Abstract
The corpus callosum (CC) is the largest set of white matter fibers connecting the two hemispheres of the brain. In humans, it is essential for coordinating sensorimotor responses, performing associative/executive functions, and representing information in multiple dimensions. Understanding which genetic variants underpin corpus callosum morphometry, and their shared influence on cortical structure and susceptibility to neuropsychiatric disorders, can provide molecular insights into the CC’s role in mediating cortical development and its contribution to neuropsychiatric disease. To characterize the morphometry of the midsagittal corpus callosum, we developed a publicly available artificial intelligence based tool to extract, parcellate, and calculate its total and regional area and thickness. Using the UK Biobank (UKB) and the Adolescent Brain Cognitive Development study (ABCD), we extracted measures of midsagittal corpus callosum morphometry and performed a genome-wide association study (GWAS) meta-analysis of European participants (combined N = 46,685). We then examined evidence for generalization to the non-European participants of the UKB and ABCD cohorts (combined N = 7,040). Post-GWAS analyses implicate prenatal intracellular organization and cell growth patterns, and high heritability in regions of open chromatin, suggesting transcriptional activity regulation in early development. Results suggest programmed cell death mediated by the immune system drives the thinning of the posterior body and isthmus. Global and local genetic overlap, along with causal genetic liability, between the corpus callosum, cerebral cortex, and neuropsychiatric disorders such as attention-deficit/hyperactivity and bipolar disorders were identified. These results provide insight into variability of corpus callosum development, its genetic influence on the cerebral cortex, and biological mechanisms related to neuropsychiatric dysfunction.
Introduction
The corpus callosum (CC) is the largest white matter tract in the human brain, facilitating higher order functions of the cerebral cortex by allowing the two hemispheres of the brain to communicate1,2. This connection is essential for coordinating sensorimotor responses, performing associative and executive functions, and representing information in multiple dimensions3,4. Most CC fibers connect corresponding left and right cortical regions of the brain, with the organization, development of axonal elongation, and myelination of callosal fibers being correlated with the rostro-caudal (front-to-back) distribution of functional areas5,6. Regional alterations in CC shape are easily assessed with neuroimaging studies, which have found local callosal abnormalities in complex neurodevelopmental and neuropsychiatric disorders6–11, such as lower anterior volumes in autism12 and lower posterior thickness in bipolar disorder13. Twin studies show up to 66% heritability for CC area14,15, and previous single-cohort studies of genetic influences on CC volume and its relationship to neuropsychiatric disorders have found heritability estimates between 22–39%16,17. Yet, the interplay between genetic variants influencing CC morphometry, the cerebral cortex, and associated neuropsychiatric disorders is not well understood.
3D magnetic resonance imaging (MRI) provides a non-invasive approach to quantify individual variations in brain regions and connections6, including the morphology of the CC, and how they are associated with brain-based traits and diseases. The midsagittal section of an anatomical brain MRI scan is able to capture the entire rostro-caudal formation of the CC, which is almost always in the field of view of 2D clinical and 3D research MRI scans alike. This 2D midsagittal representation can be segmented to offer a lower dimensional projection of the anatomical intricacies of the CC, allowing for structural measures of CC area and thickness to be computed18,19. We developed and validated a fully automated artificial intelligence based CC feature extraction tool, Segment, Measure, and AutoQC the midsagittal CC (SMACC), which we make publicly available at https://github.com/USC-LoBeS/smacc20.
Using data from the UK Biobank21 (UKB) and Adolescent Brain Cognitive Development22 (ABCD) studies, here we present results from a genome-wide association study (GWAS) meta-analysis of total area and mean thickness of the CC derived using SMACC. We also present the results for five differentiated areas based on distinguishable projections to (1) prefrontal, premotor and supplementary motor, (2) motor, (3) somatosensory, (4) posterior parietal and superior temporal, and (5) inferior temporal and occipital cortical brain regions23,24. These regions are believed to represent structural-functional coherence6. We performed a GWAS meta-analysis using two population-based cohorts, one of adolescents and another of older adults, to examine genetic influences on CC area and thickness25,26. The primary analyses were in individuals of European ancestry and the same analyses were then repeated using the data from non-European participants to assess consistency in the magnitude and direction of effect sizes. Downstream post-GWAS analyses investigated the enrichment of genetic association signals in tissue types, cell types, brain regions, and biological pathways. We examined the genetic overlap at the global and local level, using LD Score regression (LDSC)27 and Local Analysis of Variant Association (LAVA)28, respectively, and the causal genetic relationships between CC phenotypes, cortical morphometry, and related neuropsychiatric conditions.
Results
Characterization of corpus callosum shape associated loci
We conducted a GWAS of area and mean thickness of the whole corpus callosum, and five regions of the Witelson parcellation scheme (Fig. 1)23,24, using data from participants of European ancestry from the UKB (N = 41,979) and ABCD cohorts (N = 4,706). A meta-analysis of GWAS summary statistics of all CC derived metrics in UKB and ABCD was performed using METAL and the random-metal extension29,30, based on the DerSimonian-Laird random-effects model (Methods). To examine the generalizability of single nucleotide polymorphism (SNP) effects across ancestries, these same analyses were run using data from non-European participants (total N = 7,040).
Figure 1: Regions of the midsagittal corpus callosum and associated genomic loci.
An ideogram representing loci that influence total corpus callosum area, its mean thickness, and area and thickness of individual parcellations determined by the Witelson parcellation scheme in a rostral-caudal gradient (1–5). All loci are significant at the Bonferroni corrected, experiment-wide threshold of p < 6.13 x 10−9.
The GWAS meta-analysis identified 48 independent significant SNPs for total area and 18 independent SNPs for total mean thickness. Independent significant SNPs were determined in FUMA using the default threshold of r2 = 0.6, and genomic loci were determined at r2 = 0.1. This identified 28 genomic loci for total cross-sectional area, and 11 genomic loci for total mean thickness. All significant loci for total area and mean thickness showed concordance in the direction of effect between the two cohorts. There were 5 loci, all in intronic regions, each positionally mapped to genes31 that overlapped between area and mean thickness. These included IQCJ-SHIP1 (multimolecular complexes of initial axon segments and nodes of Ranvier, and calcium mediated responses)32, FIP1L1 (RNA binding and protein kinase activity)33, HBEGF (growth factor activity and epidermal growth factor receptor binding)34, CDKN2B-AS1 (involved in the NF-κB signaling pathway with diverse roles in the nervous system)35,36, and FAM107B (cytoskeletal reorganization in neural cells and cell migration/expansion)37. The genomic locus mapped to IQCJ-SHIP1 had a positive effect for total area (rs11717303, effect allele: C, effect allele frequency (EAF): 0.689, β = 4.28, s.e. = 0.51, p = 4.54 x 10−17). The same locus showed a negative effect for a different SNP on total thickness (rs12632564, effect allele: T, EAF: 0.305, β = −0.042, s.e. = 0.006, p = 2.59 x 10−12). The strongest locus for total area (rs7561572, effect allele: A, EAF: 0.532, β = −4.13, s.e. = 0.46, p = 1.98 x 10−18) was positionally mapped to the STRN gene. The strongest locus for mean thickness (rs4150211, effect allele: A, EAF: 0.265, β = −0.05, s.e. = 0.006, p = 8.20 x 10−18) was mapped to the HBEGF gene.
Loci for area overlapped between parcellations in a rostral-caudal gradient (1–5), such that: rs1122688 on the SHTN1 (or KIAA1598) gene (involved in positive regulation of neuron migration) overlapped between the genu (1) and anterior body (2); rs1268163 near the FOXO3 gene (involved in IL-9 signaling and FOXO-mediated transcription) overlapped between the posterior body (3) and isthmus (4); and rs11717303 on the IQCJ-SCHIP1 gene overlapped between the isthmus (4) and splenium (5). This gradient pattern was not observed for mean thickness. The strongest regional association was observed with splenium area (rs10901814, effect allele: C, EAF: 0.584, β = −1.69, s.e. = 0.16 p = 2.02 x 10−24) and thickness (rs11245344, effect allele: T, EAF: 0.570, β = −0.11, s.e. = 0.11, p = 6.28 x 10−22), both on the FAM53B gene. FAM53B is involved in positive regulation of the canonical Wnt signaling pathway. We observed a concordance in direction and similar magnitude effect sizes in the analyses within the data from the non-European participants. Detailed annotations and regional association plots of all genomic loci, independent significant SNPs and genes are in Supplementary Tables S1-S4 and Extended Data 1.
SNP heritability and genetic correlation between cohorts
Moderate to high genetic correlations were seen across CC phenotypes between cohorts, with rg ranging from 0.54 (s.e. = 0.27) and 0.92 (s.e. = 0.63) for area metrics, and 0.30 (s.e. = 0.16) and 0.99 (s.e. = 0.69) for thickness metrics. We used the GREML approach implemented in GCTA38,39 to estimate SNP heritability (h2SNP) for each cohort. Within the UKB, heritability values ranged for different CC phenotypes from 0.42 – 0.71, with similar results seen in the ABCD cohort (Supplementary Tables S5-S8). Total area (UKB h2SNP = 0.71, s.e. = 0.01; ABCD h2SNP = 0.74, s.e. = 0.03) and mean thickness (UKB h2SNP = 0.60, s.e. = 0.02; ABCD h2SNP = 0.77, s.e. = 0.03) showed the highest h2SNP across both cohorts. LDSC27 h2SNP estimates from the meta-analysis ranged between 0.10 (s.e. = 0.01) and 0.18 (s.e. = 0.05) for area, and 0.12 (s.e. = 0.01) and 0.16 (s.e. = 0.02) for thickness, with the area of the genu showing the highest, and area of the splenium showing the lowest h2SNP estimates. As shown in Supplementary Tables S5-S8, all LDSC RG estimates between meta-analyzed CC phenotypes were significant.
