Abstract
Increased IgG and oligoclonal bands are found in cerebrospinal fluid of humans with chronic infectious CNS disease. Studies have shown that these oligoclonal bands are antibodies directed against the agent that causes disease. Laser-capture microdissection was used to isolate individual CD38+ plasma cells from the brain of a patient with subacute sclerosing panencephalitis, and single-cell RT-PCR was used to analyze individual IgG heavy and light chains expressed by each cell. Based on overrepresented IgG sequences, we constructed functional recombinant antibodies (recombinant IgGs) and determined their specificities. Five of eight recombinant IgGs recognized measles virus, the cause of subacute sclerosing panencephalitis. These results demonstrate that overrepresented IgG sequences in postmortem brains can be used to produce functional recombinant antibodies that recognize their target antigens. This strategy can be used to identify disease-relevant antigens in CNS inflammatory diseases of unknown etiology.
Keywords: oligoclonal bands, recombinant antibodies
Increased IgG and oligoclonal bands (OGBs) are found in the cerebrospinal fluid (CSF) of humans with chronic infectious CNS diseases such as neurosyphilis, cryptococcal and tuberculous meningitis, Lyme disease, some viral meningitides, varicella zoster virus vasculopathy, and subacute sclerosing panencephalitis (SSPE), a chronic encephalitis caused by measles virus (MV). Studies in which the specificity of CSF OGBs was analyzed showed that the antibodies were directed against the agent that causes disease (1-8). For example, the OGBs in SSPE CSF and brain are antibodies directed against MV (1). These studies have led to the hypothesis that the oligoclonal IgG in the brain and CSF of patients with a chronic inflammatory CNS disease of unknown etiology, such as multiple sclerosis (MS), sarcoidosis, and Behcet's disease, is antibody directed against the agent that causes disease. Although OGBs are found in 88-100% of CSF from MS patients (9), the corresponding antigens remain unknown (10, 11). Such unanswered questions point to the need for improved techniques to identify disease-relevant antibodies and their cognate antigens and suggest the promise of such techniques in revealing the causes of inflammatory diseases with unknown etiologies.
Here, we used laser-capture microdissection (LCM) to isolate individual CD38+ plasma cells from the brain of a patient with SSPE. Single-cell RT-PCR was used to amplify individual IgG heavy (H) and light (L) chain sequences expressed by each cell. Based on overrepresented Ig sequences, we constructed functional recombinant antibodies and identified their target antigens.
Materials and Methods
SSPE Brain. Brain removed from a 14-year-old male SSPE patient 5 h after death was flash-frozen and stored at -70°C. Sections (7 μm) of frozen SSPE brain were prepared on nonplus glass slides (Fisher Scientific) at -30°C, fixed in acetone for 5 min at -20°C, and immunostained at 0°C for CD38+ cells. Briefly, sections were treated with 0.1% H2O2 for 30 sec, incubated for 2 min with PBS containing 10% goat serum and then with a 1:50 dilution of mouse anti-human CD38 antibody (DakoCytomation, DAKO) for 10 min, rinsed in PBS, and incubated for 5 min with a 1:100 dilution of horseradish peroxidase-conjugated horse anti-mouse antibody (Vector Laboratories) and 5% goat serum in PBS. After rinsing in PBS, sections were incubated for 5 min with DAB substrate (Vector Laboratories), counterstained with hematoxylin, dehydrated in nuclease-free graded alcohols to 100% ethanol, and cleared with two rinses of H20-free xylene. To preserve RNA, all aqueous solutions contained 200 units/ml RNase inhibitor (Fisher Scientific), and the total time of tissue exposure to aqueous solutions was <25 min at 0°C. Staining was visualized by light microscopy.
LCM and RT-PCR. LCM was performed on a PixCell IIe microscope (Arcturus Engineering, Mountain View, CA) with CapSure HS caps by using a pulse power of 70 mW, a 7.5-μm laser spot diameter, pulse duration of 5 ms, and target voltage of 170 mV. Individual CD38+ cells were visualized and lifted from the section onto separate caps, in sets of 12 cells per experiment. Extraction reservoirs were placed on each cap, and the assembly was stored in a tray at -20°C. After addition of 21 μl of 1× RT buffer containing 1% Nonidet P-40 and incubation at 42°C for 20 min, reverse transcription was performed by using Superscript III (Invitrogen) as described (12) but within the LCM cap reservoirs. Collection tubes were fitted onto the reservoirs, and the inverted assembly was microcentrifuged at 3,000 × g to collect reaction products. Nested PCRs were performed on 5-μl aliquots of the cDNA by using conserved 3′ IgG primers in the constant regions and pools of conserved leader and framework 1 primers from heavy chain (VH) or light chain variable region families, as described (13). PCR products were resolved on a 2% agarose gel, and appropriately sized products were excised, purified by using the MinElute Gel Extraction kit (Qiagen, Valencia, CA), and sequenced at the University of Colorado Health Sciences Cancer Center DNA Analysis and Sequencing Core. All sequences were analyzed with dnasis max software (Miraibio, Alameda, CA) and aligned to an online database VBASE from the Cambridge Center for Protein Engineering (www.mrc-cpe.cam.ac.uk). This service was used to identify the most homologous variable region germ-line segments and to determine the extent of sequence homology for all H and L chain sequences. For consistency, homologies to germ-line segments of donor DP (14) were used when applicable; otherwise, the gene locus was used to identify the most homologous germ-line segment.
