Abstract
The fundamental question of how sequence defines conformation is explicitly answered if the structures of all possible sequences of a macromolecule are determined. We present here a crystallographic screen of all permutations of the inverted repeat DNA sequence d(CCnnnN6N7N8GG), where N6, N7, and N8 are any of the four naturally occurring nucleotides. At this point, 63 of the 64 possible permutations have been crystallized from a defined set of solutions. When combined with previous work, we have assembled a data set of 37 single-crystal structures from 29 of the sequences in this motif, representing three structural classes of DNA (B-DNA, A-DNA, and four-stranded Holliday junctions). This data set includes a unique set of amphimorphic sequence, those that crystallize in two different conformations and serve to bridge the three structural phases. We have thus constructed a map of DNA structures that can be walked through in single nucleotide steps. Finally, the resulting data set allows us to dissect in detail the stabilization of and conformational variations within structural classes and identify significant conformational deviations within a particular structural class that result from sequence rather than crystal or crystallization effects.
Keywords: molecular screening
The basic principle that sequence defines the 3D structure of a macromolecule was first established in 1957 by Anfinsen (1), who showed that ribonuclease A can be reversibly denatured and renatured in solution. The exact relationship between sequence and conformation, however, remains elusive, the “protein-folding problem,” the long-coveted “Holy Grail” in protein chemistry, is yet to be solved, but not from lack of effort. The question of how sequence determines structure has been attacked by nearly every conceivable experimental and theoretical approach. The effect of sequence on the structure-stability relationship in T4 lysozyme has been extensively studied by crystallographic and thermodynamic analyses (2), whereas the propensity of single amino acids to effect formation of isolated α-helices have been studied by using host-guest peptides (3, 4). However, if the structures of all possible sequence combinations of a macromolecule are determined, then this problem is solved explicitly. We present here the results from a crystallographic screen of all possible sequence permutations within a defined inverted repeat (IR) sequence motif to construct a map of DNA structures that are available to this sequence motif.
DNA is highly polymorphic, capable of adopting a large variety of structures in crystals and solution, including right- and left-handed double helices, triple helices, and four-stranded G quartets and Holliday junctions (5, 6). The current data set of DNA structures has grown over the years, but not in a systematic manner; therefore, it has been difficult, if not impossible, to relate the structures within the framework of a common lineage of sequence or environment. Our attempt to crystallize all of the possible combinations of a defined DNA sequence motif from a common set of crystallization solutions initiated with the serendipitous findings that the sequences d(CCGGGACCGG) (7) and d(CCGGTACCGG) (8) crystallize as four-stranded Holliday junctions, the central intermediate in recombination and recombination-dependent cellular processes (9). A common ACC trinucleotide core at nucleotides N6N7N8 and an associated set of intramolecular interactions were subsequently identified that fix the junction (where the phosphoribose backbone crosses over between B-DNA duplexes) and thus allow its crystallization in these sequences (10). To search for other trinucleotides that stabilize junctions in an unbiased manner, we designed a crystallographic screen to solve the crystal structures of all 64 permutations of the sequence d(CCnnnN6N7N8GG), where N6N7N8 can be any of the four common nucleotides and nnn are specified accordingly to maintain the IR motif and thus self-complementarity of the sequences (Table 1). The sequences in this study will be referred to by the unique N6N7N8 trinucleotide motif. Although, when isolated, there are only 32 unique trinucleotides, once placed in the context of this motif, each of the 64 possible trinucleotides becomes unique. For example, the TTT sequence in this motif defines the overall sequence d(CCAAATTTGG); the AAATTT central core of this sequence is associated with highly curved B-DNA (11, 12). In contrast, the complementary AAA sequence would be found in d(CCTTTAAAGG), where the TTTAAA core is known to not show significant curvature of the DNA helix.
Table 1. Conformations observed in the single crystals of d(CCnnnN6N7N8GG).
|
N7
|
|||||
|---|---|---|---|---|---|
| N6 | G | C | A | T | N8 |
| G | A | x | x | x | G |
| B/a | b/J | B | B | C | |
| A | B | x | x | A | |
| A | B | x | b | T | |
| C | A | A | x | x | G |
| x | A/J | x | B | C | |
| - | x | x | x | A | |
| x | b | x | x | T | |
| A | x | x | x | x | G |
| B | J | B | B/J | C | |
| B | x | B | x | A | |
| b | B | b | x | T | |
| T | A | a | x | x | G |
| x | x | x | B | C | |
| x | x | x | B | A | |
| x | b | x | x | T | |
The trinucleotides N6N7N8 are presented as a triplet table, with conformations that have been determined from this screen (either new or repeats of prior structures) in bold and conformations that were determined by other groups but have not been repeated in the screen in lowercase. B-DNA structures are labeled as B, A-DNA as A, and four-stranded Holliday junction as J. Sequences that have been crystallized in the screen, but whose structures have not yet been determined, are labeled as x.