Gene-mapping and gene-set enrichment analyses
Gene-based association analysis in MAGMA40 identified 30 genes for the total area, and 34 genes for total mean thickness of the CC, with 5 genes overlapping between area and thickness (IQCJ-SCHIP1, IQCJ, BPTF, PADI2, CHIC2). The strongest association seen with area was AC007382.1 and the strongest association with mean thickness was HBEGF (Fig. 2A). There were between 15 and 31 genes for area, and between 7 and 25 genes for thickness identified within regions of the CC. Notably, IQCJ, IQCJ-SCHIP1, and STRN overlapped for all parcellations of CC area. AC007382.1 overlapped for four out of five parcellations, and STRN and PARP10 overlapped for three out of five parcellations of CC thickness (Fig. 2B, Supplementary Tables S1-S4). Enrichment of SNP heritability in 53 functional categories for each trait was determined via LDSC41. The majority of enrichment and the strongest effects across parcellations of the CC were observed in categories related to gene regulation/transcription in chromatin (Fig. 3A-B).
Figure 2: GWAS meta-analysis of midsagittal corpus callosum area and thickness.
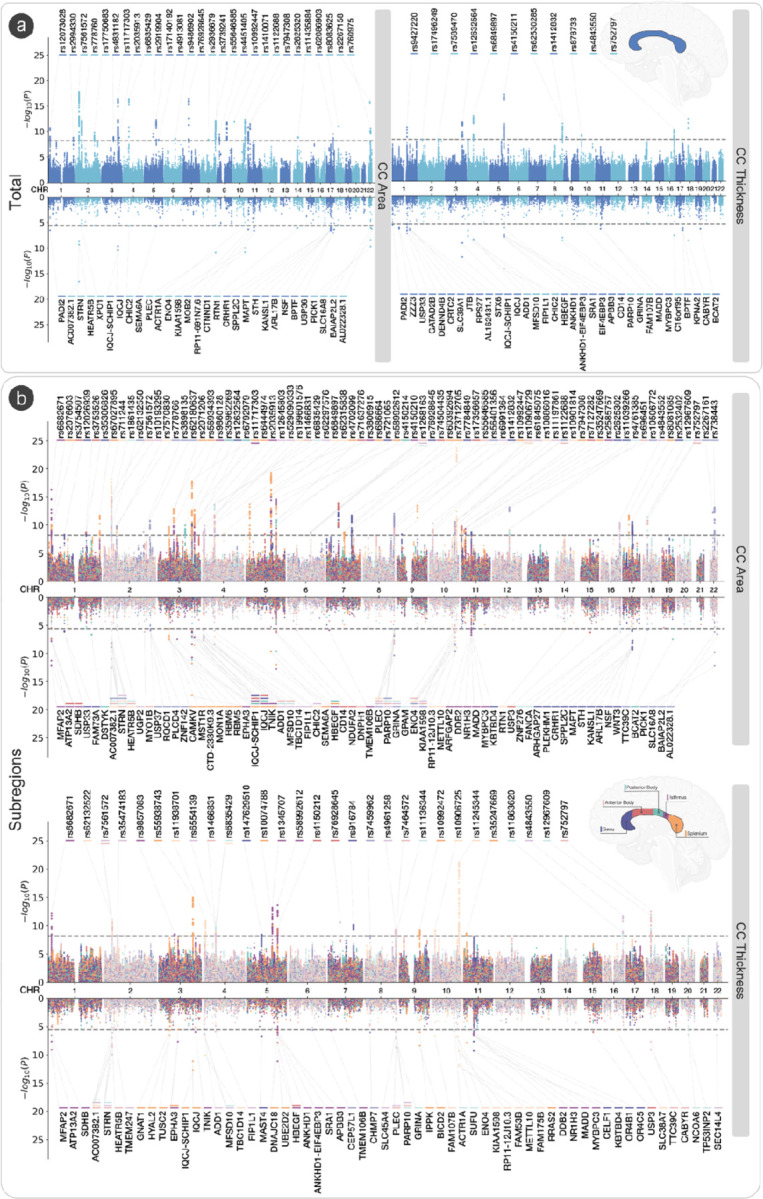
(A) Miami plot for SNPs (top) and genes (bottom) based on MAGMA gene analysis for total area and total mean thickness. (B) Miami plot for SNPs (top) and genes (bottom) based on MAGMA gene analysis for area of thickness of the CC split by the Witelson parcellation scheme23. Significant SNPs and genes are color coded by corpus callosum traits.
Figure 3: Partitioned heritability, functional annotation and enrichment of gene-sets of CC morpholog associated genetic variants.
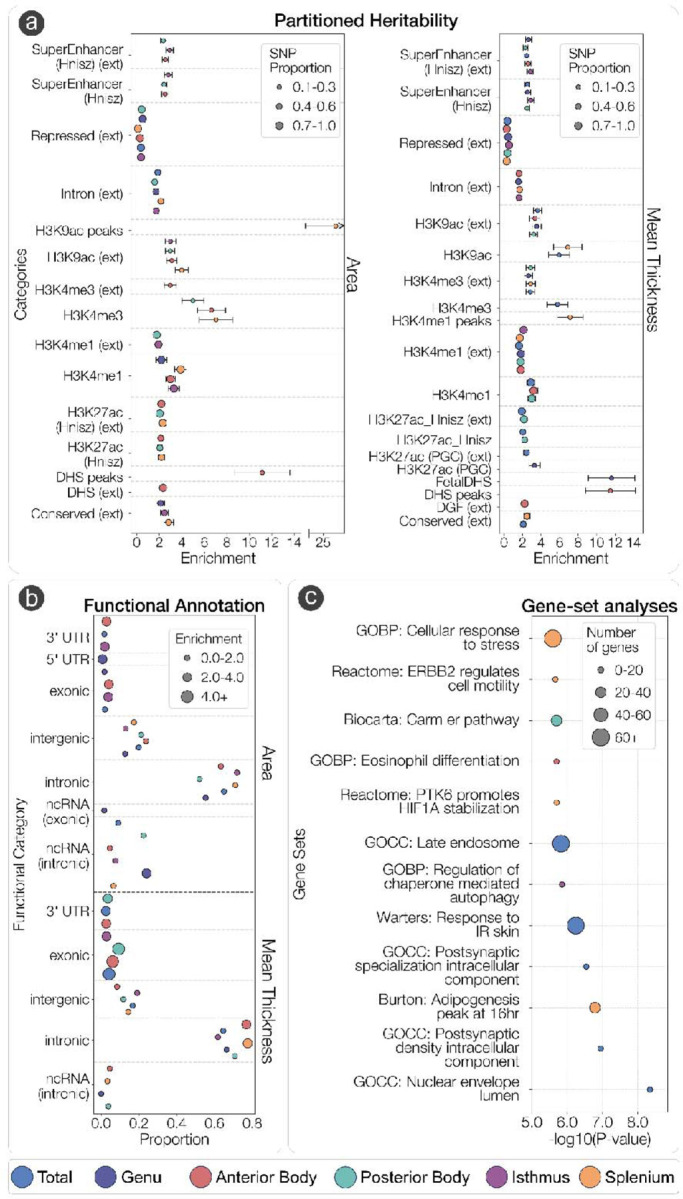
(A) Significant enrichment of SNP heritability across 53 functional categories compute by LD Score regression for area (left) and mean thickness (right). Error bars indicate 9% confidence intervals. (B) Proportion of GWAS SNPs in each functional category from ANNOVAR across each CC phenotype. (C) Significant gene-sets across CC phenotypes computed via MAGMA gene-set analysis at the Bonferroni corrected threshold of 3.23 x 10−6. GOBP: Gene-ontology biological processes, GOCC: Gene-Ontology Cellular Components.
Gene-set enrichment analyses were also completed in MAGMA (Fig. 3C). Strongest effects of significant gene sets included those involved in postsynaptic specialization for total CC area, including GO:009901 (postsynaptic specialization, intracellular component) and GO:009902 (postsynaptic density, intracellular component). A theme of signal transduction related pathways was observed for splenium area including R-HSA-6785631 (ERBB2 regulates cell motility) and R-HSA-8857538 (PTK6 promotes HIF1A stabilization). Enrichment of the “CARM1 and regulation of the estrogen receptor” was found for the posterior body thickness and is implicated transcriptional regulation via histone modifications. Enrichment of GO:1904714 (regulation of chaperone-mediated autophagy) was found for the isthmus area, which is implicated in lysosomal-mediated protein degradation. All significant results across all CC phenotypes are in Supplementary Table 18.
Tissue-specific and cell-type specific expression of corpus callosum associated genes
Gene-property enrichment analyses were completed in MAGMA with 54 tissue types from GTEx v8 and BrainSpan42,43, which includes 29 samples from individuals representing 29 different ages, as well as 11 general developmental stages. An enrichment of genes associated with isthmus thickness were expressed in the cerebellum (p(Bon) = 0.017). Area and thickness across parcellations of the CC showed an enrichment of expression of genes in the brain from early prenatal to late mid-prenatal developmental stages. An enrichment of expression of genes associated with area and thickness of the anterior body of the CC was observed in brain tissue prenatally 9 to 24 weeks post conception. Enrichment of expression of genes associated with area of the genu was observed in brain tissue 19 weeks post conception. Enrichment of expression of genes associated with total mean thickness of the CC was observed in brain tissue 19 weeks post conception. All results are shown in Supplementary Tables S19-S21. These results, along with the gene-sets involved in histone modifications, were supported by LDSC-SEG analyses using chromatin-based annotations from narrow peaks44, which showed a significant enrichment in the heritability by variants located in genes specifically expressed in DNase in the female fetal brain for total CC thickness (p(Bon) = 0.0105). Chromatin annotations showed a consistent and significant enrichment of splenium area and thickness associated variants in histone marks of the fetal brain and neurospheres (Supplementary Table S25).
Using microarray data from 292 immune cell types, area of the posterior body showed a significant enrichment in the heritability by variants located in genes specifically expressed in multiple types of myeloid cells (p(Bon) < 0.05), and area of the isthmus showed enrichment in innate lymphocytes (p(Bon) = 0.047). This further validates the aforementioned significant locus on gene FOXO3, which overlapped between the posterior body and isthmus (Supplementary Table S26).
