Abstract
It is unknown which posttranscriptional regulatory mechanisms are required for oncogenic competence. Here, we show that the LIN28 family of RNA-binding proteins (RBPs), which facilitate posttranscriptional RNA metabolism within ribonucleoprotein networks, is essential for the initiation of diverse oncotypes of hepatocellular carcinoma (HCC). In HCC models driven by NRASG12V/Tp53, CTNNB1/YAP/Tp53, or AKT/Tp53, mice without Lin28a and Lin28b were markedly impaired in cancer initiation. We biochemically defined an oncofetal regulon of 15 factors connected to LIN28 through direct mRNA and protein interactions. Interestingly, all were RBPs and only 1 of 15 was a Let-7 target. Polysome profiling and reporter assays showed that LIN28B directly increased the translation of 8 of these 15 RBPs. As expected, overexpression of LIN28B and IGFBP1-3 was able to genetically rescue cancer initiation. Using this platform to probe components downstream of LIN28, we found that 8 target RBPs were able to restore NRASG12V/Tp53 cancer formation in Lin28a/Lin28b-deficient mice. Furthermore, these LIN28B targets promote cancer initiation through an increase in protein synthesis. LIN28B, central to an RNP regulon that increases translation of RBPs, is important for tumor initiation in the liver.
Keywords: Hepatology
Keywords: Liver cancer, Translation
LIN28 proteins promote tumor initiation in hepatocellular carcinoma through their roles in protein synthesis.
Introduction
In chronically inflamed tissues subject to field cancerization, tumor initiation occurs repeatedly in different parts of the organ over long time periods. For cancer mutations to drive transformation, cells must be oncogenically competent or able to transform once the requisite mutations are established. We know that not all cells are competent because cancer mutations are often observed in nonmalignant tissues and only some cell types transform when strong cancer mutations are introduced. Oncogenic competence likely requires nonmutational processes that regulate gene expression at epigenetic, transcriptional, and posttranscriptional levels (1–3). Posttranscriptional regulation is known to involve RNA-binding proteins (RBPs), which play diverse and critical roles in RNA editing, localization, stabilization, and translation (4). While these factors are less studied in cancer because they are not as frequently mutated (5), it is clear that dysregulation of RBP expression can promote cancer growth by sustaining cell proliferation, evading apoptosis, avoiding immune surveillance, stimulating angiogenesis, and activating metastasis (6–10). There are many fundamental unanswered questions about RBPs in cancer including the following: how essential are they, and how do they operate on the molecular level?
In this study, we used the LIN28 family of RBPs to explore these questions in animal models. Lin-28 was first identified in Caenorhabditis elegans as a heterochronic factor that regulates developmental timing (11). In mammals, there are 2 Lin28 paralogs, Lin28a and Lin28b, both of which promote stem cell pluripotency, tissue growth, and carcinogenesis (12–21). Both proteins bind and inhibit the biogenesis of the Let-7 family of tumor-suppressive microRNAs, but through different mechanisms. LIN28B is more concentrated in the nucleus and is thought to block DGCR8-mediated pri- to precursor Let-7 processing, while LIN28A is localized in the cytoplasm and recruits the 3′ terminal uridylyl transferases (TUTase) named Zcchc11 or Zcchc6 (TUT4 or TUT7) (22–24). LIN28A and TUTases cooperate to uridylate Let-7 precursors, which targets them for degradation. Both LIN28A and LIN28B are also known to directly interact with thousands of other RNA targets and influence their translation into proteins (23, 25–30). LIN28 proteins, like other RBPs, reside in ribonucleoprotein (RNP) complexes, but it is unclear which individual components of these diverse RNPs are functionally important in cancer.
In general, factors that are required for tumor initiation but not progression are less understood. While factors that regulate progression represent attractive cancer drug targets, improving patient outcomes for cancers that arise from chronic tissue injury would require the suppression of repeated tumor-initiation events. To better understand requirements for RBPs in cancer initiation, we examined the earliest cancer cells. While there is no consensus cell of origin or stem cell for all liver cancer subtypes, a premalignant hepatocellular carcinoma (HCC) progenitor cell was previously shown to be capable of transformation in the setting of diethylnitrosamine (DEN) mutagenesis (31). These progenitors express CD44, aggregate, form suspension colonies in vitro, and can be isolated and transplanted based on these properties. Intriguingly, these progenitors express Lin28a and Lin28b at high levels. However, the molecular mechanisms dictating the genesis or transformation of these progenitors remain unexplored. We previously found that the deletion of Lin28a and Lin28b impaired the growth of MYC-induced pediatric hepatoblastomas in vivo, but the absence of Lin28a/Lin28b did not completely eliminate their initiation, possibly due to the high levels of MYC overexpression in that model (32). It also remains unclear how much LIN28s are required for transformation in the clinically relevant context of chronic liver disease. Furthermore, the identity of the downstream effector mechanisms associated with LIN28 proteins remains unclear. Here, we used versatile mouse models as in vivo platforms for mechanistic dissection of RBPs in cancer initiation. These studies highlight the importance of non–Let-7–dependent translational mechanisms downstream of LIN28 and the possibility of using anti-RBP approaches to prevent tumorigenesis.
Results
DEN-induced liver cancers are dependent on LIN28s.
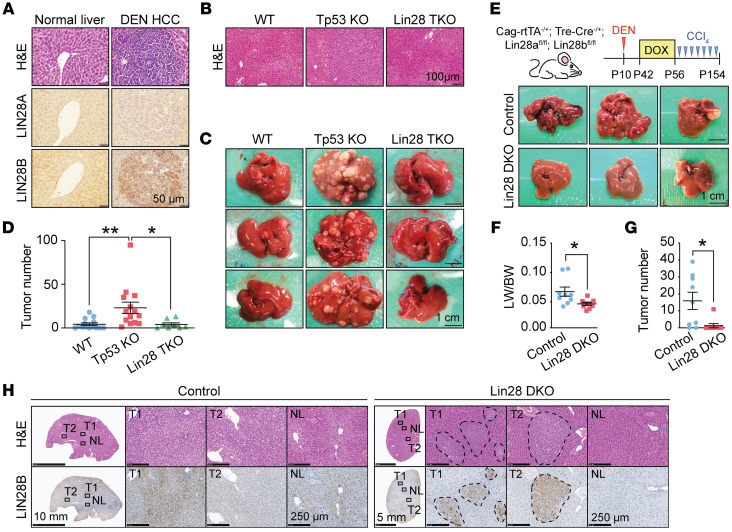
To study tumor initiation in a widely used adult HCC model, we administered DEN to FVB WT mice at 2 weeks of age. At 8 months of age, LIN28A was expressed at low levels, but LIN28B was highly expressed in microscopic hypercellular nodules observed within these livers (Figure 1A). These data suggested that both paralogs could be important for DEN-induced HCC initiation. To determine whether LIN28 proteins are required for tumor initiation, we generated liver-specific Tp53-KO mice (Albumin-Cre; Tp53fl/fl) and Lin28a/Lin28b/Tp53–triple-KO (TKO) mice (Albumin-Cre; Lin28afl/fl; Lin28bfl/fl; Tp53fl/fl). Lin28a/Lin28b/Tp53-TKO mice developed normally, had no obvious whole body or liver-associated phenotypes, and had normal liver histology (Figure 1B). Cohorts of these mice were given DEN at 2 weeks of age and aged for 10 months. Liver-tumor formation was frequent in Tp53-KO mice, but was impaired in Lin28a/Lin28b/Tp53-TKO mice (Figure 1, C and D); 64% of Tp53 WT mice, 100% of Tp53 KO mice, and 62% of Lin28a/Lin28b/Tp53 TKO mice had gross tumors, and TKO tumors were smaller than tumors from Tp53-KO mice.
Figure 1. Lin28-deficient mice are protected from DEN-driven liver tumorigenesis.
(A) H&E and IHC of LIN28A and LIN28B of DEN-induced tumor nodules and adjacent tissue from mice treated with DEN for 8 months. Images are representative of 20 individual tumors. Scale bars: 50 μm. (B) Histology images show the normal liver architecture of control (Tp53fl/fl), Tp53-KO (Albumin-Cre; Tp53fl/fl), and Lin28a/Lin28b/Tp53-TKO (Albumin-Cre; Tp53fl/fl; Lin28afl/fl; Lin28bfl/fl) mice. Scale bar: 100 μm. (C) Representative gross images of WT, Tp53-KO, and Lin28a/Lin28b/Tp53-TKO livers treated with DEN for 8 months. Scale bar: 1 cm. (D) Surface tumor numbers from WT (n = 14), Tp53-KO (n = 14), and Lin28a/Lin28b/Tp53-TKO mice (n = 8). Each dot represents 1 mouse. (E) This schematic shows DEN/CCl4 and dox administration. Representative gross images of liver from control (Cag-rtTA+/–; Lin28afl/fl; Lin28bfl/fl; n = 9) and Lin28a/Lin28b-DKO mice (Cag-rtTA+/–; TRE-Cre+/–; Lin28afl/fl; Lin28bfl/fl; n = 9) that were subjected to DEN/CCl4 administration and dox water. Scale bars: 1 cm. All the images can also be found in Supplemental Figure 1B. (F and G) Liver-to-body weight ratios (LW/BW) (F) and surface tumor numbers (G) of control (n = 9) and Lin28a/Lin28b-DKO (n = 9) mice. (H) Representative H&E images and LIN28B staining in tumors of control (n = 9) and Lin28a/Lin28b-DKO mice (n = 9). NL, normal liver; T1, tumor number 1; T2, tumor number 2. *P < 0.05; **P < 0.01.
DEN mutagen plus CCl4 chronic injury–induced HCC also required Lin28s for tumor initiation.