Production of Recombinant IgG (rIgG). H chain variable regions and full-length L chains were amplified from individual clones by using Expand High Fidelity polymerase (Roche Applied Science, Indianapolis) to incorporate restriction sites at the termini and to fuse the products to the IgG H chain or Ig L chain leader sequences, respectively. The H chain PCR product was directionally subcloned behind the CMV promoter in the modified pIgG-flag vector to express the entire human H chain, and the L chain PCR product was directionally subcloned behind the CMV promoter in the pCEP4 expression vector (G.P.O., unpublished work). After sequence verification, 7 μg of each DNA from H/L chain pairs was cotransfected into HEK293 cells (80% confluent) by using Lipofectamine 2000 (Invitrogen) and grown for 5-6 days in DMEM containing 10% dialyzed FCS. Culture supernatants were analyzed by antigen-capture ELISA to quantitate the concentration of rIgG. Plates (96-well) were coated overnight with goat anti-human IgG antibody (10 μg/ml), blocked, and incubated with dilutions of the culture supernatants from the rIgG transfections for 2 h at room temperature. After washing with PBS-0.05% Tween 20 five times for 5 min each time, bound rIgG was detected by incubation with alkaline phosphatase-conjugated goat anti-human IgG antibody (Vector Laboratories) and p-nitrophenyl phosphate for 30 min. rIgG was quantitated by monitoring in a Bio-Rad plate reader at 405 nm compared with standard amounts of purified human IgG (20-300 ng/ml). To ensure correct size and disulfide bonding, rIgG was also immunoblotted with goat anti-human IgG antibody on nonreducing SDS/PAGE gels.
Functional Assays. Each culture supernatant was used at 5-8 μg of IgG/ml to immunostain acetone-fixed coverslips of MV-infected and uninfected Vero cells as described (15). Immunostaining was also performed on coverslips of Vero cells that were transiently transfected for 48 h with 1 μg of expression constructs encoding each of five different MV proteins (phosphoprotein, hemagglutinin, nucleocapsid, fusion, or DNA polymerase), as described (16). Immunoprecipitation with 2 μg and immunoblotting with 5 μg of each rIgG per ml was performed as described (15).
Results
From frozen sections of a single SSPE brain, 110 immunostained CD38+ plasma cells were individually laser dissected, with an additional 5-10 CD38-negative cells from the surrounding neuropil, onto separate LCM caps (Fig. 1). Cells on caps were lysed with 1% Nonidet P-40, and reverse transcription was performed within the overlying collection reservoir (see Materials and Methods) by using random primers and an antisense IgG H chain constant region primer. Reaction products were centrifuged from the reservoirs, and aliquots of the product were amplified with H or L chain IgG variable region primers. Every dissected CD38+ cell was assayed for expression of variable region rearranged H chain sequences and both κ and λ L chain sequences. Approximately two-thirds of the PCRs displayed amplified PCR products that corresponded to the expected size of H and L chain variable region sequences (Fig. 2).
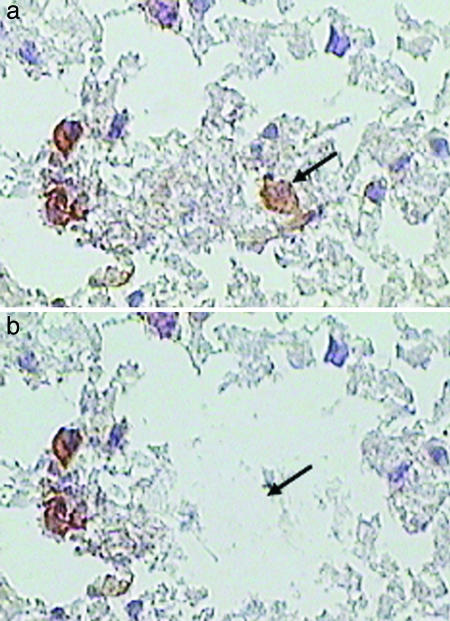
Fig. 1.