Cell-type specific analyses were performed in FUMA using data from 13 single-cell RNA sequencing datasets from the human brain. This tests the relationship between cell-specific gene expression profiles and phenotype-gene associations45. Of the 12 phenotypes tested, only total CC thickness showed significant results after going through the 3-step process using conditional analyses to avoid bias from batch effects from multiple scRNA-seq datasets. The most significant association was seen with oligodendrocytes located in the middle temporal gyrus (MTG, p(Bon) = 0.001) from the Allen Human Brain Atlas (AHBA). Oligodendrocytes (p(Bon) = 0.03) and non-neuronal cells (p(Bon) = 0.03) located in the lateral geniculate nucleus (LGN) from the AHBA also showed significant associations but were collinear (Supplementary Table S22).
LAVA-TWAS analyses28,46 (Fig. 4) of expression quantitative trait loci (eQTLs) and splicing quantitative trait loci (sQTLs) of protein-coding genes in 16 different brain, cell type, and whole blood tissues revealed the strongest eQTL associations of area and thickness with CROCC expression in whole blood for the isthmus (ρ = −0.53, p = 1.29 x 10−10). Other notable eQTL (Supplementary Table S29) findings included total CC area and isthmus area and thickness being positively associated with ATP13A2 expression in fibroblasts (ρ = 0.48, p = 1.58 x 10−7). The strongest sQTL association was a positive association observed with KANSL1 cluster 11710 in fibroblasts for genu area (ρ = 0.83, p = 1.46 x 10−14), which was the tissue type where most observed associations occurred across CC phenotypes (Supplementary Table S30). Moreover, a negative association was observed in a KANSL1 (cluster 11707) in fibroblasts for the genu area (ρ = 0.82, p = 3.11 x 10−7). An sQTL in MFSD13A (cluster 7894) in the anterior cingulate showed very strong yet opposite associations for total CC thickness (ρ = 0.42, p = 1.12 x 10−13) and total CC area (ρ = −0.44, p = 2.98 x 10−11). Other notable findings across tissue types included CRHR1 in the cortex, nucleus accumbens, and putamen, as well as UGP2 in fibroblasts, whole blood, and the putamen. No significant results from LAVA-TWAS gene-set enrichment analyses were observed after Bonferroni correction (Supplementary Tables S31-S32).
Figure 4: LAVA-TWAS analyses of corpus callosum traits with gene-expression (eQTLs) and splicing (sQTLs).
Results of local genetic correlations between CC traits and eQTLs and sQTLs from GTEx v8 using the LAVA-TWAS framework. Associations between (A) CC area and eQTLs, (B) CC thickness and eQTLs, (C) CC are and sQTLs, and (D) CC thickness and sQTLs are shown via −log10p values scaled by the direction of association (y-axis) and chromosomal location (x-axis). All significant points are colored by tissue type and labeled by CC trait. Significance thresholds for eQTLs (p < 2.01 x10−6) and sQTLs (p < 5.45 x 10−7) were determined by Bonferroni correction.
Genetic overlap of corpus callosum and cerebral cortex architecture
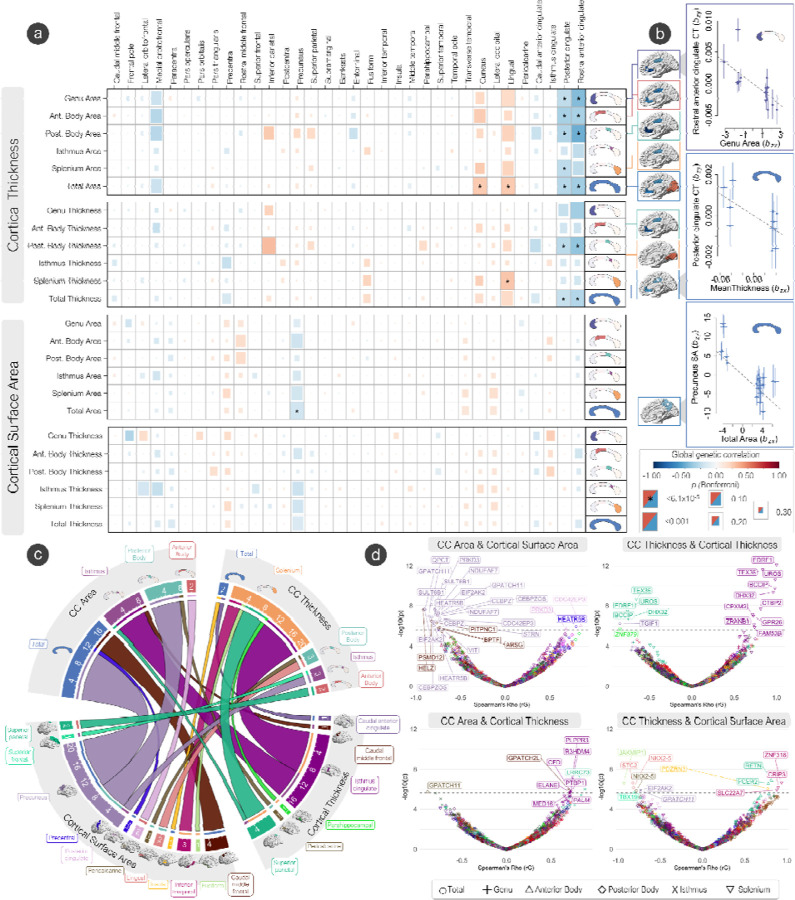
Broadly, we observed a pattern of negative genetic correlations with area and thickness of the CC with cortical thickness across regions of the cingulate cortex, but positive genetic correlations with regions’ cortical thickness across the neocortex (Fig. 5A). Specifically, we observed a significant negative genetic correlation between total area with cortical thickness of the rostral anterior cingulate (rg = −0.35, SE = 0.06) and posterior cingulate (rg = −0.28, SE = 0.06). Mean thickness was negatively genetically correlated with cortical thickness of the rostral anterior cingulate (rg = −0.29, SE = 0.06) and posterior cingulate (rg = −0.23, SE = 0.05). Positive genetic correlations were observed with cortical thickness of the lingual gyrus (rg = 0.26, SE = 0.05) and cuneus (rg = 0.27, SE = 0.06). When parcellating by the Witelson scheme, negative genetic correlations were observed for area and mean thickness with cortical thickness of regions across the cortex and the cingulate, but positive genetic correlations with regions in the occipital lobe. We also observed a significant negative genetic correlation between total area of the CC with surface area of the precuneus (rg = −0.20, SE = 0.04). (Supplementary Table S9-S10).
Figure 5: The genetic overlap of the corpus callosum and cerebral cortex.
(A) Global genetic correlations (LDSC - rG) between CC phenotypes and cerebral cortex phenotypes. The Bonferroni significance threshold was set at p 6.1 x 10−5. Surface area and cortical thickness of significant cortical regions with each CC phenotype are displayed on brain plots. (B) Of the significant global genetic correlations, significant Mendelian randomization (GSMR) results ar displayed, representing the effect of CC phenotypes on cortical phenotypes free of non-genetic confounders. (C) Chord plot displaying the number of significant bivariate local genetic correlations (LAVA) between CC and cortical phenotypes. Underlined numbers represent the total number of genes shared with that phenotype. (D) Volcano plots showing degree (−log10 p-values) and direction (rG) of local genetic correlations (LAVA) between cortical and CC phenotypes. Colors represent cortical regions labeled on the chord plot in section C. Significant genes (Bonferroni significance threshold was set at p = 2.18 x 10−6) across all phenotypes are labeled.
Genetic correlations can reflect direct causation, pleiotropy, or genetic mediation. To explore potential causal relationships between CC phenotypes and morphometry of the cerebral cortex, we ran Generalized Summary-data-based Mendelian Randomization (GSMR) analyses47 directional effect of CC phenotypes on morphometry of the cerebral cortex, but not vice-versa. (Fig. 5B, Supplementary Table S14). There was a strong negative unidirectional effect of total CC area on the precuneus surface area (bxy = −0.50, SE = 0.13, p = 0.0002), implying a greater total area and thickness of the CC results in a lower surface area of the precuneus. There was also a negative unidirectional effect of total CC mean thickness and cortical thickness of the posterior cingulate (bxy = −0.02, SE = 0.008, p = 0.02), but not vice versa. When using the Witelson parcellation scheme, there was a strong negative unidirectional effect on the area of the genu on the cortical thickness of the rostral anterior cingulate (bxy = −0.001, SE = 0.0003, p = 0.003).
Local genetic correlations of area phenotypes of the CC and surface area of the cerebral cortex with LAVA28 showed many significant negative correlations in genes between the total area and posterior body and the precuneus SA along the 2p22.2 cytogenetic band (QPCT, PRKD3, SULT6B1, NDUFAF7, EIF2AK2, HEATR5B, GPATCH11, CEBPZ, CEBPZOS, CDC42EP3, STRN, VIT) (Fig. 5C-D). Negative genetic correlations between total CC area and caudal middle frontal gyrus SA in 5 genes along the 17q24.2 cytogenetic band (HELZ, PSMD12, PITPNC1, ARSG, BPTF) were also observed. Positive local genetic correlations along the 2p22.2 cytogenetic band were observed with anterior body area and the surface area of the posterior cingulate (CDC42EP3, PRKD3), as well as total area of the CC and precentral gyrus surface area (HEATR5B).
Many negative local genetic correlations were observed with mean thickness of the splenium and cortical thickness of the superior parietal gyrus (TEX36, EDRF1, UROS, BCCIP, DHX32) and the parahippocampal gyrus (ZNF879) along the 10q26.13–10q26.2 cytogenetic bands, while positive genetic correlations were observed with isthmus cingulate cortical thickness along the 10q26.13–10q26.2 cytogenetic bands (EDRF1, TEX36, UROS, BCCIP, DHX32, CTBP2, CPXM2, GPR26, ZRANB1, FAM53B).
Area of the posterior body showed a negative local genetic correlation with pericalcarine gyrus cortical thickness (GPATCH11). Area of the isthmus showed positive local genetic correlations with the cortical thickness of the superior parietal gyrus (LRRC73), caudal middle frontal gyrus (GPATCH2L), and isthmus cingulate (PLPPR3, CFD, R3HDM4, PTBP1, ELANE, MED16, PALM) along the 19p13.3 cytogenetic band.
