Abstract
In late summer through early winter of 1998, there were several outbreaks of respiratory disease in the swine herds of North Carolina, Texas, Minnesota, and Iowa. Four viral isolates from outbreaks in different states were analyzed genetically. Genotyping and phylogenetic analyses demonstrated that the four swine viruses had emerged through two different pathways. The North Carolina isolate is the product of genetic reassortment between H3N2 human and classic swine H1N1 influenza viruses, while the others arose from reassortment of human H3N2, classic swine H1N1, and avian viral genes. The hemagglutinin genes of the four isolates were all derived from the human H3N2 virus circulating in 1995. It remains to be determined if either of these recently emerged viruses will become established in the pigs in North America and whether they will become an economic burden.
All known subtypes of influenza A viruses are found among wild avian species that serve as primary reservoirs for these agents (11). In general, an influenza virus infects only a single species; however, whole viruses may occasionally be transmitted from one species to another, and genetic reassortment between viruses from two different hosts can produce a new virus capable of infecting a third host. Avian influenza viruses are not readily introduced into humans (1), possibly because humans do not possess the α(2,3)-sialyllactose (NeuAc-2,3Gal) receptors required for attachment of the viruses to epithelial cells. However, individual viral genes can be transmitted between humans and avian species, as demonstrated by avian-human reassortant viruses that caused the 1957 and 1968 influenza pandemics (16, 33, 39). This finding suggested that an intermediate host may be needed for genetic reassortment of human and avian viruses. Pigs are considered a logical candidate for this role because they can be infected by either avian or human viruses (17, 18, 34) and because they possess both NeuAc-2,3Gal and NeuAc-2,6Gal receptors (12). In addition, there is good evidence that pigs are more frequently involved in interspecies transmission of influenza A viruses than are other animals (17, 22, 25, 30, 31).
The first cases of influenza in pigs were seen in the United States during the catastrophic human influenza pandemic of 1918–1919. Classical H1N1 swine influenza viruses are genetically and antigenically similar to the type A influenza viruses implicated in the human pandemic (27, 37, 38) and have become one of the most common causes of respiratory disease in North American pigs (38). Serologic studies of pigs in the United States in the late 1980s indicated a high (51%) prevalence of classical H1 influenza viruses, contrasted with a very low (1.1%) prevalence of H3 viruses antigenically similar to human H3 strains (6). The most recent isolation of an H3N2 swine virus in North America was in Quebec, Canada, in 1991 (2). The virus isolated is antigenically conserved and similar to 1975 human strains.
In late August of 1998, a severe outbreak of influenza-like disease occurred in a pig farm in North Carolina with 2,400 breeding sows, causing abortions and deaths among the breeding females. The sows were lethargic with high fever (104 to 107°F) and did not eat for a period of several days (exceeding 3 days for some sows). The morbidity rates approached 100%, with 7% of the breeding sows aborting their fetuses; the death rate in sows was 2%. Subsequent outbreaks, in November and December of 1998, affected pig populations in Texas, Minnesota, and Iowa. These outbreaks appeared less severe by comparison. Although the typical signs of influenza (sneezing, rough hair, and cough) were noted in greater numbers of pigs, fever was moderate and the morbidity rate was lower in Iowa. There were no reports of deaths of sows on pig farms in Minnesota or Iowa, although some mortality was reported in the Texas outbreak. Previous inoculation with an H1N1 vaccine may have alleviated some of the morbidity usually associated with influenza virus infection, but it was clearly ineffective in preventing outbreaks.
Four swine influenza viruses, including A/Swine/NorthCarolina/35922/98, A/Swine/Texas/4199-2/98, A/Swine/Minnesota/9088-2/98, and A/Swine/Iowa/8548-1/98, isolated between September and December of 1998 from influenza outbreaks in different states were used in this study. The four viruses were abbreviated as SWNC98, SWTX98, SWMN98, and SWIA98, respectively. The viruses were isolated from nasal swabs and lung tissues of sick or dead pigs and propagated in chicken embryos. All isolates were identified as belonging to the H3N2 subtype in local investigations, and this was confirmed in our laboratory.
The viral genome was sequenced as described previously (42). The sequence data were analyzed with the Wisconsin Package version 9.1-Unix (Genetics Computer Group, Madison, Wis.) and GeneDoc version 2.3.000 software (24). Phylogenetic analysis was done by the maximum parsimony method in PAUPsearch and PAUPdisplay programs of Wisconsin Package version 9.1-Unix.