LCM capture of single CD38+ plasma cells. Arrows show an individual plasma cell visualized by immunostaining SSPE brain sections for CD38+ cells (brown color) (a), which were laser-microdissected from the neuropil (b).
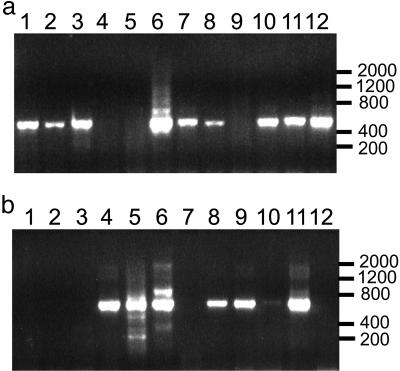
Fig. 2.
PCR amplification of H or L chain sequences from individual CD38+ cells. A representative set of 12 caps, each containing a microdissected CD38+ cell, was subjected to reverse transcription and nested PCR as described in Materials and Methods. The ≈600-bp H chain product (a) and ≈700-bp κ or λ L chain products (b) were visualized from most caps after electrophoresis in agarose gels. Size markers are indicated on the right.
Each PCR product was gel-purified and sequenced with conserved antisense primers from the constant regions of H or L chains. Table 1 lists the features of the H and L chain sequences from 65 CD38+ cells that amplified identifiable H chains, grouped according to their characteristic H chain complementarity-determining region 3 (CDR3) amino acid sequence. The VH and light chain variable region segment of each cell was also aligned to the closest germ-line segment in the VBASE database (Table 1). Among the original 110 CD38+ cells, several PCR products contained mixed VH sequences, and these cells were not examined further.
Table 1. Features of V region sequences amplified from SSPE brain CD38+ population.
| Group | CD38+ Vh CDR3 | Abundance | Germ line | Family | Percent ID | Vk CDR3 | Vλ CDR3 | Germ line | Family | Percent ID |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ALKKGEGGLRFLELYYFD | 10 | DP47 | Vh3 | 92.1 | QTWGSGMGV | Loc4b | Vλ4a | 93.7 | |
| ALKKGEGGLRFLELYYFD | 3 | DP47 | Vh3 | 92.1 | ||||||
| ALKKGEGGLRFLELYYLT | 1 | DP47 | Vh3 | 92.1 | QQNYSSPQT | DPK24 | Vκ4 | (t) | ||
| 2 | LPAAGPRSFFETYNWGMD | 8 | DP79 | Vh4 | 93.6 | AAWDDSLNAWV | DPL2 | Vλ1 | 97 | |
| LPADGPRSFFETYNNGMD | 1 | DP79 | Vh4 | 93.6 | AAWDDSLNAWV | DPL2 | Vλ1 | 97 | ||
| LPAAGPRSFFETYNWGMD | 1 | DP79 | Vh4 | 93.6 | ||||||
| 3a | IRAGAFD | 2 | DP31 | Vh3 | 95.2 | MQALQTFTF | DPK15 | Vκ2 | 99.7 | |
| 3b | IRAGAFD | 4 | DP31 | Vh3 | 95.2 | MQATQSWTF | DPK16 | Vκ2 | 98.2 | |
| IRAGAFD | 1 | DP31 | Vh3 | 95.2 | ||||||
| IRAGAFD | 1 | DP31 | Vh3 | 95.2 | (m) | |||||
| 4 | DFTSDSRGPLGWFD | 4 | DP79 | Vh4 | 93.9 | YSTDSSGDHRV | Loc3p | Vλ3 | 98.1 | |
| DFTSDSRGPLGWFD | 1 | DP79 | Vh4 | 93.9 | ||||||
| 5 | GGLAARARLVLARMD | 3 | DP63 | Vh4 | 93.8 | QQSYNTPITF | DPK9 | Vκ1 | 95.1 | |
| GGLAARARLVLARMD | 1 | DP63 | Vh4 | 93.8 | ||||||
| 6 | VRATVLTGTSMD | 2 | DP58 | Vh3 | 91.8 | GADHGSGSNFVWV | DPL22 | Vλ9 | 97.6 | |
| VRATVLTGTSMD | 1 | DP58 | Vh3 | 91.8 | ||||||
| 7 | DTGGSGSNYYHYGMD | 2 | DP10 | Vh1 | 93.2 | |||||
| DTGGSGSNYYHYGMD | 1 | DP10 | Vh1 | 93.2 | QQYNAWPPALT | DPK21 | Vκ3 | (t) | ||
| 8 | DRGGESDYDVGRGYSDHYGMD | 2 | DP71 | Vh4 | 86.9 | QQCGFSPKT | DPK22 | Vκ3 | 92.5 | |
| 9 | DQERGTILTYSDMD | 2 | DP47 | Vh3 | 95.9 | LQHNSYPHFRRR* | DPK3 | Vκ1 | 95.5 | |
| 10 | DQVPVNNWFD | 2 | DP14 | Vh1 | 95.2 | (m) | ||||
| 11 | SLTMIRGVMAFFD | 2 | DP25 | Vh1 | 87.6 | QQTYSSPSTF | DPK9 | Vκ1 | 90.9 | |
| DQVIYTGWSD | 1 | DP47 | Vh3 | 91.2 | CLYAGSTTWV | DPL10 | Vλ2 | 96.3 | ||
| GYYDSTGYKSAND | 1 | DP14 | Vh1 | 94.0 | QQTYSSPSTF | DPK9 | Vκ1 | 90.9 | ||
| LKSRIARGSYYQYFMD | 1 | DP27 | Vh2 | 93.1 | ||||||