This IR sequence motif also has the potential to adopt other DNA structures; the trinucleotide has been suggested to be the minimum motif to distinguish between the double-helical forms of B- and A-DNA (13, 14). Thus, we expected the current crystallographic screen to sample at least three different DNA structures (Fig. 1a). The IR motif, however, is limited in that it samples only those structures that are available to self-complementary sequences with Watson-Crick base pairs, thereby effectively excluding, for example, G quartets and I motifs. In addition, the nonalternating CC/GG dinucleotides at the two ends effectively exclude left-handed Z-DNA from this screen. We, therefore, consider this study as a step toward developing a set of structures under a common framework that, upon extending this motif, will eventually allow us to distinguish the effects of sequence on all possible DNA forms. For this study, we will distinguish between overall structure and the detailed conformational variations that can occur within a particular structural class.
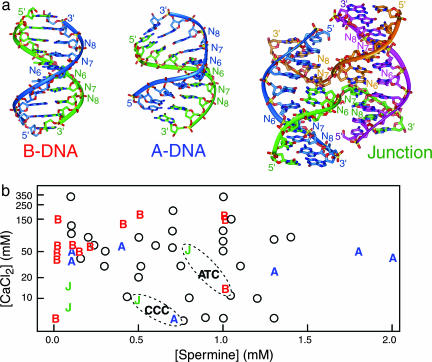
Fig. 1.
Structures from the crystallographic screen of the IR sequence d(CCnnnN6N7N8GG), where all 64 combinations of the N6N7N8 trinucleotide are sampled. (a) The conformations observed in the single crystal structures of this sequence include standard B-DNA (AGC structure shown), the altered A-DNA duplex (GGG structure shown), and the four-stranded Holliday junction (ACC structure shown). The positions of the N6N7N8 trinucleotide are labeled in each of the structures. (b) The CaCl2 and spermine concentrations yielding crystals of B-DNA (B), A-DNA (A), and junctions (J) are compared, with the concentration of CaCl2 plotted on a logarithmic scale. Open circles indicate conditions that yielded crystals, but where the conformation has not been determined. Only one label for each form is denoted in cases where crystallization conditions overlap. The conditions for crystallization of the amphimorphic sequences ATC and CCC are encompassed in ovals and labeled by the trinucleotide sequence.
The advantage of this crystallographic approach is that sequence effects on structure and conformation can be directly related to specific molecular interactions. The potential problems, however, are those purported to be inherent in DNA crystallography, including the potential that lattice interactions greatly influence or actually induce the conformation observed in the crystal (15-17). The results demonstrate that the sequence motif designed for this study is highly crystallizable and samples DNA structures and conformational variations within these structural classes broadly, apparently independent of such overt crystal lattice effects.
Materials and Methods
Deoxyoligonucleotides were synthesized with the dimethoxytrityl protecting group left intact at the 5′ terminus to facilitate purification by preparative RP-HPLC. Sequences were detritylated by treatment with 3% acetic acid and passed over a gel filtration column to yield purified DNA stocks. The DNAs were stored at -80°C as lyophilized powders and redissolved in Millipore water before use without any further purification.
All sequences were crystallized by sitting drop vapor diffusion, with setups of 10 μl of total sample drops containing 25-100 mM sodium cacodylate buffer at pH 7.0, 0-325 mM CaCl2 and 0-3 mM spermine equilibrated against 30 ml of 2-35% aqueous 2-methyl-2,4-pentanediol in the reservoir (Table 3, which is published as supporting information on the PNAS web site). These solutions are similar to previous conditions used to crystallize DNA oligomers in this laboratory (18). Each sequence was subjected to the full range of CaCl2 and spermine concentrations, with each crystal form refined individually to obtain diffraction-quality single crystals. X-ray diffraction data were collected in-house on a Rigaku (Tokyo) diffractometer with an R-AXIS IV detector or at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) and Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) synchotrons. Data were reduced by using the hkl suite of programs. Structures were solved by molecular replacement using isomorphous structures available in the Nucleic Acid Database (19) as initial models or by multiple wavelength anomalous dispersion phasing. Structures have all undergone initial refinement with addition of solvent, with nearly all Rfree values <30%. All structures have been deposited into the Protein Data Bank and the Nucleic Acid Database (Table 4, which is published as supporting information on the PNAS web site) and will be updated as they become fully refined.
