Abstract
Mitapivat, a pyruvate kinase activator, shows great potential as a sickle cell disease (SCD)-modifying therapy. The safety and efficacy of mitapivat as a long-term maintenance therapy are currently being evaluated in two open-label studies. Here we applied a comprehensive multi-omics approach to investigate the impact of activating pyruvate kinase on red blood cells (RBC) from 15 SCD patients. HbSS patients were enrolled in one of the open-label, extended studies (NCT04610866). Leukodepleted RBC obtained from fresh whole blood at baseline, prior to drug initiation, and at longitudinal timepoints over the course of the study were processed for multi-omics through a stepwise extraction of metabolites, lipids and proteins. Mitapivat therapy had significant effects on the metabolome, lipidome and proteome of SCD RBC. Mitapivat decreased 2,3-diphosphoglycerate levels, increased adenosine triphosphate levels, and improved hematologic and sickling parameters in patients with SCD. Agreement between omics measurements and clinical measurements confirmed the specificity of mitapivat on targeting late glycolysis, with glycolytic metabolites ranking as the top correlates to parameters of hemoglobin S oxygen affinity (p50) and sickling kinetics (t50) during treatment. Mitapivat markedly reduced levels of proteins of mitochondrial origin within 2 weeks of initiation of treatment, with minimal changes in reticulocyte counts. In the first 6 months of treatment there were also transient elevations of lysophosphatidylcholines and oxylipins with depletion of free fatty acids, suggestive of an effect on membrane lipid remodeling. Multi-omics analysis of RBC identified benefits for glycolysis, as well as activation of the Lands cycle.
Introduction
In red blood cells (RBC), the small molecule metabolite 2,3-diphosphoglycerate (DPG) stabilizes the deoxy conformation of hemoglobin to promote oxygen off-loading and counteract hypoxia.1 In sickle cell disease (SCD), mutation of glutamate 6 to valine in the b subunit of hemoglobin favors the polymerization of the sickle hemoglobin (HbS) upon deoxygenation.2 Therefore, elevation of 2,3-DPG in SCD is deleterious because it promotes polymerization by stabilizing HbS fibers. High levels of DPG also promote HbS polymerization by decreasing intracellular pH.3,4 DPG is an intermediate metabolite in the Rapoport-Luebering shunt off the glycolytic pathway; in the Embden-Meyerhof-Parnas glycolytic pathway, pyruvate kinase (PK) is a rate-limiting enzyme that catalyzes the second adenosine triphosphate (ATP)-generating step, in which phosphoenolpyruvate is converted to pyruvate.5 Therapeutic enhancement of the endogenous RBC PK (PKR) activity should increase the glycolytic flux, therefore leading to increases of ATP concomitantly with decreases in DPG, both of which have anti-sickling effects. Intracellular ATP is essential for the maintenance of RBC hydration and membrane integrity, which impact the pathophysiology of SCD.6
Mitapivat (AG-348, Agios Pharmaceuticals Inc, Cambridge, MA, USA) is a first-in-class, oral, allosteric activator of PK that was originally developed for treating patients with inherited PK deficiency caused by mutations in the PKLR gene. Mitapivat is approved in the USA by the Food and Drug Administration for the treatment of hemolytic anemia in adults with PK deficiency, and in the European Union by the European Medicines Agency and in Great Britain by the Medicines and Healthcare Products Regulatory Agency for the treatment of PK deficiency in adults. Its ability to enhance the activity of wild-type PK subsequently led to clinical trials of mitapivat in other hemolytic anemias, including thalassemia and SCD. Indeed, proof-of-concept for activating PK as a therapeutic approach was established in two independent studies of mitapivat, a phase I, open-label, multiple dose ascending study7 (NCT04000165) and a phase II, open-label study8 (www.trialregister.nl NL8517). In both studies, mitapivat improved hematologic parameters, increased ATP and decreased DPG levels with decreased sickling.7, 8 The safety and efficacy of mitapivat as a long-term maintenance therapy for patients with SCD are currently being evaluated in both studies. In the present study, we apply a comprehensive multi-omics approach9-11 to investigate the impact of activating PK on RBC from SCD patients on mitapivat therapy in the NCT04610866 extended study. The rationale for these omics analyses was to test the metabolic effects of mitapivat on late glycolysis and other pathways, including ATP synthesis, and redox status of the sickle RBC cytosol and membrane (lipidome). At the same time, proteomics analyses afforded the opportunity to monitor changes in PK levels, while also assessing the impact on the proteome and (ATP-dependent) phosphoproteome as a whole.
Methods
Study design and preparation of blood samples
This study evaluated one of the exploratory endpoints in an open label phase I/II study (NCT04610866), i.e., the longterm safety and tolerability of mitapivat. The study was approved by the National Heart, Lung, and Blood Institute Institutional Review Board and was performed in accordance with the Declaration of Helsinki.12 Blood samples for ex-vivo studies were obtained from 15 patients with HbSS enrolled in the study. All the patients were adult (age >18 years) with confirmed SCD (HbSS) and a baseline hemoglobin between 7.1-10.5 g/dL, with no recent blood transfusions, erythropoietin therapy, or changes in SCD-specific therapies including hydroxyurea and L-glutamine.13 All patients started mitapivat at a dose of 50 mg twice daily, escalating after 4 weeks to 100 mg twice daily; dose adjustments were performed for reasons of safety and tolerability, as per the Principal Investigator’s discretion. At the time of data cutoff (March 23, 2023), all 15 patients had completed the core period of 24 weeks (visit 6, V6), 14 patients had completed 48 weeks (V8), ten patients had completed 72 weeks (V10) and six patients had completed 92 weeks (V12). RBC obtained from fresh whole blood in EDTA at baseline (V1, prior to drug initiation) and longitudinal timepoints were collected over the course of the study. After centrifuging 6 mL of whole blood at 800 g for 10 minutes at room temperature, the plasma was removed and the RBC pellets were resuspended by adding 3 mL of phosphate-buffered saline. To obtain leukodepleted RBC, the resuspended RBC were subjected to a leukodepletion process using a NEO High-Efficiency Leukocyte Reduction Filter for RBC (Haemonetics, PA, USA). Samples were flash-frozen in ethanol and dry ice, and kept frozen at -80°C until analysis. PKR activity was measured as described elsewhere.14,15 In total, 150 (6x12, 4x10, 4x8 and 1x6) timepoint samples were analyzed (Figure 1A). Sickling kinetics were measured by counting the fraction of sickled red cells as a function of time in a 384-well plate using a machine learning method while slowly deoxygenating cells with nitrogen to 5% oxygen in the oxygen pressure- and temperature-controlled humidified chamber of a Biotek “Lionheart FX” automated microscope system (Agilent Technologies).16 The t50 is the time at which 50% of the cells are sickled. Oxygen dissociation curves were measured with a Hemox Analyzer (TCS Scientific Corp, PA, USA). Briefly PKR activity was measured by a coupled enzyme system with lactate dehydrogenase (LDH) in which the pyruvate produced by PKR was reduced to lactate with the concomitant oxidation of NADH to NAD. The progress of the reaction was followed by a change in the oxidation state of the cofactor spectrophotometrically at 340 nm. PKR protein levels were determined by Mesoscale Assay (MesoScale Discovery) goat anti-PKLR antibody (Aviva) and mouse anti-PKLR antibody (Abcam). SULFO-TAG goat anti-mouse (Mesoscale Discovery) was used as the detection antibody.