To address the question of the genetic composition and origin of the causative viruses in the four influenza outbreaks, we partially sequenced each of the eight viral gene segments. Nucleotide sequences ranging from 595 to 1,385 bases of each gene were compared with those available in GenBank to determine the extent of homology (Table 1). The four swine viruses isolated in 1998 (abbreviated as SW98 viruses) could be separated on the basis of closest genetic relatedness. For the earlier isolate, SWNC98, the two genes encoding the hemagglutinin (HA) and neuraminidase (NA) surface glycoproteins and one internal gene encoding PB1 are most closely related to the corresponding genes from recent human H3N2 influenza viruses (98 to 99% homology). The other five internal genes of SWNC98 have the highest homology with genes of the classic H1N1 swine lineage (93 to 98%). For the remaining three viruses, the HA, NA, and PB1 genes belong to the same lineage as those from SWNC98 (97 to 99% homology), while the NP, M, and NS genes originated from the classical swine lineage (98% homology). Interestingly, two internal genes from these isolates, PB2 and PA, are closely related to the genes of avian influenza A viruses (93 to 97% homology). Since only a few PB2 and PA gene sequences from recently isolated avian viruses were available for comparison with GenBank data, the actual homologies between swine and avian PB2 and PA genes may be closer than the estimates reported here.
TABLE 1.
Genetic homology of the swine viruses isolated in the United States
| Gene segment | Region compared (nt)b | SWNC98
|
SWTX98, SWMN98, and SWIA98
|
Avg % homology overalla | ||||
|---|---|---|---|---|---|---|---|---|
| Lineage | Virus with the greatest homology | % Homology | Lineage | Virus with the greatest homology | Avg % homology | |||
| PB2 | 44–1,600 | Swine | A/Swine/TN/24/77 (H1N1) | 93 | Avian | A/Seal/MA/133/82 (H4N5) | 93 | 82 |
| PB1 | 1,088–2,236 | Human | A/Miyagi/29/95 (H3N2) | 98 | Human | A/Miyagi/29/95 (H3N2) | 99 | 98 |
| PA | 25–620 | Swine | A/WI/4755/94 (H1N1) | 94 | Avian | A/Swine/HK/81/78 (H3N2) | 97 | 81 |
| HA | 78–1,061 | Human | A/Finland/363/95 (H3N2) | 98 | Human | A/Finland/339/95 (H3N2) | 98 | 96 |
| NP | 34–1,024 | Swine | A/MD/12/91 (H1N1) | 98 | Swine | A/Swine/IA/17672/88 (H1N1) | 98 | 96 |
| NA | 26–1,411 | Human | A/Shiga/20/95 (H3N2) | 99 | Human | A/Shiga/25/97 (H3N2) | 99 | 97 |
| M | 52–964 | Swine | A/WI/4755/94 (H1N1) | 95 | Swine | A/WI/4755/94 (H1N1) | 98 | 93 |
| NS | 22–866 | Swine | A/Swine/IA/17672/88 (H1N1) | 97 | Swine | A/Swine/IA/17672/88 (H1N1) | 98 | 98 |
SWNC98 versus SWTX98, SWMN98, and SWIA98.
nt, nucleotide.
Thus, the swine H3N2 viruses isolated during the 1998 outbreaks are genetic reassortants incorporating gene segments of different origins. SWNC98 was generated by reassortment between a recent human virus and a classical swine virus, whereas each of the remaining three isolates arose from reassortment events among recent H3N2 human, classic swine H1N1, and avian influenza A viruses.
