Abstract
Local base stacking and conformational mobility play a major role in the structure and function of nucleic acids. We have recently shown that the low-energy CD spectrum of 2-aminopurine (2-AP), i.e., the CD spectral region above 300 nm, can be used to monitor conformational changes in polynucleotides at or near mono- and dinucleotide 2-AP residues that replace adenine residues in DNA and RNA. Here, we extend this technique to pyrrolo-cytosine (PC), a fluorescent analogue of cytosine. The low-energy CD spectrum of a PC dinucleotide in dsDNA exhibits an exciton couplet with two bands of opposite sign centered at 350 nm. This signal is characteristic of base stacking between adjacent PC residues in a right helical conformation. Isolated PC nucleotide residues inserted into polynucleotide chains also display chirality that reflects the asymmetric environment of their sequence context. Thus, we show that the low-energy CD spectra of C(PC)A and A(PC)C sequences in dsDNA have opposite signs. It appears that the measurement of the low-energy CD spectra of PC residues will usefully complement 2-AP measurements by serving to characterize the local conformations and dynamics of nucleic acids near C residues and G-C base pairs.
Keywords: base stacking, dinucleotide conformations, exciton coupling, nucleic acid conformations, spectroscopy
Circular dichroism (CD) spectroscopy is commonly used to investigate the conformation of nucleic acids. However, conformational environments of particular nucleotide residues within DNA or RNA cannot be studied by this method because the spectroscopic signals of these residues are buried in the total CD of the polynucleotide. This limitation has been partially overcome by using the low-energy CD of 2-aminopurine (2-AP) residues at defined sites within DNA and RNA (1). The successful use of this technique with 2-AP is a prototype investigation for the present study of another nucleic acid base analogue, pyrrolo-cytosine (PC), that can probe G-C sequences.
2-AP is a structural isomer of adenine in which the amino group has been moved from position 6 to position 2 of the purine ring. It has a low-energy electronic transition at 305 nm that is fluorescent (2, 3). Base stacking causes a red shift in the absorbance maximum of this transition (2, 4). 2-AP behaves in other ways like adenine. In particular, the insertion of 2-AP residues opposite T or U residues in duplex DNA or RNA does not significantly change the conformation or stability of these structures (4, 5) (unpublished results), and furthermore, 2-AP nucleoside triphosphates can serve as substrates for DNA and RNA polymerases in vivo and in vitro (6-11).
DNA molecules that contain 2-AP residues exhibit two types of low-energy CD spectra (1). The first is due to exciton coupling between two stacked 2-AP residues (12) in a right helical conformation, such as in B-form dsDNA. This CD signal is characterized by a positive CD band located at a wavelength above the maximum UV absorbance of 2-AP and a negative band below this wavelength. The low-energy tail of the large oligonucleotide CD signal near 270 nm can mask the negative band; thus, in practice, a positive exciton band above 320 nm is often the most reliable indication of exciton coupling between 2-AP residues. The second type of signal, a single CD band centered at the wavelength of maximum UV absorbance, is characteristic of 2-AP in a context without exciton coupling. This type of signal has been observed for single 2-AP residues or for pairs of 2-AP residues that are not stacked.
Using this method, we have seen local conformational changes in polynucleotides caused by a number of variables, including thermal melting of dsDNA, conformational changes such as B-form to A-form conversions, and the denaturing effects of ethanol (1). We are currently investigating protein-induced RNA conformational changes that are implicated in regulating transcription in E. coli and phage λ by site-specifically placing 2-AP residues in the loop of an RNA hairpin, boxB (unpublished results); binding of cognate protein to boxB produces CD changes that reflect, for example, stacking of nonadjacent bases in the loop and extrusion of the intervening base. Hence, the low-energy CD of site-specifically placed 2-AP residues within nucleic acids provides a useful way to study biochemical processes that depend on local DNA and RNA conformations.