The Albumin-Cre–driven model has limitations that prevent a clear understanding of how impactful an anti-LIN28 therapeutic strategy might be in patients with chronic liver damage and HCC. First, it is an embryonic KO and not a model of inducible Lin28 loss in adults. Second, it is liver specific and does not assess potential extrahepatic toxicities associated with a whole-body Lin28 loss. Finally, the above DEN-only model did not involve chronic liver injury, which is integral to cirrhosis pathogenesis and the ensuing development of HCC. To address these limitations, we developed an inducible genetic deletion model that would accurately mimic complete LIN28A and LIN28B inhibition in the entire animal. We generated uninducible control (TRE-Cre; Lin28afl/fl; Lin28bfl/fl) and inducible whole-body Lin28a/Lin28b double-KO (DKO) mice (CAG-Rtta; TRE-Cre; Lin28afl/fl; Lin28bfl/fl), which allowed us to delete Lin28a and Lin28b in all cells of the adult mouse upon doxycycline (dox) administration (Figure 1E). At 2 weeks of age (P14), a single dose of DEN was injected to induce liver mutagenesis. At 6 weeks of age, dox was given to all mice for 2 weeks, which caused deletion of Lin28a/Lin28b in all cells of experimental mice but had no genetic impact on control mice (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI165734DS1). To introduce chronic injury, we initiated chronic carbon tetrachloride (CCl4) twice a week for 12 weeks starting at 10 weeks of age, which caused repeated centrilobular hepatocyte necrosis, accompanied by inflammation and fibrosis.
Almost all gross liver tumors were prevented in the setting of dual Lin28 deletion (Figure 1, E–G, and Supplemental Figure 1B). Seven of nine control mice developed grossly detectable tumors of the liver surface, while only 1 of 9 Lin28a/Lin28b-DKO mice developed tumors (Figure 1G). We asked whether the tumors that arose in the Lin28a/Lin28b-DKO background had escaped Lin28b deletion, and indeed we detected LIN28B expression in most tumors that formed in the setting of Lin28a/Lin28b deletion (Figure 1H). These data indicated that LIN28 expression was critical for cancer development in the DEN alone and in the DEN+CCl4 models that integrate mutagenesis, chronic tissue damage, and fibrosis. Next, we asked whether LIN28s might be affecting tumor development through influences on tissue injury or fibrogenesis. In addition to the DEN+CCl4 model, we also subjected WT and Lin28a/Lin28b-DKO mice to 12 or 20 weeks of long-term CCl4 injury without DEN (Supplemental Figure 1C). Even after 20 weeks of CCl4, there was no induction of Lin28a/Lin28b expression in WT liver tissues (data not shown), and there were no differences in the amount of fibrosis-related gene expression between WT and Lin28a/Lin28b-DKO mice (Supplemental Figure 1, C and D). We also examined Lin28 expression levels in livers from mice fed Western diets for 6 months. While these mice developed fatty liver disease and had increased fibrogenic gene expression (Supplemental Figure 1E), there was no increase in Lin28a or Lin28b expression (Supplemental Figure 1F). In human livers, LIN28A and LIN28B overexpression were observed in HCCs, but not in nonmalignant liver tissues from stage 4 fibrosis patients (Supplemental Figure 1, G–J). Collectively, these data suggest that blocking LIN28s impaired cancer formation but did not affect other histologic processes associated with chronic liver disease.
NRASG12V activation and Tp53 loss cause a multifocal mixed HCC/cholangiocarcinoma.
It is challenging to dissect the underlying molecular mechanisms associated with mouse models of cancer, in part due to the low-throughput nature of mouse genetics. To dissect LIN28-associated mechanisms in vivo, we needed more rapid models that could also mimic the genetics of the chronic injury models used above. We previously examined the genetic drivers of DEN cancers. Sequencing of 242 of the most commonly mutated genes in human and murine HCCs in 50 individual DEN tumors (33) identified recurrent, mutually exclusive mutations in Egfr (Phe254Ile), Hras (Gln61Arg), and Braf (Val637Glu). These data suggested that DEN tumor transformation depends on at least one of these EGFR-RAS-MAPK–activating mutations. In line with these findings, an increase of p-ERK, a core effector of RAS signaling, was detected in DEN tumors and premalignant lesions (Supplemental Figure 2). To determine whether isolated RAS pathway activation and Tp53 deletion could recapitulate the DEN-induced HCC models, we tested a model of liver cancer driven by NRASG12V activation. We used hydrodynamic transfection (HDT) to introduce a transposable vector containing a constitutively active NRASG12V into liver-specific Tp53-KO mice. This was sufficient to drive multifocal carcinomas within 2–3 weeks (Supplemental Figure 3A) (34, 35). Individual NRASG12V-driven tumors contained both HCC and cholangiocarcinoma (CCA) histologies (Supplemental Figure 3B), a mixed entity that is frequently seen clinically. Regions of CCA stained for CK19 and EPCAM, while regions of HCC expressed high levels of IGF2 and AFP, and both histological types were MYC positive (Supplemental Figure 3, B and C). This NRASG12V/Tp53 model was advantageous because the driver lesions were defined and because the rate of tumorigenesis was shortened from 8 months to under 1 month.
Within NRASG12V/Tp53 cancers, we found high LIN28B and modest LIN28A expression in a subset of cancer cells, corroborated by quantitative PCR (qPCR) on bulk tumor samples (Supplemental Figure 3, C and D). We also saw increased expression of IGF2BP1, IGF2BP2, and IGF2BP3, oncofetal proteins known to be regulated by the LIN28/Let-7 pathway (Supplemental Figure 3, C and D). To determine whether the earliest lesions that ultimately gave rise to these tumors expressed LIN28A or LIN28B, we examined the hypercellular lesions that arose shortly after HDT. These small lesions were detected as early as 12 days after HDT near the pericentral veins or in zone 3 of liver lobules (Supplemental Figure 3E). We hypothesized that these small 5 - to 10-cell diameter lesions could contain the cells of origin for this HCC model. Similar to the DEN-induced HCC progenitors, these lesions expressed IGF2BP3 and were LIN28B but not LIN28A positive (Supplemental Figure 3E), suggesting a greater importance for LIN28B in tumor initiation.
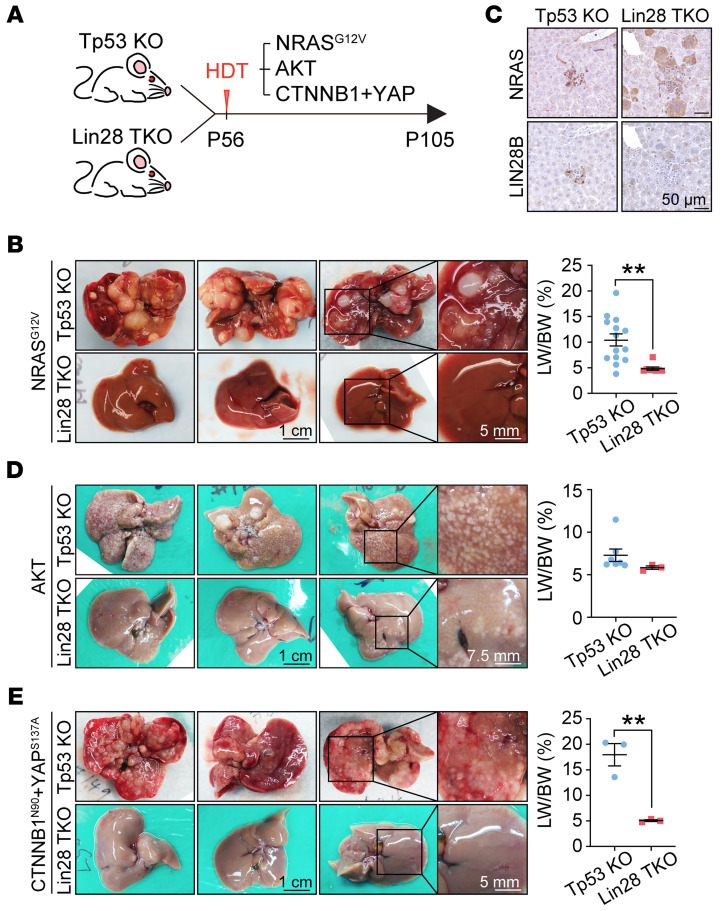
To determine whether LIN28 proteins are also required for NRASG12V/Tp53-specific liver cancer development, we generated the same NRASG12V activation model in the context of liver-specific Lin28a/Lin28b/Tp53-TKO mice (Figure 2A). Single Tp53-KO mice succumbed to tumorigenesis within 7 weeks, but Lin28a/Lin28b/Tp53-TKO mice did not develop any cancers even 3 months after HDT (Figure 2B). This demonstrated an even greater requirement for LIN28 proteins in the NRASG12V/Tp53 model when compared with the DEN model. To confirm that HDT plasmids were successfully delivered, we showed that NRASG12V was expressed in the nonmalignant livers of Lin28a/Lin28b/Tp53-TKO mice (Figure 2C). As expected, LIN28B expression was observed in Tp53-KO but not in Lin28a/Lin28b/Tp53-TKO mice (Figure 2C). These results showed that mice without LIN28s are almost completely protected from tumor development in this model, suggesting that LIN28s are required for oncogenic competence and HCC initiation. To determine whether this was an NRASG12V-specific effect, we also performed similar experiments with CTNNB1N90+YAPS127A+Tp53-KO and AKT+Tp53-KO HDT models (Figure 2, D and E). HCC formation, though not completely blocked, was dramatically reduced in these other models. These observations suggested that LIN28 proteins are critical gatekeepers for transformation driven by multiple oncogenic pathways.
Figure 2. Lin28a and Lin28b are both required for liver cancer development.
