Abstract
The structure of a genetic network is uncovered by studying its response to external stimuli (input signals). We present a theory of propagation of an input signal through a linear stochastic genetic network. We found that there are important advantages in using oscillatory signals over step or impulse signals and that the system may enter into a pure fluctuation resonance for a specific input frequency.
Keywords: systems biology, synthetic biology, stochastic processes
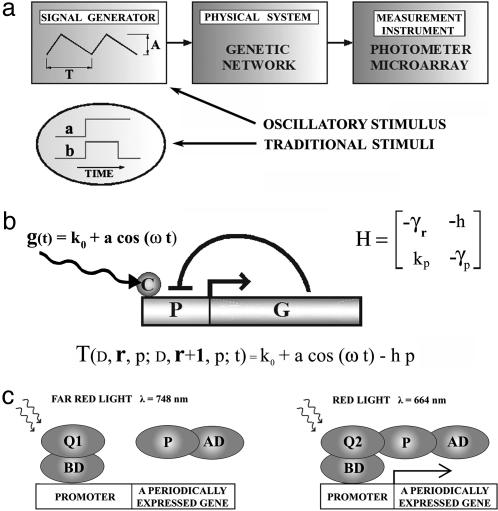
The nature of a physical system is revealed through its response to external stimulation. The stimulus is imposed on the system, and its effects are then measured (Fig. 1a). This approach is widely used in biology: a cell culture is perturbed with a growth factor, a heat shock, etc. The data measured contain the initial information encoded into the stimulus and the information about the intrinsic characteristics of the system. The more parameters the experimentalist can adjust to craft the perturbation stimulus, the more information about the system can be revealed. In recent years we have witnessed a tremendous increase in measurement capabilities (e.g., microarray and proteomic technologies, better reporter genes). However, the success of the systems approach to molecular biology depends not only on the measurement instruments, but also on an effective design and implementation of the input stimulus, which has not been thoroughly explored. Traditionally, two types of time-dependent stimuli are at work in molecular biological experiments (1, 2). For example, a step stimulus is obtained when at one instant of time a growth factor is added to the medium (Fig. 1a, graph a). The stimulus from graph b in Fig. 1a is a superposition of two step stimuli. The investigator can control the height of the step stimulus (the concentration of the growth factor) or the time extension of the heat shock. The cells respond to these stimuli only transiently. The response is dampened after some time and becomes harder to detect because of noise. To overcome the noise, the concentration of the stimulus is typically increased to the point where the strength of the stimulus raises far above its physiological range.
Fig. 1.
Genetic networks stimulated by signal generators. (a) Genetic network response depends on the type of the applied stimulus. (b) An autoregulatory network. The gene G is under the influence of a cofactor C that rhythmically modulates the activity of the promoter P. The matrix H contains the parameters that dictate the transition probabilities of the stochastic model. The transition probability per unit time from r to r + 1 mRNA molecules, T(D, r, p; D, r + 1, p; t), is modulated by the oscillatory signal generator. The DNA, D, and the protein, p, do not change in this transition. (c) The gene is turned on with a red light pulse of wavelength λ = 664 nm. With a far red light of wavelength λ = 748 nm the gene is turned off. Adapted from ref. 5. AD, activation domain; BD, binding domain; Q, a protein that changes its form upon light exposure, from Q1 to Q2 and back; P, a protein that interacts only with form 2 of protein Q (Q2).
We propose to implement a molecular switch at the level of gene promoter and use it to impose an oscillatory stimulus. In the absence of experimental noise, any stimulus can be used to determine the input-output properties of a genetic network. However, in the presence of experimental noise, oscillatory input has many advantages: (i) the measurements can be extended to encompass many periods so the signal-to-noise ratio can be dramatically improved; (ii) the measurement can start after transient effects subside, so that the data become easier to incorporate into a coherent physical model; and (iii) an oscillatory stimulus has more parameters (period, intensity, slopes of the increasing and decreasing regimes of the stimulus) than a step stimulus. As a consequence, the measured response will contain much more quantitative information. Experimental results from neuroscience prove that oscillatory stimulus can modulate the mRNA expression level of genes. For example, the c-fos transcription level in cultured neurons is enhanced 400% by an electrical stimulus at 2.5 Hz and reduced by 50% at 0.01 Hz (3). Also, the mRNA levels of cell recognition molecule L1 in cultured mouse dorsal root ganglion neurons change if the frequency of the electric pulses is varied. The expression level of L1 decreases significantly after 5 days of 0.1-Hz stimulation but not after 5 days of 1-Hz stimulation (4). To extend the oscillatory approach to other types of cells, a two-hybrid assay (5) can be used to implement a molecular periodic signal generator (Fig. 1c). The light switch is based on a molecule (phytochrome in ref. 5) that is synthesized in darkness in the Q1 form. When the Q1 form absorbs a red light photon (wavelength 664 nm) it is transformed into the form Q2. When Q2 absorbs a far red light (wavelength 748 nm) the molecule Q goes back to its original form, Q1. The transitions take milliseconds. The protein P interacts only with the Q2 form, thus recruiting the activation domain to the target promoter. In this position, the promoter is open and the gene is transcribed. After the desired time has elapsed, the gene can be turned off by a photon from a far red light source. Using a sequence of red and far red light pulses the molecular switch can be periodically opened and closed.