| SADTSTAYYGLD | 1 | DP47 | Vh3 | 96.6 | LQDYNYPLTF | DPK3 | Vκ1 | 99.2 | ||
| STGTDYYSYYMD | 1 | DP73 | Vh5 | 86.6 | YSTDTSGNFRV | Loc3p | Vλ3 | 99.2 | ||
| EGQLALDQYYYYYMD | 1 | DP50 | Vh3 | 96.3 | NSYTSISTVV | DPL11 | Vλ2 | 93.6 | ||
| DRTGYTSFLFD | 1 | DP31 | Vh3 | 90.0 | SSYAGRNKGYV | DPL12 | Vλ2 | 96 | ||
| DPEEQWLADYFD | 1 | DP47 | Vh3 | 97.6 | MQATQSWTF | GTWDSSLSARV | DPL5 | Vλ1 | 98.9 | |
| VEVGPNEDFYMD | 1 | DP88 | Vh1 | 90.1 | QQSYSFPWTF | DPK9 | Vκ1 | 89.4 | ||
| EVAGGADIEVVPAAIGVDYHYGMD | 1 | DP79 | Vh4 | 97.3 | QSADSSGSYKV | (t) |
Each line identifies the CDR3 amino acid sequence and prevalence of each distinct H chain clone, the germ-line family, most homologous germ-line segment, percent identity to the closest germ-line segment, and the associated L chain amplification for that clone. *, an in-frame stop codon; (m), a mixed sequence that could not be further analyzed; (t), a truncated sequence.
Overall, 11 distinct groups of overrepresented VH-region sequences were identified from 55 of the 65 CD38+ cells analyzed (83%). Sequences in the remaining 10 CD38 + cells were encountered only once. Variable region sequences comprising the largest plasma cell clone were amplified from 14 individual CD38+ cells (22% of the entire repertoire), and the second-largest clone contained 10 identical H chain sequences from CD38+ cells (15% of the repertoire). All of these H chain clones were functional rearrangements of V(D)J segments, and virtually all demonstrated significant mutation from their corresponding germ-line VH segment (86-98% identity). Most of the H chain clones also expressed a distinct functional κ or λ L chain (Table 1), usually in the absence of a visible PCR product for the complementary λ or κ PCR in that cell. The L chain variable regions were also categorized by their characteristic CDR3 segments and were mutated from the most homologous germ-line light chain variable region segment but generally to a lesser extent than seen for the matching H chains. L chains could not be amplified from several of the cells or from any of the CD38+ cells comprising Group 10. In one plasma cell of Group 1, repeated attempts failed to amplify the predicted λ L chain sequence expressed in 10 of the other Group 1 plasma cells. Instead, a rearranged κ L chain sequence was detected. In several other cells, multiple functionally rearranged L chain sequences were encountered. Of the 65 cells in this analysis, ≈15% had PCR products containing more than one L chain sequence. For example, the single CD38+ cell identified by the H chain CDR3-DPEEQWLADYFD expressed both a κ and λ L chain sequence. Several other cells either expressed in-frame stop codons resulting in a nonfunctional L chain (clone 9), did not contain a PCR-detectable L chain (clone 10), or contained a single unverified L chain sequence among all of the cells in clone 7. In most dissected CD38+ cells where more than one L chain variable region was amplified, a dominant L chain was identifiable, and the authentic L chain was identified by the presence of the same H/L pair in other cells. Interestingly, four of the eight cells with an identical H chain in clone 3 (IRA-GAFD) expressed the same κ L chain derived from the DPK16 germ-line segment, whereas two other cells expressed a distinct κ L chain derived from the DPK15 germ line.