(A) Schematic for HDT of transposons in Tp53-KO and Lin28a/Lin28b/Tp53-TKO mice. (B) Representative gross images (left) and liver-to-body weight ratios (right) of Tp53-KO (n = 14) and Lin28a/Lin28b/Tp53-TKO (n = 8) mice receiving transposons carrying NRASG12V for 7 weeks (P105). Scale bars: 1 cm; 5 mm (right panels). (C) IHC shows NRAS and LIN28B expression in early lesions of Tp53-KO and Lin28a/Lin28b/Tp53-TKO mice that had NRASG12V injected 2 weeks prior (P70). Scale bars: 50 μm. (D) Representative gross images (left) and liver-to-body weight ratios (right) of Tp53-KO (n = 7) and Lin28a/Lin28b/Tp53-TKO (n = 3) mice receiving transposons carrying AKT for 7 weeks (P105). Scale bars: 1 cm; 7.5 mm (right panels). (E) Representative gross images (left) and liver-to-body weight ratios (right) of Tp53-KO (n = 3) and Lin28a/Lin28b/Tp53-TKO (n = 3) mice receiving transposons carrying CTNNB1N90 and YAPS137A for 7 weeks (P105). Scale bars: 1 cm; 5 mm (right panels). **P < 0.01.
Using Lin28-deficient mouse models to define bypass and rescue pathways.
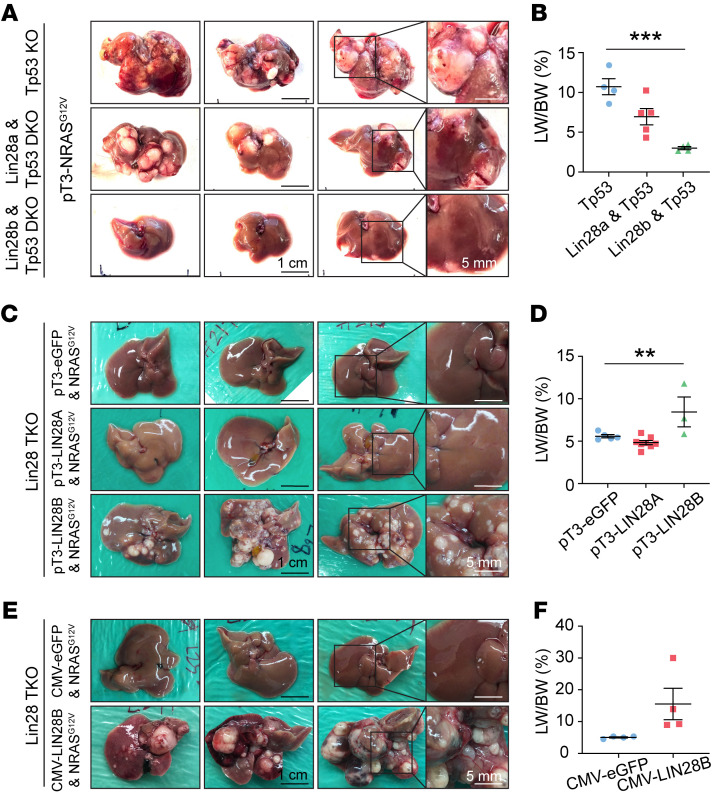
The rapid carcinogenesis and the stark phenotypic difference between Tp53-KO and Lin28a/Lin28b/Tp53-TKO mice facilitated mechanistic dissection in vivo. To first determine whether Lin28a or Lin28b was the more critical component, we genetically segregated mice into ones that had either Lin28a or Lin28b deletion but not both along with Tp53 deletion (Albumin-Cre; Lin28afl/fl; Tp53fl/fl or Albumin-Cre; Lin28bfl/fl; Tp53fl/fl). After NRASG12V activation in each of these models, we saw reduced tumorigenesis in both Lin28a/Tp53- and Lin28b/Tp53-DKO models (Figure 3, A and B), but neither model showed the degree of tumor prevention that was seen when both paralogs were deleted. This indicated that each gene was partially responsible for tumor formation, with Lin28b having a larger effect. We then asked whether overexpression of a single Lin28 paralog in tandem with NRASG12V activation could rescue the Lin28a/Lin28b/Tp53-TKO model. LIN28B overexpression was able to rescue HCC (Figure 3, C and D). In contrast, neither GFP nor LIN28A overexpression was able to rescue HCC. The observation that LIN28A did not rescue was consistent with the lack of LIN28A expression in early cancer lesions. These results indicated LIN28A was required but not sufficient to initiate NRASG12V/Tp53 liver cancers.
Figure 3. Lin28b is indispensable for NRASG12V/Tp53-driven liver cancer development.
(A) Representative gross images of Tp53-KO (Albumin-Cre; Tp53fl/fl; n = 4), Lin28a/Tp53-DKO (Albumin-Cre; Tp53fl/fl; Lin28afl/fl; n = 5), and Lin28b/Tp53-DKO (Albumin-Cre; p53fl/fl; Lin28bfl/fl; n = 4) mice that received NRASG12V by HDT for 7 weeks. Scale bars: 1 cm; 5 mm (right panels). (B) Liver-to-body weight ratios for A. One-way ANOVA was performed. (C) Representative gross images of Lin28a/Lin28b/Tp53-TKO mice that received NRASG12V in combination with pT3-eGFP (n = 5), pT3-LIN28A (n = 8), or pT3-LIN28B (n = 3) for 7 weeks. Scale bars: 1 cm; 5 mm (right panels). (D) Liver-to-body weight ratios for C. One-way ANOVA was performed. (E) Representative gross images of Lin28a/Lin28b/Tp53-TKO mice that received NRASG12V combined with pCMV-eGFP (n = 4) or pCMV-LIN28B (n = 4) for 7 weeks. Scale bars: 1 cm; 5 mm (right panels). (F) Liver-to-body weight ratios for E. **P < 0.01; ***P < 0.001.
To determine whether LIN28B was necessary for cancer maintenance in addition to initiation, we also examined survival in Lin28a/Lin28b/Tp53-TKO mice with either transient or continuous overexpression of LIN28B. We injected pT3-NRASG12V but not LIN28B into Tp53-KO mice as the control HCC model. This experiment resulted in cancers with ongoing endogenous Lin28a/Lin28b expression since the Tp53-KO mice are WT for Lin28a and Lin28b. Transient Lin28b overexpression in Lin28a/Lin28b/Tp53-TKO mice was achieved with a LIN28B plasmid without transposon integration sequences (pcDNA3.1-CMV-hLIN28B). Continuous LIN28B overexpression in Lin28a/Lin28b/Tp53-TKO mice was achieved with a LIN28B plasmid with transposon integration sequences (pT3-LIN28B). We reasoned that the lack of transposon integration would allow for initiation, but there would be no persistent LIN28B to drive long-term tumor growth after initiation. The first observation was that NRASG12V plus transient LIN28B overexpression in Lin28a/Lin28b/Tp53-TKO mice could lead to lethal tumors (Figure 3, E and F) with survival similar to that of the control model (Supplemental Figure 4A), indicating that only a brief burst of LIN28B expression was needed to initiate HCC. Two months after HDT, no LIN28B protein expression was seen in the transient rescue group (Supplemental Figure 4B), supporting the observation that LIN28B is required only for initiation. The second major observation is that continuous overexpression of LIN28B did not lead to more aggressive tumor progression compared with the other 2 models, suggesting the lack of a maintenance requirement for LIN28B. We also tested this in vitro by perturbing LIN28B expression in Huh7 and SNU308 cells. LIN28B knockdown in both Huh7 and HCC53N cells showed no influence on cell proliferation (Supplemental Figure 4, C and D), and overexpression in SNU308 did not cause a marked increase in proliferation (Supplemental Figure 4E). Together, these data indicate that LIN28B is not required for tumor maintenance in some HCCs exhibiting LIN28B overexpression.
We then used this in vivo system to define the functional importance of LIN28B’s mRNA targets. By exploiting the difference between the Tp53-KO and the Lin28a/Lin28b/Tp53-TKO mice, we attempted to identify genetic suppressors of the Lin28a/Lin28b deletion phenotype. To determine whether known targets of LIN28A/LIN28B would be able to rescue cancer initiation in Lin28a/Lin28b-deficient mice, we tested Igf2bp1, Igf2bp2, and Igf2bp3. Each of these genes is highly expressed in embryos and in human HCCs but not in adult livers (Supplemental Figure 3D and Supplemental Figure 5A) and thus they are considered “oncofetal” RBPs (36, 37). Igf2bp mRNAs are known to be stabilized by LIN28 binding and downregulated by Let-7 (38). We found that the coinjection of either Igf2bp1, Igf2bp2, or Igf2bp3 in tandem with NRASG12V efficiently rescued cancers in Lin28a/Lin28b/Tp53-TKO mice (Supplemental Figure 5B). We reasoned that because these downstream effectors could “suppress” the TKO phenotype, other molecular effectors might also be revealed in a similar way.
LIN28s bind to RBPs and their mRNAs in an oncofetal regulon.