There are four input parameters that can be varied: the period (T), the time separation between the pulses (s), and the amplitude (A) of the red and the far red pulses. The mRNA concentration profile will depend on these parameters and can be measured with high-throughput technology (6). Protein levels also will depend on the input signal. The proteins can be recorded with 2D PAGE analysis or MS. If one single gene product is targeted, than a real-time luminescence recording can be used (7). A periodic generator can be used to investigate biological systems for which the mRNA and protein concentrations naturally oscillate in time. An example of such a system is the circadian clock that drives a 24-h rhythm in living organisms from human to cyanobacteria. The core oscillator is a molecular machinery based on an autoregulatory feedback loop involving a set of key genes (Bmal, Per1, Per2, Per3, Cry1, Cry2, etc.) (8). Experimental procedures used to elucidate the clock mechanism are based on measuring the circadian wheel-running behavior of mice under normal light/dark cycles or in constant darkness (dark/dark) conditions. Experimental evidence demonstrates that laws of quantitative nature govern the molecular clock. For example (9), the internal clock of Cry1 mutants have a free-running (i.e., dark/dark conditions) period of 22.51 ± 0.06 h, which is significantly lower than the period of 23.77 ± 0.07 h for WT mice. In contrast, a Cry2 mutant has a significantly higher period of 24.63 ± 0.06 h. In light/dark conditions, both mutants follow the 24-h period of the entrained light cycles. A double Cry1,2 mutant is arrhythmic in dark/dark conditions and follows a 24-h rhythm in light/dark conditions. To explain these experimental values we suggest using a light-switchable generator to drive the expression level of Cry1 and Cry2 and measure the dynamics of transcription and the translation for the rest of the key clock genes. Another application of the periodic generator is to modulate a constitutively expressed gene by superimposing an oscillatory profile on top of its flat level. Then, the genes that show a modulation with a frequency equal to the generator's frequency will be detected by a microarray experiment. Why is this approach different from the one where a step stimulus is used? Because the frequency of the generator is not an internal parameter of the biological system. The genes that interact with the driven gene will be modulated by the input frequency. The rest of the genes will have different expression profiles dictated by the internal parameters of the biological system. This point of view is supported by our findings (6) that the circadian clock (which is an endogenous periodic signal generator) propagates its output to only 8-10% of the transcriptome in mice peripheral tissues (liver or heart). In contrast to the oscillatory input, when a step stimulus is applied, all of the expression profiles are dictated by the internal parameters of the biological system. Except for the height of the step stimulus (the dose of the factor applied) there is no external parameter implemented into the input signal. As such, it is difficult to separate those genes that directly respond to the input signal and consequently avoid artifacts. With the applications described in mind, we study the propagation of an input signal through a stochastic genetic network.
The Response of a Stochastic Genetic Network to an Input Stimulus
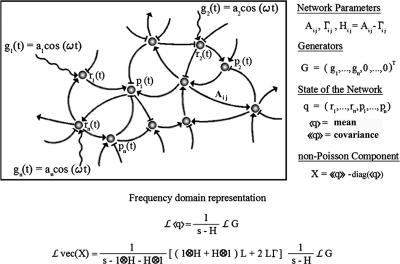
The effects of an oscillatory input have been studied on specific biological systems by using models based on differential equations (10-12). The stochastic character is embedded into these equations as an exterior additive term. In contrast, we compute the generator's effects on the mean and fluctuation of the gene products with a stochastic model (13-15). In this way, the generated stimulus and the noisy nature of the cell are entangled in the stochastic genetic model. For a network of n genes (Fig. 2) the state of a cell is described by the mRNA and protein molecule numbers: q = (r1,..., rn, p1,..., pn). We assume that, during any small time interval Δt, the probability for the production of a molecule of the ith type is 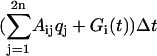 , i.e., qi is increased by 1 with the above probability. The function Gi(t) represents the time-varying input signal and modulates the mRNA production only: G = (g1(t),..., gn(t), 0,..., 0)T (the superscript T is the transposition operation that transforms G into a column vector for notational convenience in what follows). The parameter Aij represents the influence of the jth type of molecule on the production rate of a molecule of the ith type. Similarly, there is a matrix of parameters Γij governing the degradation rates of the molecules. For simplicity, we assume that the input stimulus directly affects only the production rates. The mean μ = 〈q〉 and the covariance matrix ν = 〈〈q〉〉 = 〈〈(q - 〈q〉) (q - 〈q〉)T〉 of the state q are driven by the generator G.