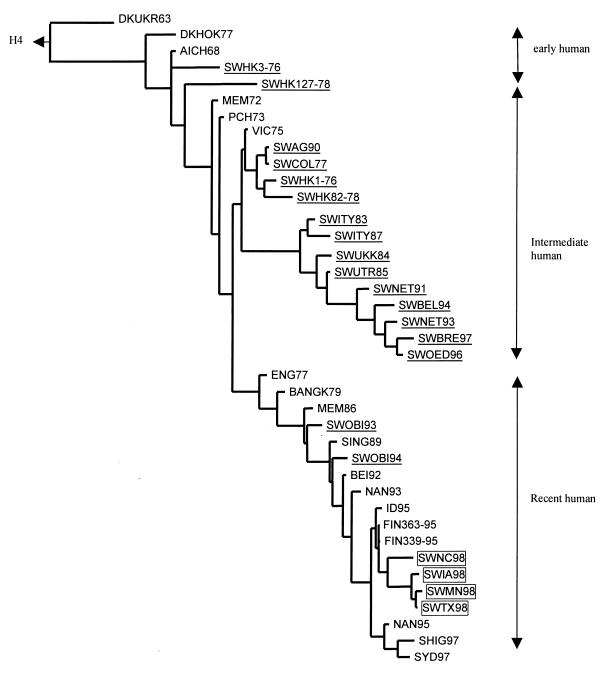
To identify more precisely the host or hosts in which the SW98 viruses originated, we constructed phylogenetic trees, using the HA1 region of the HA gene and partial sequences of the PB2 and NP genes. To learn whether the swine H3N2 viruses isolated in the United States are related to the H3 swine virus isolated previously, we used nucleotide sequences of representative swine viruses isolated at different times and in different regions of the world to construct an HA1 tree. As shown in Fig. 1, the swine H3 HA genes can be segregated into three lineages, including early human strains, intermediate human strains, and recent human strains. This grouping differs somewhat from that reported by Nerome et al. (23). The swine viruses of the early human lineage seem to be derived from the human strain A/Aichi/2/68. The intermediate human-derived swine viruses, which have been isolated continuously in Europe since 1983 (3), were closely related to A/Victoria/3/75-like viruses and appeared to have established a separate lineage. The recent human lineage includes two swine viruses isolated in 1993 and 1994 in Japan, A/Swine/Obihiro/1/93 and A/Swine/Obihiro/1/94 (13, 14), as well as the four SW98 viruses isolated in the United States. It is apparent from the phylogenetic tree that the HAs of the SW98 isolates were derived from neither the recent Japanese swine viruses nor the A/Swine/Ange-Gardien/150/90 (SWAG90) virus isolated in Canada (2). Rather, they are all descendants of human viruses circulating in 1995.
FIG. 1.
Phylogenetic tree of HA1 nucleotide sequences of the H3 hemagglutinin. The tree was generated by the maximum parsimony method in the PAUPsearch and PAUPdisplay programs of the Wisconsin Package version 9.1-Unix (Genetics Computer Group) and is rooted to the H4 sequence of A/Duck/Czechoslovakia/56 (H4N6). Horizontal lines are proportional to the numbers of nucleotide substitutions between branch points. Swine viruses are underlined, and the SW98 viruses are boxed. Abbreviations: DKUKR63, A/Duck/Ukraine/1/63 (H3N8); DKHOK77, A/Duck/Hokkaido/5/77 (H3N?); AICH68, A/Aichi/2/68 (H3N2); MEM72, A/Memphis/102/72 (H3N2); PCH73, A/Port Chalmers/1/73 (H3N2); VIC75, A/Victoria/3/75 (H3N2); ENG77, A/England/321/77 (H3N2); BANGK79, A/Bangkok/1/79 (H3N2); MEM86, A/Memphis/6/86 (H3N2); SING89, A/Singapore/12/89 (H3N2); BEI92, A/Beijing/32/92 (H3N2); NAN93, A/Nanchang/12/93 (H3N2); NAN95, A/Nanchang/933/95 (H3N2); FIN363-95, A/Finland/363/95 (H3N2); FIN339-95, A/Finland/339/95 (H3N2); ID95, A/Idaho/4/95 (H3N2); SHIG97, A/Shiga/25/97 (H3N2); SYD97, A/Sydney/05/97 (H3N2); SWHK1-76, A/Swine/Hong Kong/1/76 (H3N2); SWHK3-76, A/Swine/Hong Kong/3/76 (H3N2); SWCOL77, A/Swine/Colorado/1/77 (H3N2); SWHK82-78, A/Swine/Hong Kong/82/78 (H3N2); SWHK127-78, A/Swine/Hong Kong/127/78 (H3N2); SWITY83, A/Swine/Italy/309/83 (H3N2); SWUKK84, A/Swine/Ukkel/1/84 (H3N2); SWUTR85, A/Swine/Utrecht/4/85 (H3N2); SWITY87, A/Swine/Italy/635/87 (H3N2); SWAG90, A/Swine/Ange-Gardien/150/90 (H3N2); SWNET91, A/Swine/Netherlands/784/91 (H3N2); SWNET93, A/Swine/Netherlands/L2/93 (H3N2); SWOBI93, A/Swine/Obihiro/1/93 (H3N2); SWOBI94, A/Swine/Obihiro/1/94 (H3N2); SWBEL94, A/Swine/Belgium/231/94 (H3N2); SWOED96, A/Swine/Oedenrode/7C/96 (H3N2); SWBRE97, A/Swine/Breugel/97 (H3N2).