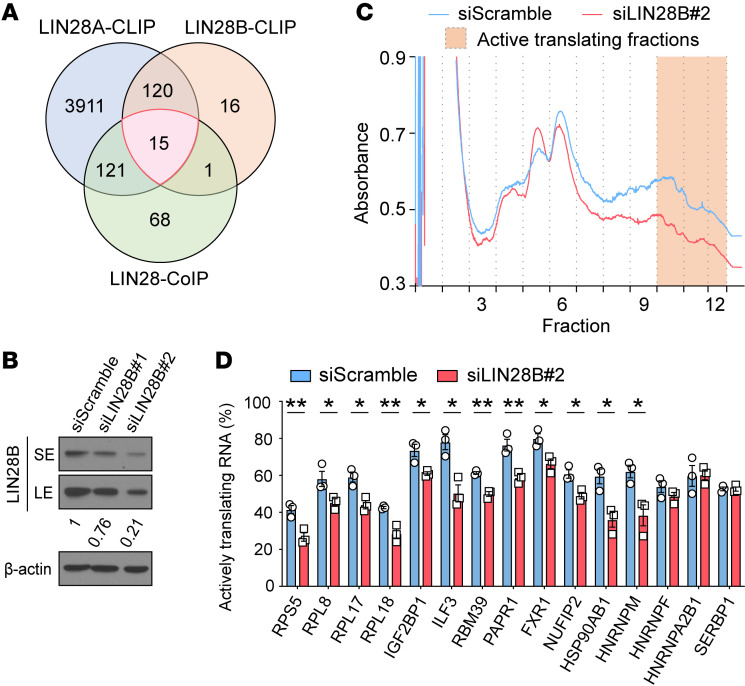
To biochemically define mechanisms by which LIN28s promote cancer, we investigated RBP networks involving LIN28 proteins. We hypothesized that there is a LIN28-associated RNA regulon or a group of functionally related mRNAs and proteins whose expression is coordinated through direct interactions. LIN28s interact with thousands of mRNA targets, so we sought to identify the subset of these interactions that might have a larger functional impact. In addition, LIN28 proteins have protein cofactors that likely augment LIN28 functionality. We reasoned that LIN28s could enhance the translation of mRNAs that ultimately become protein cofactors in a feed-forward loop. We sought to identify cofactors that also interact with their own mRNA transcripts by integrating LIN28-mRNA and LIN28-proteomic interaction data sets. We performed LIN28 co-IP and mass spectrometry (MS) on lysates from embryonic stem cells and leukemia cell lines with high levels of LIN28A or LIN28B expression (Supplemental Table 1). We then analyzed published eCLIP data for LIN28A and LIN28B from embryonic stem cells and HEK293T cells (Supplemental Tables 2 and 3) (38–41). This eCLIP data identified mRNAs bound and regulated by LIN28. The intersection of these protein and mRNA interaction data sets revealed 15 factors that bind to LIN28A or LIN28B as both proteins and mRNAs (Figure 4A and Supplemental Table 4). Using Western blotting (WB) and RNA IP qPCR (RIP-qPCR), we confirmed that all 15 targets interacted with LIN28B as mRNAs and proteins (Supplemental Figure 6, A and B). Interestingly, all 15 were RBPs or ribosomal proteins (Supplemental Table 5) and 14 of 15 targets were not likely to be regulated by Let-7 because they do not have Let-7–binding sites in their 3′ UTRs based on TargetScan (https://www.targetscan.org/vert_80/) analysis (only Igf2bp1 is Let-7 regulated). According to TCGA transcriptomic data, all 15 were overexpressed in human HCCs as compared with nonmalignant liver tissues (Supplemental Figure 6C), supporting oncogenic functionality for this RNA regulon.
Figure 4. LIN28B promotes the translation of RBP target mRNAs.
(A) Venn diagram shows 15 factors that bind to LIN28 proteins as both proteins and mRNAs. See gene names in Supplemental Tables 1–4. (B) WB analysis shows LIN28B expression 48 hours after siRNA treatment in Huh7 cells. SE, short exposure; LE, long exposure. The numbers below the boxes show relative intensity. (C) Polysome profiling shows total translational activity in Huh7 cells with LIN28B siRNA knockdown compared with control siRNA–treated cells. The graph shows a representative profile from 3 replicate experiments. (D) RT-qPCR analysis of LIN28 targets show the percentage of active translating mRNA fraction in control and LIN28B knockdown Huh7 cells. Data include 3 biological replicates. *P < 0.05; **P < 0.01.
Given the relationship between LIN28s and these RBPs, we hypothesized that LIN28s might directly regulate the stability or expression of their mRNAs. Three possible mechanisms of LIN28 regulation were tested: (a) transcriptional activation, (b) mRNA stabilization, and (c) translational enhancement. To distinguish among these, we performed LIN28B loss- and gain-of-function experiments in 2 human liver cancer cell lines that do and do not express LIN28B (Huh7 and SNU308, respectively) (Figure 4B and Supplemental Figure 7A). Since LIN28B has been reported to act as a transcription factor and is expressed in the nucleus (42, 43), we asked whether these targets were transcriptionally regulated by LIN28B. Neither LIN28B siRNA knockdown nor retroviral overexpression influenced target mRNA levels (Supplemental Figure 7, A and B). Second, we tested the idea that LIN28B could increase target mRNA stability through direct interactions, as suggested previously (38, 44). To examine mRNA stability, we inhibited transcription with actinomycin D after LIN28B knockdown or overexpression. These perturbations did not change relative mRNA levels for any target except for SERBP1 (Supplemental Figure 7, C and D). These data provided little evidence for direct LIN28B effects on mRNA transcription or stabilization.
LIN28B promoted the translation of target genes through protein-mRNA interactions.
Next, we tested to determine whether LIN28B regulates the translation of some of these targets. Previously, it was shown that LIN28 proteins bring mRNAs to polysomes, resulting in increased translational efficiency (25, 41, 45–47). To more broadly identify mRNA targets whose translation is increased with the presence of LIN28B, we performed polysome analysis of control and LIN28B siRNA cell lines. LIN28B knockdown in Huh7 resulted in an overall reduction of polysome abundance (Figure 4C). After LIN28B knockdown, 12 of the 15 oncofetal regulon mRNAs (RPL8, RPL17, RPL18, RPS5, ILF3, IGF2BP1, RBM39, PARP1, HSP90AB1, FXR1, NUFIP2, HNRNPM) redistributed to less active polysome fractions, indicating reduced association with actively translating ribosomes (Figure 4D). Three targets (HNRNPF, HNRNPA2B1, SERBP1) showed no change in polysome distribution (Figure 4D), indicating that not all of LIN28B’s interacting mRNAs are regulated from a translational standpoint.
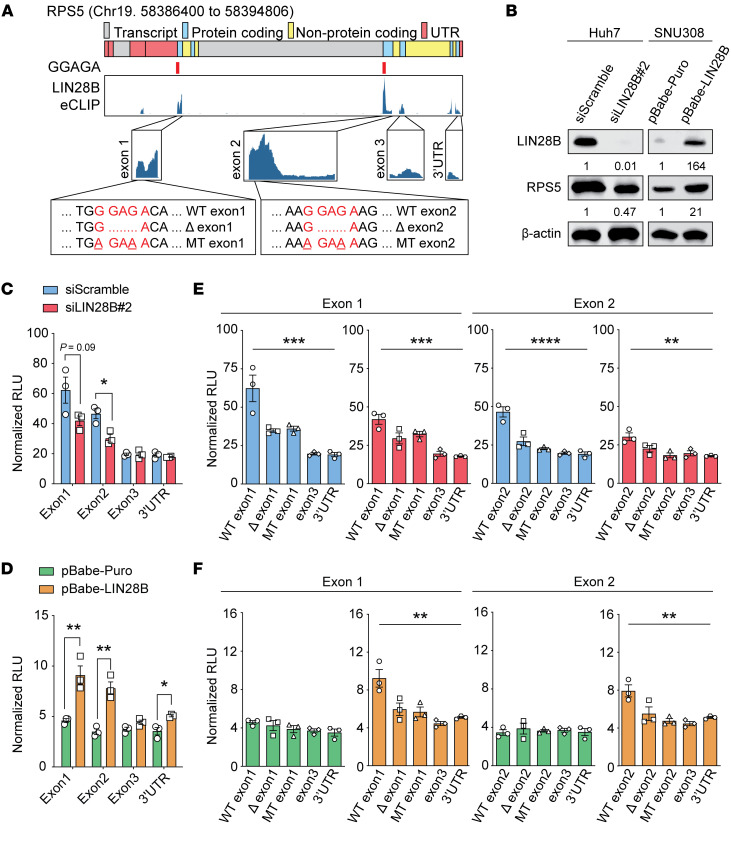
To further determine whether LIN28B directly regulates target mRNAs found in polysomes, we established dual-luciferase reporter assays to probe translational regulation. For ribosomal protein S5 (RPS5) mRNA, we identified direct LIN28B-binding regions through examination of eCLIP-Seq data from HepG2, an HCC cell line (48). LIN28B binds to exons 1 and 2 of RPS5 mRNA (Figure 5A), so these LIN28B-binding regions were cloned into the renilla luciferase reporter 3′ UTR. Because eCLIP data show minimal LIN28B binding to exon 3 or the 3′ UTR of RPS5, we used these as negative control sequences. We transfected these reporters into Huh7, an HCC cell line with high LIN28B (Figure 5B). RPS5 exon 1 and exon 2 reporters showed increased luciferase activity when compared with exon 3 or 3′ UTR reporters; LIN28B knockdown specifically reduced luciferase only for exon 1 and exon 2 reporters (Figure 5C). We then introduced the same reporters into SNU308, a liver cancer cell line with little LIN28B expression. As expected, we did not see a marked difference between exon 1/2 and exon 3/3′ UTR reporters, presumably because LIN28B was not present to affect translation. Consistent with this, exon 1/2 reporter activity in SNU308 increased after LIN28B overexpression (Figure 5D). These data indicated that the presence of LIN28B was critical for increasing the translation of LIN28-responsive reporters. Because LIN28B is known to bind a consensus GGAGA motif (41), we identified such motifs in exon 1 and 2 and generated reporters in which these GGAGA motifs were deleted or mutated. The mutated reporters showed decreased luciferase activity compared with WT reporters in Huh7 cells and SNU308 cells overexpressing LIN28B, but not in SNU308 without LIN28B expression (Figure 5, E and F). We also observed similar findings for interleukin enhancer-binding factor 3 (ILF3) reporters (Supplemental Figure 8, A–F). To more directly demonstrate mRNA binding by LIN28B, we overexpressed FLAG-LIN28B and an equal mixture of luciferase constructs containing the WT or 2 mutant motifs. FLAG-LIN28B pulldown led to the enrichment of luciferase mRNA containing the WT binding motif. Both the deletion and mutant binding motifs were not as enriched (Supplemental Figure 8G). Together, these data show that LIN28B binds to specific sequences within target mRNAs, resulting in increased translation.
Figure 5. LIN28B regulates RPS5 translation through direct mRNA binding.