, i.e., qi is increased by 1 with the above probability. The function Gi(t) represents the time-varying input signal and modulates the mRNA production only: G = (g1(t),..., gn(t), 0,..., 0)T (the superscript T is the transposition operation that transforms G into a column vector for notational convenience in what follows). The parameter Aij represents the influence of the jth type of molecule on the production rate of a molecule of the ith type. Similarly, there is a matrix of parameters Γij governing the degradation rates of the molecules. For simplicity, we assume that the input stimulus directly affects only the production rates. The mean μ = 〈q〉 and the covariance matrix ν = 〈〈q〉〉 = 〈〈(q - 〈q〉) (q - 〈q〉)T〉 of the state q are driven by the generator G.
Fig. 2.
Response of a stochastic genetic network to an oscillatory input. The Laplace transform L changes the dynamic variable from time to frequency. In the vec(X) all of the elements of matrix X are arranged in a column vector.
The transfer of the signal from the generator through the genetic network to the output measured data is encapsulated in a set of transfer matrices. Specifically, let H = A - Γ and denote the Laplace transforms of μ and G byLμ and LG. Here and in what follows, μ and G are represented as column vectors. The connection between the mean and the generators is given by formula 1, which is typical for a deterministic linear system. However, the genetic system is stochastic and the measure of the intrinsic noise is quantified by the covariance matrix ν. The effect of the stimulus generators is most transparent if we split ν in a Poisson and a non-Poisson component: ν = diag(μ) + X. Here diag(μ) represents a matrix with the components of the vector μ on its diagonal, with all of the other terms being zero. For a Poisson process, X = 0 and thus the term diag(μ) is called the Poisson component of ν. The non-Poisson component X = ν - diag(μ) can be expressed in terms of the generators (Appendix and Supporting Text, which is published as supporting information on the PNAS web site):
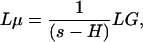 |
[1] |
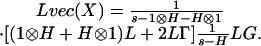 |
[2] |
The vec(X) is a vector constructed from the matrix X by stripping the columns of X one by one and stacking them one on top of each other in vec(X). We emphasize here that the time variation of the generators G in 2 can take any form and is not bounded to be periodic or a step stimulus.
There are three matrices that transfer the information from the generators to the non-Poisson component, Lvec(X) = M3M2M1LG. The first, M1 = (s - H)-1, is the same as the transfer matrix for the mean. The second, M2 = (1 ⊗ H + H ⊗ 1)L + 2LΓ, breaks the symmetry between the degradation and production parameters that are otherwise hidden in the matrix H = A - Γ. The ⊗ is the Kronecker product of two matrices. The matrix L (with elements 0 and 1) is the lifting matrix from dimension of the mean (2n values) to the dimension of vec(X) (4n2 values). The third matrix is M3 = (s - 1 ⊗ H - H ⊗ 1)-1. If λi is the eigenvalue of H then all combinations λi + λj are the eigenvalues of 1 ⊗ H + H ⊗ 1. Thus M3 represents the analog of M1 in the space of covariance variables.
For a step stimulus, these eigenvalues are of primal importance: the measured signal is a superposition of components with different eigenvalues and has a complicated mathematical expression. However, for a periodic stimulus, the frequency of the external generator is the important parameter. This frequency is fixed by the experimentalist not by the biological system. Only the phase and the amplitude of the output signal depends on the system's eigenvalues and the mathematical form is less cumbersome then for the step stimulus. The input-output relations, 1 and 2, were derived from the master equation written for the probability of the states of the genetic network. Thus, we must specify the initial conditions for the probability of the states. These conditions refer here to states for which one molecular component vanishes (qi = 0, for one i). The input-output relations, 1 and 2, are independent of these boundary states if the Γ matrix is diagonal. A diagonal Γ matrix was used in ref. 13, and we will use it also in the example that follows. Tools developed in the field of system identification can be used to create models for the networks under study (16). The difference between the system identification classical models and a genetic network is that the latter is a stochastic process by nature, whereas the former are deterministic models with a superimposed noise from external sources. However, the formulas that describe the relations between the mean and covariance of the stochastic process and the input signals, 1 and 2, are of the same general nature as those used in system identification theory (16). In the next section we will use 1 and 2 to analyze one of the most fundamental regulatory motifs in a genetic network: an autoregulatory gene that acts upon itself through a negative feedback (17-19). The fluctuation can drive this biological system out of its equilibrium state (20).