Omics analyses
The omics methods and statistical analyses are reported extensively in the Online Supplementary Methods - Extended. Metabolomics, lipidomics and proteomics analyses were performed as previously described.17-19 The statistical analysis was conducted using MetaboAnalyst v 5.0 and RStudio (v.4.2.3). Biorender (https://www.biorender.com/) was used to generate summary vignettes.
Results
Mitapivat had significant effects on the sickle red blood cell metabolome, lipidome and proteome
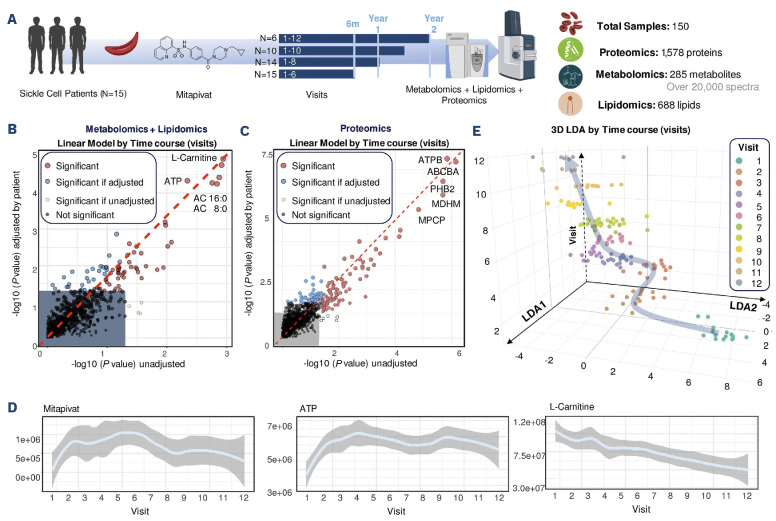
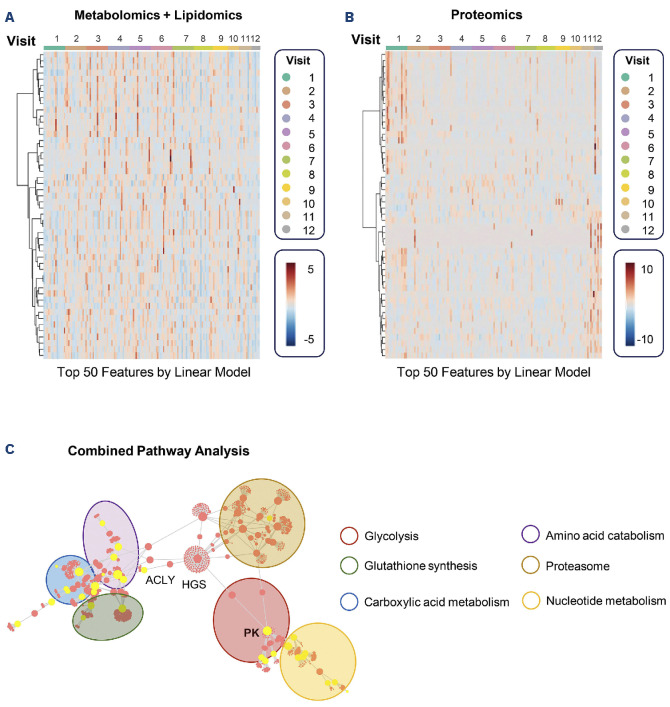
We performed two separate analyses of the data collected on the longitudinal samples. We first analyzed all 150 samples available from all visit timepoints 1-12 (V1 to V12) - a breakdown of biological replicates available for each timepoint is shown in Figure 1A. We then analyzed 90 samples for all 15 patients up to V6 (Figure 1A). All raw omics data and elaborations are provided in Online Supplementary Table S1, including complete blood counts from the patients at the timepoints analyzed in the study. Unsupervised analyses of multi-omics data were performed via repeated measure analysis of variance (ANOVA) and linear models of combined metabolomics and lipidomics data (Figure 1B), and proteomics data (Figure 1C), respectively. These analyses identified molecules associated with mitapivat treatment, either when testing for unadjusted variables, or upon adjustments for confounders such as patient-specific responses. Mitapivat levels were detected via mass spectrometry in the leukodepleted RBC, suggesting successful drug delivery (Figure 1D). Of note, ATP and L-carnitine levels were identified as the top two metabolites with the strongest positive and negative responses, respectively, across all patients throughout the whole duration of the study (Figure 1D). First, we performed supervised analysis of combined multi-omics data via linear discriminant analysis (LDA). Figure 1E shows the results were plotted based on the top two major components (LDA1 and LDA2 – x and y, respectively), while discriminating the samples across visits along the z axis. Plotting the same results using LDA3 as a sample clustering factor for the z axis revealed patient-specific responses to the treatment, with a confounded, yet still observable clustering of the samples by visit number (Online Supplementary Figure S1A). This patient-specific heterogeneity can be at least in part explained by the heterogeneity of mitapivat levels, in accordance with the design of the clinical protocol, as detected by mass spectrometry (Online Supplementary Figure S1B). Overall, the temporal trends of omics responses to mitapivat treatment across visits was evident, as highlighted by heatmap representations, especially when focusing on the top 50 metabolites/lipids (Figure 2A) or proteins (Figure 2B) by LDA (Online Supplementary Table S1). Similar results were obtained by time-series ANOVA, when focusing on the patients for whom analyses at all time points were available. Such trends are highlighted by line plot representations of selected top responding omics results (Online Supplementary Figure S1C) and are further illustrated by the heatmap representation of time series measurements across all samples at all visits (without the filter for the top significant features (Online Supplementary Figure S1D).
Figure 1.