Results
To study how sequence defines the structure and conformation of DNA, we attempted to determine the crystal structures of all 64 permutations of the IR sequence d(CCnnnN6N7N8GG), where N6, N7, and N8 are any of the four naturally occurring nucleotides. At this point in the study, a remarkable 63 of the possible 64 sequences (the lone exception being CGA) have been crystallized from a common set of crystallization solutions, indicating that this sequence motif is readily crystallizable and thus ideal for this type of exhaustive structural screen. We currently have solved the structures of 23 sequences from the screen and, along with similar structures from previous studies on sequences in this same motif, have a total of 29 sequences for the study. Crystals that were previously solved and have been reproduced by using the current solutions are included in the 23. The screen itself yielded 24 additional structures from 21 previously unreported sequences. Although incomplete, the current structures from the screen are sufficient to define a set of general rules that show how individual nucleotides distinguish between various DNA structures and affect their detailed conformations.
Structural Classes. The structures resulting from this screen fall into three classes: the right-handed double-helical forms of B- and A-DNA and four-stranded Holliday junction (Fig. 1a). Not surprisingly, a majority of these sequences (17, including 5 from previous reports) crystallize as B-DNA duplexes, 7 form only A-DNA, and 1 sequence (ACC) forms only the junction (Table 1). A unique aspect of the results, however, is that a set of amphimorphic sequences (those that crystallize as two different structures) have been identified that link each of these structural phases. They include ATC, which crystallized as both B-DNA and junction, and CCC, which crystallized as both A-DNA and junction under the crystallization conditions of the screen. In addition, GCC, which was previously reported as B-DNA (20), was crystallized under our conditions as a junction (18) (the B form was crystallized with Mg2+, whereas the junction was with Ca2+ cations). These amphimorphic sequences, therefore, sit at the interfaces between the junction and the two duplex DNA forms. Finally, GGC, which was previously reported as A-DNA (21), was crystallized as B-DNA in the current study (the difference being the alcoholic precipitant used to crystallize the A-DNA form) and represents a sequence at the B-A interface.
Crystallization Conditions. In comparing crystallization solutions, we see first that double-stranded A- and B-DNAs crystallize across nearly the entire range of divalent and polyvalent cations (Fig. 1b). B-DNAs are seen to crystallize at lower spermine concentrations, but, interestingly, at higher calcium (II) concentrations than A-DNAs. Still, all sequences were subjected to the entire range of solutions in this screen and, therefore, there was no bias toward any structural form designed into the experiment. Four-stranded junctions generally crystallize under lower Ca+2 solutions (<15 mM) than the B-DNAs. A comparison of the crystallization solutions of the amphimorphic sequences, however, confirm our expectations that high concentrations of divalent cations are required to shield the negative electrostatic potential at the phosphates of the compact stacked-X junction (22) and to prevent migration of the junction along the DNA strands (23) of a given sequence. ATC was seen to form junctions at higher concentrations of Ca2+ and B-DNA duplexes at lower cation concentrations. Interestingly, CCC forms a junction at higher Ca2+ concentration but is A-DNA at lower concentrations. Thus, for any particular sequence, the junction is stabilized by higher concentrations of divalent cations, as expected for B-DNA (22), but we now see that this is also true for the A-DNA duplex. We note, however, that in both cases, the duplex and junction forms can coexist at the intermediate Ca2+ crystallization solutions.
A comparison of the two forms of GCC shows that Ca2+ is more effective than Mg2+ at stabilizing the junction over B-DNA [it should be noted that ACC crystallizes as a junction with either Ca2+ or Mg2+ (7, 8)]. Finally, GGC is seen to be induced by alcohol to form A-DNA. Although one expects alcohols to favor the A-form (24), this study directly implicates alcoholic precipitants on the structural class in crystals. Thus, the solution conditions that favor crystallization of each conformation are generally consistent with what has been observed for the behavior of DNA in solution.