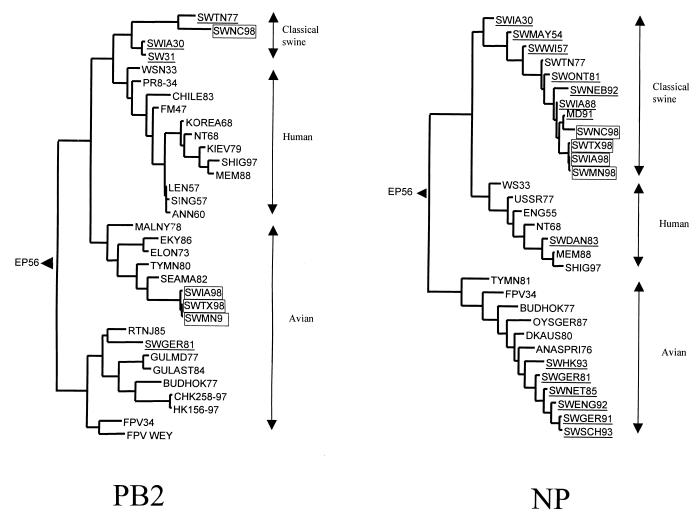
Phylogenetic analysis of the PB2 and NP genes showed a clear division of the SW98 viruses into different lineages (Fig. 2). In the PB2 tree, swine viruses are separated into a classical swine lineage and an avian lineage. SWNC98 belongs to the classical swine lineage, while the remaining three SW98 viruses cluster together on the North American avian branch. In the NP tree, the swine viruses are divided into three lineages, including classical swine and human-like and avian-like lineages. All of the NP genes from the SW98 viruses belong to the classical swine lineage. SWNC98 is more closely related to A/MD/12/91 than to the remaining SW98 isolates.
FIG. 2.
Phylogenetic trees for partial PB2 and NP sequences of influenza A viruses. Nucleotide residues 44 to 1,600 of all PB2 genes and residues 34 to 1,024 of all NP genes were analyzed. The trees were generated by the method described in the legend to Fig. 1. Abbreviations for the PB2 tree: SWIA30, A/Swine/Iowa/15/30 (H1N1); SW31, A/Swine/1976/31 (H1N1); SWTN77, A/Swine/Tennessee/24/77 (H1N1); SWGER81, A/Swine/Germany/2/81 (H1N1); WSN33, A/WSN/33 (H1N1); PR8-34, A/Puerto Rico/8/34 (H1N1); FM47, A/Fort Monmouth/1/47 (H1N1); LEN57, A/Leningrad/134/17/57 (H2N2); SING57, A/Singapore/1/57 (H2N2); ANN60, A/Ann Arbor/6/60 (H2N2); KOREA68, A/Korea/426/68 (H2N2); NT68, A/NT/60/68 (H3N2); KIEV79, A/Kiev/59/79 (H1N1); CHILE83, A/Chile/1/83 (H1N1); MEM88, A/Memphis/8/88 (H3N2); SHIG97, A/Shiga/25/97 (H3N2); FPV34, A/Chicken/FPV/Rostock/34 (H7N1); FPVWEY, A/Chicken/FPV/Weybridge/34 (H7N7); CHK258-97, A/Chicken/Hong Kong/258/97 (H5N1); HK156-97, A/Hong Kong/156/97 (H5N1); BUDHOK77, A/Budgerigar/Hokkaido/1/77 (H4N6); GULMD77, A/Gull/Maryland/704/77 (H13N6); MALNY78, A/Mallard/New York/6750/78 (H2N2); RTNJ85, A/Ruddy Turnstone/New Jersey/47/85 (H4N6); TYMN80, A/Turkey/Minnesota/833/80 (H4N2); SEAMA82, A/Seal/Massachusetts/133/82 (H4N5); GULAST84, A/Gull/Astrakhan/227/84 (H13N6); ELON73, A/Equine/London/1416/73 (H7N7); EKY86, A/Equine/Kentucky/2/86 (H3N8). Abbreviations for the NP tree; SWIA30, A/Swine/Iowa/15/30 (H1N1); SWMAY54, A/Swine/May/54 (H1N1); SWWI57, A/Swine/Wisconsin/1/57 (H1N1); SWTN77, A/Swine/Tennessee/24/77 (H1N1); SWONT81, A/Swine/Ontario/2/81 (H1N1); SWIA88, A/Swine/Iowa/17672/88 (H1N1); SWNEB92, A/Swine/Nebraska/1/92 (H1N1); MD91, A/Maryland/12/91 (H1N1); WS33, A/Wilson-Smith/33 (H1N1); USSR77, A/USSR/90/77 (H1N1); ENG55, A/England/19/55 (H1N1); NT68, A/NT/60/68 (H3N2); SWDAN83, A/Swine/Dandong/9/83 (H3N2); MEM88, A/Memphis/2/88 (H3N2); SHIG97, A/Shiga/25/97 (H3N2); TYMN81, A/Turkey/Minnesota/1661/81 (H1N1); FPV34, A/Chicken/FPV/Rostock/34 (H7N1); BUDHOK77, A/Budgerigar/Hokkaido/1/77 (H4N6); OYSGER87, A/Oystercatcher/Germany/87 (H1N1); DKAUS80, A/Duck/Australia/749/80 (H1N1); ANASPRI76, A/Anas Acuta/Primorje/695/76 (H2N3); SWHK93, A/Swine/Hong Kong/168/93 (H1N1); SWGER81, A/Swine/Germany/2/81 (H1N1); SWNET85, A/Swine/Netherlands/12/85 (H1N1); SWENG92, A/Swine/England/195852/92 (H1N1); SWGER91, A/Swine/Germany/8533/91 (H1N1); SWSCH93, A/Swine/Schleswig-Holstein/1/93 (H1N1).