Mean thickness of the posterior body showed negative local genetic correlations with the surface area of the lingual gyrus (STC2, NKX2–5, 5q35.2) and pericalcarine gyrus (NKX2–5). Mean thickness of the isthmus showed negative local genetic correlations with the precuneus (EIF2AK2, GPATCH11, 2p22.2) and superior frontal gyrus (TBX19) surface area. Total mean thickness of the CC showed a positive genetic correlation with surface area of the insula (PDZRN3). The anterior body mean thickness showed positive local genetic correlations with surface area of the superior parietal gyrus (RETN, FCER2). Splenium mean thickness showed positive genetic correlations with inferior temporal gyrus surface area (ZNF318, CRIP3, SLC22A7) along the 6p21.1 cytogenetic band.
Genetic overlap of corpus callosum and associated neuropsychiatric phenotypes
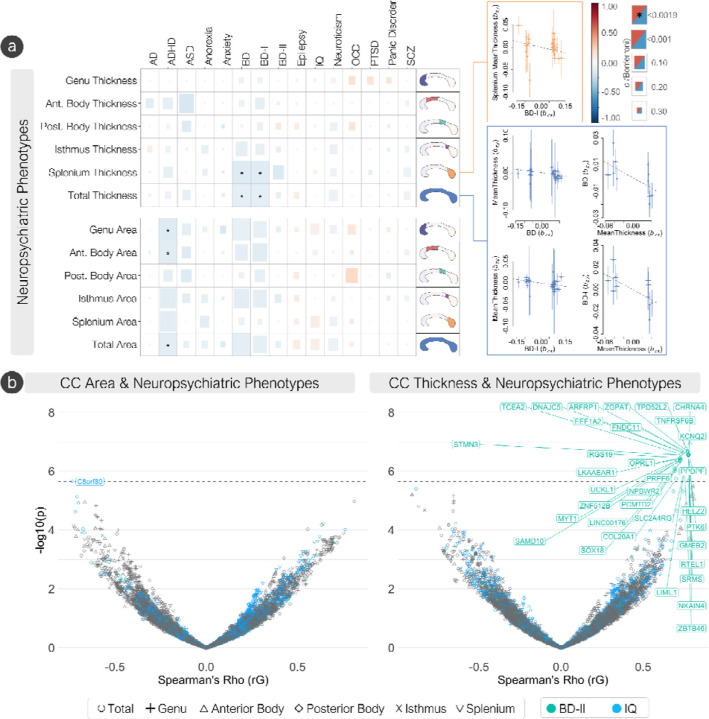
We observed a significant genetic correlation (Fig. 6A, Supplementary Table S11) between total CC area and ADHD (rg = −0.11, SE = 0.03), bipolar disorder (BD, rg = −0.10, SE = 0.03), and bipolar I disorder (BD-I, rg = −0.10, SE = 0.03). Total mean thickness was genetically correlated with BD (rg = −0.10, SE = 0.03) and BD-I (rg = −0.10, SE = 0.03). When analyzing the regional Witelson parcellations, the area of the genu was genetically correlated with ADHD risk (rg = −0.13, SE = 0.03), and the mean thickness of the splenium was genetically correlated with risk for BD (rg = −0.13, SE = 0.03) and BD-I (rg = −0.12, SE = 0.03).
Figure 6: The genetic overlap of the corpus callosum and neuropsychiatric phenotypes.
(A) Global genetic correlations between CC traits and neuropsychiatric phenotypes. The Bonferroni significance threshold was set at p = 0.0019. Of the significant global genetic correlations, significant Mendelian randomization (GSMR) results ar displayed, representing the effect of CC phenotypes on neuropsychiatric phenotypes free of non-genetic confounders. (B) Volcano plots showing degree (−log10 p-values) and direction (rG) of local genetic correlations (LAVA) between neuropsychiatric and CC phenotypes. Phenotypes with significant associations are colored (IQ an bipolar II disorder). Significant genes (Bonferroni significance threshold was set at p = 2.23 x 10−6) across all neuropsychiatric phenotypes. AD: alzheimer’s disease, ADHD: attention deficit hyperactivity disorder, ASD: autism spectrum disorder, BD: bipolar disorder, BD-I: bipolar I disorder, BD-II: bipolar II disorder, IQ: intelligence quotient, OCD: obsessive-compulsive disorder, PTSD: post-traumatic stress disorder, SCZ: schizophrenia.
GSMR analyses showed causal bidirectionality of genetic liability of BD (bxy = −0.06, SE = 0.02, p = 0.006) and BD-I (bxy = −0.05, SE = 0.02, p = 0.003) on total mean thickness of the CC, and mean thickness of the CC on BD (bxy = −0.19, SE = 0.08, p = 0.01) and BD-I (bxy = −0.23, SE = 0.09, p = 0.02). When using the Witelson parcellation, GSMR analyses showed causal directionality of genetic liability of BD-I on mean thickness of the splenium (bxy = −0.09, SE = 0.04, p = 0.01), but not vice versa (Fig. 6A, Supplementary Table S15).
Local genetic correlations with LAVA28 (Fig. 6B, Supplementary Table S17) showed 34 positive local genetic correlations between thickness of the posterior body and bipolar II disorder (BD-II) along the 20q13.33 cytogenetic band (top 5 genes being KCNQ2, TPD52L2, TNFRSF6B, ZGPAT, ARFP1), and one negative local genetic correlation between total CC area and IQ (C8orf89).
Discussion
We performed a GWAS meta-analysis of corpus callosum morphometry using our artificial intelligence based extraction tool, SMACC, from 46,685 individuals using UKB and ABCD. The majority of studies investigating the genetic influence via candidate genes on CC structure and development have been conducted using various animal models and post-mortem human studies6. Given the difference of the human CC compared to animal models6, this study provides genome-wide insight into human variation and genes that influence the human CC in vivo.
We show the genetic architecture of the CC is highly polygenic, and specific genetic variants influence CC subregions along a rostral-caudal gradient. Five loci that were positionally mapped to genes were identified to influence both total area and mean thickness of the CC (IQCJ-SHIP1, FIP1L1, HBEGF, CDKN2B-AS1, and FAM107B). IQCJ-SHIP1 had the strongest effect across total area and mean thickness, implicating mechanisms such as conduction of action potentials in myelinated cells via organizing molecular complexes at the nodes of Ranvier and axon initial segments, calcium mediated responses, as well as axon outgrowth and guidance48. The strongest locus for total area was mapped to the STRN gene. STRN has been heavily implicated in the Wnt signaling pathway, which controls the expression of genes that are essential for cell proliferation, survival, differentiation, and migration via transcription factors49–51. The HBEGF gene was the strongest locus for total mean thickness, implicating mechanisms in early development. HBEGF expression is localized in the ventricular zone and cortical layers during development52, and has been implicated in regulating cell migration via chemoattractive mechanisms52. Significant enrichment of heritability of total mean thickness in various histone marks from chromatin data (ATAC-seq) of the fetal brain and cortex derived primary cultured neurospheres, significant tissue expression in the brain 19-weeks post conception, as well as enrichment of gene sets involving regulation of histone modification, suggests genetic variants in regions of open chromatin and transcriptional activity regulation in early development are key mechanisms underlying CC morphometry. When histones are acetylated, they become more negatively charged. This negative charge repels the negatively charged DNA, causing the DNA to be “pushed away” from the histones. This loosening of the DNA-histone complex makes it easier for transcription factors to access the DNA and initiate transcription53.
Parcellation of the CC into the five regions defined by the Witelson scheme allowed for further refinement and genetic understanding of its morphometry in a rostral-caudal gradient. Our results provide insight as to which molecular mechanisms influence this functionally defined gradient (i.e. prefrontal, premotor/supplementary motor, primary motor, primary sensory, and parietal/temporal/occipital)24. An overlap of genetic loci along the most anterior (genu and anterior body, SHTN1) and most posterior (isthmus and splenium, IQCJ-SCHIP1) regions of the CC, along with splenium heritability enrichment of in histone chromatin marks of the fetal brain and dorsolateral prefrontal cortex, implicates regulation of neuron migration and action potential conduction. But the overlap of the FOXO3 along the area of the posterior body and isthmus implicates IL-9 signaling and FOXO-mediated transcription responsible for triggering apoptosis54. Only the posterior body and isthmus showed heritability enrichment in immune cells including myeloid cells and innate lymphocytes. The thinning of the CC (along the posterior body and isthmus) occurs in a functional gradient connecting the somatosensory and parietal association areas of the brain6,55,56. This follows activity dependent pruning by functional area6, where somatosensory circuits are pruned in early development in an experience dependent context57. As immune cells are increasingly being recognized as key players in brain maturation and neurodevelopment58, our results suggest IL-9 mediating a neuroprotective effect in the CC during the cell dieback phase58,59, and may play a significant role in posterior CC morphometry. LAVA-TWAS results showed another potential mechanism of isthmus pruning via expression of ATP13A2 in fibroblasts, and splicing of genes involved in NF-κB signaling60. ATP13A2 is involved in lysosomal-mediated apoptosis61, suggesting such regulation of fibroblast mediated growth of callosal projections62. This is also supported by the current discovery of enrichment of genes related to isthmus area in the “regulation of chaperone mediated autophagy pathway”, which may influence isthmus morphometry.