Fluctuation Resonance
Four parameters characterize the system: the feedback strength A12 = -h, the translation rate A21 = kp, and two degradation rates, Γ11 = γr,Γ22 = γp. The gene regulation is under the control of its own protein product and the protein activity is modulated by a cofactor. The cofactor is driven by a periodic light-switchable generator g(t) = k0 + acos(ωt) (Fig. 1a). Before the generator is applied, the transcription rate is equal with k0 and the system is in a steady state. Through the transfer matrices, 1 and 2, the light generator will impose a periodic evolution of the mean and covariance matrix for mRNA and protein product. We denote the mean mRNA by 〈r(t)〉 and the mean number of protein by 〈p(t)〉. We will concentrate on the protein number in what follows. After the transients are gone, 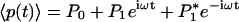 , that is the protein number will oscillate with an amplitude P1 on top of a baseline P0; here * represents complex conjugation. The fluctuation of the protein number, 〈〈p(t)〉〉, differs from the mean number by a quantity that we denoted by Xpp(t): 〈〈p(t)〉〉 = 〈p(t)〉 + Xpp(t). For a pure Poisson process, 〈〈p(t)〉〉 = 〈p(t)〉. Thus the term Xpp(t) represents the deviation from a Poisson process. If there is some information about the genetic system that can be uncovered by measuring not only the mean but also the covariance matrix, then this information is hidden only in the non-Poisson component Xpp(t). The quantity Xpp(t) is not interesting only from a statistical point of view but also from a dynamical one. The equation for the time evolution of 〈〈p(t)〉〉 takes its most simple form if it is written for Xpp(t). That is, the time dependence of the mean value must be subtracted from the time evolution of 〈〈p(t)〉〉. Similar to the mean value, the non-Poisson component of the fluctuation will oscillate in time,
, that is the protein number will oscillate with an amplitude P1 on top of a baseline P0; here * represents complex conjugation. The fluctuation of the protein number, 〈〈p(t)〉〉, differs from the mean number by a quantity that we denoted by Xpp(t): 〈〈p(t)〉〉 = 〈p(t)〉 + Xpp(t). For a pure Poisson process, 〈〈p(t)〉〉 = 〈p(t)〉. Thus the term Xpp(t) represents the deviation from a Poisson process. If there is some information about the genetic system that can be uncovered by measuring not only the mean but also the covariance matrix, then this information is hidden only in the non-Poisson component Xpp(t). The quantity Xpp(t) is not interesting only from a statistical point of view but also from a dynamical one. The equation for the time evolution of 〈〈p(t)〉〉 takes its most simple form if it is written for Xpp(t). That is, the time dependence of the mean value must be subtracted from the time evolution of 〈〈p(t)〉〉. Similar to the mean value, the non-Poisson component of the fluctuation will oscillate in time, 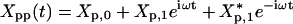 with complex amplitude Xp,1. The relative strength of the fluctuation versus the mean value can be described by using the Fano factor (13): 〈〈p(t)〉〉/〈p(t)〉 = 1 + Xpp(t)/〈p(t)〉. For oscillatory inputs, the response of the network is best described in frequency domain rather than in time. In frequency domain, as an analog of the Fano factor we consider the ratio of the amplitude of Xpp(t) versus the amplitude of 〈p(t)〉.
with complex amplitude Xp,1. The relative strength of the fluctuation versus the mean value can be described by using the Fano factor (13): 〈〈p(t)〉〉/〈p(t)〉 = 1 + Xpp(t)/〈p(t)〉. For oscillatory inputs, the response of the network is best described in frequency domain rather than in time. In frequency domain, as an analog of the Fano factor we consider the ratio of the amplitude of Xpp(t) versus the amplitude of 〈p(t)〉.
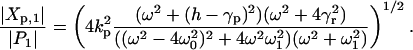 |
[3] |
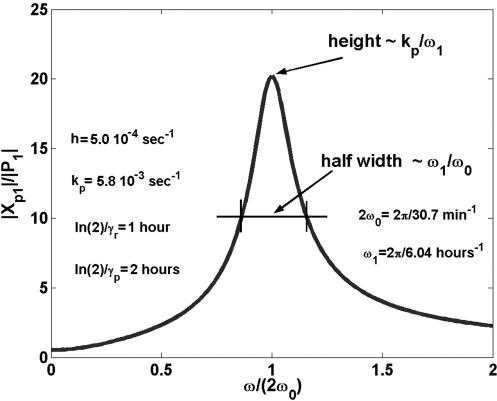
Here ω1 = γr + γp. The complex amplitudes Xp,1 and P1 depend on the input frequency, and therefore resonance phenomena can be detected in the system. If the light-switchable generator oscillates with double the natural frequency ω20 = hkp + γrγp, that is, ω = 2ω0 we find a state of resonance for fluctuation and not for the mean (Fig. 3).
Fig. 3.
Fluctuation resonance. The amplitude Xp,1 of the non-Poisson component is much higher than the amplitude of the mean protein number, P1, at ω = 2ω0.
For ω = 2ω0 the system will be in a pure fluctuation resonance. In such a situation the molecular noise can drive the cell out of its equilibrium state, which can have dramatic consequence on the cell fate. Our model being linear cannot cover the entire phenomena that accompanies a system whose state is close to resonance. However, a linear model suggests the existence of pure fluctuation resonance. At fluctuation resonance, the deviation from a Poissonian process is high. The oscillation amplitude for protein fluctuation is much greater then the amplitude of the mean. Experimental results (21) show that typical values for the ratio kp/γr are 40 for lacZ and 5 for lacA. These values suggest that there are natural conditions for a strong height fluctuation resonance (Fig. 3). However, for a sharp fluctuation resonance (small half width), we need h > γr or γp, a condition that does not appear in all genetic networks. It is through the experimental study that we will clarify how some biological systems can sustain fluctuation resonance and others can not. Besides resonance, the frequency response provides other insights into the structure of the autoregulatory system. The parameters of the system can be read from the measured data. The frequency response of the mean values behave like the response of a classical linear system to input signals. The new aspects are those related to fluctuations. Like Xpp(t) and Xrr(t), the correlation coefficient between the mRNA and protein number will oscillate in time: 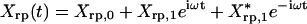 with amplitude Xrp,1. Taking the ratios of the amplitudes
with amplitude Xrp,1. Taking the ratios of the amplitudes 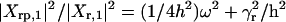 ,
, 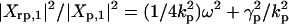 , we observe that all four parameters of the system can be estimated from the slopes and the intercepts of the above ratios as a function of ω2. Detail formulas for each amplitude are given in Supporting Text.