Crystal Lattices. The crystals for which we have diffraction data (43 sequences) fall into 15 unique space groups (including 7 of the 14 Bravais lattice types) and 27 associated crystal forms, demonstrating that the structures of this sequence motif are not restricted by the crystal lattice, but are free to assume a variety of crystal forms (Table 4). In addition, the two amphimorphic sequences ATC and CCC have crystal forms that are specific for their respective conformations, but that these crystal forms can coexist under certain conditions. Furthermore, at least one space group (monoclinic C2) is common for both B-DNA and the junction, and a second (orthorhombic P212121) is seen for both A-DNA and B-DNA. Our supposition, therefore, is that each sequence adopts a crystal lattice that can accommodate the particular structure(s) formed in the crystallization solution. Thus, the crystal lattice serves less as a tyrant (15, 16) here than as an experimental facilitator that leaves no doubt concerning the structure(s) of each sequence.
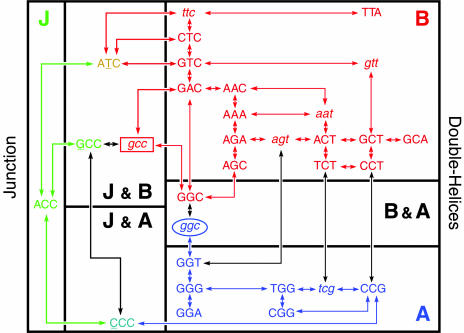
Crystallographic Map of DNA Structural Space. The data set resulting from this screen defines a phase map that relates DNA structures to sequence and environment (Fig. 2). We can walk through this map of DNA structures in single-nucleotide steps starting with the ACC sequence, the core trinucleotide that uniquely forms the Holliday junction. A transition of the central C of ACC to T generates the amphimorphic ATC sequence that sits at a junction/B-DNA interface that depends on the concentration of divalent cations. The transition from ACC to GCC defines a similar interface, but one that depends on the type of divalent cation. To fully enter the B-phase, additional transitions or transversions to convert ATC to (G/C/T)TC, or GCC to GCT or GTC are required. Alternatively, the transversion of ACC to CCC yields an amphimorphic sequence at the junction/A-DNA interface. The trinucleotide is pulled further into the fully A-DNA phase by systematic transitions and transversions from CCC that lead toward GGG, the classic A-DNA trinucleotide. Finally, the interchange between A- and B-DNA duplexes is seen to occur through the amphimorphic sequence GGC in a solvent-dependent manner or, more directly, from GGT to AGT, which is consistent with the understanding that A/T favors B-DNA (25).
Fig. 2.
Map of DNA structure space sampled by crystals of d(CCnnnN6N7N8GG). The map is divided into three specific structural classes (labeled B for B-DNA, A for A-DNA, and J for junctions) and the interfaces between each conformational phase. The sequences in uppercase letters define those that have been uniquely solved or reproduced in the current study, while those in lowercase letters are structures from previous studies, but not reproduced here. The rectangle around GCC indicates that the structure is induced by a change in divalent cations (from Ca2+ to Mg2+). Similarly, the oval around GGC indicates that the A form is induced by alcohol. Arrows trace paths through the conformational map as the N6N7N8 trinucleotide undergoes single-nucleotide transitions or transversions. These are not unique paths, but show one set of consistent single-nucleotide steps through the conformational space.
Discussion
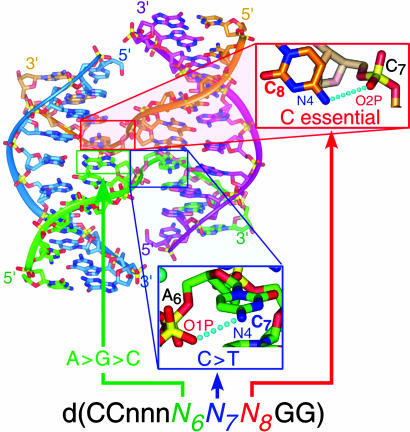
This study started with the goal of identifying trinucleotides in the d(CCnnnN6N7N8GG) sequence motif that form four-stranded DNA Holliday junctions, and indeed the screen has done that. The trinucleotides N6N7N8 = ACC, GCC, ATC, and CCC that are now identified as junction-forming are associated with specific interactions observed in the four-stranded complex. One unique aspect of the study is that each phase of the structure map is linked by amphimorphic sequences, which allows us to delineate the effects of single nucleotides at each position of the trinucleotide core on the stability of the junction relative to both A- and B-DNA duplexes. The parent ACC trinucleotide has been shown to stabilize DNA junctions in the presence of various cations (26), with G·A mismatches (7), in drug cross-linked constructs (27), and with the terminal C·G base pairs of decanucleotide motif replaced by T·A base pairs (26). Thus, ACC is defined as the most stabilizing of the junction-forming trinucleotides. A common feature of all of the junction trinucleotides is the cytosine at the N8 position. The current study shows that not all NNC type trinucleotides form junctions; thus, the cytosine at N8 appears to be essential, but not sufficient to define a junction, which can directly be attributed to the hydrogen bond from the cytosine N4 amino to the phosphate oxygen at the junction crossover (Fig. 3). We note, however, that this interaction can be partially replaced by an analogous Br···O halogen bond, as in the structure of the ACbr5U junction (18, 28).