Alterations of the metabolome in sickle red blood cells from patients on treatment with mitapivat. (A) Overview of the clinical study. Fifteen sickle cell patients (SS genotype) were enrolled in this clinical trial, with all 15 patients being treated for 6 months, 14 for a whole year and six for up to 2 years (visit 12). Red blood cell (RBC) samples underwent multi-omics characterization. (B, C) Linear model analysis of metabolomics and lipidomics data (B) or proteomics data (C) identified molecules associated with the treatments, either unadjusted (x axis) or adjusted by patient-specific responses (y axis). Highlighted metabolites (B) or proteins (C) represent the variables with the highest weights across linear discriminant analysis 1 (LDA1). (D) Line plots of mitapivat, ATP and carnitine, the very drug being administered, along with the levels of the top metabolites affected by the treatment. In light blue, median metabolite levels across all samples, while range intervals are shown in light gray. Data are shown as peak area abundance (arbitrary unit on the y axis), while the x axis represents visits 1-12. (E) LDA identified two major components (LDA1 and LDA2 – x and y, respectively) discriminating samples across visits (z axis).
Pathway analyses of combined multi-omics data identified multiple sub-networks of metabolites/proteins involved in glycolysis (PKR – KPYR) representing one of the nodes with the highest betweenness centrality (Figure 2C). Additional pathways significantly affected by mitapivat included proteins of mitochondrial origin and carboxylic acids of the Krebs cycle, amino acid catabolism, especially glutaminolysis and glutathione synthesis and tryptophan/ kynurenine metabolism, nucleoside metabolism and proteasome components.
Figure 2.
Heatmap and network analysis of top metabolites and lipids or proteins affected by mitapivat treatment in sickle red blood cells. (A, B) The top 50 metabolites/lipids (A) and proteins (B) (based on linear discriminant analyses) affected by mitapivat treatment are shown as a function of time (visits). A full list of these features is provided in Online Supplementary Table S1. (C) Merged data from these analyses were uploaded to Omicsnet to perform combined pathway analyses.
Mitapivat treatment significantly affects the levels of mitochondrial proteins
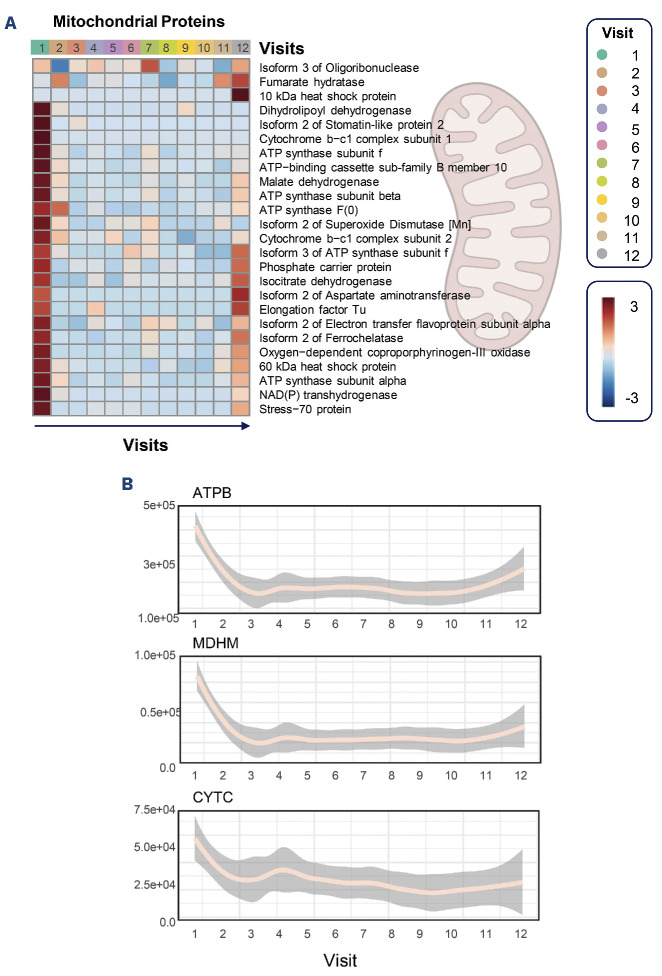
The most consistent and important finding in the proteome was the significant depletion of proteins of mitochondrial origin in leukodepleted RBC immediately after 2 weeks of mitapivat treatment at V2 (Figure 3A). Several components of the mitochondrial electron transport chain (e.g., ATPB) or other key cytosolic enzymes (e.g., mitochondrial malate dehydrogenase – MDHM) with roles in apoptosis (e.g., cytochrome c – CYTC) were rapidly depleted within 2 weeks of initiating mitapivat and their levels remained low in most patients for the whole duration of the study (Figure 3B). This effect appeared to be lost for a subset of mitochondrial proteins (especially components of complex V ATP synthase; Krebs cycle enzymes isocitrate dehydrogenase and fumarate hydratase, aspartate aminotransferase and mitochondrial elongation factors) by visit 12 (Figure 3A), but this could be due to the small sample size. Of note, these results could not be explained by changes in reticulocyte counts, since there were minimal changes in reticulocyte levels throughout the duration of the study (Online Supplementary Table S1; Online Supplementary Figure S2A). Correlation of omics data to complete blood counts did not highlight a significant association between proteins of mitochondrial origin and reticulocyte, platelet or white blood cell counts (Online Supplementary Figure S2B). Only the levels of MDH cytosolic (MDHC) and mitochondrial (MDHM) subunits were negatively and positively correlated with reticulocyte counts, though they ranked 315 and 317, respectively, in the list of omics correlates to this complete blood count parameter (Online Supplementary Figure S2B). Despite the drop in mitochondrial proteins immediately after the first visits, the levels of corresponding carboxylic acids were transiently elevated between visits 5-6 after 24 weeks of mitapivat treatment (Online Supplementary Figure S3A). As an internal validation of the omics results and caveat in the interpretation of the data, it is worth noting that thrombospondin 1 (TSP1) and platelet factor 4 (PLF4) ranked among the top three positive correlates to platelet counts (Online Supplementary Figure S2B).
Figure 3.
Impact of mitapivat treatment on red blood cell residual mitochondrial proteins. (A) Heatmap of median values of peak areas for proteins identified despite the gene ontology classification as proteins of mitochondrial origin or localization. (B) Selected line plots for the most significantly affected members of this group through the course of the study.