rIgGs were synthesized from representative clones of 8 of the 11 overrepresented clones (clones 1, 2, 3A and 3B, 4, 5, 8, and 11). To construct rIgGs, the H chain variable region sequence of each clone was fused to a common human IgG leader sequence by using PCR amplification, and the product was directionally cloned into pIgG1-Flag, derived from the mammalian expression vector pCEP-4 containing the cloned IgG1 constant region domains with a Flag peptide at the C terminus (G.P.O., unpublished work). Full-length L chains were similarly subcloned into the pCEP4 expression vector. H and L chains for each of the clones were transiently cotransfected into HEK293-EBNA cells, and the culture supernatants collected after 6 days were used for structural and functional analyses. Each of the recombinant antibodies was intact IgG, as verified by immunoblotting on nonreducing SDS/PAGE gels (data not shown). IgG concentrations were quantitated by antigen-capture ELISA, and those at low concentrations were concentrated to levels of 5-15 μg/ml. All IgGs were examined in functional assays.
Five of the eight rIgGs produced from the SSPE brain stained MV-infected but not uninfected Vero cells (Fig. 3). Although the same H chain in clone 3 was associated in two distinct populations with different κ L chains (clones 3A and 3B), only the rIgG from clone 3B stained MV-infected cells at the concentrations tested. Each of the five rIgGs also stained Vero cells transiently transfected with the MV nucleocapsid gene but not the MV hemagglutinin or fusion protein genes (Fig. 4); none of the rIgGs stained Vero cells transfected with the MV phosphoprotein or DNA polymerase genes (data not shown). rIgG from clone 3B immunostained cells in frozen sections of SSPE brain but not normal human brain (Fig. 5). The rIgG could be distinguished from endogenous human IgG by using the Flag epitope. Clones 1 and 2 also stained SSPE brain (data not shown).
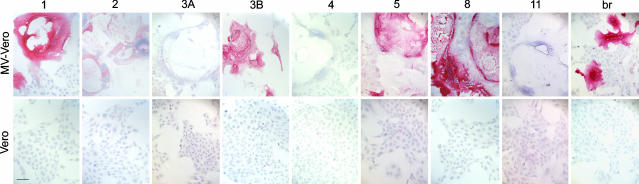
Fig. 3.
Immunostaining of MV infected cells. Five of the eight overrepresented IgG sequences, when expressed as rIgGs, stained MV-infected but not uninfected Vero cells. IgG extracted from SSPE brain (br) also stained MV-infected but not uninfected cells. Positive staining (red) was visualized by New Fuschin and counterstaining with hematoxylin. (Bar, 100 μm.)
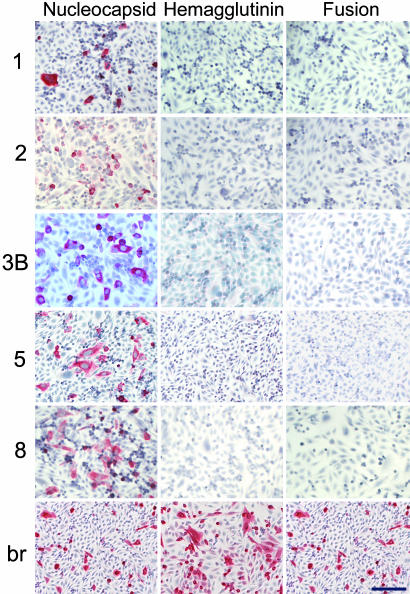
Fig. 4.
Immunostaining of individual MV proteins by rIgGs. rIgG (5-8 μg/ml) stained Vero cells transiently transfected with the MV nucleocapsid gene but not cells transfected with the MV fusion protein or hemagglutinin genes. IgG (2 μg/ml) extracted from an SSPE brain (br) stained cells transiently transfected with all three MV genes. Positive staining (red) was visualized by New Fuschin and counterstaining with hematoxylin. (Bar, 100 μm.)
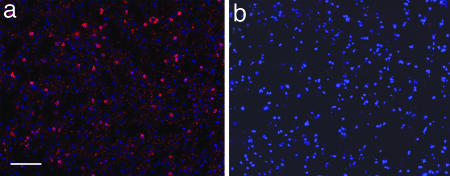
Fig. 5.
Clone 3B rIgG stains SSPE brain. rIgG (5 μg/ml prepared from clone 3B) stained sections of SSPE (a) but not normal (b) human brain. Positive staining (red) was visualized by Texas red immunofluorescence, and nuclei were visualized after DAPI staining (blue). (Bar, 100 μm.)