Fig. 3.
Correlating sequence effects to atomic interactions in junctions. The interactions that are identified as being important for fixing the junction in ACC are shown in the insets. General rules for junction-forming sequences are noted in green, red, and blue for the nucleotides N6, N7, and N8, respectively. The inset for the cytosine C8 to phosphate of N7 is rotated relative to the orientation of the overall structure.
In addition, all of the current junction-forming trinucleotides have a pyrimidine at the central N7 position. Comparing ACC with the amphimorphic ATC trinucleotide, however, indicates that a cytosine is more effective than thymine at stabilizing the junction. There is a potential interaction between the N4 amino group of cytosine and a phosphate oxygen on the same strand but at the DNA duplex across the junction. This interaction, however, is longer (3.2-3.5 Å) than would be expected for an effective hydrogen bond. In addition, we see that the methyl group of the ATC structure is oriented directly toward and is within 4.1-4.5 Å of the oxygen atoms of this same phosphate. This observation suggests that the stabilizing effect of the pyrimidine base is primarily electrostatic, with the N4 amino group of cytosine being more effective than the methyl group of thymine as a counter to the phosphate oxygen. Thus, the general sequence rule is NCC > NTC in forming the junction for electrostatic reasons.
With a single exception, N6 of the N6N7N8 trinucleotide is a purine, with A > G in stabilizing the junction. This order is evident from the observation that ATC is amphimorphic and capable of forming a junction, but GTC forms B-DNA. The exception to a purine at N6 is CCC. Again, the results indicate that a cytosine at N6 is less stabilizing to the junction than either A or G through the argument that CCC is amphimorphic and CTC is B-DNA. Thus, the series at the N6 nucleotide is A > G > C.
It is interesting that the amphimorphic CCC sequence crystallizes as a duplex in the A form, but as a four-stranded junction with arms that adopt the B-DNA structure. This finding suggests that, at least for DNA, the four-stranded junction favors B-type double helices even if the sequence has a strong propensity for A-DNA. Again, this results from the hydrogen-bonding interaction of the cytosine at N8 that is required to stabilize the junction in the IR motif (this interaction would not be available with a deep major groove that one would expect with A-DNA arms).
The conformation map shows that A-DNA is associated with the trinucleotide motifs GGN, NGG, and CC(C/G). B-DNA is favored as individual C/G base pairs of these A-DNA triplets are replaced by T/A base pairs (Fig. 2). When applying previous trinucleotide rules to distinguish A-DNA from B-DNA, those derived from calculations of hydrophobic surfaces (14) correctly predicted 16/24 (67%) of the sequences crystallized as A- or B-DNA, whereas those from experimental alcohol titrations (13) correctly predicted 21/28 (75%) sequences (not all trinucleotides are represented in the respective scales). The relatively poor showings reflect the fundamental differences between these two former methods for predicting A- and B-DNA and the design of the current experiment. The hydrophobicity scale was derived from a structural data set that was highly variable in how the sequences were crystallized (relying on the available structures at the time) and included a large number of lattice distorted A-DNAs. On the other hand, the alcohol titration scale was determined spectroscopically as the amount of trifluoroethanol required to induce a B- to A-DNA transition. The current study, in contrast, relates the two conformations through sequences that are explicitly determined from nearly identical crystallization solutions. Thus, the structures derived by the crystallographic screen provide a potential means to derive a set of rules to predict the sequence formation of A- and B-DNA from a consistent data set.