(A) eCLIP data show LIN28B-binding regions on RPS5 mRNA. Red marks under the gene indicate the location of consensus LIN28B-binding motifs (GGAGA). Deletion (Δ) and mutation (MT) of consensus motifs were introduced to prevent LIN28B binding. Exon 1, 2, 3, and 3′ UTR sequences were inserted into the Renilla 3′ UTR region. (B) WB analysis of RPS5 and LIN28B in Huh7 and SNU308 cell lines. Numbers below the boxes show relative intensities. (C) Renilla luciferase activity promoted by RPS5 exon 1, exon 2, exon 3, and 3′ UTR reporters in LIN28B siRNA knockdown (n = 3) versus control Huh7 cells (n = 3). (D) Renilla luciferase activity promoted by RPS5 exon 1, exon 2, exon 3, and 3′ UTR reporters in LIN28B overexpression (n = 3) versus control SNU308 cells (n = 3). (E and F) Renilla luciferase activity promoted by WT RPS5 sequences compared with deletion and mutation containing reporters in control (blue) and LIN28B siRNA knockdown (red) Huh7 cells (n = 3) (E) and in control overexpression (green) and LIN28B overexpression (orange) SNU308 cells (n = 3) (F). One-way ANOVA was performed. **P < 0.01; ***P < 0.001; ****P < 0.0001.
LIN28 target genes can rescue tumorigenesis by increasing protein synthesis in Lin28-deficient mice.
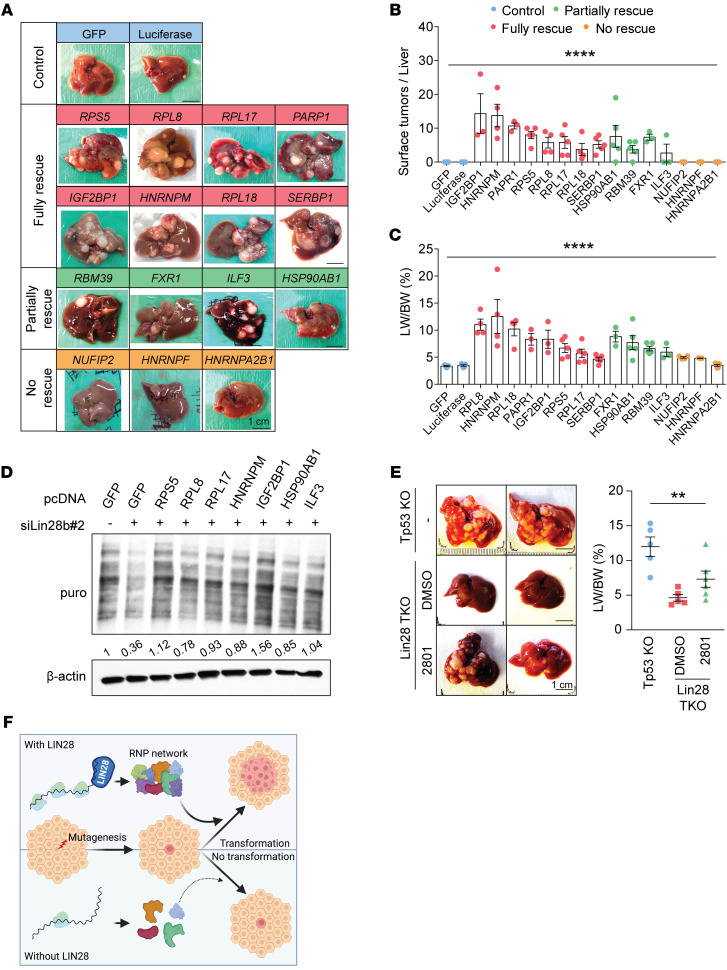
We tested the functional importance of these RNA regulon components by attempting to rescue HCC in the Lin28a/Lin28b/Tp53-TKO model. NRASG12V, SB100, and GFP injected into TKO mice were not able to initiate any tumorigenesis. To ensure that the lack of tumorigenesis was not due to GFP-related effects, we also showed that a construct containing Luciferase did not rescue tumorigenesis (n = 3; Figure 6A and Supplemental Figure 9). Next, we asked whether overexpressing LIN28 target genes would be able to rescue HCC in TKO mice. To do this, we individually coinjected 15 transposons containing full-length cDNAs of the RBP targets along with NRASG12V and SB100 into TKO mice, then harvested livers 7 weeks after HDT. In contrast with GFP or Luciferase, 8 of 15 LIN28 targets could cause full rescues of HCCs in TKO mice (Figure 6A) and 4 of 15 showed partial rescues (Figure 6, B and C, and Supplemental Figure 9). A full rescue was defined as all mice regaining grossly visible tumors on the liver surface, while a partial rescue was defined as only a subset of mice regaining visible tumors. Interestingly, the genes with the strongest rescue effects were also those that were subject to translational regulation by LIN28B (Figure 4D), while 2 of 3 genes (HNRNPF and HNRNPA2B1) that did not rescue were not translationally affected by LIN28. This use of transposons containing GFP or Luciferase also ensured that the rescue of tumorigenesis was not simply due to the random integration of transposons into cancer-driving loci (Figure 6A and Supplemental Figure 9).
Figure 6. A subset of LIN28’s RBP targets can rescue tumorigenesis in Lin28a/Lin28b/Tp53-TKO mice in part through increases in protein synthesis.
(A) Representative gross images of Lin28a/Lin28b/Tp53-TKO mice receiving NRASG12V in combination with pT3-RBPs (n > 3), pT3-eGFP (n = 5), or pT3-Luciferase (n = 5) for 7 weeks. Scale bars: 1 cm. All the images can also be found in Supplemental Figures 5B and 9. (B) Surface tumor number for A. One-way ANOVA was performed. (C) Liver-to-body weight ratios for A. One-way ANOVA was performed. (D) WB analysis quantified OP-puro labeling protein in Huh7 cells with Lin28b knockdown plus target overexpression. Number below the box shows relative intensity. (E) Representative gross images of Tp53-KO (Albumin-Cre; Tp53fl/fl; n = 5) and Lin28a/Lin28b/Tp53-TKO (Albumin-Cre; Tp53fl/fl; Lin28afl/fl; Lin28bfl/fl) mice that received NRASG12V by HDT for 7 weeks. TKO mice were subjected to BAZ2A inhibitors (n = 6) or DMSO (n = 5) once per week starting 3 days after HDT. Scale bars: 1 cm. One-way ANOVA was performed. (F) Schematic shows the importance of LIN28 proteins for tumor initiation. The image was designed and drawn using BioRender. **P < 0.01; ****P < 0.0001.
We asked how these targets rescued tumorigenesis in the absence of LIN28 proteins. We observed that LIN28 proteins regulated protein synthesis since the loss of Lin28a/Lin28b reduced polysome abundance (Figure 4C). In support of a broad influence on protein synthesis, we performed an additional assay to measure global protein synthesis rates. O-propargyl-puromycin (OP-puro) is an analog of puromycin that is incorporated into newly translated proteins. OP-puro is used to measure newly synthesized proteins. Cycloheximide was used as a strong control that almost completely inhibited translation. We observed that LIN28B siRNA knockdown decreased OP-puro fluorescence signaling in Huh7 cells (Supplemental Figure 10A). WB analysis also showed a similar finding (Supplemental Figure 10B). Just because LIN28B promoted global protein synthesis rates does not mean that this is the means through which cancer initiation is promoted. We sought to determine whether an increase in protein synthesis is sufficient for cancer initiation. One set of findings that supports a role for protein synthesis is that when we rescue tumorigenesis with target gene overexpression in vivo, the corresponding in vitro overexpression of these targets also showed an increase in global protein synthesis (Figure 6D).
To functionally test whether protein synthesis is sufficient to rescue the effects of Lin28a/Lin28b loss, we used an orthogonal method to increase protein synthesis in the absence of LIN28 proteins. We enhanced translation through the inhibition of BAZ2A/B proteins. BAZ2A and BAZ2B are chromatin-remodeling proteins that suppress ribosomal protein and ribosomal DNA transcription. Genetic or pharmacological inhibition of BAZ2A/B proteins leads to increased protein synthesis and liver regeneration (49). A small molecule called GSK2801 promotes increases in protein synthesis through bromodomain inhibition of BAZ2A/B proteins (50). GSK2801 rescued tumor development in Lin28a/Lin28b/Tp53-TKO mice. Four out of six mice receiving GSK2801 developed visible tumors on the liver, while 0 of 4 vehicle-treated TKO livers developed tumors (Figure 6E). These data show that increases in translation, in part mediated through LIN28, are important for liver tumor initiation caused by NRASG12V/Tp53.
Discussion
The processes that regulate oncogenic competence and tumor initiation are especially important for cancers associated with chronic inflammatory conditions, such as cirrhosis. While LIN28s are also important for pediatric hepatoblastoma progression (32) and HCC cell line growth in vitro (51), the degree to which they are required for initiation of adult HCC in animal models is not clear. Here, we showed that the lack of LIN28A and LIN28B had no impact on liver development or function, but largely abrogated tumor initiation driven by powerful oncogenes such as NRAS, CTNNB1, YAP, and AKT. The phenotypic magnitude of these observations, paired with the fact that LIN28 proteins are dispensable in adults, suggests that they could be attractive targets for liver cancer prevention. It is fortuitous that these particular requirements for oncogenic competence are not apparently essential for normal adult physiology or regeneration.
The stark nature of the Lin28a/Lin28b deletion phenotype made it easier to dissect the underlying molecular mechanisms associated with LIN28-driven cancers. LIN28s are well known to inhibit Let-7 miRNA biogenesis and function. However, the non–Let-7 target genes that exert important oncogenic effects are comparably less investigated. We hypothesized that LIN28s help to organize a ribonuclear protein regulon involving many other RNAs and RBPs that are required for oncogenic competence. We showed that LIN28s interact with a large number of mRNAs and proteins within cancer cells and most of these interactions did not occur through Let-7, given the lack of Let-7–binding sites within target mRNAs. Instead, LIN28s directly bind the mRNAs of these targets, bring them to polyribosomes, and increase their protein synthesis rates (Figure 6F).