In this article, we extend the low-energy CD technique to PC, to probe specific sequences of nucleic acids that are not accessible to 2-AP. PC is a fluorescence analog of cytosine (Fig. 1) with an excitation maximum at 350 nm and an emission maximum at 460 nm (13-16). Fluorescence quenching of site-specifically placed PC residues has previously been used to study local conformations in the nucleic acid components of complexes of T7 RNA polymerase (14, 15) and HIV-1 reverse transcriptase (17).
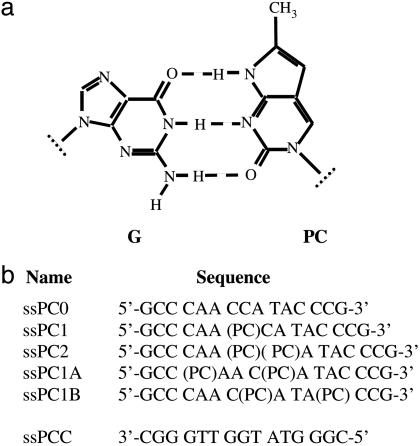
Fig. 1.
Structure of a guanine-PC base pair (a), and sequences and nomenclature of ss oligonucleotides used in this study (b). ssPCC is the complementary strand used to form the ds oligonucleotides dsPC0, dsPC1, dsPC2, dsPC1A, and dsPC1B.
PC has not been as extensively studied as 2-AP, and its effects on nucleic acid structure and stability are less well known. As far as we have been able to determine, PC incorporation by DNA replication in vivo has not been investigated, and the mutagenicity of this analogue is unknown. The PC deoxyribonucleoside triphosphate can be used in PCRs (16), and a recent study has shown that, in vitro, reverse transcriptase-catalyzed DNA-dependent DNA synthesis is not affected by the incorporation of PC residues into the template strand (17). It seems that this template-directed synthesis takes place with high fidelity, because the probability of nucleotide misincorporation directed by either a C or a PC residue located in the template strand is approximately the same.
Furthermore, the RNase H activity of HIV-1 reverse transcriptase properly cleaves DNA-RNA substrate with PC in the DNA strand, suggesting that replacing C residues with PC introduces no gross structural deformation of the oligonucleotide (17). Likewise, substitution of PC for C has little effect on the thermal stability of dsDNA oligonucleotides (13, 16). Although further studies of the differences between PC and C are clearly needed, the available data indicate that PC residues can be considered, at least to a first approximation, as the functional equivalent of cytosine residues.
PC absorbs at 350 nm (13, 14), and one would expect adjacent PC residues in a B-form DNA conformation to show exciton bands centered at this wavelength (18, 19). Less interference from the CD of the naturally occurring bases is expected for a CD spectrum centered at 350 nm than for 2-AP, which absorbs near 320 nm in dsDNA (1). Thus, the use of PC residues should permit extension of our low-energy CD methods to the investigation of G-C sequences.
To examine this possibility, we have measured the spectroscopic properties of single-stranded (ss) and double-stranded (ds) 15-mer oligonucleotides containing PC residues at various positions (Fig. 1b). The oligonucleotide PC2 has two adjacent PC residues and was used to study the effects of base stacking on CD. Other polynucleotide sequences were chosen to permit us to investigate the effect of sequence changes on the CD of an isolated PC chromophore. The oligonucleotide PC1 has a residue in an A(PC)C sequence and PC1A has two PC residues in a C(PC)A sequence, whereas PC1B has one residue in each sequence.
Materials and Methods
Oligonucleotides containing PC (Fig. 1) were synthesized and PAGE-purified by Qiagen. The sequences of all of the oligonucleotides used were identical, except for the substitution of C by PC. Unless otherwise stated experiments were carried out at 20°C in 20 mM sodium phosphate buffer containing 100 mM NaCl and 0.1 mM EDTA (pH 7.5) and made with deionized water (Barnstead). Concentration units are mol (ss or ds) of oligonucleotide per liter.
To avoid complications due to secondary structure within the complementary ss oligonucleotide ssPCC (see below), concentrations were calculated from the A260 values of ss oligonucleotides at 80°C, using the recently determined extinction coefficients of Cavaluzzi et al. (20) and a molar extinction coefficient of 4,000 M-1·cm-1 for PC-monophosphate (John Randolph, Glen Research, personal communication). Thermal denaturation experiments were carried out by raising the temperature in the cuvette by 2°/min, and melting was monitored by UV absorbance at 260 nm, using a Cary 3E spectrometer.