, we observe that all four parameters of the system can be estimated from the slopes and the intercepts of the above ratios as a function of ω2. Detail formulas for each amplitude are given in Supporting Text.
The Spectrum, the Experimental Noise, and the Importance of the Input Stimulus
We described the use of a periodic signal to decipher a genetic network. Traditionally, a step stimulus is used in biology for pathway detection (i.e., adding a growth factor to the culture). From the response to a step stimulus we can extract, in principle, the parameters of the system. The natural question is then: why should we generate a periodic stimulus when there is already a step stimulus in use? Seeking an answer, we notice that the measured data in our studied example can be expressed as a sum of exponentially decaying functions, e-λt, if a step stimulus was used (Supporting Text). For a periodic input, the response contains only exponentials with imaginary argument, eiωt. Mathematically, the main difference between exponentials with real arguments, e-λt, and those with imaginary arguments, eiωt, is that with the former we cannot form an orthogonal basis of functions, whereas such a basis can be formed with the latter. If we depart from our example, we can say that, in general, the response of the network to a step input will be a sum of components that are not orthogonal on each other. The time dependence of these nonorthogonal components can be more complex than an exponential function; they can contain polynomials in time or decaying oscillations, depending on the position in the complex plane of eigenvalues of the transfer matrix H. In constrast, the permanent response obtained from a periodic input is a sum of Fourier components that form an orthogonal set. Orthogonal components are much easier to separate than nonorthogonal ones. This mathematical difference explains the advantage of using oscillatory inputs. However, an argument can be made that increasing the number of replicates will be enough to separate the step response from noise. In what follows we study how many replicates are needed to successfully fight the experimental noise. We will show that fewer replicates are needed if the genetic network is probed with an oscillatory generator rather than a step signal. To keep the argument simple, we will study the difficulty of separating nonorthogonal components for a network for which the response to a step stimulus is a sum of decaying exponentials. The argument can be extended to other types of nonorthogonal components, but this line of thought falls outside the scope of this article. The measured data being a superposition of exponential terms can be written as:
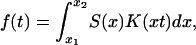 |
[4] |
with K(xt) = e-ixt for the periodic response and K(xt) = e-xt for the step stimulus. The spectral function S(x) depends on the network's parameters and the type of the input signal. For example, the spectrum of the autoregulatory system for a periodic input is S(x) = S0δ(x) + S1δ(x - iω) + S1*δ(x + iω), where δ(x) is the Dirac delta function. The coefficients S0, S1 take specific values if the spectrum refers to mean mRNA, proteins, or their correlations. For example, for the protein fluctuation:
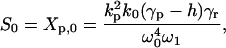 |
[5] |
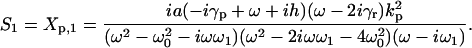 |
[6] |
A detailed description of the spectrum for an autoregulatory network is given in Supporting Text. For oscillatory inputs that are not pure cosine function and for more complicated networks, the spectrum is more complex, but is still connected with the measured data like in 4. The spectrum S(x) carries information about the parameters of the genetic network and can be recovered from the data f(t). The network's parameter can be estimated from the spectrum once a model of the network is chosen. Our goal is to show that the spectrum obtained from an oscillatory input signal is much less distorted by the experimental noise than the spectrum obtained from a step input. Laboratory measurements are samples of f(t) at N discrete time points. Given a finite number N of measured data points, f1,..., fN, the spectrum for the periodic case S(x) can only be approximated as a weighted sum of N terms (Supporting Text): 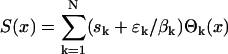 . Each term,
. Each term, 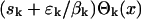 , contains a function
, contains a function 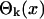 that does not depend on the measured data and the weights sk + εk/βk that are computed from the measured data f1,..., fN. In the absence of experimental noise, εk = 0, all N coefficients sk can be computed from the measured data. When experimental noise is present, εk ≠ 0, what we compute from measured data is sk + εk/βk, and we cannot separate sk from it because we do not know the actual value for εk. The best we can do is to use only those terms for which sk > εk/βk, so the effect of the distortion on sk is not large. Unfortunately, the distortion increases as βk gets smaller, which actually happens when k increases. A term can be recovered from noise if βk-1 < sk/εk. Usually, this relation is valid for k = 1... Jp, with Jp being the last term that can be recovered. A similar relation holds for the exponential case, with αk instead of βk and Je instead of Jp. It is desirable that both cutoffs (Jp, Je) be as close as possible to the number of sampled points, N. The striking difference between the two cases is that the cutoff Jp is much larger then the cutoff Je. This difference is a consequence of the fact that the numbers αk decrease exponentially to 0 (22), whereas βk stays close to 1 for many k before eventually dropping close to zero (23). This huge difference between αk and βk has its origin in the fact that the set of functions of time, exp(-λt), indexed by λ, do not form an orthogonal set, whereas the functions exp(iωt), indexed by ω, are orthogonal.
