakeling, A. E., Guy, S. P., Woodburn, J. R., Ashton, S. E., Curry, B. J., Barker, A. J. & Gibson, K. H. (2002) Cancer Res. 62, 5749–5754. [PubMed] [Google Scholar]
- 5.Moyer, J. D., Barbacci, E. G., Iwata, K. K., Arnold, L., Boman, B., Cunningham, A., DiOrio, C., Doty, J., Morin, M. J., Moyer, M. P., et al. (1997) Cancer Res. 57, 4838–4848. [PubMed] [Google Scholar]
- 6.Sordella, R., Bell, D. W., Haber, D. A. & Settleman, J. (2004) Science 305, 1163–1167. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein, I. B. (2002) Science 297, 63–64. [DOI] [PubMed] [Google Scholar]
- 8.Fukuoka, M., Yano, S., Giaccone, G., Tamura, T., Nakagawa, K., Douillard, J., Nishiwaki, Y., Vansteenkiste, J., Kudoh, S., Rischin, D., et al. (2003) J. Clin. Oncol. 12, 2237–2246. [DOI] [PubMed] [Google Scholar]
- 9.Kris, M. G., Natale, R. B., Herbst, R. S., Lynch, T. J., Prager, D., Belani, C. P., Schiller, J. H., Kelly, K., Spiridonidis, H., Sandler, A., et al. (2003) J. Amer. Med. Assoc. 290, 2149–2158. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Soler, R., Chachoua, A., Hammond, L. A., Rowinsky, E. K., Huberman, M., Karp, D., Rigas, J., Clark, G. M., Santabarbara, P. & Bonomi, P. (2004) J. Clin. Oncol. 22, 3238–3247. [DOI] [PubMed] [Google Scholar]
- 11.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, N. & Sawyers, C. L. (2001) Science 293, 876–880. [DOI] [PubMed] [Google Scholar]
- 12.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117–125. [DOI] [PubMed] [Google Scholar]
- 13.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D. & Sawyers, C. L. (2004) Science 305, 399–401. [DOI] [PubMed] [Google Scholar]
- 14.Chen, L. L., Trent, J. C., Wu, E. F., Fuller, G. N., Ramdas, L., Zhang, W., Raymond, A. K., Prieto, V. G., Oyedeji, C. O., Hunt, K. K., et al. (2004) Cancer Res. 64, 5913–5919. [DOI] [PubMed] [Google Scholar]
- 15.Tamborini, E., Bonadiman, L., Greco, A., Albertini, V., Negri, T., Gronchi, A., Bertulli, R., Colecchia, M., Casali, P. G., Pierotti, M. A., et al. (2004) Gastroenterology 127, 294–299. [DOI] [PubMed] [Google Scholar]
- 16.Cools, J., DeAngelo, D. J., Gotlib, J., Stover, E. H., Legare, R. D., Cortes, J., Kutok, J., Clark, J., Galinsky, I., Griffin, J. D., et al. (2003) New Engl. J. Med. 348, 1201–1214. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, S., Boggon, T. J., Dayaram, T., Janne, P. A., Kocher, O., Meyerson, M., Johnson, B. E., Eck, M. J., Tenen, D. G. & Halmos, B. (2005) N. Engl. J. Med. 352, 786–792. [DOI] [PubMed] [Google Scholar]
- 18.Pao, W., Miller, V. A., Politi, K. A., Riely, G. J., Somwar, R., Zakowski, M. F., Kris, M. & Varmus, H. (2005) PLoS Med. 2, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blencke, S., Ullrich, A. & Daub, H. (2003) J. Biol. Chem. 278, 15435–15440. [DOI] [PubMed] [Google Scholar]
- 20.Wiley, H. S. (2003) Exp. Cell Res. 284, 78–88. [DOI] [PubMed] [Google Scholar]
- 21.Amann, J., Kalyankrishna, S., Massion, P. P., Ohm, J. E., Girard, L., Shigematsu, H., Peyton, M., Juroske, D., Huang, Y., Stuart Salmon, J., et al. (2005) Cancer Res. 65, 226–235. [PubMed] [Google Scholar]
- 22.Tracy S., Mukohara, T., Hansen, M., Meyerson, M., Johnson, B. E. & Janne, P. A. (2004) Cancer Res. 64, 7241–7244. [DOI] [PubMed] [Google Scholar]
- 23.Koizumi, F., Shimoyama, T., Taguchi, F., Saijo, N. & Nishio, K. (March 10, 2005) Int. J. Cancer, 10.1002/ijc.20985. [DOI] [PubMed]
- 24.Rabindran, S. K., Discafani, C. M., Rosfjord, E. C., Baxter, M., Floyd, M. B., Golas, J., Hallett, W. A., Johnson, B. D., Nilakantan, R., Overbeek, E., et al. (2004) Cancer Res. 64, 3958–3965. [DOI] [PubMed] [Google Scholar]
- 25.Tsou, H., Overbeek-Klumpers, E. G., Hallett, W. A., Reich, M. F., Floyd, M. B., Johnson, B. D., Michalak, R. S., Nilakantan, R., Discafani, C., Golas, J., et al. (2005) J. Med. Chem. 48, 1107–1131. [DOI] [PubMed] [Google Scholar]
- 26.Torrance, C. J., Jackson, P. E., Montgomery, E., Kinzler, K. W., Vogelstein, B., Wissner, A., Nunes, M., Frost, P. & Discafani, C. M. (2000) Nat. Med. 6, 1024–1028. [DOI] [PubMed] [Google Scholar]
- 27.Discafani, C. M., Carroll, M. L., Floyd, M. B., Jr., Hollander, I. J., Husain, Z., Johnson, B. D., Kitchen, D., May, M. K., Malo, M. S., Minnick, A. A., Jr., et al. (1999) Biochem. Pharmacol. 57, 917–925. [DOI] [PubMed] [Google Scholar]