The topographic organization of the CC correlates with the homotopic bilateral regions of the cortex it is known to connect5. A variety of genetically regulated principal mechanisms influence CC neuronal and glial proliferation, neuronal migration and specification, midline patterning, axonal growth and guidance, and post-guidance refinement to homotopic analogs in the cortex63,64. Our results suggest potential genetic mechanisms contributing to callosal-cortical organization. We show an overall negative global genetic correlation of CC phenotypes with the cortical thickness of the cingulate and surface area of the posterior parietal cortices, including a unidirectional negative effect of genu area on rostral anterior cingulate thickness, and total area on precuneus surface area free of any non-genetic confounders. Positive global genetic correlations of total CC area and splenium thickness with cortical thickness in the occipital cortex were also observed. Local genetic correlations of the CC were observed throughout the cerebral cortex, most pronounced with total CC area and splenium thickness. Notable findings included numerous genes in the chr2p22 cytogenetic band showing negative correlations between total CC and posterior body area with precuneus surface area, including the significant STRN gene observed across all CC phenotypes, further implicating the Wnt signaling pathway and dendritic calcium signaling in the context of neurodevelopment65,66. Within this cytogenetic band, HEATR5B was also positively genetically associated with precentral gyrus surface area. Opposing genetic effects were observed between splenium thickness with isthmus cingulate thickness (i.e. positive) vs. superior parietal cortex thickness (i.e. negative) in genes in the chr10q26.13 cytogenetic band. Clinical phenotypes associated with the central nervous system due to copy number variations of chr10q26.13 include abnormal cranium development, global developmental delay and learning difficulties, and neuropsychiatric manifestations including ADHD, impulsivity or autistic behaviors67–69. This provides a novel testable hypothesis for functional follow up studies, as alterations in the isthmus cingulate and superior parietal cortex have been observed in large-scale studies of various neurodevelopmental disorders70. Positive genetic associations in the chr19p13.3 cytogenetic band were observed between the isthmus area and isthmus cingulate cortical thickness, which has been implicated with microcephaly, ventriculomegaly and developmental delay71,72.
Our results demonstrate opposing genetic relationships between CC phenotypes and thickness of the cingulate cortex (negative) vs the neocortex (positive), which suggests a strong genetic component underlying the development of the CC via pioneer axons and chemotaxis. Developmentally, pioneer axons emerge in the cingulate and project their axons across the midline using guidance cues. A large portion of these callosal projections are pruned and myelinated in an activity dependent manner, such that axonal remodeling is highly dependent on correlated neural activity in the cortex6,73–75. The strongest local genetic correlation supporting this finding was observed between total mean thickness of the CC and rostral anterior cingulate thickness on TGIF1. As TGIF1 is implicated in holoprosencephaly (i.e. where the brain fails to develop two hemispheres), forebrain development via alterations in the Sonic Hedgehog (SHH) pathway, and disruption of axonal guidance via chemoattractive mechanisms76,77, these results provide a potential genetic localization for functional follow-up. The isthmus cingulate, in relation to the isthmus and splenium, was the only cingulate region showing positive local genetic correlations, providing further evidence of distinct molecular mechanisms (e.g. immune-mediated apoptosis and regulation of callosal projections) compared to the rest of the corpus callosum underlying its structure and development.
Abnormalities of the CC have also been associated with various neurological/neuropsychiatric disorders6. The negative global genetic correlations observed in CC area with ADHD and CC thickness with bipolar disorder, indicate that the allelic differences resulting in smaller CC area and thickness are partly shared with those resulting in a greater risk for ADHD and bipolar disorder, respectively. Positive local genetic correlations heavily implicated the 20q13.33 cytogenetic band in the relationship between posterior body thickness and bipolar II disorder, providing a plausible neurobiological mechanism underlying an observed genetic risk at this locus78–80, and an observed morphological difference in the CC13,81,82. A negative genetic correlation at the C8orf89 locus, known to have biased expression in the testis83,84, was observed between total CC area and IQ. Strong evidence suggests the high similarity in gene expression and proteome between the brain and testes are due to involvement in the speciation process85.
In summary, this work identifies genome-wide significant loci of morphometry of the overall corpus callosum and its sectors, convergence on biological functions, tissues and cell types, as well as the genetic overlap with the cerebral cortex and neuropsychiatric conditions.
Methods
Artificial intelligence corpus callosum extraction and segmentation with SMACC
Data Preprocessing:
All UKB participants completed a 31-minute neuroimaging protocol using a Siemens Skyra 3 Tesla scanner and a 32-channel head coil in one of three MRI scanning locations. All 3D structural T1-weighted brain scans were acquired using the following parameters: 3D MPRAGE, sagittal orientation, in-plane acceleration factor = 2, TI/TR = 880/2000 ms, voxel resolution = 1 x 1 x 1 mm, acquisition matrix = 208 x 256 x 256 mm. All scans were pre-scan normalized using an on-scanner bias correction filter. More details of the imaging protocols may be found in the following reference papers86,87.
All ABCD participants completed a neuroimaging protocol in one of three scanner types at 21 different sites88. The Siemens Prisma had the following parameters for the T1-weighted scans: TI/TR = 1060/2500 ms, TE = 2.88 ms, voxel resolution = 1 x 1 x 1 mm, acquisition matrix = 176 x 256 x 256, flip angle = 8 degrees. The Philips Achieva Ingenia had a TI/TR = 1060/6.31 ms, voxel resolution = 1 x 1 x 1 mm, acquisition matrix = 225 x 256 x 256 mm and a flip angle = 8 degrees. The GE MR750 had a TI/TR = 1060/2500 ms, TE = 2 ms, voxel resolution = 1 x 1 x 1 mm, acquisition matrix = 208 x 256 x 256, and a flip angle = 8 degrees.
All T1w MRIs were registered to MNI15289–91 1mm space with 6 degrees of freedom using FSL’s flirt92 command.
SMACC development and UNet training:
Mid-sagittal T1w, T2w, and FLAIR images from UK Biobank21, PING93, HCP94, and ADNI195 were used for training the UNet model for CC segmentation. Individual study scanner parameters can be found in their respective references. The demographic information for the datasets used to create the UNet model is shown in Supplementary Table 31. Augmentation of image data is a common procedure in deep learning to prevent model overfitting and improve model accuracy96. All the images were downsampled by a factor of 2, 3, 4 and 5 along the sagittal axis and then upsampled back to original size using MRtrix’s mrgrid command to include low resolution images in the training97. To include lower resolution T1w images resembling older or clinical data in training, all the images were harmonized using a fully unsupervised deep-learning framework based on a generative adversarial network (GAN)98 to a subject from the ICBM dataset90. Images were also rotated clockwise in increments of 15 degrees and then resized to 256*256. Black boxes were randomly added to the images to imitate partial agenesis cases. Supplementary Figure 1 shows some T1w augmented images that were the input training images for the UNet model.
UNet Implementation:
A Tensorflow implementation of UNet99 was trained on 80% of the images for 250 epochs until the difference between the intersection over union (IOU) after consecutive iterations was less than 1x10−4. The U-Net architecture is structured with a contracting pathway and an expansive pathway. The contracting pathway repeatedly performs two 3x3 convolutions (without padding), with each convolution followed by a rectified linear unit (ReLU) and a 2x2 max pooling operation. At each stage in the expansive pathway the feature map is upsampled followed by a 2x2 convolution which reduces the feature channels by half. Then, the corresponding cropped feature map from the contracting pathway is concatenated, and two 3x3 convolutions are applied, with each one followed by a ReLU. We used the following training parameters: 1x10−4 learning rate and an Adam optimizer100. The rest of the data was used for validation. The midsagittal CC (midCC) was initially segmented using image processing techniques101 on subjects from ADNI1 (N=1032, 54–91 years), PING (N=1178, 3–21 years), HCP (N=963, 22–37 years) and UKB (N=190, 45–81 years). These masks were then visually verified and manually edited by neuroanatomical experts which served as the ground truth. To evaluate the model, the area of overlap between the predicted segmentation and the ground truth was calculated.
CC shape metrics extracted with SMACC:
SMACC provides outputs of global and regional shape metrics extracted from the corpus callosum segmentation, including area, thickness, length, perimeter and curvature. The regional shape metrics were based on a 5 compartment version of the Witelson atlas23,24. The Witelson atlas is composed of the (1) genu, (2) anterior midbody, (3) posterior midbody, (4) isthmus, and (5) splenium. The metrics used for the GWAS analysis were area and mean thickness of the total CC and all of the parcellations of the Witelson atlas. The thickness is defined as the distance in the inferior-superior direction between the top and bottom of the contour and at every point along the length of the segment, then averaged across the region of interest. The total area is the summation of the number of voxels with intensity value greater than 0.5 in the segmentation.
Corpus callosum segmentation quality control (QC) with SMACC:
To ensure that segmentations were of appropriate quality without having to manually assess all output images, which eventually may scale to hundreds of thousands of scans, we included an automated quality control (QC) assessment into SMACC. The regional and global metrics were used as inputs to the machine learning models detailed below for automatic binary classification of segmentations as Pass or Fail. CC segmentations from SMACC were manually assessed across multiple datasets by neuroanatomical experts. This included data from UKB (N=12,902, aged 45–81 years), ADNI1 (N=724, aged 54–91 years), PING (N=857, 3–21 years) and HCP (N=615, 22–37 years), all of which served as the ground truth for QC model building. All data was split 80/20 for training/testing.
Figure 7 gives the overview and the flow of SMACC. Several architectures including a 3-layer sequential neural network with 42 neurons, 22 in the second layer, and 11 in the third layer; a wide & deep neural network with 80 neurons in the first 3 layers and 40 in the last 3 layers, XGBoost classifier and an ensemble model were tested to classify the segmentations from the UNet as pass or fail. The ensemble model consisted of XGBoost, k-nearest neighbors (KNN), support vector classifier (SVC), logistic regression, and a random forest classifier. The results from all the classifiers in the ensemble model were combined using a majority voting classifier. All the models were compared using metrics including precision, recall, F1 score and Area Under the Curve (AUC). Supplementary Table 34 shows the performance of different models based on the shape metrics extracted from the CC segmentations.
Figure 7: Segment, Measure, and AutoQC the midsagittal CC (SMACC) pipeline -.