that does not depend on the measured data and the weights sk + εk/βk that are computed from the measured data f1,..., fN. In the absence of experimental noise, εk = 0, all N coefficients sk can be computed from the measured data. When experimental noise is present, εk ≠ 0, what we compute from measured data is sk + εk/βk, and we cannot separate sk from it because we do not know the actual value for εk. The best we can do is to use only those terms for which sk > εk/βk, so the effect of the distortion on sk is not large. Unfortunately, the distortion increases as βk gets smaller, which actually happens when k increases. A term can be recovered from noise if βk-1 < sk/εk. Usually, this relation is valid for k = 1... Jp, with Jp being the last term that can be recovered. A similar relation holds for the exponential case, with αk instead of βk and Je instead of Jp. It is desirable that both cutoffs (Jp, Je) be as close as possible to the number of sampled points, N. The striking difference between the two cases is that the cutoff Jp is much larger then the cutoff Je. This difference is a consequence of the fact that the numbers αk decrease exponentially to 0 (22), whereas βk stays close to 1 for many k before eventually dropping close to zero (23). This huge difference between αk and βk has its origin in the fact that the set of functions of time, exp(-λt), indexed by λ, do not form an orthogonal set, whereas the functions exp(iωt), indexed by ω, are orthogonal.
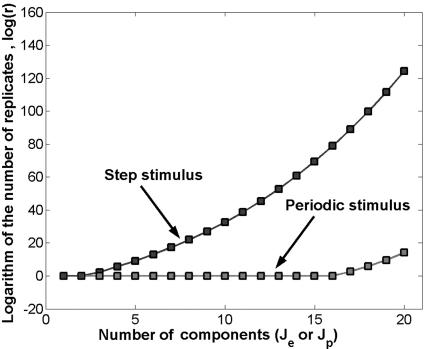
In theory, however, we can still hope that a step stimulus can deliver good estimates if the noise εk is reduced by using r replicates (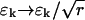 ). This is not the case. Fig. 4 represents the number of replicates needed to recover the component Je or Jp if the signal-to-noise ratio (SNR) is 10 (
). This is not the case. Fig. 4 represents the number of replicates needed to recover the component Je or Jp if the signal-to-noise ratio (SNR) is 10 (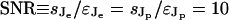 ). The number of replicates grows very fast in the exponential case (for SNR = 10 and N = 20 we need 269 replicates for the fourth spectral component), whereas in the periodic case, the number of replicates stays low for many spectral components (only for the 17th component it raises to 14, with SNR = 10 and N = 20). See also Fig. 5, which is published as supporting information on the PNAS web site.
). The number of replicates grows very fast in the exponential case (for SNR = 10 and N = 20 we need 269 replicates for the fourth spectral component), whereas in the periodic case, the number of replicates stays low for many spectral components (only for the 17th component it raises to 14, with SNR = 10 and N = 20). See also Fig. 5, which is published as supporting information on the PNAS web site.
Fig. 4.
The number of replicates needed to recover a given spectral component is shown.
Conclusions and Discussions
We studied the response of a linear stochastic genetic network to an input stimulus (signal). We provide a general formula that relates the mean and covariance matrix of mRNAs and proteins to the input generators. The particular type of periodic signals was studied in detail for an autoregulatory system. We found that fluctuation resonance can manifest in such systems. Besides interesting physical phenomena that can be detected with a periodic signal, the oscillatory input is useful for experimental noise rejection. We compared two experimental designs: one that uses a step stimulus as a perturbation and another that uses a periodic input. We concluded that the response of the genetic network to a periodic stimulus is much easier to detect from noise than the response of the same network to a step stimulus. This conclusion applies whenever the response of the network to an oscillatory input is a sum of Fourier components and can be the case for many nonlinear networks. However, the input-output relations, 1 and 2, apply only to a linear stochastic model. A linear model is a good approximation around a steady state of the genetic network. A genetic network is a nonlinear system and can have several steady states. If the signal generator does not vary in time, the genetic network will be characterized by one of these steady states. When the signal generator starts to oscillate with an amplitude that doesn't drive the network faraway from its steady state, the linear model is a good approximation. For large amplitudes, the nonlinear effects start to be important, and at some values of the generator's amplitude, the network will jump close to a different steady state. Such nonlinear behaviors cannot be described by a linear model. Also, the parameters that describe the network are supposed to be constant in time. This approximation is valid if the changes in the network parameters are slow with respect to the changes produced by the oscillatory input signals. The input frequency should be chosen so that the system can be considered with constant coefficients for the elapsed time of measurements. Also, the period of oscillations must be less than the trend effects caused by growth, apoptosis, etc. Besides biological effects that span large intervals of time, experimental artifacts, like medium evaporation, can superimpose a trend on the measured profile. The input period should be less than the time characteristics of these trends. These trends will impose a limit for the lower range of the input frequencies. The response to oscillations also depends on the time characteristics of the system under study. If the system has a high damping factor, the high frequencies will be strongly attenuated and the output signal is not measurable. With all of these restrictions, the experimentalist still has the freedom to work in a frequency band, a freedom not present in the step stimulus.