A key question is how might LIN28 promote translation in general, rather than just the translation of its target genes. Interestingly, many of LIN28’s target proteins are involved in ribosome biogenesis and RNA metabolism, suggesting that the targets of LIN28 might themselves exert broad influences on protein synthesis. For example, RPS5, RPL17, RPL18, and RPL8 are each ribosomal components. Because LIN28 increases the protein production of these ribosomal components, there could be a resulting feed-forward loop that increases global protein synthesis. If LIN28B was only binding nontranslation-related proteins that were not involved in protein synthesis, then one would only see a specific increase in the synthesis of those proteins, but not a global increase in protein synthesis. Indeed, a previous study also showed interactions between LIN28B and general protein synthesis machinery in cancer (29). The interactions between LIN28 and ribosomal components are conserved in different cancer types, including neuroblastoma. In these ways, the reactivation of an oncofetal RBP network involving LIN28 proteins is essential for malignant transformation. A major goal in cancer prevention would be to eliminate or target genes that are required for cancer development, but which have no essential physiological functions. Our study highlights a group of oncofetal genes that might be required during embryogenesis, unnecessary in normal adult tissues, but then required again for cancer initiation.
Methods
Sex as a biological variable.
For the DEN-induced HCC model and the CCl4-induced chronic injury model, sex was not considered as a biological variable. For HDT-induced HCC models, only males were used due to higher and faster rates of cancer development.
Mice.
Albumin-Cre and Tp53fl/fl mice were purchased from The Jackson Laboratory and backcrossed more than 15 generations into the FVB/N inbred mouse strain to generate Tp53-KO (Albumin-Cre+/–; Tp53fl/fl) mice. Conditional Lin28a/Lin28b-DKO mice (Cag-rtTA–/+; TRE-Cre–/+; Lin28afl/fl; Lin28bfl/fl) were reported previously (52, 53). Lin28a/Tp53-DKO (Albumin-Cre; Tp53fl/fl; Lin28afl/fl), Lin28b/Tp53-DKO (Albumin-Cre; Tp53fl/fl; Lin28bfl/fl), and Lin28a/Lin28b/Tp53-TKO (Albumin-Cre; Tp53fl/fl; Lin28afl/fl; Lin28bfl/fl) mice were generated by crossing Tp53 to Lin28a and Lin28b double-floxed mice. DEN (MilliporeSigma) was diluted in saline and injected intraperitoneally at age P15 at a dose of 25 μg/g. CCl4 (MilliporeSigma) was diluted 1:10 in corn oil (MilliporeSigma) and injected once per week. To induce fatty liver disease, mice were fed with Western diet containing 21.1% fat, 41% sucrose, and 1.25% cholesterol by weight (Envigo, TD.120528) and high sugar water (23.1 g/L D-fructose (Sigma-Aldrich, F0127) and 18.9 g/L D-glucose (Sigma-Aldrich, G8270) as described previously (54).
Antibodies.
Antibodies used were as follows: NRAS (Abcam, AB77392, IHC), p-ERK (CST, 9101, IHC), LIN28B (Proteintech, 16178-1-AP or CST 5422, IHC and WB), LIN28A (CST, 8641, IHC), c-MYC (MilliporeSigma, M4439, IHC), RPS5 (Abcam, AB210745, WB), IGF2BP1 (Abcam, AB166798, IHC), IGF2BP2 (Abcam, AB124930, IHC), IGF2BP3 (Proteintech, 14642-1-AP, IHC), CK19 (DSHB, TROMA-III, IHC), EpCAM (CST, 14452, IHC), and β-actin (CST 4970, WB).
Cell culture.
Huh7 and SNU308 cells were provided by Helen Hobbs’s lab at University of Texas Southwestern. The HCC53N cell line was previously generated by our lab from FVB mice with NRASG12V and Tp53 deleted liver cancers. Huh7 cells were cultured in DMEM with 10% FBS and penicillin-streptomycin. SNU308 cells were cultured in DMEM with 10% heat-inactivated FBS and penicillin-streptomycin. All cells were cultured at 37°C in a 5% CO2 incubator. For transfections, Huh7 cells (2.5 × 105) were transfected with 25 pmol siRNA (Life Technologies) in 6-well plates by using Lipofectamine RNAiMAX and OptiMEM (Life Technologies), as described in the manufacturer’s instructions. To generate SNU308 cells with LIN28B overexpression, cells were transfected in a 10 cm dish using Lipofectamine 3000 and OptiMEM, then selected with puromycin for 1 week to establish stable overexpression clones.
Protein extraction and WB analysis.
Cells were lysed using RIPA lysis buffer supplemented with Complete Protease Inhibitor (Roche) and subsequent 20% amplitude sonication for 5 seconds. Lysates were cleared by 14,000 g centrifugation at 4°C for 15 minutes. Equivalent lysates were separated by SDS-PAGE and electro-transferred onto polyvinylidene difluoride membranes (Fisher Scientific). Following blocking in 5% nonfat dry milk in PBST for 30 minutes, membranes were incubated in primary antibodies overnight. Horseradish-peroxidase–conjugated secondary antibodies diluted 1:5,000 in 5% nonfat milk were used. Bands were visualized with SuperSignal West Pico or Femto substrate kits (Thermo Fisher). The following commercial primary antibodies supplemented with 0.02% sodium azide were used for immunoblot analysis: NRAS, LIN28B, RPS5, RPL8 (Abcam, AB155136), RPL17 (Abcam, AB155781), RPL18 (Abcam, AB241988), IGF2BP1, ILF3 (Abcam, AB92355), RBM39 (Sigma-Aldrich, HPA001591), PARP1 (Abcam, AB191217), FXR1 (Abcam, AB155124), SERBP1 (Thermo Fisher Scientific, TA800699), HNRNPF (Invitrogen, PA5-79382), HNRNPM (Invitrogen, MA1-34981), HNRNPA2B1 (Invitrogen, PA5-34939), HSP90AB1 (Invitrogen, MA1-10372), puromycin (Thermo Fisher Scientific, MABE343MI), and β-actin.
Molecular biology and cloning.
The transposon plasmids used for in vivo tumorigenesis rescue studies were modified from pT3-Ef1ɑ-GW, which was provided by Xin Chen at UCSF (San Francisco, California, USA). Full-length cDNAs including GFP, Luciferase, RPS5, RPL8, RPL17, RPL18, SERBP1, PARP1, IGF2BP1, RBM38, FXR1, ILF3, HSP90AB1, NUFIP2, HNRNPM, HNRNPF, and HNRNPA2B1 were purchased from Horizon Discovery and used to replace the CcdB gene using the gateway LR recombination reaction (Invitrogen, 11791100). Sequences were then confirmed by Sanger sequencing.
HDT.
SB100, pT3-MYC, and pT3-CTNNB1 transposon plasmids were obtained from Xin Chen at UCSF. All mice were injected at approximately 6 weeks of age, when their body weights were at least 20 g. HDT plasmids were suspended in 2 mL of saline and administered via tail-vein injection over 7 seconds. A 10:1 mass ratio of combined HDT plasmids to SB100 transposase plasmid was used.
Histology and IHC.
Tissues were fixed in 4% paraformaldehyde for 16 to 24 hours and embedded in paraffin. Tissue sections were deparaffinized with xylene and rehydrated with a graded series of ethanol (100%, 95%, 80%, and 50% ethanol and distilled water), followed by 2 washes of 5 minutes each in PBS with 0.05% Tween-20 (PBST). Antigen retrieval was performed for 20 minutes in sodium citrate buffer (10 mM at pH 6) at 90–100°C, followed by a wash with PBST for 5 minutes. Tissue sections were then incubated for 10 minutes in 3% (vol/vol) hydrogen peroxide in methanol to block endogenous peroxidase activity. Sections were then washed for 5 minutes in PBST and blocked at 25°C for 1 hour using 2% normal goat serum, 2% BSA, and 0.1% Triton X-100 in PBS. Tissue sections were then incubated in a humidified chamber overnight at 4°C with primary antibody (1/200 in TBST). Sections were subsequently washed with PBST (3× for 5 minutes each) and incubated at 25°C for 1 hour with a secondary antibody. After washing with PBST (3× for 5 minutes each), sections were incubated with ready-to-use streptavidin peroxidase (Lab Vision) for 10 minutes at 25°C and then color was developed using the DAB Kit (Vector Laboratories). Sections were counterstained with hematoxylin. The following primary antibodies were used: NRAS, p-ERK, LIN28B, LIN28A, c-MYC, IGF2BP1, IGF2BP2, IGF2BP3, CK19, and EpCAM.
RNA extraction and RT-qPCR.