The midsagittal slice from a participant registered to MNI space with 6 degrees of freedom serves as an input to the UNet architecture used for the midsagittal corpus callosum segmentation. The Witelson atlas was used for segmenting the CC into five different regions. Global and subregion metrics (thickness and area-shown in green) were extracted from the segmentation. The thickness (black arrow) is defined as the distance in the inferior-superior direction between the top and bottom of the contour, after reorientation to standard space, at every point along the length of the segment, then average across the region of interest. These metrics serve as input for the ensemble machine learning model used for labelin CC segmentations as having passed or failed quality control (QC). Abbreviations: Montreal Neurological Institute - MNI, CC - corpus callosum, ML - Machine Learning, KNN - K Nearest Neighbors, SVC - Support Vector Classifier
SMACC vs FreeSurfer:
Comparing SMACC and FreeSurfer via Dice scores with respect to manual masks:
For assessing the accuracy of the SMACC compared to the ground truth and compared to the commonly used tool FreeSurfer102, we ran the SMACC pipeline on 30 subjects from the Hangzhou Normal University (HNU) test-retest dataset103,104. Each subject in this dataset was scanned with a full brain T1w MRI 10 times within a period of 40 days, for a total of 300 scans. All 300 scans had also been manually segmented by a neuroanatomical expert to serve as the ground truth. Segmentations from SMACC and FreeSurfer v7.1 were compared to manual segmentations using the Dice overlap coefficient. The average Dice coefficient between automated CC masks from SMACC and ground truth segmentations was 0.94 across all scans. The average Dice score between FreeSurfer CC segmentations and manual masks was 0.82. The Dice score was consistently higher for all the subjects using SMACC. Supplementary Figure 2 and Supplementary Table 35, show a few midCC segmentations obtained from SMACC compared to FreeSurfer.
ICC for SMACC:
To assess test-retest reliability of SMACC the intraclass correlation (ICC) scores were calculated. Average ICC values for thickness and area of the Witelson parcellations and the total CC were greater than 0.9 and are shown in Supplementary Figure 3.
Study cohorts
U.K. Biobank:
The UK Biobank (UKB) is a large population level cohort study conducting longitudinal deep phenotyping of around 500,000 participants in the United Kingdom (UK) aged between 40–69 at recruitment. All participants provided informed consent to participate. The North West Centre for Research Ethics Committee (11/NW/0382) granted ethics approval for the UK Biobank study21. We used genotype data from UKB released in May 2018. The data was collected from 489,212 individuals, and 488,377 of those individuals passed quality control checks by UKB. The genotypes were then imputed using two reference panels: the Haplotype Reference Consortium (HRC) reference panel and a combined reference panel of the UK10K and 1000 Genomes projects Phase 3 (1000G) panels21. There were 8,422,770 SNPs following quality control (QC) of the data which included having a genotyping call rate (SNPs missing in individuals) of greater than 95%, removing variants with a minor allele frequency less than 0.01 (1%), removing variants with Hardy-Weinberg equilibrium p-values less than 1e-6, and removing individuals with greater than three standard deviations away from the mean heterozygosity rate. To determine European ancestry in UKB, the ENIGMA MDS protocol (https://enigma.ini.usc.edu/protocols/genetics-protocols/) was completed using 10 components. The mean and standard deviations of the first and second genetic components of individuals who were classified as Utah residents with Northern and Western European ancestry from the CEPH collection (CEU) from the HapMap 3 release were then calculated. Individuals in UKB who were within a distance of 0.0101 on components 1 and 2 were classified as of European ancestry (N = 41,979). The MDS plot of individuals included in the analysis overlaid over the HapMap 3 population is available in Supplementary Figure 4.
ABCD:
The Adolescent Behavioral Cognitive Development (ABCD) study is the largest study in the United States (USA) following adolescent children starting from 9 years of age through adolescence with deep phenotyping including neuroimaging and genotyping using the Smokescreen™ Genotyping array consisting over over 300,000 SNPs88,105,106. Only neuroimaging from baseline (ages 9–10) were used. Following imputation using the ENIGMA protocol107 with the European 1000 Genomes Phase 3 Version 5 reference panel, phased using Eagle version 2.3108, and the QC process as described in the UKB cohort, a total of 4,706 European ancestry children, and 5,683,360 SNPs were included. To determine European ancestry in ABCD, the methods described for the UKB were completed. The MDS plot of individuals included in the analysis overlaid over the HapMap 3 population is available in Supplementary Figure 5.
We also analyzed non-European ancestry individuals to examine the generalization of the observed effects across ancestries. Using the aforementioned methods, we included 1504 individuals from the UKB and 5536 individuals from ABCD.
GWAS meta-analysis of corpus callosum morphometry
Genome-wide association analysis (GWAS) for UKB and ABCD separately for all CC phenotypes were completed via a linear whole-genome ridge regression model using REGENIE, allowing for the control of genetic relatedness109. Covariates included age, sex, age*sex interaction, and the first 10 genetic principal components. A two-step REGENIE analysis was completed with the following parameters. For step 1, the entire dataset was used with a block size of 1000 and leave-one-out-chromosome validation109. Step 2 was completed with a threshold for minor allele count of 5, a block size of 1000, and otherwise default parameters.
A meta-analysis of GWAS summary statistics of all CC derived metrics in UKB and ABCD were conducted using METAL software and the random-metal extension29,30, based on the random-effects model. A random-effects model was chosen since the effect sizes of SNPs on the corpus callosum has the potential to be different between the UKB and ABCD cohorts due to age. White matter volume is known to increase through childhood and start decreasing in middle adulthood110, which may result in different genetic effect sizes being observed. We opted to conduct a meta-analysis instead of using a two-stage discovery-replication approach because Skol et al. have shown that this method is more powerful, despite using more stringent significance levels for multiple correction111, and is common practice in the literature112–114. Percent variance (R2) explained by each significant SNP was calculated using the approach described in Rietvield et al115. The R2 of each variant j was calculated via:
where and are the minor and major allele frequencies, is the estimated effect of the variant within the meta-analysis and 2 is the estimated variance of the trait (for which we used the pooled variance of the trait across UKB and ABCD. In order to determine the number of independent traits, matrix spectral decomposition was computed using matSpD in R on the phenotypic correlations between CC traits using the method proposed by Li and Ji116,117. This resulted in 8.16 effective independent variables, and a significance threshold of p = 5 x 10−8/8.16 = 6.13 x 10−9. Meta-analyses were also completed for non-European individuals.
Heritability and genetic correlations within and between cohorts
To determine SNP heritability (h2SNP) tagged from SNPs used in the analysis, we used the GREML approach implemented in GCTA38,39, while adjusting for the same covariates as in the GWAS. The SNP heritability (h2SNP) from LDSC,27 was also computed, which estimates heritability casually explained by common reference SNPs. Genetic correlations between the UKB and ABCD cohorts for area and thickness of each parcellation of the CC defined by the Witelson scheme, and total CC were completed using LDSC27. Between cohort heterogeneity of h2SNP should not be considered unusual, as the genetic influence observed on the corpus callosum has the potential to be different between the UKB and ABCD cohorts due to age - white matter volume is known to increase through childhood and start decreasing in middle adulthood110, as well as the smaller sample size in ABCD making it harder for LDSC to detect polygenic effects118.
Gene-mapping and gene enrichment analyses
Genetic variants (SNPs) were mapped to genes using information about genomic position, expression quantitative trait loci (eQTL) information, and 3D chromatin interaction mapping as implemented in FUMA v1.5.2 with the experiment-wide significance threshold (p = 6.13 x 10−9)119. Pathway enrichment analyses using the results from the full meta-analyses with no pre-selection of genes via MAGMA v1.0840 gene-set analysis in FUMA. Genes located in the MHC region were excluded (hg19: chromosome 6: 26Mb - 34Mb). There were 19,021 gene sets from MSigDB v7.0120 (Curated gene sets: 5500, GO terms: 9996), and 9 other data resources including KEGG, Reactome and Biocarta (https://www.gsea-msigdb.org/gsea/msigdb/collection_details.jsp#C2). MAGMA uses gene-based P-values to identify genes that are more strongly associated with a phenotype than would be expected by chance. MAGMA then applies a competitive test to compare the association of genes in a gene set to the association of genes outside of the gene set. This allows MAGMA to identify gene sets that are enriched for association signals. MAGMA corrects for a number of confounding factors, such as gene length and size of the gene set, to ensure that the results are not due to chance. A gene-based association analysis (GWGAS) in MAGMA was completed using the full summary statistics for each trait from METAL. Corrections for multiple comparisons were completed using the Bonferroni approach.
To determine whether genes associated with CC morphometry cluster into biological functions, tissue types, or specific cell types, we used the full results of the meta-analyzed genome-wide association studies (GWAS) rather than prioritizing genes. Pathway analysis as described above was completed.
We performed gene-property and gene-set analysis using the MAGMA software on 54 tissue types from the GTEx v8 database and BrainSpan42,43, which includes 29 samples from individuals representing 29 different ages of brains, as well as 11 general developmental stages.
Single cell RNA-sequencing data sets used in the cell-type specific analyses included the human developmental and adult brain samples from the PsychENCODE consortium121, human brain samples of the middle temporal gyrus and lateral geniculate nucleus from the Allen Brain Atlas122, human brain samples using DroNc-seq123, two datasets of human prefrontal cortex brain samples across developmental stages which show per cell type average across different ages, and per cell type per age average expression124, two datasets of human brain samples with and without fetal tissue125, human brain samples from the temporal cortex126, and human samples from the ventral midbrain from 6–11 week old embryos127. A 3-step workflow is implemented in FUMA to determine association between cell-type specific expression and CC morphometry-gene association supported by multiple independent datasets, which has been extensively described45. All tests were corrected using the Bonferroni approach.
Partitioned heritability of meta-analysis results by cell and tissue type with LDSC
Partitioned heritability analysis was completed to estimate the amount of heritability explained by annotated regions of the genome41,44. We tested for enrichment of CC h2 of variants located in multiple tissues and cell types using the LDSC-SEG approach, with all analyses being corrected for the FDR44. Annotations indicating specific gene expression in multiple tissues/cell types from the Genotype-Tissue Expression (GTEx) project and Franke lab were downloaded from https://alkesgroup.broadinstitute.org/LDSCORE/LDSC_SEG_ldscores/. We also downloaded 489 tissue-specific chromatin-based annotations from narrow peaks for six epigenetic marks from the Roadmap Epigenomics and ENCODE projects128,129. These annotations were downloaded from the URL mentioned above. This would allow us to either verify or identify new findings from the gene expression analysis from an independent source using a different type of data. Finding new patterns of chromatin enrichment can help us to understand how genes are regulated. For example, if we find that a particular epigenetic mark is enriched in a region of the genome that is associated with a specific gene in a specific tissue type, this could suggest that the gene is regulated by that epigenetic mark in that specific tissue type. Gene expression data from the Immunological Genome (ImmGen) project130, which contains microarray data on 292 immune cell types from mice, was used to test immune cell-type-specific enrichments. Data was downloaded from the aforementioned link.