The five MV-staining rIgGs also recognized a single 60-kDa species in immunoblots of MV-infected Vero cells and SSPE brain, corresponding to the size of the nucleocapsid protein. Polyclonal IgG extracted from an SSPE brain recognized this species as well as other MV proteins in infected cell lysates. None of the rIgGs or the extracted polyclonal IgG reacted with uninfected cell lysates or normal human brain (Fig. 6a and data not shown). Both rIgG from clones 3A and 3B, differing only in their distinct L chains, recognized the 60-kDa species in lysates of SSPE brain to different degrees (Fig. 6b). rIgGs from clones 1, 2, and 3b also immunoprecipitated a single 60-kDa protein from lysates of MV-infected Vero cells but not from uninfected cells (Fig. 6c). Although all were used at the same concentration and at the same conditions, the rIgGs precipitated the 60-kDa species with varying intensity, with rIgG 2 reacting only weakly.
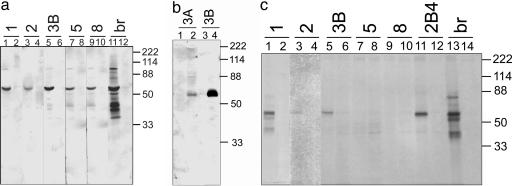
Fig. 6.
Immunoblotting and immunoprecipitation of MV by rIgG. (a) Each rIgG (3 μg/ml) was applied to lysates of MV-infected (lanes 1, 3, 5, 7, 9, and 11) or uninfected (lanes 2, 4, 6, 8, 10, and 12) Vero cells. In MV-infected lysates, each of the rIgGs that stained MV-infected cells also recognized a 60-kDa band corresponding to the size of the MV nucleocapsid protein; IgG (3 μg/ml) extracted from SSPE brain (br) recognized multiple MV proteins in MV-infected cells (lane 11) but not in uninfected cells (lane 12). (b) Two rIgGs from the clone 3 H chain, expressing two distinct κ L chains, both recognized a 60-kDa band in SSPE brain (lanes 2 and 4) but not in normal human brain (lanes 1 and 3); at the same concentration, the 3B rIgG reacted more intensely than the 3A rIgG. (c) Two micrograms of rIgGs 1, 2, 3B, 5, and 8, or a monoclonal antibody (2B4) directed against the MV nucleocapsid protein (15) or IgG extracted from SSPE brain (br) were used to immunoprecipitate pairs of 35S-labeled lysates of MV-infected (lanes 1, 3, 5, 7, 9, 11, and 13) or uninfected (lanes 2, 4, 6, 8, 10, 12, and 14) Vero cells. rIgG from clones 1, 2, and 3B immunoprecipitated a major species of 60 kDa from MV-infected lysates (lanes 1, 3, and 5), whereas the SSPE brain-extracted IgG precipitated additional bands from the MV-infected lysates (lane 13). Antibody 2B4 also immunoprecipitated the 60-kDa species from MV-infected (lane 11) but not uninfected (lane 12) lysates. Molecular mass standards are indicated on the right.
Discussion
By using a laser-capture technique to isolate individual CD38+ plasma cells from SSPE brain and single-cell RT-PCR to analyze the IgG repertoire expressed by these cells, we identified clonal populations that contained features of somatic mutation and a targeted antibody response. The over-represented IgG sequences were expressed as functional recombinant antibodies (rIgGs), and most reacted with MV, the cause of SSPE. Our determination of the specific reactivity of rIgGs is consistent with and extends studies in other chronic infectious CNS disorders in which the oligoclonal IgG was shown to be antibody directed against the agent that causes disease (reviewed in ref. 10). The strategies and techniques developed herein have the potential to identify the causative antigen in chronic inflammatory CNS diseases of unknown etiology, such as multiple sclerosis, sarcoidosis, and Behcet's disease, particularly because multiple analyses of IgGs in MS brain plaques and CSF have revealed features of a targeted antibody response (17-22).
Compared with earlier analysis of RNA extracted from a large piece of SSPE brain (16), the LCM approach provides the advantage of fidelity of recombinant antibodies produced by accurate pairing of H and L chains produced by a single plasma cell. For example, when the clone 1 H chain in this study was examined earlier in combination with different L chains (16), the antibody stained MV-infected cells but did not identify a MV protein in immunoblots or cells transfected with vectors expressing MV proteins. However, when the clone 1 H chain was combined with its authentic L chain, the rIgG not only stained MV-infected cells but also recognized the MV nucleocapsid protein in immunoblots and transfected cells (Figs. 4 and 6).