Mitapivat significantly promotes PKR activity and boosts DPG consumption and ATP production in human sickle red blood cells in vivo
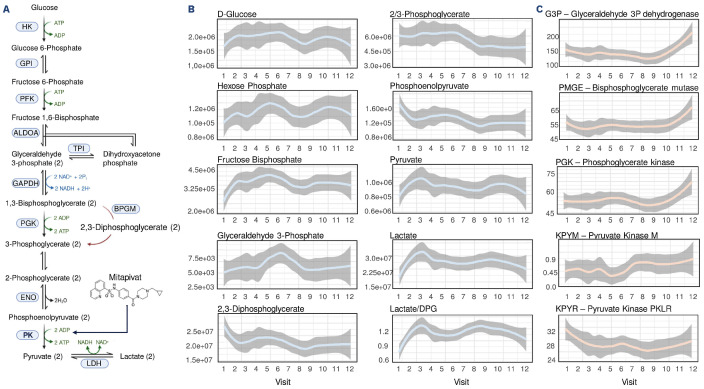
Mechanistically, mitapivat was designed to stabilize the active conformation of PKR (also known as KPYR), thus boosting late glycolysis, with concomitant consumption of DPG and generation of ATP (Figure 4A). Consistent with the proposed mechanism of mitapivat, our results confirmed the elevation of glycolytic metabolites upstream to DPG (hexose phosphate – isomers, fructose bisphosphate and glyceraldehyde 3-phosphate), concomitant with the reduction of DPG, phosphoglycerate (isomers) and phosphoenolpyruvate downstream to DPG. Of note, while our mass spectrometry-based approach does not distinguish between the 1,3- and 2,3-DPG isomers, the latter being by far the more abundant in mature RBC, the spectrometry results were in strong agreement with standard clinical measurements of 2,3-DPG via enzymatic assays (see the correlation analyses below). The end products of glycolysis in RBC, pyruvate and lactate were both significantly elevated after visits 2-3 (2-3 months’ interval after initiation of mitapivat treatment) (Figure 4B). Some of these changes were consistent with elevations in the levels of multiple glycolytic enzymes, especially after visit 10 (at 72 weeks of treatment), suggestive of additional phenomena favoring glycolysis beyond PKR activation. Of note, the levels of PKR (KPYR, Figure 4C) were not found to increase with treatment, with minor albeit significant decreases after the first visit and levels that remained constant after that timepoint. No significant changes were observed in other PK isoforms (KPYM – pyruvate kinase M), with elevations in other glycolytic enzymes (glyceraldehyde 3-phosphate dehydrogenase, bisphosphoglycerate mutase, phosphoglycerate kinase) (Figure 4C) but not hexokinase (HK1) or KPYR/HK1 ratios (Online Supplementary Figure S1C) after visit 11. Also of note, the mass spectrometry-based metabolic measurements of increased ATP (Figure 1D) and decreased DPG (Figure 4B) were independently validated via standard CLIA-certified clinical chemistry assays (see correlation analyses below). Elevation of lactate and consumption of DPG were also consistent with the enzymatic assay-based detection of PKR activity.
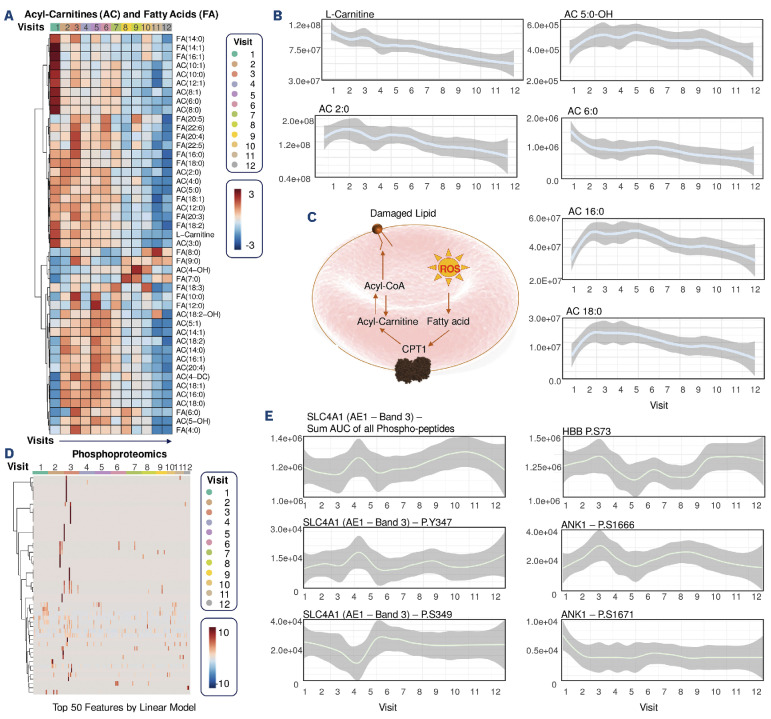
Beyond glycolysis: mitapivat treatment significantly reduces sickle red blood cell acyl-carnitines, induces transient increases in pentose phosphate pathway and amino acid levels
The metabolic pathways that were most significantly affected by mitapivat other than glycolysis were acyl-carnitine and free fatty acid metabolism (Figure 5). The levels of almost all free fatty acids decreased over the course of the treatment, while those of almost all acyl-carnitines transiently increased between visits 2 and 6, to decrease again afterwards, suggesting a stabilization of the Lands cycle pathway of damaged membrane lipid remodeling (Figure 5A-C). This consideration is in part supported by the lipidomics data, showing transient elevations of oxylipins and bile acids within the same time window (Online Supplementary Figure S3C). Of note, all very-long chain (especially C20 series) acyl-carnitines showed a strong positive correlation with RBC counts, suggesting an association between RBC numbers (but not size – mean corpuscular volume and hemoglobin concentration) with the acyl-carnitine pool throughout the study (Online Supplementary Figure S2B).
Consistent with a transient membrane remodeling phenotype at visits 2-3, altered phosphoproteomics profiles were observed – especially for the structural proteins band 3 (SLC4A1) and ankyrin (ANK1), as well as functional proteins (HBA1, HBG, HBB in the heatmap in Figure 5D, HBB P.S73 in Figure 5E), transporters (the monocarboxylate transporter SLC16A1; the excitatory amino acid transporter SLC1A3) and the modulatory proteins adducin 1 and 2 (Figure 5D). However, trends diverged for different residues, with transient elevation in phospho-Y347 of SLC4A1 corresponding to decreases in the levels of the neighboring phospho-S349 residue (Figure 5E). Similar observations held true for ANK1, with elevated phospho-S1666 accompanied by parallel decreases in phospho-S1671 (Figure 5E).