A different line of thought emerges when it comes to analyzing whether the oscillatory method can be scaled to large networks. Experimentally, using high-throughput measurements (microarray and proteomic tools), a large set of gene products can simultaneously be measured. The experimentalist is searching for a pathway that is controlled by a gene. Using oscillatory signals to stimulate the desired gene, the time variation of the downstream genes will contain in its spectrum the input frequency, so these genes will be detected. Moving the signal generator along the pathway, more and more local patches of the network will be uncovered. The global view of the network will consist of all of these patches connected together. The theoretical framework for connecting a set of patches is unclear at present. Experimentally, however, we verified that a source of oscillations propagates into a large genetic network (6). Specifically, a microarray experiment was conducted on mice entrained for 2 weeks on a 24-h period of light/dark signals. The periodic input signal was not implemented at the level of gene promoter; it was an exterior periodic source of light that entrained the internal clock of the cell. After entrainment, and in complete darkness, the output signals (mRNA) were measured every 4 h for 2 days with an Affymetrix (Santa Clara, CA) platform. From ≈6,000 expressed genes in heart, ≈500 showed a mRNA that oscillates within a 24-h period. The same results were reported in ref. 24. The next step is to implement the generator at the gene promoter level and measure the spread of the input signal into the network.
Given the advantages of a periodic stimulus presented above, we believe that the experimental implementation of a periodic generator at the promoter level will prove fruitful in the study of genetic networks.
Supplementary Material
Acknowledgments
We thank Kai-Florian Storch for valuable discussions about the experimental design of the periodic generator, Charles J. Weitz for pointing out to us the Cry1,2 mutant experiments, and the reviewers for their comments. Our work was created and developed independently of ref. 25, which was brought to our attention by one reviewer in December 2004. O.L. is grateful to his colleagues at the Center for Biotechnology and Genomic Medicine for their encouragement. This work was supported in part by National Institutes of Health Grant 1R01HG02341 and National Science Foundation Grant DMS-0090166.
Appendix
The genetic network is described by a linear stochastic network (13-15). The network is driven by using signal generators placed inside the promoters of a subset of genes that are part of the network. For a gene we will denote by D, r, and p the number of DNA, mRNA, and protein molecules, respectively, per cell. We consider r,p to be variables but D to be a constant, and we normalize it to D = 1. The state of a cell that contains n active genes is specified by: q̃ = (D1, D2,..., Dn, r1, r2,..., rn, p1, p2,..., pn). The genetic state is changing in time; for a short transition time, dt, only one q̃i changes its value, and this new value can be either q̃i + 1 or q̃i - 1. We consider in this article a linear stochastic genetic network characterized by the following transition probabilities: 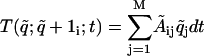 ,
, 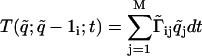 . Here q̃ is the initial state and 1i is a vector of length M with all elements 0 except the one in the position i that is 1. The time variation of the generators that drives gene expression is encapsulated in the matrix Ãij, which governs the production of different molecules. The matrices Ãij and
. Here q̃ is the initial state and 1i is a vector of length M with all elements 0 except the one in the position i that is 1. The time variation of the generators that drives gene expression is encapsulated in the matrix Ãij, which governs the production of different molecules. The matrices Ãij and 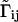 consist of four submatrices, corresponding to splitting the state q̃ into two subgroups. One subgroup contains only the DNA states (D1,..., Dn) and the other subgroup contains the protein and mRNA states q = (r1, r2,..., rn, p1, p2,..., pn),
consist of four submatrices, corresponding to splitting the state q̃ into two subgroups. One subgroup contains only the DNA states (D1,..., Dn) and the other subgroup contains the protein and mRNA states q = (r1, r2,..., rn, p1, p2,..., pn),
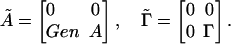 |
[7] |
The generator submatrix Gen has a special form. It is a 2n × n matrix and locates the position of the generators in the genetic network: Genij = gi(t)δij, i = 1...2n, j = 1,...n. Each gene promoter is driven by one generator gi(t), i = 1,..., n, which will influence the mRNA production of gene i. The same mRNA production can be influenced by the protein concentration, and this feedback effect is described by the elements of the 2n × 2n matrix A (7). The structure of the matrix A is a consequence of the topology of the genetic network. The equation for the probability P(q̃, t) of the network to be in the state q̃ at time t is: 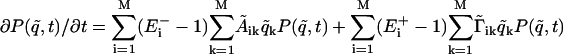 , where the shift operators
, where the shift operators 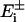 are given by
are given by 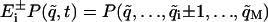 .