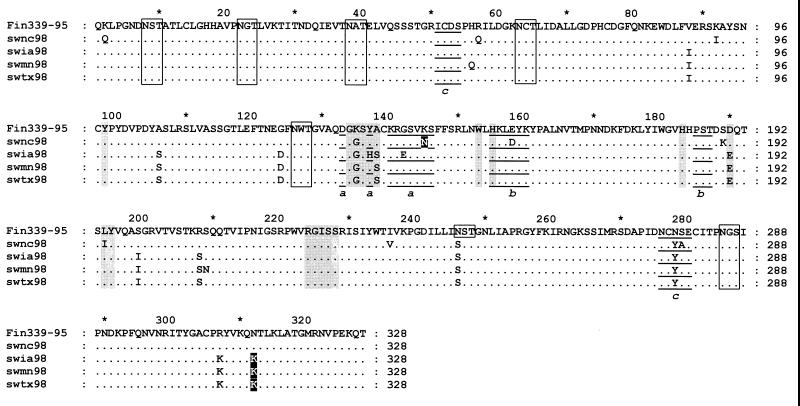
The HA1 sequences of A/Finland/339/95 in GenBank showed the closest homology with the HA1s of the SW98 viruses. Because of the difficulty in finding the actual precursor of the latter viruses, we used the sequence of MDCK-grown A/Finland/339/95 (26) as prototype to determine the mutations that may have occurred after the introduction of the HA gene from humans to pigs (Fig. 3). Twelve to 14 amino acid differences were found between the prototype virus and the SW98 isolates, resulting in an overall sequence identity of 95.7 to 96.3%. To minimize the possibility that the amino acid changes in the SW98 viruses resulted from sequence differences between the actual precursor and the A/Finland/339/95 prototype, we aligned the 10 most closely related sequences from GenBank with the deduced amino acid sequences of the SW98 viruses (data not shown). This comparison revealed two of the changes: one at position 145, found in 8 of 10 related human viruses, and another at position 312, found in only 1 of 10 related human viruses. These mutations may have been inherited from the precursor virus and should not be regarded as changes occurring after human-to-pig transmission (Fig. 3).
FIG. 3.
Alignment of HA1 amino acid sequences of SW98 viruses with that of A/Finland/339/95 (H3N2). The residues in open boxes are potential glycosylation sites; shaded residues denote the receptor-binding site; residues in reverse type represent amino acid changes that may have been derived from a human virus precursor (see text); underlined residues are antigenic sites (lowercase letters indicate discrete antigenic sites).
The majority of differences between the prototype and SW98 HA1s occurred in regions other than the proposed antigenic sites (5, 21, 40); indeed, the percentage of mutations in these sites ranged from 10 to 27%. Six potential glycosylation sites (Asn-X-Ser/Thr) are conserved at positions 8, 22, 38, 63, 126, and 285 in the HA1 domains of the SW98 and A/Finland/339/95 viruses. Owing to an Asn246 to Ser246 substitution, the SW98 viruses lost one potential glycosylation site at position 246. Compared with A/Finland/339/95, the SW98 viruses have several amino acid changes in the receptor-binding sites of their HA1 domains: SWNC98 has a single mutation (Lys135 to Gly135), while each of the remaining isolates has three (Lys135 to Gly135, Ala138 to Ser138, and Asp190 to Glu190). SWIA98 has an additional residue replacement in its receptor-binding site (Tyr137 to His137). The amino acid sequence of the SW98 viruses at residues 226 to 228, a site known to affect receptor-binding properties (29), was Ile-Ser-Ser, typical of human influenza virus strains circulating in 1995 (19, 20, 26).