Liver total RNA was isolated using TRIzol reagent (Invitrogen, 15596018) followed by purification using the RNeasy Mini Kit (QIAGEN). For reverse-transcription qPCR (RT-qPCR), cDNA synthesis was performed with 1 μg of total RNA using iScript RT Supermix (Bio-Rad, 1708840) in a total of 20 μl volume per reaction. To measure mRNA expression, each cDNA sample (20 μl) was diluted to 200 μl, and 5 μl was combined with primers and iTaq Universal SYBR Green Supermix (Bio-Rad, 172-5121) in a total of 12 μl volume of reaction. The mRNA levels were normalized to β-actin expression. The following RT-qPCR primers were used: hLIN28A (forward: 5′-GAGCATGCAGAAGCGCAGATCAAA; reverse: 5′-TATGGCTGATGCTCTGGCAGAAGT), hLIN28B (forward: 5′-GCCCCTTGGATATTCCAGTC; reverse: 5′-TGACTCAAGGCCTTTGGAAG), hGAPDH (forward: 5′-ATGGCCTTCCGTGTTCCT; reverse: 5′-CAGGCGGCACGTCAGAT), hVIM (forward: 5′-TGTCCAAATCGATGTGGATGTTTC; reverse: 5′-TTGTACCATTCTTCTGCCTCCTG), hCOL1A1 (forward: 5′-GATTCCCTGGACCTAAAGGTGC; reverse: 5′-AGCCTCTCCATCTTTGCCAGCA), hACTA2 (forward: 5′-AAAAGACAGCTACGTGGGTGA; reverse: 5′-GCCATGTTCTATCGGGTACTTC), hRPS5 (forward: 5′-ATGACCGAGTGGGAGACAG; reverse: 5′-GCTTTGCGGAAGCGTTTGG), hRPL8 (forward: 5′-AAGGGCATCGTCAAGGACATC; reverse: 5′-CAGCTCCGTCCGCTTCTTAAA), hRPL17 (forward: 5′-GAACCCCACGAAATCATGCAA; reverse: 5′-TGAACACGAAGATTGGAACCTC), hRPL18 (forward: 5′-ATGTGCGGGTTCAGGAGGTA; reverse: 5′-CTGGTCGAAAGTGAGGATCTTG), hIGF2BP1 (forward: 5′-GCGGCCAGTTCTTGGTCAA; reverse: 5′-TTGGGCACCGAATGTTCAATC), hILF3 (forward: 5′-AGCATTCTTCCGTTTATCCAACA; reverse: 5′-GCTCGTCTATCCAGTCGGAC), hRBM39 (forward: 5′-CAATGCTTGAGGCTCCTTACA; reverse: 5′-TCCGTTCCTTACTTTTGCTTCTC), hPARP1 (forward: 5′-CGGAGTCTTCGGATAAGCTCT; reverse: 5′-TTTCCATCAAACATGGGCGAC), hFXR1 (forward: 5′-GAGAAGACGGTATGGTTCCATTT; reverse: 5′-AGGCGTTCCATTCTTAGCTGT), hNUFIP2 (forward: 5′-GGTGAACTAAACGGTAATGCTGG; reverse: 5′-GCTAGTGTCTACAACTTGCTGG), hSERBP1 (forward: 5′-ATTTGACGACGAATCGGACCC; reverse: 5′-GTTCTTGCGGTCTTTCTGGGA), hHNRNPF (forward: 5′-CTGCTCTGTTGAGGACGTG; reverse: 5′-CCTGCCCTCTCTAGTGTAGATG), hHNRNPM (forward: 5′-GCGGCGACGGAGATCAAAA; reverse: 5′-CTCATTCTGAGCAGGTCGTTC), hHNRNPA2B1 (forward: 5′-ATTGATGGGAGAGTAGTTGAGCC; reverse: 5′-AATTCCGCCAACAAACAGCTT), hACTB (forward: 5′-AGAAGGATTCCTATGTGGGCG; reverse: 5′-CATGTCGTCCCAGTTGGTGAC), mAfp (forward: 5′-CTGGCGATGGGTGTTTAGAA: reverse: 5′-GCCTGAGAGTCCATACTTGTTAG), mIgf2 (forward: 5′-TACCTCTCAGGCCGTACTT; reverse: 5′-ACTGTCTCCAGGTGTCATATTG), mLin28a (forward: 5′-AGGCGGTGGAGTTCACCTTTAAGA; reverse: 5′-AGCTTGCATTCCTTGGCATGATGG), mLin28b (forward: 5′-TTTGGCTGAGGAGGTAGACTGCAT; reverse: 5′-ATGGATCAGATGTGGACTGTGCGA), mIgfbp1 (forward: 5′-ATCAGCCCATCCTGTGGAAC; reverse: 5′-TGCAGCTAATCTCTCTAGCACTT), mIgfbp2 (forward: 5′-CAGACGCTACGCTGCTATCC; reverse: 5′-CCCTCAGAGTGGTCGTCATCA), mIgfbp3 (forward: 5′-CGCCCCACTTACAATGGGAG; reverse: 5′-CTGCCGTTTCCGAATCCGT), mVim (forward: 5′-ACCGCTTTGCCAACTACAT: reverse: 5′-TTGTCCCGCTCCACCTC), mActa2 (forward: 5′-GAGAAGCCCAGCCAGTCG; reverse: 5′-ATCTTTTCCATGTCGTCCCAGTTG), mCol1a1 (forward: 5′-TTCTCCTGGCAAAGACGGACTCAA; reverse: 5′-AGGAAGCTGAAGTCATAACCGCCA), and mGapdh (forward: 5′-ACCACAGTCCATGCCATCAC; reverse: 5′-TCCACCACCCTGTTGCTGTA).
Luciferase assays.
LIN28B-binding sites in the human RPS5, ILF3, IGF2BP1, HSP90AB1, and HNRNPM genes plus approximately 250 bp of flanking sequence were amplified and ligated into the XbaI site of pGL3-control (Promega). Mutagenesis was performed by reamplifying each fragment with primers containing the correct mutations. Mutated fragments were then reintroduced into the pGL3 3′ UTR reporter plasmid using the In-Fusion HD Cloning Kit (Clontech). Twenty-four hours after transfection, cells were lysed and assayed for firefly and renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to renilla luciferase activity to obtain relative luciferase activity. All transfections were performed in triplicate, and 3 experimental trials were performed.
Polysome profiling.
Sucrose gradients were prepared right before use in Beckman ultracentrifuge tubes as described previously (55). One day prior to the experiment, gradients were allowed to diffuse for 16 hours at 4°C. The next day, 20–40 × 106 Huh7 cells were trypsinized and washed 2× with ice-cold PBS containing 100 μg/mL cycloheximide. After the second wash, PBS was discarded and cell pellets resuspended in 750 μL of Polysome Extraction Buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1% NP-40 in distilled water) containing cycloheximide, protease inhibitor cocktail, and RNAse inhibitors. Cells were lysed for 10 minutes on ice and sheared through a 27.5-gauge needle 3–4 times. The lysates were centrifuged at 15,000 g for 5 minutes at 4°C and the supernatant lysate RNA concentration was quantified by NanoDrop (Thermo Fisher Scientific). Equal amounts of lysate (500–600 μg RNA) were loaded across all gradients. The gradients were centrifuged at 35,000 g for 2 hours at 4°C and run on a fractionator machine (Bio-Rad) to visualize and collect polysome fractions. Each collected fraction was mixed with 3× volume of 100% ethanol and 20 μg glycogen carrier and incubated overnight at −20°C. The next day, fractions were centrifuged at 20,000 g for 30 minutes at 4°C to precipitate RNA pellets. Pellets were dried for 20 minutes at room temperature, resuspended in 100 μL Nanopure water (Thermo Fisher Scientific) and 350 μL RNeasy RLT lysis buffer, and loaded onto RNeasy columns (QIAGEN). The RNeasy Kit was used to isolate RNA; then cDNA synthesis and RT-qPCR were performed, and 20 ng of Luciferase mRNA control (Promega, L4561) was added to each fraction prior to RNA extraction to control for variability in total RNA in fractions during RNA isolation and reverse transcription. Fractions associated with fewer than 3 ribosomes were grouped together (poorly translated mRNAs), and fractions with more than 3 ribosomes were grouped together (efficiently translated mRNAs). RT-qPCR was used to quantify mRNA levels in each group. Experiments on all groups were performed in triplicate, and multiple experimental trials were performed.
MS.
FLAG-LIN28B and associated proteins were immunoprecipitated with ANTI-FLAG M2 affinity agarose beads according to the manufacturer’s protocol (MilliporeSigma, catalog A2220). Coprecipitated proteins were separated on a 4%–20% polyacrylamide gel (Bio-Rad) and visualized using the Bio-Safe Coomassie Stain (Bio-Rad). Multiple bands covering most protein sizes were excised and treated with dithiothreitol and iodoacetamide. Proteins were digested in gel. Resulting peptides were gel extracted and analyzed by liquid chromatography–MS (LC-MS) as described previously (56). Peptide matches were filtered by mass accuracy, tryptic state, XCorr, and confirmed by manual inspection.
OP-puro labeling.
For puromycin labeling of newly synthesized protein in cells, after 48 hours of siRNA transfection, 1 μM OP-puro was added to cells for 1 hour. For Western quantification, cells were then washed with cold PBS and lysed with RIPA. For immunocytochemistry analysis, cells were fixed in 4% formaldehyde for 10 minutes, then permeabilized with 0.05% Triton X-100.
Immunofluorescence.
Cells were fixed in 4% PFA for 10 minutes, then permeabilized with 0.5% Triton X-100 for 10 minutes. Cells were then blocked with 5% BSA for 1 hour. After blocking, cells were incubated with primary antibodies overnight. Then cells were washed 3× with PBS, followed by secondary antibody incubation for 1 hour. Nuclei were stained with Hoechst before mounting to a microscope slide. Images were captured using confocal microscopy (Zeiss) and assembled using Fiji ImageJ software (version 2.0.0-rc-43/1.51q). The following antibodies were used: LIN28B, puromycin, anti-mouse Alexa Fluor 488 (Life Technologies, A21131), and anti-rabbit Alexa Fluor 594 (Life Technologies, A21207).
IP.
To examine the interactions between LIN28B protein and target proteins, FLAG-LIN28B constructs were transfected into HEK-293T cells grown in 6-well plates at 80% confluence. One day after transfection, cells were trypsinized and seeded in 10 cm dishes. Cells were harvested when they reached 80% confluence using Pierce RIPA buffer. Anti-FLAG M2 agarose beads (Sigma-Aldrich, A2220, 25 μl of the slurry beads per sample) were washed with Wash Buffer (Thermo Fisher Scientific, 88828, buffer A supplemented with 200 mM NaCl) added to the extracts and rotated overnight at 4°C. After incubation, beads were washed with Wash Buffer 3× and directly boiled with 1× protein loading buffer for 10 minutes at 95°C. The supernatants of the boiled samples were resolved by SDS-PAGE and analyzed by WB. The following antibodies were used: FLAG (CST, 14793S) and rabbit-IgG (CST, 2729).
Statistics.