In the same time window (visit 2-6 – i.e., until 6 months of treatment), all nucleotides and, even more strikingly, all free amino acids (especially glutamine, arginine, methionine) increased before decreasing again to levels comparable to pre-treatment levels by visit 10 – suggestive perhaps of altered intake from the bloodstream or increased proteolysis to remove damaged protein components, consistent with the elevation in proteasomal proteins and the increased availability of ATP to fuel energy-dependent proteasomal activity (Online Supplementary Figure S3B and S3D). In this view, it is interesting to note that after visit 2 a remarkable elevation in glutaminolysis was accompanied by a delayed elevation in the levels of the key antioxidant reduced glutathione (GSH) only by visit 4 (Online Supplementary Figure S3E). Of note, kynurenine levels correlated with those of mitochondrial proteins, showing a depletion after visit 2, increases at visits 3 and 4, followed by a decrease and then an increase again at visit 10 in a subset of patients (driving increases of the median of the line plot in Online Supplementary Figure S3E).
Figure 4.
Impact of mitapivat on sickle red blood cell glycolysis. (A) Overview of glycolysis, showing the reaction catalyzed by red cell pyruvate kinase (PKR, Uniprot name KPYR) - the target of mitapivat. (B, C) Line plots for mass spectrometry-based measurements of peak areas of glycolytic metabolites and enzymes during the course of the study.
Figure 5.
Sickle red blood cell membrane remodeling after mitapivat treatment. (A, B) Acyl-carnitines and free fatty acids are significantly affected by mitapivat treatment. (A) Heatmap of the median values of each metabolite in these pathways across all subjects for up to 2 years of treatment (visit 12). (B) Highlight of acyl-carnitine depletion during the course of the treatment, especially free and saturated fatty acid-conjugated acyl-carnitines, (C) with an of the pathway overview. (D) Longitudinal phosphoproteomics analyses suggest a transient increase in protein phosphorylation at visits 2-3. (E) Significant changes in band 3 (SLC4A1), hemoglobin beta (HBB) and ankyrin (ANK1) were observed, with diverging trends at different S/T/Y residues).
Changes to the acyl-carnitine, free fatty acid and oxylipin pools were accompanied by widespread changes in the lipidome in almost all classes (Online Supplementary Figure S4A). Indeed, all classes but diacylglycerols increased transiently after visit 2 and decreased afterwards (similar to the trends described above for the amino acids). Of all these changes, the most notable was the transient elevation of lyosphospholipids (especially lysophosphatidylcholines– LPC) and sphingomyelins (especially SM) (Online Supplementary Figure S4).
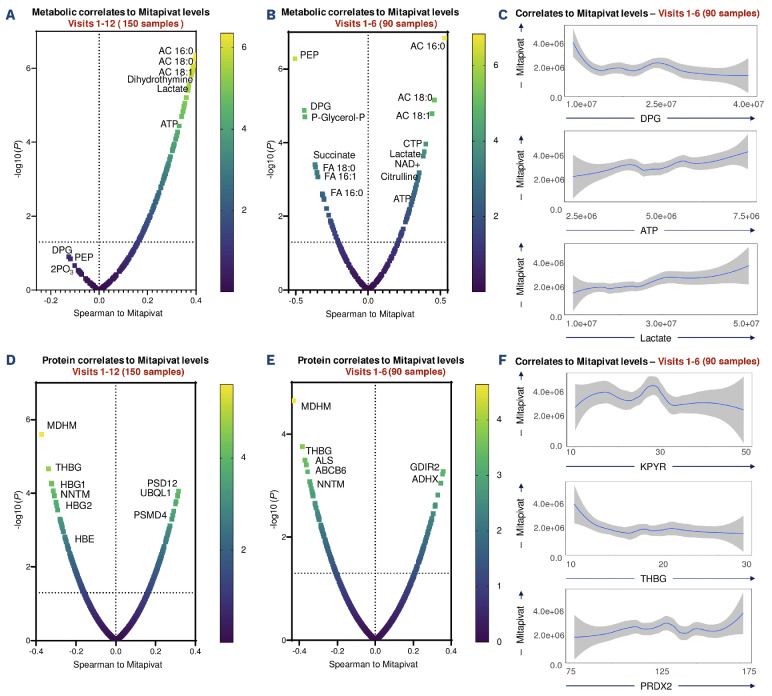
Omics correlates to mass spectrometry-detected mitapivat levels in HbSS red blood cells
After cataloging the changes in the metabolome, lipidome and proteome over the course of the study, we then correlated omics findings to functional measurements, based on two groups of data for all 15 patients: all timepoints V1-V12 (n=150 samples) and for timepoints V1 to V6 (n=90 samples). Mass spectrometry-based measurements of mitapivat levels in RBC confirmed a strong positive association between mitapivat levels and glycolysis, with lactate and ATP ranking among the most significant positive correlates, and DPG and phosphoenolpyruvate (PEP) as the top negative correlates, confirming the specificity of the treatment (Figure 6A-C). Mitapivat levels were positively associated with the most abundant acyl-carnitines, especially palmitoyl (AC 16:0), stearoyl (AC 18:0) and oleyl (AC 18:1), confirming a potential link between membrane lipid remodeling and mitapivat (Figure 6A, B). Similarly, mitapivat levels were negatively associated with free fatty acids and succinate. At the protein level, mitapivat was strongly negatively associated with multiple mitochondrial proteins, especially mitochondrial malate dehydrogenase (MDHM) (Figure 6D, E). On the other hand, we observed a positive association with key RBC antioxidant enzymes such as peroxiredoxin 2 (PRDX2) (Figure 6F).
Figure 6.
Omics correlates to mass spectrometry-detected mitapivat in sickle red blood cells. (A-F) Metabolite (A-C) and protein (D-F) correlates to mitapivat levels in red blood cells from patients with sickle cell disease during a 2-year period of treatment (visits 1-12; N=6) or just within the first 6 months (N=15). Volcano plots indicate Spearman correlations (x axis) and -log10 of related P values. (C and F) Line plots for selected metabolites (C) and proteins (F), with mitapivat levels (independent variable) shown on the y axis upon 90 degree rotation of the original graph for ease of visualization.