LAVA - TWAS
We used the LAVA-TWAS framework to investigate the relationship between CC traits and gene expression in brain tissues, fibroblasts, lymphocytes, and whole blood from the GTEx consortium (v8)83 in all protein coding genes, as it has ability to model the uncertainty of eQTL effects compared to other commonly used TWAS approaches, which have been shown to be prone to high type-I errors (false positives), and provides a directly interpretable effect size in the rG estimate46. Analyses were performed on all protein coding genes (N = 18,380) between all CC phenotypes and eQTLs/sQTLs for each tissue. Genotype data from the European sample of the 1000 Genomes (phase 3) project131 was used to estimate SNP LD for LAVA. For each eQTL/sQTL that had a significant genetic signal for both the CC phenotype and cortical phenotype (univariate p-values less than 1 x 10−4), the local bivariate genetic correlation between the two was estimated and tested. All LAVA-TWAS results were corrected using the Bonferroni approach. Following TWAS, trait specific enrichment analysis via a Fisher’s exact test of the top 1% of genes, to evaluate overrepresentation in 7,246 MSigDB v6.2132 gene sets and gain insight into biological pathways, was conducted. Gene sets were subset such that they must have consisted of at least one of the top 1% of genes, to avoid testing gene-sets with no significantly associated genes. All enrichment testing for eQTLs and sQTLs was performed with Bonferroni correction.
Global and local genetic correlations with cortical morphometry and mendelian randomization
The CC develops in such a manner that callosal projections are over-produced then refined during development. The majority of cortical projections are refined during postnatal stages and are under the influence of guidance cues6. As many genes are responsible for callosal axon guidance, we sought to investigate the genetic relationship between our derived CC traits and the genetic architecture of the human cerebral cortex6. We used LDSC to determine the global genetic correlation between area and thickness of the total and parcellated regions of the corpus callosum, and the GWAS summary statistics of each globally corrected region-of-interest of the cerebral cortex from the ENIGMA-3 GWAS133. We performed bi-directional Mendelian Randomization analyses to investigate if significant genetic correlations observed could be driven by genetic causal relationships between an exposure (e.g., area and thickness of different regions of the CC) and outcome (e.g., regional surface area & cortical thickness). Analyses were performed with summary statistics using GSMR47. All analyses were corrected using the Bonferroni approach. To capture potential local shared genetic effects across the genome, we ran LAVA28 for all protein coding genes (N = 18,380) between all CC phenotypes and surface area and cortical thickness of regions in the ENIGMA3 GWAS. Genotype data from the European sample of the 1000 Genomes (phase 3) project131 was used to estimate SNP LD for LAVA. For each gene that had a significant genetic signal for both the CC phenotype and cortical phenotype (univariate p-values less than 1 x 10−4), the local bivariate genetic correlation between the two was estimated and tested. All results were corrected using the Bonferroni approach.
Global and local genetic correlations with neuropsychiatric conditions and mendelian randomization
Abnormalities of the corpus callosum have also been heavily implicated in several neurological and neuropsychiatric conditions such as autism spectrum disorders (ASDs), ADHD, bipolar disorder, schizophrenia, visual impairments and epilepsy9,56,134–141. We used LDSC to determine the global genetic correlation between area and thickness of the total and parcellated regions of the corpus callosum, and 15 neuropsychiatric traits. Mendelian randomization analysis, and local genetic correlations were run as done for the brain cortical phenotypes.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (Grant Nos. R01 MH1340004 and R01 AG059874 [to NJ], National Science Foundation Graduate Research Fellowship Program (Grant No. 2020290241 [to RRB], the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), and UK Biobank (Resource Application No. 11559). SEM was supported by NHMRC grants APP1172917 and APP1158127.
Footnotes
Code Availability
The code and model used to extract the corpus callosum and its metrics is available at https://github.com/USC-LoBeS/smacc/.
Ethics Declarations
Competing Interests
No authors have any other conflicts of interest to disclose.
Data Availability
This work is a meta-analysis. Upon publication, the full meta-analytic summary statistics will be made available in ENIGMA-Vis142.
References
- 1.Fame R. M., MacDonald J. L. & Macklis J. D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 34, 41–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenlon L. R. & Richards L. J. Contralateral targeting of the corpus callosum in normal and pathological brain function. Trends Neurosci. 38, 264–272 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Paul L. K. Developmental malformation of the corpus callosum: a review of typical callosal development and examples of developmental disorders with callosal involvement. J. Neurodev. Disord. 3, 3–27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown W. S., Jeeves M. A., Dietrich R. & Burnison D. S. Bilateral field advantage and evoked potential interhemispheric transmission in commissurotomy and callosal agenesis. Neuropsychologia 37, 1165–1180 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Caminiti R. et al. Diameter, length, speed, and conduction delay of callosal axons in macaque monkeys and humans: comparing data from histology and magnetic resonance imaging diffusion tractography. Journal of Neuroscience 33, 14501–14511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De León Reyes N. S., Bragg-Gonzalo L. & Nieto M. Development and plasticity of the corpus callosum. Development 147, (2020). [DOI] [PubMed] [Google Scholar]
- 7.Piras F. et al. Corpus callosum morphology in major mental disorders: a magnetic resonance imaging study. Brain Commun 3, fcab100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeulen C. L., du Toit P. J., Venter G. & Human-Baron R. A morphological study of the shape of the corpus callosum in normal, schizophrenic and bipolar patients. J. Anat. 242, 153–163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unterberger I., Bauer R., Walser G. & Bauer G. Corpus callosum and epilepsies. Seizure 37, 55–60 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Zhao G. et al. A Comparative Multimodal Meta-analysis of Anisotropy and Volume Abnormalities in White Matter in People Suffering From Bipolar Disorder or Schizophrenia. Schizophr. Bull. 48, 69–79 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L. et al. Alterations in white matter microarchitecture in adolescents and young adults with major depressive disorder: A voxel-based meta-analysis of diffusion tensor imaging. Psychiatry Res Neuroimaging 323, 111482 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Valenti M. et al. Abnormal Structural and Functional Connectivity of the Corpus Callosum in Autism Spectrum Disorders: a Review. Review Journal of Autism and Developmental Disorders 7, 46–62 (2020). [Google Scholar]
- 13.Videtta G. et al. White matter modifications of corpus callosum in bipolar disorder: A DTI tractography review. J. Affect. Disord. 338, 220–227 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Scamvougeras A., Kigar D. L., Jones D., Weinberger D. R. & Witelson S. F. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci. Lett. 338, 91–94 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Woldehawariat G. et al. Corpus callosum size is highly heritable in humans, and may reflect distinct genetic influences on ventral and rostral regions. PLoS One 9, e99980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell M. L. et al. Distributed genetic effects of the corpus callosum subregions suggest links to neuropsychiatric disorders and related traits. Acta Neuropsychiatr. 1–8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S.-J. et al. The genetic architecture of the corpus callosum and its genetic overlap with common neuropsychiatric diseases. J. Affect. Disord. 335, 418–430 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Joshi S. H. et al. Statistical shape analysis of the corpus callosum in Schizophrenia. Neuroimage 64, 547–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luders E., Thompson P. M. & Toga A. W. The development of the corpus callosum in the healthy human brain. J. Neurosci. 30, 10985–10990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadewar S. P. et al. A Comprehensive Corpus Callosum Segmentation Tool for Detecting Callosal Abnormalities and Genetic Associations from Multi Contrast MRIs. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2023, 1–4 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkow N. D. et al. The conception of the ABCD study: From substance use to a broad NIH collaboration. Dev. Cogn. Neurosci. 32, 4–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witelson S. F. Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain 112 (Pt 3), 799–835 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Hofer S. & Frahm J. Topography of the human corpus callosum revisited—Comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage 32, 989–994 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Panizzon M. S. et al. Distinct Genetic Influences on Cortical Surface Area and Cortical Thickness. Cereb. Cortex 19, 2728–2735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler A. M. et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 53, 1135–1146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan B. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werme J., van der Sluis S., Posthuma D. & de Leeuw C. A. An integrated framework for local genetic correlation analysis. Nat. Genet. 54, 274–282 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemani G. Explodecomputer/random-Metal: Adding Random Effects Model. (Zenodo, 2022). doi: 10.5281/ZENODO.6974695. [DOI] [Google Scholar]
- 31.Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin P.-M. et al. Schwannomin-interacting protein-1 isoform IQCJ-SCHIP-1 is a late component of nodes of Ranvier and axon initial segments. J. Neurosci. 28, 6111–6117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann I., Martin G., Friedlein A., Langen H. & Keller W. Human Fip1 is a subunit of CPSF that binds to U rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626–626 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oyagi A. & Hara H. Essential roles of heparin-binding epidermal growth factor-like growth factor in the brain. CNS Neurosci. Ther. 18, 803–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song C., Qi Y., Zhang J., Guo C. & Yuan C. CDKN2B-AS1: An indispensable long non-coding RNA in multiple diseases. Curr. Pharm. Des. 26, 5335–5346 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Kaltschmidt B. & Kaltschmidt C. NF-κB in the nervous system. Cold Spring Harb. Perspect. Biol. 1, a001271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakajima H. & Koizumi K. Family with sequence similarity 107: A family of stress responsive small proteins with diverse functions in cancer and the nervous system (Review). Biomed Rep 2, 321–325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Lee S. H., Goddard M. E. & Visscher P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leeuw C. A., Mooij J. M., Heskes T. & Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finucane H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Consortium GTEx et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finucane H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe K., Umićević Mirkov M., de Leeuw C. A., van den Heuvel M. P. & Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Leeuw C., Werme J., Savage J. E., Peyrot W. J. & Posthuma D. On the interpretation of transcriptome-wide association studies. PLoS Genet. 19, e1010921 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papandréou M.-J. et al. CK2-regulated schwannomin-interacting protein IQCJ-SCHIP-1 association with AnkG contributes to the maintenance of the axon initial segment. J. Neurochem. 134, 527–537 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Liu J. et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther 7, 3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munji R. N., Choe Y., Li G., Siegenthaler J. A. & Pleasure S. J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 31, 1676–1687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chenn A. & Walsh C. A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Caric D. et al. EGFRs mediate chemotactic migration in the developing telencephalon. Development 128, 4203–4216 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Zentner G. E. & Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 20, 259–266 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Huang H. & Tindall D. J. Dynamic FoxO transcription factors. J. Cell Sci. 120, 2479–2487 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Aboitiz F., Scheibel A. B., Fisher R. S. & Zaidel E. Fiber composition of the human corpus callosum. Brain Res. 598, 143–153 (1992). [DOI] [PubMed] [Google Scholar]