In addition to the structural map, the structures from this crystallographic screen also allow us to relate sequence to conformational variations that are important for recognition and function of each structural class (nine such parameters from Table 5, which is published as supporting information on the PNAS web site, are summarized in Table 2). A comparison of the base pair and base step parameters of the structures in this study shows that A-DNAs are conformationally homogenous compared with B-DNA. We would not draw this conclusion from a comprehensive analysis of all A-DNA crystal structures available in the Nucleic Acid Database (19). Although the A-DNA structures detailed here show some sequence-dependent variations in their double-helical conformations, they do not vary dramatically from the canonical form. We attribute this finding to the consistency in the current data set of A-DNA structures, where solution conditions and sequence end effects have been controlled.
Table 2. Helical parameters for structures from the crystallographic screen of the sequence motif d(CCnnnN6N7N8GG).
| Rotational parameters,°
|
Translational parameters, Å
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Structures | Helical twist | Propeller twist | Tilt | Roll | Buckle | Rise | Slide | X displacement | Zp |
| B-DNA | |||||||||
| <Base pairs> | 34.7 (15.5) | −12.0 (8.1) | −0.62 (10.9) | 1.74 (7.98) | −0.23 (8.57) | 3.30 (0.4) | 0.66 (1.02) | 0.53 (1.56) | −0.67 (0.59) |
| <Structures> | 35.6 (1.2) | −11.6 (2.8) | −0.05 (0.52) | 2.18 (1.51) | −0.31 (2.62) | 3.31 (0.06) | 0.71 (0.40) | 0.49 (0.48) | −0.68 (0.24) |
| GCA | 38.2* | −12.9 | −0.43 | −0.12* | −4.94* | 3.39 | 1.32* | 1.43* | −0.9 |
| Junctions | |||||||||
| <Base pairs> | 37.5 (4.3) | −11.2 (8.1) | −0.14 (4.69) | 1.90 (4.47) | −1.16 (6.09) | 3.40 (0.23) | 1.63 (1.11) | 2.11 (1.89) | −0.97 (1.07) |
| <Structures> | 37.5 (0.33) | −11.2 (3.0) | −0.14 (0.65) | 1.90 (0.52) | −1.16 (2.93) | 3.40 (0.03) | 1.63 (0.11) | 2.11 (0.13) | −0.97 (0.14) |
| A-DNA | |||||||||
| <Base pairs> | 30.4 (4.2) | −7.82 (8.47) | 0.62 (3.91) | 6.88 (5.69) | −0.57 (7.78) | 3.30 (0.24) | −1.74 (0.31) | −4.47 (1.25) | 2.27 (0.39) |
| <Structures> | 30.4 (0.8) | −7.82 (3.57) | 0.62 (0.67) | 6.88 (1.71) | −0.57 (2.42) | 3.30 (0.06) | −1.74 (0.15) | −4.47 (0.25) | 2.27 (0.15) |
| Amphimorphic junction structures | |||||||||
| ATC (B-DNA) | 36.8 | −17.3 | −0.06 | −0.08 | 7.35 | 3.34 | −0.07 | −0.16 | −0.26 |
| ATC (Junction-LS) | 37.8 | −13.0 | 0.47 | 1.53 | 0.91 | 3.34 | 1.63 | 1.99 | −1.08 |
| ATC (Junction-HS) | 37.2 | −16.5 | 0.20 | 2.28 | 0.37 | 3.38 | 1.77 | 2.11 | −1.15 |
| CCC (Junction) | 38.0 | −14.2 | −1.28 | 2.15 | −7.19 | 3.37 | 1.60 | 2.60 | −0.97 |
| CCC (A-DNA) | 30.5 | −8.09 | −0.29 | 7.1 | 0.83 | 3.24 | −1.76 | 13.66 | 2.38 |
The rotational and translational parameters that characterize the helical conformations of nucleic acid structures [as defined (31) and calculated by the program 3dna (32)] are compared for the mean values of all base pairs [<Base pairs> (standard deviations)] and as means averaged across the structures [<Structure> (standard deviation of mean)] for structures in the screen that are B-DNA, four-stranded junctions, and A-DNA.
Values that fall at least 1 SD outside the mean of the average structural class.
The B-DNA duplexes and junction arms show similar sequence-dependent conformational variation, with the mean and degree of variability of each helical parameter being very similar between the two structural forms. This finding supports the model that the arms of the junction mirror the properties of B-DNA in general (29). When the helical parameters for the amphimorphic sequence ATC are compared between the junction and B-DNA structures, the helical arms of the junction are seen to be more typical, in many respects, of standard B-DNA than the actual B-DNA structure for this sequence. For example, the B form of ATC shows significant buckling within and slide and roll between stacked base pairs from the average B-DNA duplex of the data set. In contrast, the arms of the junction more or less fall into the norms of the B-DNA structures, with the exception that they are slightly overwound and show a positive slide and a displacement of the phosphate group (ZP) that is more negative than B-DNA duplexes. It is clear from an analysis of the overall data set, however, that these features are particular to the crossover of the DNA strands of the junctions.