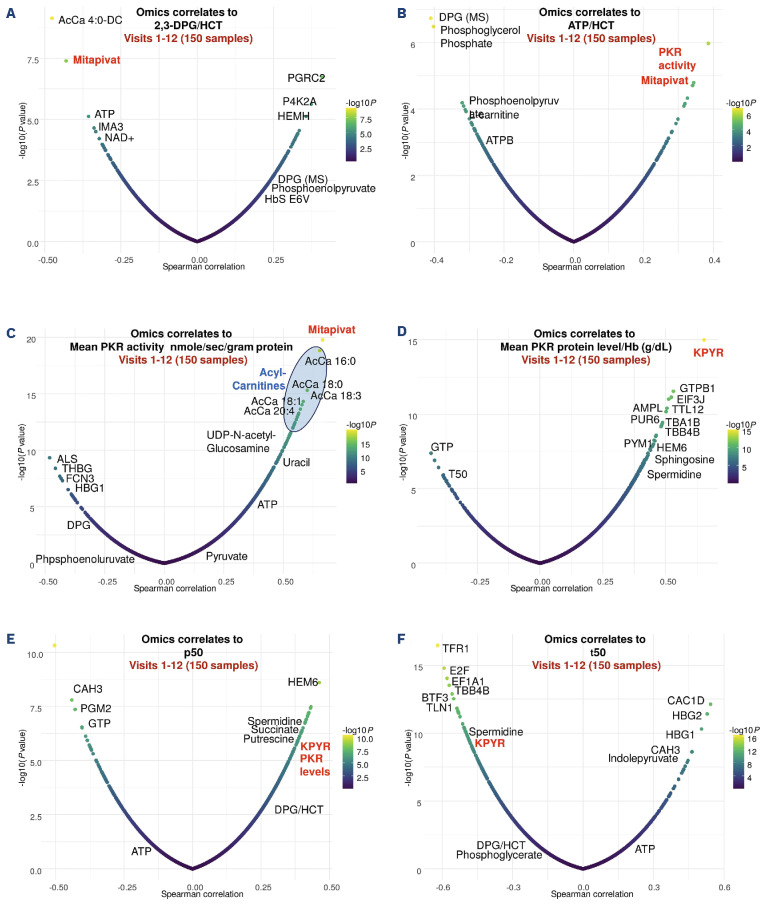
Unsupervised analyses confirm a significant association between mitapivat levels, PKR activity, DPG consumption and ATP levels
Clinical measurements of DPG corrected for hematocrit were strongly negatively associated with omics-measured mitapivat and ATP levels (Figure 7A) while ATP corrected for hematocrit was positively associated with omics-measured PKR activity and mitapivat levels (Figure 7B). Clinical measurements of PKR activity also positively correlated with elevations of almost all acyl-carnitines and mitapivat (Figure 7C). Serving as an internal control for the quality of the proteomics data, KPYR (PKR protein) levels measured by mass spectrometry were identified as the top overall positive correlate to PKR levels measured in the clinical arm of the study (Figure 7D). A decrease in p50 and an increase in t50 are both anti-sickling effects; increased HbS oxygen affinity (decreased p50) indicates less very low affinity polymer in red cells, while increased t50 indicates longer delay times, allowing more cells to escape the microcirculation before HbS polymerizes and makes RBC less flexible. Elevations in the levels and activity of KPYR mass-spectrometry measurements were positively associated with increases in p50 (along with succinate, as well as spermidine and putrescine) (Figure 7E), polyamines whose levels were positively correlated to reticulocyte counts (Online Supplementary Figure S2B) and negatively associated with sickling time (t50) (Figure 7F), suggestive of functional implications of these omics changes upon mitapivat treatment.
Discussion
Clinical studies have shown that RBC in patients with SCD have elevated levels of DPG and a functional deficiency of ATP, alterations which have also been observed in more recent metabolomic studies.9-11 Recently, we reported13 that mitapivat is well-tolerated in patients with SCD, having beneficial effects on several hematologic parameters: above all, the mean hemoglobin at 24 weeks increased significantly from baseline (mean increase: 1.38 g/dL, standard deviation: 0.88 g/dL; P<0.0001), with minor changes in fetal hemoglobin percentages.13 Here we describe results from the first multi-omics analysis of RBC from patients with SCD being treated with mitapivat therapy for up to 2 years. We confirm the specificity of mitapivat, a PK activator, using a combination of metabolomics, proteomics, lipidomics and correlation to clinical measurements of DPG and ATP levels, PK protein and activity levels, HbS oxygen affinity (p50), and sickling kinetics (t50). We leverage state-of-the-art high-throughput approaches,9,10,20-26 which we had recently used to investigate the metabolome of subjects with sickle cell trait and SCD,11,27,28 and patients with PK deficiency.7,29,30 Our measurements confirmed that the increased RBC ATP levels are sustained during a time window of 2 years in this study. Through a combination of multi-omics approaches we show that elevated ATP levels are indeed associated with elevation in pools of reduced glutathione (glutathione synthesis is an ATP-dependent process)31 and, above all, activation of the Lands cycle, a pathway that relies on acyl-carnitine pools to restore oxidatively damaged lipids.21 These findings are interesting in that they are aligned with similar observations from metabolomics studies on dried blood spots from SCD patients on mitapivat treatment from the SCORE trial.32 In this context, it is worth noting that our data suggest the presence of a transition period in which increased ATP availability is associated with depletion of free carnitine, transient increases in lysophospholipids and declines in free fatty acids, and alterations of ATP-dependent membrane protein phosphorylation33 profiles (e.g., SLC4A1, ANK1), suggestive of ongoing membrane lipid remodeling within the (<120-day) lifespan of the sickle RBC originally exposed to the drug at the beginning of the treatment. Our results also suggest that replenishing depleted carnitine pools via exogenous supplementation (dietary or – better delivered – intravenously administered) of L-carnitine34 could be a testable intervention to complement the mitapivat regimen under evaluation in this study, at least at initiation of treatment within the first 6 months. It appears plausible that the initial lipidomics profile is a readout of existing, circulating, irreversibly damaged sickle RBC that will need to be completely removed from the bloodstream before a full representation of the benefits of increased ATP for mature erythrocytes derived from de novo erythropoiesis when already exposed to mitapivat could be manifested. In this regard, it is worth noting that very-long chain acyl-carnitine levels were associated with total RBC counts, but not reticulocyte, platelet or white blood cell counts, nor to other RBC-related parameters (mean corpuscular volume, mean corpuscular hemoglobin, hematocrit, etc.). An alternative explanation consistent with the data is that an extension in RBC lifespans upon mitapivat treatment would account for the generally lower carnitine pools, since carnitine reservoirs are consumed as RBC age in the circulation.35
Figure 7.
Multi-omics findings correlate to functional readouts on red blood cells from patients with sickle cell disease on mitapivat treatment for 2 years. Correlates (Spearman) are shown for 2,3-DPG and ATP upon normalization to hematocrit (A, B), PKR activity and levels (C, D), p50 (E) and sickling time - t50 (F). Whole blood levels of ATP and 2,3-DPG were measured using a validated liquid chromatography tandem mass spectrometry assay with lower limit of quantitation at 50.0 μg/mL and converted to intracellular concentrations by dividing by the hematocrit (as a fraction).