.
We need the time evolution equations for mRNAs and proteins: μi = 〈qi〉 and νij = 〈qiqj〉 - 〈qi〉〈qj〉, i, j = 1,..., 2n. In matrix notation, for the column vector μ and for the matrix X with elements given by Xij = νij - δijμi we obtain:
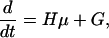 |
[8] |
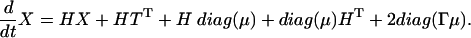 |
[9] |
Here HT is the transpose matrix of H = A - Γ and diag(μ) has nonzero elements only on the principal diagonal: diag(μ)ij = δijμi. Using the Laplace transform, the solution to 8 is 1. Eq. 9 is a matrix equation. To solve this equation we first transform it to an equation where the unknown is a column vector. The transformation needed is X → vec(X), where the column vector vec(X) contains the columns of the matrix X one on top of the other, starting with the first column and ending with the last column. The vec mapping has the useful property that vec(HX) = (1 ⊗ H)vec(X), vec(XH) = (HT ⊗ 1)vec(X), where 1 is the unit matrix and A ⊗ B is the tensor product of matrices A and B. The column vector vec(diag(μ)) can be expressed in terms of the column vector μ: vec(diag(μ)) = Lμ, were L is a lift matrix from a space of dimension of μ to the square of this dimension: L = (P1,..., P2n)T, (Pk)ij = δikδjk. The solution to 9 takes the form 2.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gardner, T. S., di Bernardo, D., Lorenz, D. & Collins, J. J. (2003) Science 301, 102-105. [DOI] [PubMed] [Google Scholar]
- 2.Vance, W., Arkin, A. & Ross, J. (2002) Proc. Natl. Acad. Sci. USA 99, 5816-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng, H. Z., Fields, R. D. & Nelson, P. G. (1993) J. Neurosci. Res. 35, 459-467. [DOI] [PubMed] [Google Scholar]
- 4.Itoh, K., Stevens, B., Schachner, M. & Fields, R. D. (1995) Science 270, 1369-1372. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu-Sato, S., Huq, E., Tepperman, J. M. & Quail, P. H. (2002) Nat. Biotechnol. 20, 1041-1044. [DOI] [PubMed] [Google Scholar]
- 6.Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H. & Weitz, C. J. (2002) Nature 417, 78-83. [DOI] [PubMed] [Google Scholar]
- 7.Izumo, M., Johnson, C. H. & Yamazaki, S. (2003) Proc. Natl. Acad. Sci. USA 100, 16089-16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reppert, S. M. & Weaver, D. R. (2002) Nature 418, 935-941. [DOI] [PubMed] [Google Scholar]
- 9.van der Horst, G. T. J., Muijtjens, M., Kobayashi, K., Takano, R., Kanno, S., Takao, M., de Wit, J., Verkerk, A., Eker, A. P. M., vanLeenen, D., et al. (1999) Nature 398, 627-630. [DOI] [PubMed] [Google Scholar]
- 10.Smolen, P., Baxter, D. A. & Byrne, J. H. (1998) Am. J. Physiol. 43, C531-C542. [DOI] [PubMed] [Google Scholar]
- 11.Hasty, J., Dolnik, M., Rottschafer, V. & Collins, J. (2002) Phys. Rev. Lett. 88, 148101. [DOI] [PubMed] [Google Scholar]
- 12.Simpson, M. L., Cox, C. D. & Sayle, G. S. (2003) Proc. Natl. Acad. Sci. USA 100, 4551-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thattai, M. & van Oudenaarden, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swain, P. S., Elowitz, M. B. & Siggia, E. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12795-12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Kampen, N. G. (1992) Stochastic Processes in Physics and Chemistry (North-Holland, Amsterdam).
- 16.Ljung, L. (1999) System Identification-Theory for the User (1999) (Prentice-Hall, Upper Saddle River, NJ), 2nd Ed.
- 17.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799-804. [DOI] [PubMed] [Google Scholar]
- 18.Becskei, A. & Serrano, L. (2000) Nature 405, 590-593. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld, N., Elowitz, M. & Alon, U. (2002) J. Mol. Biol. 323, 785-793. [DOI] [PubMed] [Google Scholar]
- 20.Isaacs, F., Hasty, J., Cantor, C. & Collins, J. (2003) Proc. Natl. Acad. Sci. USA 100, 7714-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennell, D. & Riezman, H. (1977) J. Mol. Biol. 114, 1-21. [DOI] [PubMed] [Google Scholar]
- 22.Bertero, M., Brianzi, P. & Pike, E. R. (1985) Proc. R. Soc. London A 398, 23-44. [Google Scholar]
- 23.Slepian, D. (1983) SIAM Rev. 25, 379-393. [Google Scholar]
- 24.Panda, S., Antoch, M.P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S. & Hogenesch, J. B. (2002) Cell 109, 307-320. [DOI] [PubMed] [Google Scholar]
- 25.Gadgil, C., Lee, C.-H. & Othmer, H. G. (2005) Bull. Math. Biol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.