We observed approximately equal numbers of CD20+ B cells and CD38+ plasma cells in the SSPE brain (data not shown) but restricted laser capture to CD38+ plasma cells, because these cells likely produce most of the IgG. In our repertoire of 65 isolated plasma cells, 11 H chain clones were expressed multiple times, most of which amplified with a single functional κ or λ L chain, and 10 H chain clones were encountered once. One of the expressed L chain clones contained an in-frame stop codon and was nonfunctional (clone 9). Several explanations are possible: (i) our primers may not have amplified a second functional L chain from these cells; (ii) capture of a nearby B cell with the CD38+ plasma cell might have revealed a nonfunctional L chain from the B cell indistinguishable from the authentic L chain; (iii) the nonfunctional L chain might reflect a failed rearrangement early in B cell development; or (iv) to escape deletion (23), a self-reactive B cell might undergo receptor editing by expressing a newly recombined L chain. Although we did not detect a functional L chain in this clone, additional evidence for receptor editing may be found in the Group 3 clone, where the same H chain in different cells was associated with expression of distinct κ L chains and might account for differing affinities of the 3A and 3B rIgGs for their cognate antigen (see below).
Five of the eight rIgGs constructed from overrepresented H/L chain clones recognized MV in multiple functional assays. The rIgGs stained MV-infected cells in tissue culture and in SSPE brain and also recognized denatured MV antigens in immunoblots of MV-infected cell lysates and SSPE brain. Immunoblotting with two rIgGs (3A and 3B) revealed the same 60-kDa MV protein, even though only the 3B construct stained MV-infected cells and SSPE brain at the concentrations tested. This finding suggests a differential affinity of rIgGs for the same MV antigen and supports the notion that L chain usage influences affinity more than the specificity of antigen binding (24).
It is interesting that all of the functional rIgGs in this study reacted to the MV nucleocapsid. No immediate explanation is apparent, particularly because the serum and CSF of patients with SSPE contain antibodies to many MV proteins (25, 26). Possibly, some of the rIgGs that did not stain MV-infected cells were low-affinity antibodies to other MV proteins and will require further study. The high percentage of virus-specific plasma cell clones identified thus far in this brain (60% of clones tested) compares favorably with a model for CNS demyelination produced by mouse hepatitis virus, in which ≈30% of plasma cells recruited to the CNS are virus-specific (27).
The demonstration that rIgG constructed from single plasma cells of postmortem SSPE brain react to the disease-causing agent points to the applicability of such techniques in identifying disease-relevant antigens in other inflammatory CNS diseases of unknown cause.
Acknowledgments
We thank Dr. Jeffrey Bennett, University of Colorado Health Sciences Center, for providing advice during this work; Marina Hoffman for editorial review; and Cathy Allen for preparing the manuscript. The SSPE brain was kindly provided by the National Neurological Research Specimen Bank, Veterans Affairs Medical Center (Los Angeles). This work was supported by National Institutes of Health Grants NS41549 (to M.P.B.) and NS32623 (to D.H.G., M.P.B., and G.P.O.).
Author contributions: M.P.B., G.P.O., and D.H.G. designed research; M.P.B. and K.M.K. performed research; M.P.B., G.P.O., A.M.R., P.R.R., and C.D.C. contributed new reagents/analytic tools; M.P.B., K.M.K., A.M.R., P.R.R., and D.H.G. analyzed data; and M.P.B., K.M.K., G.P.O., and D.H.G. wrote the paper.
Abbreviations: CDR3, complementarity-determining region 3; OGB, oligoclonal bands; LCM, laser-capture microdissection; MV, measles virus; rIgG, recombinant IgG; SSPE, subacute sclerosing panencephalitis; CSF, cerebrospinal fluid; L, light; H, heavy; VH, heavy chain variable.