The same methods described above were used to analyze the NA sequences of the SW98 viruses. The SW98 NA sequences were most closely related to those of A/Shiga/20/95 and A/Shiga/25/97 in GenBank. When the deduced amino acid sequences of the SW98 viruses were aligned with that of MDCK-grown A/Shiga/25/97 (19) (data not shown), 9 to 13 amino acid differences were found. Five of the changes were reckoned as derivations from the precursor human virus by comparison with 12 of the most closely related sequences from GenBank. The residues in the enzymatic site (7) are conserved among the A/Shiga/25/97 and SW98 viruses. Eight potential glycosylation sites were observed at positions 61, 70, 86, 146, 200, 234, 329, and 402 in the NA protein of A/Shiga/25/97. Six of them are conserved in the SW98 viruses; however, the potential glycosylation site at position 329 was lost in these viruses through the substitution of Ser331 for Arg331. The SWTX98, SWMN98, and SWIA98 viruses lack a potential glycosylation site, due to substitution of Asp402 for Asn402, that is present in the SWNC98 isolate.
Pigs appear to have a relatively weak species-specific barrier against infection by avian as well as human influenza A viruses (32, 38). Avian influenza viruses were introduced into pigs in Asia and Europe (9, 10, 17, 31), and genetic reassortment between human-like and avian-like or swine viruses occurred in pigs (4, 35). Our genetic analysis of the influenza viruses isolated from pigs in 1998 provides compelling evidence of interspecies transmission of human and avian viruses to pigs and of genetic reassortment among the human, swine, and avian influenza A viruses. To our knowledge, this is the first reported case in which the genome of an influenza virus consisted of gene segments from three different viral lineages.
An avian H1N1 influenza virus was transmitted to European pigs in the early 1980s (31, 32) and replaced the classical H1N1 strain that had been circulating in these animals since 1976 (3, 8). In 1983 to 1985, genetic reassortment took place between the avian-like and human-like H3N2 swine virus that had been transmitted to European pigs in the mid-1970s (3, 4). The reassortant strain, which possess “surface” genes of human origin and “internal” genes of avian origin, later replaced the human-like H3N2 virus (3). The H3N2 reassortant with avian internal genes has apparently superseded the antigenically related human-like strain. Successful adaptation of the reassortants with avian-derived internal genes suggests that these viruses might have a growth advantage in pigs compared with classical swine and human-derived viruses. Our finding also suggests that combination of gene segments from human, avian, and classical swine influenza viruses creates a favorable genetic background for transmission and replication of the hybrid virus in pigs. Indeed, the “triple” reassortant viruses described here caused influenza outbreaks in at least three states, in contrast to the human-swine isolate (SWNC98), which appears to have infected pigs in North Carolina only.
Amino acid sequence analysis revealed multiple changes in the surface proteins of the SW98 viruses, compared with prototype sequences. This finding could reflect laboratory adaptation of the viruses to growth in eggs, rather than adaptation to pigs, since previous studies have documented both antigenic and sequence differences in the hemagglutinin between MDCK-grown and egg-grown human viruses (15, 26, 28). Egg-adaptation substitutions for human viruses usually occur at residues 137 (Y to C), 156 (E to K), 186 (S to I), 226 (L to Q), 248 (T to I), and 276 (T to I) (15, 26, 28). Comparison of A/Finland/339/95 virus grown in eggs versus that grown in MDCK cells revealed only one amino acid difference, I194, in the HA1 domain (26). Thus, since the SW98 viruses also had a change to I at position 194 and lacked any mutations commonly associated with virus growth in eggs, we suggest that most of the amino acid changes observed did not result from virus propagation in eggs.
Although several amino acid changes in the receptor-binding pocket of SW98 viruses are duplicated in human viruses (19, 20, 26), others (H137 and S138) are unique. Since the affinity requirement for HA binding differs according to the host species, it is not surprising that some residues in the receptor-binding site were replaced after interspecies transmission of the SW98 viruses. The relatedness of the SW98 viruses to the human H3N2 strains isolated in 1995 was further emphasized by the presence of the same amino acid sequences (Ile-Ser-Ser) at residues 226 to 228 in the receptor-binding site (19, 20, 26).