Fluorescence spectra were taken with a Jobin Yvon Fluorolog spectrometer, and the results are reported as counts per second. Samples placed into a 10 × 4-mm cuvette with the long path-length oriented in the direction of the incident light, were excited at 353 nm, and emission spectra were recorded at 20°C. Maximum emission was observed at 455 nm for all experiments. OD at 355 nm was <0.05, and inner filter corrections were not required.
CD spectra were taken between 500 and 195 nm on a Jasco J-720 CD spectrometer equipped with a thermostated cell holder. The results presented represent the average of 8-20 spectra and are plotted as Δε, (the difference in molar extinction coefficient of left and right circularly polarized light) per mol of residue (19). CD values per mol of nucleotide at wavelengths below 300 nm [units, liter (mol nt)-1·cm-1] were compared with CD values per mol of PC residue above 300 nm [units, liter (mol PC)-1·cm-1].
Results
Preparation and Characterization of Oligonucleotides that Contain PC Residues. Solutions of 7 μM concentrations of ss deoxyribooligonucleotides (units, mol of oligonucleotide per liter; optical densities of ≈1 OD at 260 nm) were divided into two portions. One was used to characterize the ss oligonucleotides, and the other was mixed with the complementary ss oligonucleotide to form dsDNA complexes. Low-energy absorbance of PC residues at 350 nm, with an intensity of 2% of the OD at 260 nm per PC residue for ss oligonucleotides, could be clearly observed without interference from the absorbance of the naturally occurring bases.
The ds nature of the polynucleotide was confirmed by thermal melting, which was monitored by changes in the UV absorbance at 260 nm. The melting profile was cooperative in nature, as expected for a melting process that involves a ds- to ssDNA phase transition. The Tm of the unmodified construct without PC, dsPC0, was 59°C, whereas the Tm of dsPC1 and dsPC2 were 1°C and 2°C higher, respectively. These results show, as previously reported (13, 16), that the substitution of C residues by PC has little effect on the overall stability of dsDNA molecules.
The fluorescence properties of PC are known and have been used to probe local conformational changes induced by proteins in polynucleotides that contain PC (14, 15, 17). The fluorescence intensity of ssPC2 per mol of PC was one-quarter of that seen with ssPC1 or ssPC1A. The lower intensity observed for the ss oligonucleotide with two adjacent PC residues, compared with that seen for isolated PC residues, may reflect quenching due to base stacking interactions between PC residues in ssPC2 (see below). The fluorescence signal further decreased when ss oligonucleotides were titrated with the complementary oligonucleotide ssPCC. Upon formation of the ds complex, 86% quenching was observed for dsPC1, and 80% quenching was observed for dsPC2, relative to the initial fluorescence signal of their constituent ss oligonucleotides.
To eliminate possible unwanted effects of variable oligonucleotide sequence, the ss oligonucleotides chosen for these studies were identical, except for substitution of C residues by PC at various positions (Fig. 1b). As a result, ssPCC, the complementary ss oligonucleotide used to make all of the ds oligonucleotides, contains eight guanosine residues, and several findings suggest that this ss molecule may possess significant secondary structure. Thus, we observed that the value of A260 for ssPCC increased by 10% when the temperature was changed from 15°C to 80°C, and the resulting melting transition was cooperative with a Tm of 57°C. In contrast, broad thermal transitions were observed for the other ss oligonucleotides, with total increases in A260 of only 3-5%. In addition, the CD spectrum of the ssPCC oligonucleotide construct showed peaks at 269 and 289 nm. Similar CD spectra have been previously reported for guanine-rich ss oligonucleotides and attributed to the formation of higher-order oligonucleotide structures (21).