The data in most panels reflect multiple experiments performed on different days using mice derived from different litters. In all experiments, mice were not excluded from analysis after the experiment was initiated unless the mice died. Data and error bars indicate mean and SEM. Statistical tests used are noted in the figure legends. Unless otherwise stated in the methods or figure legends, 2-tailed Student’s t tests (2-sample equal variance) were used to test the significance of differences between 2 groups. For time-course–expression experiments, 1-way ANOVA was performed and the significance shown was compared with the initial time point. For experiments involving 2 groups and different time points, 2-way ANOVAs were used, and the significance was compared with the means of each group at different time points. Variation is indicated using SEM and presented as mean ± SEM. Statistical significance was defined as P < 0.05.
Study approval.
All mice were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee at University of Texas Southwestern under protocol APN 2015-101118. For human tissue samples, all patients gave consent under Institutional Review Board STU062013-063 or STU092013-010.
Data availability.
All TCGA data used in this study were obtained through the NIH National Cancer Institute (NCI) (https://portal.gdc.cancer.gov/). All data supporting the findings are available within the article, supplemental materials, or Supporting Data Values files. Other data can be obtained upon request.
Author contributions
MHH, YW, and HZ conceived the project, performed the experiments, and wrote the manuscript. JMY, LL, XS, LHN, XW, and YHL assisted with experiments and mouse husbandry. XL performed bioinformatic analyses. SKK and CB contributed to conceptualization of the project. JTP and GQD performed LIN28B co-IP experiments and MS.
Supplementary Material
Acknowledgments
We thank C. Lewis (University of Texas Southwestern Tissue Procurement Service) and J. Shelton (University of Texas Southwestern Histopathology Core) for histopathology. We thank Joshua Mendell’s lab (University of Texas Southwestern) for assistance with polysome profiling and luciferase reporter assays and Helen Hobbs’s lab for HCC cell lines. This study was in part supported by a sponsored research agreement with Redona Therapeutics. HZ was also supported by the Pollack Foundation, NIH/NCI R01 grants (CA190525, AA028791, DK125396), a Cancer Prevention & Research Institute of Texas (CPRIT) New Investigator grant, and an Emerging Leader Award from the Mark Foundation For Cancer Research (21-003-ELA). The Zhu lab is supported by an Innovation Award from the Moody Medical Research Institute.
Version 1. 06/14/2024
In-Press Preview
Version 2. 08/01/2024
Electronic publication
Footnotes
Conflict of interest: HZ had a sponsored research agreement with Redona Therapeutics (formerly Twentyeight-Seven Therapeutics) that supported part of this work. HZ is a cofounder of Quotient Therapeutics and Jumble Therapeutics and is an advisor for Newlimt, Alnylam Pharmaceuticals, and Chroma Medicines. HZ receives research support from Chroma Medicines and owns stock in Ionis and Madrigal Pharmaceuticals. GQD holds equity in and has received consulting fees from Redona.
Copyright: © 2024, Hsieh et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2024;134(15):e165734.https://doi.org/10.1172/JCI165734.
Contributor Information
Yonglong Wei, Email: weiyl@ynu.edu.cn.
Lin Li, Email: Lin2.Li@utsouthwestern.edu.
Liem H. Nguyen, Email: liem.nguyen@utsouthwestern.edu.
Yu-Hsuan Lin, Email: yu-hsuan.lin@utsouthwestern.edu.
Xuxu Sun, Email: xuxu.sun@shsmu.edu.cn.
Xun Wang, Email: xun.wang@utsouthwestern.edu.
Xin Luo, Email: xin.luo@utsouthwestern.edu.
Sarah K. Knutson, Email: sarah.k.knutson@gmail.com.
Christina Bracken, Email: chrissy.brack@gmail.com.
George Q. Daley, Email: george.daley@childrens.harvard.edu.
John T. Powers, Email: john.powers@austin.utexas.edu.
Hao Zhu, Email: hao.zhu@utsouthwestern.edu.
References
- 1.Baggiolini A, et al. Developmental chromatin programs determine oncogenic competence in melanoma. Science. 2021;373(6559):eabc1048. doi: 10.1126/science.abc1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 3.Macaluso M, et al. Genetic and epigenetic alterations as hallmarks of the intricate road to cancer. Oncogene. 2003;22(42):6472–6478. doi: 10.1038/sj.onc.1206955. [DOI] [PubMed] [Google Scholar]
- 4.Zhao BS, et al. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelamraju Y, et al. Mutational landscape of RNA-binding proteins in human cancers. RNA Biol. 2018;15(1):115–129. doi: 10.1080/15476286.2017.1391436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira B, et al. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3(7):506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Q, et al. RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat Rev Cancer. 2017;17(6):381–392. doi: 10.1038/nrc.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvinge H, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rooij LPMH, et al. Post-transcriptional regulation in hematopoiesis: RNA binding proteins take control. Biochem Cell Biol. 2019;97(1):10–20. doi: 10.1139/bcb-2017-0310. [DOI] [PubMed] [Google Scholar]
- 10.Gebauer F, et al. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 11.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Shyh-Chang N, et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155(4):778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt EJ, et al. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development. 2012;139(24):4514–4523. doi: 10.1242/dev.083279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Farzaneh M, et al. Concise review: LIN28/let-7 signaling, a critical double-negative feedback loop during pluripotency, reprogramming, and tumorigenicity. Cell Reprogram. 2017;19(5):289–293. doi: 10.1089/cell.2017.0015. [DOI] [PubMed] [Google Scholar]
- 17.Balzeau J, et al. The LIN28/let-7 pathway in cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovnicki J, et al. LIN28B promotes the development of neuroendocrine prostate cancer. J Clin Invest. 2020;130(10):5338–5348. doi: 10.1172/JCI135373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang LD, et al. The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia. 2015;29(6):1320–1330. doi: 10.1038/leu.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12(4):395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornton JE, et al. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18(10):1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piskounova E, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Qiu C, et al. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38(4):1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakar NY, et al. RELB alters proliferation of human pluripotent stem cells via IMP3- and LIN28-mediated modulation of the expression of IGF2 and other cell-cycle regulators. Stem Cells Dev. 2015;24(16):1888–1900. doi: 10.1089/scd.2014.0587. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. Lin28 mediates radiation resistance of breast cancer cells via regulation of caspase, H2A.X and Let-7 signaling. PLoS One. 2013;8(6):e67373. doi: 10.1371/journal.pone.0067373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, et al. Lin28 promotes dental pulp cell proliferation via upregulation of cyclin-dependent proteins and interaction with let-7a/IGF2BP2 pathways. Biomed Pharmacother. 2019;113:108742. doi: 10.1016/j.biopha.2019.108742. [DOI] [PubMed] [Google Scholar]
- 29.Missios P, et al. LIN28B alters ribosomal dynamics to promote metastasis in MYCN-driven malignancy. J Clin Invest. 2021;131(22):10:e145142. doi: 10.1172/JCI145142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tummala R, et al. Lin28 induces resistance to anti-androgens via promotion of AR splice variant generation. Prostate. 2016;76(5):445–455. doi: 10.1002/pros.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He G, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155(2):384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen LH, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell. 2014;26(2):248–261. doi: 10.1016/j.ccr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018;44(4):447–459. doi: 10.1016/j.devcel.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184(4):912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molina-Sánchez P, et al. Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology. 2020;159(6):2203–2220. doi: 10.1053/j.gastro.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajimura S, et al. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci U S A. 2005;102(4):1240–1245. doi: 10.1073/pnas.0407443102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood TL, et al. Expression of the IGFBP-2 gene in post-implantation rat embryos. Development. 1992;114(1):59–66. doi: 10.1242/dev.114.1.59. [DOI] [PubMed] [Google Scholar]
- 38.Hafner M, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19(5):613–626. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graf R, et al. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 2013;10(7):1146–1159. doi: 10.4161/rna.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48(2):195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao T, et al. LIN28B regulates transcription and potentiates MYCN-induced neuroblastoma through binding to ZNF143 at target gene promotors. Proc Natl Acad Sci U S A. 2020;117(28):16516–16526. doi: 10.1073/pnas.1922692117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Y, et al. Lin28a binds active promoters and recruits tet1 to regulate gene expression. Mol Cell. 2016;61(1):153–160. doi: 10.1016/j.molcel.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su J, et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene. 2021;40(39):5814–5828. doi: 10.1038/s41388-021-01978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samsonova A, et al. Lin28, a major translation reprogramming factor, gains access to YB-1-packaged mRNA through its cold-shock domain. Commun Biol. 2021;4(1):359. doi: 10.1038/s42003-021-01862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin J, et al. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39(9):3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng S, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29(3):496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 48.Van Nostrand EL, et al. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 2020;21(1):90. doi: 10.1186/s13059-020-01982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apaydin MS, et al. Stochastic roadmap simulation for the study of ligand-protein interactions. Bioinformatics. 2002;18 Suppl 2:S18–S26. doi: 10.1093/bioinformatics/18.suppl_2.S18. [DOI] [PubMed] [Google Scholar]
- 50.Stolfi RL, Martin DS. Chemotherapeutic activity of L-histidinol against spontaneous, autochthonous murine breast tumors. Chemotherapy. 1990;36(6):435–440. doi: 10.1159/000238801. [DOI] [PubMed] [Google Scholar]
- 51.Newman JM, Linke R. Chinese immigrant food habits: a study of the nature and direction of change. R Soc Health J. 1982;102(6):268–271. doi: 10.1177/146642408210200613. [DOI] [PubMed] [Google Scholar]
- 52.Shinoda G, et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells. 2013;31(8):1563–1573. doi: 10.1002/stem.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuchida T, et al. Corrigendum to “A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer” [J Hepatol 69 (2018) 385-395] J Hepatol. 2018;69(4):988. doi: 10.1016/j.jhep.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gandin V, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp. 2014;(87):51455. doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All TCGA data used in this study were obtained through the NIH National Cancer Institute (NCI) (https://portal.gdc.cancer.gov/). All data supporting the findings are available within the article, supplemental materials, or Supporting Data Values files. Other data can be obtained upon request.