There are, however, specific sequences that fall well outside the standard conformational variations of the B-DNA double helix. The overall structure of GCA, for example, deviates significantly from standard B-DNA, with five of the nine parameters in Table 2 falling at least 1 SD from the mean values for the overall structures of this class. In particular, the duplex is highly overwound (at ≈9.4 bp per turn as estimated from the helical twist), with the stacked base pairs showing significant roll, buckling, and slide. The experimental design of the study indicates that these variations are defined by the sequence rather than by the crystal lattice or crystallization conditions. Interestingly, the sequence TGCGCA is the repeating binding motif for at least one eukaryotic promoter (30) and, therefore, such conformational perturbations may play a role in protein recognition.
In summary, a crystallographic data set of DNA structures is being assembled from a well defined sequence motif and a relatively consistent set of crystallization solutions. This set allows us to correlate sequence and environment with structural classes and conformational variability within structural classes. Thus, it is clear that the strategy of broadly sampling structures by crystallographic screening of a specific sequence motif directly defines the effects of sequence on macroscopic behavior at the level of detailed molecular interactions. Although currently limited to DNA structures of self-complementary sequences, the results of the study show that the basic premise is correct: if the structures of all permutations of a molecule can be determined, the sequence effects on the overall structure and the details of their conformation are explicitly known.
Supplementary Material
Acknowledgments
We thank members of the Karplus laboratory at Oregon State University and Dr. J. Vargason for help with crystallographic problems. This work was supported by National Institutes of Health Grant R1GM62957A and National Science Foundation Grant MCB0090615. X-ray diffraction facilities were supported by the Proteins and Nucleic Acids Facility Core of the Environmental Health Sciences Center at Oregon State University (National Institute on Environmental Health Sciences Grant ES00210) and the Murdock Charitable Trust. Several DNA oligomers were synthesized in the Center for Gene Research and Biotechnology at Oregon State University. The BioCARS Sector 14 beamline (Argonne National Laboratory, Argonne, IL) is supported by the National Institutes of Health, National Center for Research Resources under Grant RR07707, and the Advanced Photon Source was supported by the U.S. Department of Energy under Contract W-31-109-Eng-38. The Advanced Light Source at Lawrence Berkeley National Laboratory is supported by the U.S. Department of Energy under Contract DE-AC03-76SF00098.
Author contributions: F.A.H. and P.S.H. designed research; F.A.H., A.T., Z.J.R.J., M.H., D.R., J.W., and E.C. performed research; F.A.H., A.T., and Z.J.R.J. contributed new reagents/analytic tools; F.A.H., Z.J.R.J., M.H., and P.S.H. analyzed data; and F.A.H. and P.S.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: IR, inverted repeat.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1ZEZ, 1ZF4, 1ZF3, 1ZFC, 1ZF2, 1ZF1, 1ZEW, 1ZFM, 1ZFH, 1ZFF, 1ZF5, 1ZFE, 1ZF7, 1ZFG, 1ZF0, 1ZFB, 1ZEX, 1ZEY, 1ZFA, 1ZF9, 1ZF8, and 1ZF6).