Glycosylation of viral antigens masks and unmasks antigenic sites (5, 36, 40, 41) and therefore is an important process in the generation of new viruses. Clear evidence exists for the importance of carbohydrates in modulating antigenicity: for example, a new glycosylation site in the HA molecule prevented antibody binding and viral neutralization in a mutant of A/Aichi/1/68 (36). Alignment of H3 proteins shows that human viruses have acquired seven potential glycosylation sites in their HA1 domains, whereas avian viruses have only four potential glycosylation sites (data not shown). This suggests that the presence of glycosylation sites may correlate with immunologic pressure in the host. Avian viruses are subject to less immunologic pressure than mammalian viruses and therefore do not require as much carbohydrate to mask their antigenic sites. We speculate that the swine viruses isolated in 1998 lost several potential glycosylation sites in their HAs and NAs after they were introduced from humans into pigs, in part because of the weaker selection pressure exerted by the latter host.
Emergence of the H3N2 reassortant influenza viruses in the pigs of the United States raises further concerns about whether the viruses will become established in the pigs in North America and whether they will become an economic burden. It is worth considering the inclusion of H3N2 inactivated virus in the swine vaccination regimen. This event also emphasized the need for surveillance of pig populations in North America for evidence of continuous circulation of this virus in pigs.
Nucleotide sequence accession numbers.
All sequences used in this study were sent to GenBank, and the accession numbers are AF153232 to AF153263.
Acknowledgments
We thank Gary Walch, USDA, Belleville, Tex., for field investigation and providing the virus from Texas, David Walker and Lijuan Zhang for excellent technical assistance, and John Gilbert for editorial assistance.
These studies were supported by Public Health Service grants AI29680 and AI95357 and Cancer Center Support (CORE) grant CA-21765 from the National Institutes of Health and by the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 2.Bikour M H, Frost E H, Deslandes S, Talbot B, Weber J M, Elazhary Y. Recent H3N2 swine influenza virus with haemagglutinin and nucleoprotein genes similar to 1975 human strains. J Gen Virol. 1995;76:697–703. doi: 10.1099/0022-1317-76-3-697. [DOI] [PubMed] [Google Scholar]
- 3.Campitelli L, Donatelli I, Fonim E, Castrucci M R, Fabiani C, Kawaoka Y, Krauss S, Webster R G. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. doi: 10.1006/viro.1997.8514. [DOI] [PubMed] [Google Scholar]
- 4.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 5.Caton A J, Brownlee G G, Yewdell J W, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 6.Chambers T M, Hinshaw V S, Kawaoka Y, Easterday B C, Webster R G. Influenza viral infection of swine in the United States 1988–1989. Arch Virol. 1991;116:261–265. doi: 10.1007/BF01319247. [DOI] [PubMed] [Google Scholar]
- 7.Colman P M, Varghese J N, Laver W G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 8.Donatelli I, Campitelli L, Castrucci M R, Ruggieri A, Sidoli L, Oxford J S. Detection of two antigenic subpopulations of A (H1N1) influenza viruses from pigs: antigenic drift or interspecies transmission? J Med Virol. 1991;34:248–257. doi: 10.1002/jmv.1890340410. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Shortridge K F, Krauss S, Li P H, Kawaoka Y, Webster R G. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70:8041–8046. doi: 10.1128/jvi.70.11.8041-8046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinshaw V S, Alexander D J, Aymard M, Bachmann P A, Easterday B C, Hannoun C, Kida H, Lipkind M, MacKenzie J S, Nerome K, Schild G C, Scholtissek C, Senne D A, Shortridge K F, Skehel J J, Webster R G. Antigenic comparisons of swine-influenza-like H1N1 isolates from pigs, birds and humans: an international collaborative study. Bull World Health Organ. 1984;62:871–878. [PMC free article] [PubMed] [Google Scholar]
- 11.Hinshaw V S, Webster R G. The natural history of influenza A viruses. In: Beare A S, editor. Basic and applied influenza research. Boca Raton, Fla: CRC Press; 1982. pp. 79–104. [Google Scholar]
- 12.Ito T, Kida H, Kawaoka Y. Receptors of influenza A viruses: implication for the role of pigs in the generation of pandemic human influenza viruses. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza. Vol. 3. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 516–519. [Google Scholar]
- 13.Katsuda, K., M. Imai, T. Shirahata, H. Kida, and H. Goto. 1996. Unpublished data.