References
- 1.Vandvik, B., Norrby, E., Nordal, H. J. & Degre, M. (1976) Scand. J. Immunol. 5, 979-992. [DOI] [PubMed] [Google Scholar]
- 2.Vandvik, B., Norrby, E., Steen-Johnson, J. & Sensvold, K. (1978) Eur. Neurol. 17, 13-22. [DOI] [PubMed] [Google Scholar]
- 3.Porter, K. G., Sinnamon, D. G. & Gillies, R. R. (1977) Lancet 1, 1262. [DOI] [PubMed] [Google Scholar]
- 4.Coyle, P. K. & Wolinsky, J. S. (1981) Ann. Neurol. 9, 557-562. [DOI] [PubMed] [Google Scholar]
- 5.Vartdal, F., Vandvik, B., Michaelsen, T. E., Loe, K. & Norrby, E. (1981) Ann. Neurol. 11, 35-40. [DOI] [PubMed] [Google Scholar]
- 6.ter Meulen, V., Stephenson, J. R. & Kreth, H. W. (1983) Comp. Virol. 18, 105-159. [Google Scholar]
- 7.Martin, R., Martens, U., Sticht-Groh, V., Dorries, R. & Kruger, H. (1988) J. Neurol. 235, 229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgoon, M. P., Hammack, B. N., Owens, G. P., Maybach, A. L., Eikelenboom, M. J. & Gilden, D. H. (2003) Ann. Neurol. 54, 459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattson, D. H., Roos, R. P. & Arnason, B. G. W. (1981) Ann. Neurol. 9, 34-41. [DOI] [PubMed] [Google Scholar]
- 10.Gilden, D. H., Devlin, M. E., Burgoon, M. P. & Owens, G. P. (1996) Multiple Sclerosis 2, 179-183. [DOI] [PubMed] [Google Scholar]
- 11.Burgoon, M. P., Gilden, D. H. & Owens, G. P. (2004) Front. Biosci. 9, 786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie, A. M., Gilden, D. H., Williamson, R. A., Burgoon, M. P., Yu, X., Helm, K., Corboy, J. R. & Owens, G. P. (2004) J. Immunol. 173, 649-656. [DOI] [PubMed] [Google Scholar]
- 13.Owens, G. P., Ritchie, A. M., Burgoon, M. P., Williamson, R. A., Corboy, J. R. & Gilden, D. H. (2003) J. Immunol. 171, 2725-2733. [DOI] [PubMed] [Google Scholar]
- 14.Cook, G. P. & Tomlinson, I. M. (1995) Immunol. Today 16, 237-242. [DOI] [PubMed] [Google Scholar]
- 15.Burgoon, M. P., Williamson, R. A., Owens, G. P., Ghausi, O., Bastides, R. B., Burton, D. R. & Gilden, D. H. (1999) J. Neuroimmunol. 94, 204-211. [DOI] [PubMed] [Google Scholar]
- 16.Burgoon, M. P., Owens, G. P., Smith-Jensen, T., Walker, D. & Gilden, D. H. (1999) J. Immunol. 163, 3496-3502. [PubMed] [Google Scholar]
- 17.Qin, Y., Duquette, P., Poole, R. & Antel, J. (1998) J. Clin. Invest. 102, 1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo, M., Dono, M., Gazzola, P., Roncella, S., Valetto, A., Chiorazzi, N., Mancardi, G. L. & Ferrarini, M. (2000) J. Immunol. 164, 2782-2789. [DOI] [PubMed] [Google Scholar]
- 19.Owens, G. P., Kraus, H., Burgoon, M. P., Smith-Jensen, T., Devlin, M. E. & Gilden, D. H. (1998) Ann. Neurol. 43, 236-243. [DOI] [PubMed] [Google Scholar]
- 20.Owens, G. P., Burgoon, M. P., Anthony, J., Kleinschmidt-DeMasters, B. K. & Gilden, D. H. (2001) Clin. Immunol. 98, 258-263. [DOI] [PubMed] [Google Scholar]
- 21.Baranzini, S. E., Jeong, M. C., Butoni, C., Murray, R. S., Bernard, C. C. A. & Oksenberg, J. R. (1999) J. Immunol. 163, 5133-5144. [PubMed] [Google Scholar]
- 22.Smith-Jensen, T., Burgoon, M. P., Anthony, J., Kraus, H., Gilden, D. H. & Owens, G. P. (2000) Neurology 54, 1227-1232. [DOI] [PubMed] [Google Scholar]
- 23.Kouskoff, V. & Nemazee, D. (2001) Life Sci. 69, 1105-1113. [DOI] [PubMed] [Google Scholar]
- 24.Marks, J. D., Griffiths, A. D., Malmqvist, M., Clackson, T. P., Bye, J. M. & Winter, G. (1992) Biotechnology (N.Y.) 10, 779-783. [DOI] [PubMed] [Google Scholar]
- 25.Mehta, P. D., Kane, A. & Thormar, H. (1977) J. Immunol. 118, 2254-2261. [PubMed] [Google Scholar]
- 26.Pohl-Koppe, A., Kaiser, R., Meulen, V. T. & Liebert, U. G. (1995) Clin. Diagn. Virol. 4, 135-147. [DOI] [PubMed] [Google Scholar]
- 27.Tschen, S. I., Bergmann, C. C., Ramakrishna, C., Morales, S., Atkinson, R. & Stohlman, S. A. (2002) J. Immunol. 168, 2922-2929. [DOI] [PubMed] [Google Scholar]