References
- 1.Anfinsen, C. B. (1973) Science 181, 223-230. [DOI] [PubMed] [Google Scholar]
- 2.Matthews, B. W. (1996) FASEB J. 10, 35-41. [DOI] [PubMed] [Google Scholar]
- 3.Ramshaw, J. A., Shah, N. K. & Brodsky, B. (1998) J. Struct. Biol. 122, 86-91. [DOI] [PubMed] [Google Scholar]
- 4.Persikov, A. V., Ramshaw, J. A., Kirkpatrick, A. & Brodsky, B. (2000) Biochemistry 39, 14960-14967. [DOI] [PubMed] [Google Scholar]
- 5.Lebrun, A. & Lavery, R. (1997) Curr. Opin. Struct. Biol. 7, 348-354. [DOI] [PubMed] [Google Scholar]
- 6.Minsky, A. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 317-342. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz-Lombardia, M., Gonzalez, A., Eritja, R., Aymami, J., Azorin, F. & Coll, M. (1999) Nat. Struct. Biol. 6, 913-917. [DOI] [PubMed] [Google Scholar]
- 8.Eichman, B. F., Vargason, J. M., Mooers, B. H. & Ho, P. S. (2000) Proc. Natl. Acad. Sci. USA 97, 3971-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Y. & West, S. C. (2004) Nat. Rev. Mol. Cell Biol. 5, 937-944. [DOI] [PubMed] [Google Scholar]
- 10.Hays, F. A., Watson, J. & Ho, P. S. (2003) J. Biol. Chem. 278, 49663-49666. [DOI] [PubMed] [Google Scholar]
- 11.Stefl, R., Wu, H., Ravindranathan, S., Sklenar, V. & Feigon, J. (2004) Proc. Natl. Acad. Sci. USA 101, 1177-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marini, J. C., Levene, S. D., Crothers, D. M. & Englund, P. T. (1982) Proc. Natl. Acad. Sci. USA 79, 7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolstorukov, M. Y., Ivanov, V. I., Malenkov, G. G., Jernigan, R. L. & Zhurkin, V. B. (2001) Biophys. J. 81, 3409-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basham, B., Schroth, G. P. & Ho, P. S. (1995) Proc. Natl. Acad. Sci. USA 92, 6464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiGabriele, A. D., Sanderson, M. R. & Steitz, T. A. (1989) Proc. Natl. Acad. Sci. USA 86, 1816-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson, R. E., Goodsell, D. S. & Neidle, S. (1994) Proc. Natl. Acad. Sci. USA 91, 3579-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramakrishnan, B. & Sundaralingam, M. (1993) J. Biomol. Struct. Dyn. 11, 11-26. [DOI] [PubMed] [Google Scholar]
- 18.Hays, F. A., Vargason, J. M. & Ho, P. S. (2003) Biochemistry 42, 9586-9597. [DOI] [PubMed] [Google Scholar]
- 19.Berman, H. M., Olson, W. K., Beveridge, D. L., Westbrook, J., Gelbin, A., Demeny, T., Hsieh, S. H., Srinivasan, A. R. & Schneider, B. (1992) Biophys. J. 63, 751-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinemann, U., Alings, C. & Bansal, M. (1992) EMBO J. 11, 1931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer-Jung, C., Moras, D. & Timsit, Y. (1998) EMBO J. 17, 2709-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duckett, D. R., Murchie, A. I., Diekmann, S., von Kitzing, E., Kemper, B. & Lilley, D. M. (1988) Cell 55, 79-89. [DOI] [PubMed] [Google Scholar]
- 23.Panyutin, I. G., Biswas, I. & Hsieh, P. (1995) EMBO J. 14, 1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprecher, C. A., Baase, W. A. & Johnson, W. C., Jr. (1979) Biopolymers 18, 1009-1019. [DOI] [PubMed] [Google Scholar]
- 25.Peticolas, W. L., Wang, Y. & Thomas, G. A. (1988) Proc. Natl. Acad. Sci. USA 85, 2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorpe, J. H., Gale, B. C., Teixeira, S. C. & Cardin, C. J. (2003) J. Mol. Biol. 327, 97-109. [DOI] [PubMed] [Google Scholar]
- 27.Eichman, B. F., Mooers, B. H., Alberti, M., Hearst, J. E. & Ho, P. S. (2001) J. Mol. Biol. 308, 15-26. [DOI] [PubMed] [Google Scholar]
- 28.Auffinger, P., Hays, F. A., Westhof, E. & Ho, P. S. (2004) Proc. Natl. Acad. Sci. USA 101, 16789-16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichman, B. F., Ortiz-Lombardia, M., Aymami, J., Coll, M. & Ho, P. S. (2002) J. Mol. Biol. 320, 1037-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano, R., Mita, T. & Matsui, T. (1992) Biochim. Biophys. Acta 1132, 49-57. [DOI] [PubMed] [Google Scholar]
- 31.Olson, W. K., Bansal, M., Burley, S. K., Dickerson, R. E., Gerstein, M., Harvey, S. C., Heinemann, U., Lu, X. J., Neidle, S., Shakked, Z., et al. (2001) J. Mol. Biol. 313, 229-237. [DOI] [PubMed] [Google Scholar]
- 32.Lu, X. J. & Olson, W. K. (2003) Nucleic Acids Res. 31, 5108-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.