- 14.Katsuda K, Shirahata T, Kida H, Goto H. Antigenic and genetic analyses of the hemagglutinin of influenza viruses isolated from pigs in 1993. J Vet Med Sci. 1995;57:1023–1027. doi: 10.1292/jvms.57.1023. [DOI] [PubMed] [Google Scholar]
- 15.Katz J M, Wang M, Webster R G. Direct sequencing of the HA gene of influenza (H3N2) virus in original clinical samples reveals sequence identity with mammalian cell-grown virus. J Virol. 1990;64:1808–1811. doi: 10.1128/jvi.64.4.1808-1811.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaoka Y, Krauss S, Webster R G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 18.Kundin W D. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature. 1970;228:857. doi: 10.1038/228857a0. [DOI] [PubMed] [Google Scholar]
- 19.Lindstrom S E, Hiromoto Y, Nerome R, Omoe K, Sugita S, Yamazaki Y, Takahashi T, Nerome K. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindstrom S, Sugita S, Endo A, Ishida M, Huang P, Xi S H, Nerome K. Evolutionary characterization of H3N2 influenza viruses: novel changes in the receptor binding domain. Arch Virol. 1996;141:1346–1355. doi: 10.1007/BF01718836. [DOI] [PubMed] [Google Scholar]
- 21.Lubeck M D, Gerhard W. Topological mapping antigenic sites on the influenza A/PR/8/34 virus hemagglutinin using monoclonal antibodies. Virology. 1981;113:64–72. doi: 10.1016/0042-6822(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 22.Mancini G, Donatelli I, Rozera C, Arangio Ruiz G, Butto S. Antigenic and biochemical analysis of influenza “A” H3N2 viruses isolated from pigs. Arch Virol. 1985;83:157–167. doi: 10.1007/BF01309913. [DOI] [PubMed] [Google Scholar]
- 23.Nerome K, Kanegae Y, Shortridge K F, Sugita S, Ishida M. Genetic analysis of porcine H3N2 viruses originating in southern China. J Gen Virol. 1995;76:613–624. doi: 10.1099/0022-1317-76-3-613. [DOI] [PubMed] [Google Scholar]
- 24.Nicholas K B, Nicholas H B, Jr, Deerfield D W. GeneDoc: analysis and visualization of genetic variation. EMBnew News. 1997;4:14. [Google Scholar]
- 25.Ottis K, Sidoli L, Bachmann P A, Webster R G, Kaplan M M. Human influenza A viruses in pigs: isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch Virol. 1982;73:103–108. doi: 10.1007/BF01314719. [DOI] [PubMed] [Google Scholar]
- 26.Pyhala R, Ikonen N, Haanpaa M, Kinnunen L. HA1 domain of influenza A (H3N2) viruses in Finland in 1989–1995: evolution, egg-adaptation and relationship to vaccine strains. Arch Virol. 1996;141:1033–1046. doi: 10.1007/BF01718607. [DOI] [PubMed] [Google Scholar]
- 27.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha E P, Xu X, Hall H E, Allen J R, Regnery H L, Cox N J. Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses. J Gen Virol. 1993;74:2513–2518. doi: 10.1099/0022-1317-74-11-2513. [DOI] [PubMed] [Google Scholar]
- 29.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 30.Rota P A, Rocha E P, Harmon M W, Hinshaw V S, Sheerar M G, Kawaoka Y, Cox N J, Smith T F. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989;27:1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholtissek C, Burger H, Bachmann P A, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 32.Scholtissek C, Ludwig S, Fitch W M. Analysis of influenza A virus nucleoproteins for the assessment of molecular genetic mechanisms leading to new phylogenetic virus lineages. Arch Virol. 1993;131:237–250. doi: 10.1007/BF01378629. [DOI] [PubMed] [Google Scholar]
- 33.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 34.Schultz U, Fitch W M, Ludwig S, Mandler J, Scholtissek C. Evolution of pig influenza viruses. Virology. 1991;183:61–73. doi: 10.1016/0042-6822(91)90118-u. [DOI] [PubMed] [Google Scholar]
- 35.Shu L L, Lin Y P, Wright S M, Shortridge K F, Webster R G. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology. 1994;202:825–833. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 36.Skehel J J, Stevens D J, Daniels R S, Douglas A R, Knossow M, Wilson I A, Wiley D C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci USA. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 38.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster R G, Laver W G. Studies on the origin of pandemic influenza. I. Antigenic analysis of A 2 influenza viruses isolated before and after the appearance of Hong Kong influenza using antisera to the isolated hemagglutinin subunits. Virology. 1972;48:433–444. doi: 10.1016/0042-6822(72)90054-2. [DOI] [PubMed] [Google Scholar]
- 40.Wiley D C, Wilson I A, Skehel J J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 41.Wilson I A, Ladner R C, Skehel J J, Wiley D C. The structure and role of the carbohydrate moieties of influenza virus haemagglutinin. Biochem Soc Trans. 1983;11:145–147. [PubMed] [Google Scholar]
- 42.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]