One of the most striking and unexpected findings was the decrease in RBC mitochondrial proteins within 2 weeks of exposure to mitapivat treatment. Although some immature reticulocytes could have been retained in the leukodepleted RBC, we utilized the same technique for leukodepletion that was used in a previous study.36 Furthermore, the mitapivat treatment was not associated with significant changes in reticulocyte counts in this study, and mitochondrial protein levels were also not significantly associated with reticulocyte counts (with the exception of MDH isoforms). Of note, we previously showed that mature RBC accounted for the major source of mitochondrial DNA (detected by polymerase chain reaction-based assay) in the leukodepleted RBC. Multiple groups have reported that mature sickle red cells abnormally retain mitochondria that contribute to sickle inflammatory pathology in various ways – as a source of cell-free mitochondrial DNA that acts as a damage-associated molecular pattern and by generation of reactive oxygen species.36-38 Functionally, we had previously suggested a link between the accumulation of mitochondrial metabolites such as succinate and the stabilization of transcription factors such as the hypoxia-inducible factor 1α and its downstream targets including the pro-inflammatory cytokine interleukin 1β.39 Succinate levels are a predictor of cardiovascular function and exercise intolerance both in murine models and patients with SCD.26 Here it is interesting to observe that early decreases of proteins of potential mitochondrial origin in the mature erythrocytes were associated with transient elevations and then decreases of carboxylic acids such as succinate. In this regard, we and others previously associated elevation of carboxylic acids, as well as kynurenine levels to activation of cGAS-STING-interferon signaling not just upon viral infection, but also upon age- or disease-related elevations of circulating mitochondrial DNA and RNA.40-42 In the context of SCD, we have recently associated RBC and plasma kynurenine levels to poor cardiorenal function and outlook in this patient population,9-11 as well as to elevation in basal levels of hemolysis and osmotic fragility in older healthy blood donors with higher body mass indices.43 Transient elevation of kynurenine in the earliest visits during the trial was followed by ultimate declines below the initial, pre-treatment levels, suggesting a potential beneficial effect of mitapivat on this pathway. There are several points to note regarding this study. First of all, here we performed a longitudinal study in SCD patients, which is more powerful than cross-sectional studies that include healthy controls, as it enables direct monitoring of the impact of mitapivat in a patient-specific fashion, while controlling for factors like the complexity of a disease such as SCD. However, similar omics studies are necessary to understand the impact of mitapivat in healthy controls, and to determine whether mitapivat-associated changes promote a phenotypic change towards a “healthy RBC” omics profile. While samples were buffy coat-depleted and filtered to remove residual platelets and leukocytes, it is not possible to exclude cell-extrinsic effects on the RBC metabolome, which is indeed influenced by metabolite uptake from plasma, as is the case for kynurenine.43
Altogether, muti-omics investigations confirmed the direct benefits of activating PK in SCD, i.e., increasing ATP and decreasing DPG, clearly correlating with functional measurements of oxygen affinity (p50) and sickling kinetics (t50). An increase in ATP does, however, have additional beneficial effects, one of these being an almost immediate reduction in protein and metabolic markers of retention of mitochondria in the mature RBC. Other beneficial effects shown in changes in the metabolomics-lipid-proteomics profile provide insights into a potential series of pathways that warrants further mechanistic testing in the future. Our data also suggest that, on top of glycolytic metabolites, mitochondrial proteins could be a useful biomarker for monitoring response of biochemical efficacy of activating PK as a therapeutic approach in SCD.
Supplementary Material
Funding Statement
Funding: Clinical research was supported by the intramural divisions of National Heart, Lung, and Blood Institute (NHLBI) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institutes of Health (NIH). AD’A was supported by funds from the NHLBI (R01HL146442, R01HL149714, R01HL148151, R01HL161004). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Data-sharing statement
All the raw data generated in this study are available in Online Supplementary Table S1.
References
- 1.D’Alessandro A, Earley EJ, Nemkov T, et al. Genetic polymorphisms and expression of Rhesus blood group RHCE are associated with 2,3-bisphosphoglycerate in humans at high altitude. Proc Natl Acad Sci U S A. 2024;121(1):e2315930120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sunshine HR, Hofrichter J, Ferrone FA, Eaton WA. Oxygen binding by sickle cell hemoglobin polymers. J Mol Biol. 1982;158(2):251-273. [DOI] [PubMed] [Google Scholar]
- 3.Hofrichter J, Ross PD, Eaton WA. Supersaturation in sickle cell hemoglobin solutions. Proc Natl Acad Sci U S A. 1976;73(9):3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg MA, Husson MA, Bunn HF. Participation of hemoglobins A and F in polymerization of sickle hemoglobin. J Biol Chem. 1977;252(10):3414-3421. [PubMed] [Google Scholar]
- 5.Nemkov T, Stephenson D, Earley E, et al. Biological and genetic determinants of red blood cell glycolysis. bioRxiv. Sept 11, 2023. doi:10.1101/2023.09.11.55725 [preprint, not peer-reviewed]. [Google Scholar]
- 6.Adebiyi MG, Manalo JM, Xia Y. Metabolomic and molecular insights into sickle cell disease and innovative therapies. Blood Adv. 2019;3(8):1347-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu JZ, Conrey A, Frey I, et al. A phase 1 dose escalation study of the pyruvate kinase activator mitapivat (AG-348) in sickle cell disease. Blood. 2022;140(19):2053-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk MJ, Rab MAE, van Oirschot BA, et al. Safety and efficacy of mitapivat, an oral pyruvate kinase activator, in sickle cell disease: a phase 2, open-label study. Am J Hematol. 2022;97(7):E226-E229. [DOI] [PubMed] [Google Scholar]
- 9.Sun K, D’Alessandro A, Ahmed MH, et al. Structural and functional insight of sphingosine 1-phosphate-mediated pathogenic metabolic reprogramming in sickle cell disease. Sci Rep. 2017;7(1):15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darghouth D, Koehl B, Madalinski G, et al. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood. 2011;117(6):e57-e66. [DOI] [PubMed] [Google Scholar]
- 11.D’Alessandro A, Nouraie SM, Zhang Y, et al. In vivo evaluation of the effect of sickle cell hemoglobin S, C and therapeutic transfusion on erythrocyte metabolism and cardiorenal dysfunction. Am J Hematol. 2023;98(7):1017-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. [DOI] [PubMed] [Google Scholar]