- 56.Aboitiz F. & Montiel J. One hundred million years of interhemispheric communication: the history of the corpus callosum. Braz. J. Med. Biol. Res. 36, 409–420 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Faust T. E., Gunner G. & Schafer D. P. Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 22, 657–673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zengeler K. E. & Lukens J. R. Innate immunity at the crossroads of healthy brain maturation and neurodevelopmental disorders. Nat. Rev. Immunol. 21, 454–468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renault V. M. et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu T., Zhang L., Joo D. & Sun S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 17023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Veen S. et al. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 578, 419–424 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Smith K. M. et al. Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat. Neurosci. 9, 787–797 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Pânzaru M.-C. et al. Genetic heterogeneity in corpus callosum agenesis. Front. Genet. 13, 958570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paul L. K. et al. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat. Rev. Neurosci. 8, 287–299 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Castets F. et al. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 134, 1051–1062 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartoli M., Monneron A. & Ladant D. Interaction of calmodulin with striatin, a WD-repeat protein present in neuronal dendritic spines. J. Biol. Chem. 273, 22248–22253 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Yatsenko S. A. et al. Identification of critical regions for clinical features of distal 10q deletion syndrome. Clin. Genet. 76, 54–62 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Vera-Carbonell A. et al. Clinical comparison of 10q26 overlapping deletions: delineating the critical region for urogenital anomalies. Am. J. Med. Genet. A 167A, 786–790 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Lin S. et al. Chromosome 10q26 deletion syndrome: Two new cases and a review of the literature. Mol. Med. Rep. 14, 5134–5140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson P. M. et al. ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry 10, 100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palumbo P. et al. Clinical and molecular characterization of a de novo 19p13.3 microdeletion. Mol. Cytogenet. 9, 40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swan L. & Coman D. Ocular Manifestations of a Novel Proximal 19p13.3 Microdeletion. Case Rep. Genet. 2018, 2492437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Innocenti G. M. & Price D. J. Exuberance in the development of cortical networks. Nat. Rev. Neurosci. 6, 955–965 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Gavrish M. et al. Molecular mechanisms of corpus callosum development: a four-step journey. Front. Neuroanat. 17, 1276325 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwards T. J., Sherr E. H., Barkovich A. J. & Richards L. J. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain 137, 1579–1613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taniguchi K. et al. Genetic and Molecular Analyses indicate independent effects of TGIFs on Nodal and Gli3 in neural tube patterning. Eur. J. Hum. Genet. 25, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okada A. et al. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369–373 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Fullerton J. M., Donald J. A., Mitchell P. B. & Schofield P. R. Two-dimensional genome scan identifies multiple genetic interactions in bipolar affective disorder. Biol. Psychiatry 67, 478–486 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Li Y. et al. Genome-wide methylome analyses reveal novel epigenetic regulation patterns in schizophrenia and bipolar disorder. Biomed Res. Int. 2015, 201587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park N. et al. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol. Psychiatry 9, 1091–1099 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Sarrazin S. et al. Corpus callosum area in patients with bipolar disorder with and without psychotic features: an international multicentre study. J. Psychiatry Neurosci. 40, 352–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang F. et al. Abnormal corpus callosum integrity in bipolar disorder: a diffusion tensor imaging study. Biol. Psychiatry 64, 730–733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uhlén M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Matos B., Publicover S. J., Castro L. F. C., Esteves P. J. & Fardilha M. Brain and testis: more alike than previously thought? Open Biol. 11, 200322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller K. L. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alfaro-Almagro F. et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hagler D. J. et al. Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage 202, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazziotta J. C., Toga A. W., Evans A., Fox P. & Lancaster J. A probabilistic atlas of the human brain: Theory and rationale for its development. Neuroimage 2, 89–101 (1995). [DOI] [PubMed] [Google Scholar]
- 90.Mazziotta J. et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1293–1322 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mazziotta J. et al. A four-dimensional probabilistic atlas of the human brain. J. Am. Med. Inform. Assoc. 8, 401–430 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jenkinson M., Bannister P., Brady M. & Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Jernigan T. L. et al. The Pediatric Imaging, Neurocognition, and Genetics (PING) Data Repository. Neuroimage 124, 1149–1154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Van Essen D. C. et al. The WU-Minn Human Connectome Project: An overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petersen R. C. et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 74, 201–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shorten C. & Khoshgoftaar T. M. A survey on image data augmentation for deep learning. J. Big Data 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tournier J.-D. et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 202, 116137 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Liu M. et al. Style Transfer Generative Adversarial Networks to Harmonize Multi-Site MRI to a Single Reference Image to Avoid Over-Correction. bioRxiv 2022.09.12.506445 (2022) doi: 10.1101/2022.09.12.506445. [DOI] [Google Scholar]
- 99.Ronneberger O., Fischer P., Brox T. & Navab N. Medical image computing and computer-assisted intervention–MICCAI 2015. Cham: Springer. [Google Scholar]
- 100.Kingma D. P. & Ba J. Adam: A Method for Stochastic Optimization. arXiv [cs.LG] (2014). [Google Scholar]
- 101.Zhu A. H. et al. Robust automatic corpus callosum analysis toolkit: mapping callosal development across heterogeneous multisite data. in 14th International Symposium on Medical Information Processing and Analysis vol. 10975 177–184 (SPIE, 2018). [Google Scholar]
- 102.Fischl B. FreeSurfer. Neuroimage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gorgolewski K. J. et al. A high resolution 7-Tesla resting-state fMRI test-retest dataset with cognitive and physiological measures. Sci Data 2, 140054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zuo X.-N. et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data 1, 140049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baurley J. W., Edlund C. K., Pardamean C. I., Conti D. V. & Bergen A. W. Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17, 145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uban K. A. et al. Biospecimens and the ABCD study: Rationale, methods of collection, measurement and early data. Dev. Cogn. Neurosci. 32, 97–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stein J. L. et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 44, 552–561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loh P.-R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mbatchou J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 110.Bethlehem R. A. I. et al. Brain charts for the human lifespan. Nature 604, 525–533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Skol A. D., Scott L. J., Abecasis G. R. & Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006). [DOI] [PubMed] [Google Scholar]
- 112.Demontis D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. (2023) doi: 10.1038/s41588-022-01285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nagel M. et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 50, 920–927 (2018). [DOI] [PubMed] [Google Scholar]
- 114.Kim J. J. et al. Multi-ancestry genome-wide association meta-analysis of Parkinson’s disease. Nat. Genet. 56, 27–36 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rietveld C. A. et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li J. & Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Nyholt D. R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74, 765–769 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.rkwalters & Palmer D. Nealelab/UKBB_ldsc: v2.0.0 (Round 2 GWAS Update). (2022). doi: 10.5281/zenodo.7186871. [DOI] [Google Scholar]
- 119.Watanabe K., Taskesen E., Van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liberzon A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hodge R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Habib N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhong S. et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018). [DOI] [PubMed] [Google Scholar]
- 125.Darmanis S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U. S. A. 112, 7285–7290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hochgerner H. et al. STRT-seq-2i: dual-index 5’ single cell and nucleus RNA-seq on an addressable microwell array. Sci. Rep. 7, 16327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.La Manno G. et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heng T. S. P., Painter M. W. & Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 131.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liberzon A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grasby K. L. et al. The genetic architecture of the human cerebral cortex. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luders E. et al. Associations between corpus callosum size and ADHD symptoms in older adults: The PATH through life study. Psychiatry Res Neuroimaging 256, 8–14 (2016). [DOI] [PubMed] [Google Scholar]
- 135.Lau Y. C. et al. Autism traits in individuals with agenesis of the corpus callosum. J. Autism Dev. Disord. 43, 1106–1118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tao B. et al. Characteristics of the corpus callosum in chronic schizophrenia treated with clozapine or risperidone and those never-treated. BMC Psychiatry 21, 538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saar-Ashkenazy R. et al. White-matter correlates of anxiety: The contribution of the corpus-callosum to the study of anxiety and stress-related disorders. Int. J. Methods Psychiatr. Res. e1955 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Westerhausen R. et al. The corpus callosum as anatomical marker of intelligence? A critical examination in a large-scale developmental study. Brain Struct. Funct. 223, 285–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Di Paola M. et al. The structure of the corpus callosum in obsessive compulsive disorder. Eur. Psychiatry 28, 499–506 (2013). [DOI] [PubMed] [Google Scholar]
- 140.Kitayama N. et al. Morphologic alterations in the corpus callosum in abuse-related posttraumatic stress disorder: a preliminary study. J. Nerv. Ment. Dis. 195, 1027–1029 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saar-Ashkenazy R. et al. Breakdown of Inter-Hemispheric Connectivity Is Associated with Posttraumatic Symptomatology and Memory Impairment. PLoS One 11, e0144766 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shatokhina N. et al. ENIGMA-Vis: A Web Portal to Browse, Navigate & Visualize Brain Genome-Wide Association Studies (GWAS). Biol. Psychiatry 89, S136 (2021). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This work is a meta-analysis. Upon publication, the full meta-analytic summary statistics will be made available in ENIGMA-Vis142.