- 13.Conrey A, Frey I, Asomaning N, et al. Long-term safety and efficacy of mitapivat, an oral pyruvate kinase activator, in adults with sickle cell disease: extension of a phase 1 dose escalation study. Blood. 2023;142(Suppl 1):273. [Google Scholar]
- 14.Beutler E, Blume KG, Kaplan JC, Löhr GW, Ramot B, Valentine WN. International Committee for Standardization in Haematology: recommended methods for red-cell enzyme analysis. Br J Haematol. 1977;35(2):331-340. [DOI] [PubMed] [Google Scholar]
- 15.Beutler E. Red Cell Metabolism: A Manual of Biochemical Methods. 3rd edition. Orlando (FL): Grune & Stratton; 1984. [Google Scholar]
- 16.Metaferia B, Cellmer T, Dunkelberger EB, et al. Phenotypic screening of the ReFRAME drug repurposing library to discover new drugs for treating sickle cell disease. Proc Natl Acad Sci U S A. 2022;119(40):e2210779119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemkov T, Reisz JA, Gehrke S, Hansen KC, D’Alessandro A. High-throughput metabolomics: isocratic and gradient mass spectrometry-based methods. Methods Mol Biol. 2019;1978:13-26. [DOI] [PubMed] [Google Scholar]
- 18.Reisz JA, Zheng C, D’Alessandro A, Nemkov T. Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics. Methods Mol Biol. 2019;1978:121-135. [DOI] [PubMed] [Google Scholar]
- 19.Thomas T, Stefanoni D, Dzieciatkowska M, et al. Evidence of structural protein damage and membrane lipid remodeling in red blood cells from COVID-19 patients. J Proteome Res. 2020;19(11):4455-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song A, Wen AQ, Wen YE, et al. p97 dysfunction underlies a loss of quality control of damaged membrane proteins and promotes oxidative stress and sickling in sickle cell disease. FASEB J. 2022;36(5):e22246. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, Bogdanov M, Zhang Y, et al. Hypoxia-mediated impaired erythrocyte Lands’ cycle is pathogenic for sickle cell disease. Sci Rep. 2016;6:29637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Dai Y, Wen J, et al. Detrimental effects of adenosine signaling in sickle cell disease. Nat Med. 2011;17(1):79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dembélé KC, Mintz T, Veyrat-Durebex C, et al. Metabolomic profiling of plasma and erythrocytes in sickle mice points to altered nociceptive pathways. Cells. 2020;9(6):1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato GJ, Steinberg MH, Gladwin MT. Intravascular hemolysis and the pathophysiology of sickle cell disease. J Clin Invest. 2017;127(3):750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribeiro PR, Teixeira RdS, Souza AR, et al. Blood plasma metabolomics of children and adolescents with sickle cell anaemia treated with hydroxycarbamide: a new tool for uncovering biochemical alterations. Br J Haematol 2021;192(5):922-931. [DOI] [PubMed] [Google Scholar]
- 26.Cendali FI, Nemkov T, Lisk C, et al. Metabolic correlates to critical speed in murine models of sickle cell disease. Front Physiol. 2023;14:1151268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemkov T, Skinner S, Diaw M, et al. Plasma levels of acylcarnitines and carboxylic acids correlate with cardiovascular and kidney function in subjects with sickle cell trait. Front Physiol. 2022;13:916197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Alessandro A, Nouraie SM, Zhang Y, et al. Metabolic signatures of cardiorenal dysfunction in plasma from sickle cell patients as a function of therapeutic transfusion and hydroxyurea treatment. Haematologica. 2023;108(12):3418-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glenthøj A, van Beers EJ, Al-Samkari H, et al. Mitapivat in adult patients with pyruvate kinase deficiency receiving regular transfusions (ACTIVATE-T): a multicentre, open-label, singlearm, phase 3 trial. Lancet Haematol. 2022;9(10):e724-e732. [DOI] [PubMed] [Google Scholar]
- 30.Al-Samkari H, Galactéros F, Glenthøj A, et al. Mitapivat versus placebo for pyruvate kinase deficiency. N Engl J Med. 2022;386(15):1432-1442. [DOI] [PubMed] [Google Scholar]
- 31.D’Alessandro A, Anastasiadi AT, Tzounakas VL, et al. Red blood cell metabolism in vivo and in vitro. Metabolites. 2023;13(7):793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dijk MJ, van der Veen S, Rab MAE, et al. Untargeted metabolomics on dried blood spots of patients with sickle cell disease treated with the pyruvate kinase activator mitapivat. Blood. 2022;140(Suppl 1):21-23. [Google Scholar]
- 33.Bardyn M, Crettaz D, Rappaz B, et al. Phosphoproteomics and morphology of stored human red blood cells treated by protein-tyrosine-phosphatases inhibitor. Blood Adv. 2023;8(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu P, Chen C, Zhang Y, et al. Erythrocyte transglutaminase-2 combats hypoxia and chronic kidney disease by promoting oxygen delivery and carnitine homeostasis. Cell Metab. 2022;34(2):299-316.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Alessandro A, Key A, Amireault P, et al. Genetic regulation of carnitine metabolism controls lipid damage repair mechanisms and hemolytic propensity of human red blood cells during aging in vivo and in vitro. Blood. 2023;142(Suppl 1):4032. [Google Scholar]
- 36.Tumburu L, Ghosh-Choudhary S, Seifuddin FT, et al. Circulating mitochondrial DNA is a proinflammatory DAMP in sickle cell disease. Blood. 2021;137(22):3116-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriconi C, Dzieciatkowska M, Roy M, et al. Retention of functional mitochondria in mature red blood cells from patients with sickle cell disease. Br J Haematol. 2022;198(3):574-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jagadeeswaran R, Lenny H, Vazquez B, et al. The abnormal presence of mitochondria in circulating red blood cells cause an increased oxygen consumption rate, ROS generation and hemolysis in patients with sickle cell disease. Blood. 2017;130(Suppl 1):2237.29170191 [Google Scholar]
- 39.Tannahill GM, Curtis AM, Adamik J, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zecchini V, Paupe V, Herranz-Montoya I, et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature. 2023;615(7952):499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giordano AMS, Luciani M, Gatto F, et al. DNA damage contributes to neurotoxic inflammation in Aicardi-Goutières syndrome astrocytes. J Exp Med. 2022;219(4):e20211121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unali G, Crivicich G, Pagani I, et al. Interferon-inducible phospholipids govern IFITM3-dependent endosomal antiviral immunity. EMBO J. 2023;42(10):e112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemkov T, Stephenson D, Erickson C, et al. Regulation of kynurenine metabolism by blood donor genetics and biology impacts red cell hemolysis in vitro and in vivo. Blood. 2024;143(5):456-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw data generated in this study are available in Online Supplementary Table S1.