The presence of secondary structure in ssPCC has two consequences for our study. First, to determine oligonucleotide concentration, it is necessary to take into account the effects of this structure on the observed UV absorbance (see Materials and Methods). In addition, we note that the ssPCC oligonucleotide shows an apparent Tm that is similar to the values reported above for the ds oligonucleotides. Consequently, it appears that the formation of secondary structure within ssPCC could potentially compete with ds oligonucleotide formation. However, we were able to show that this competition does not affect our experiments. In particular, we found that the interaction between the two oligonucleotides was rapid; no time-dependent fluorescent changes were observed during titration of the fluorescent oligonucleotide with ssPCC. Furthermore, the binding of the complementary strand to the fluorescent oligonucleotides was stoichiometric. Hence, in our studies, the formation of secondary structure in ssPCC noted above does not appear to interfere with the formation of duplex molecules.
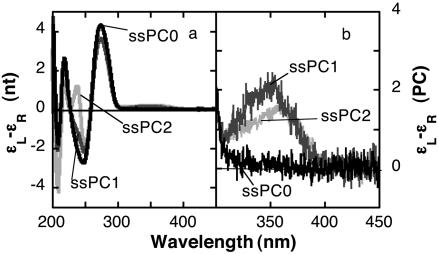
Low-Energy CD of Oligonucleotides That Contain PC Residues. We subsequently investigated the CD spectra of oligonucleotides with PC residues in various positions. The CD peak at 275 nm decreased from 4.3 [liters (mol nt)-1·cm-1] in ssPC0 to 3.6 [liters (mol nt)-1·cm-1] in ssPC1 and ssPC2 (Fig. 2). The low-energy CD spectra of ssPC1 consisted of a single band at 350 nm, which is the position of the PC absorption maximum. The low-energy band of the spectrum obtained with the ssPC2 oligonucleotide was somewhat less intense and shifted to 360 nm. This red shift in the CD maximum could be a consequence of appearance of a high-energy peak due to stacking between PC bases in a fraction of the ssPC2 population.
Fig. 2.
High-energy (a) and low-energy (b) CD spectra of 7 μM ss oligonucleotides. Reaction conditions were as described in Materials and Methods.
The CD spectra of the ds oligonucleotides dsPC0, dsPC1, and dsPC2 showed a peak at 265 nm and a shoulder at 288 nm. The intensity of the CD band at 265 nm was Δε = 3.2 [liters (mol nt)-1·cm-1] for dsPC0, and the intensity of this band decreased to 2.2 and 1.6 [liters (mol nt)-1·cm-1] in dsPC1 and dsPC2, respectively.
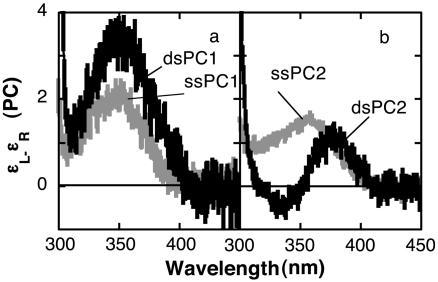
Transition from ss to ds oligonucleotide produced local conformational changes at the PC residues that affected the low-energy CD spectrum. Formation of dsPC1 from ssPC1 increased the intensity of the 350-nm band that was observed in the ss oligonucleotide by a factor of 1.8 but did not change the wavelength of maximum intensity (Fig. 3a). Hence, the low-energy CD spectrum of dsPC1 has its maximum intensity at the same wavelength as the absorption spectrum of this transition, as expected in the absence of exciton coupling. We infer that the observed CD reflects the chiral placement of adjacent nucleotides near the PC residue, and that this placement changes with base stacking.
Fig. 3.
Low-energy CD spectra of 7 μM ss oligonucleotide and 3.5 μM ds oligonucleotides. (a) PC1. (b) PC2. Concentration units are mol oligonucleotide per liter. CD units are liters (mol PC)-1·cm-1.
In contrast, the low-energy CD spectrum of dsPC2 exhibits two bands of opposite sign centered on the absorption maximum (Fig. 3b). The spectrum has a peak at 380 nm and a trough at 335 nm, and crossed the baseline at 352 ± 2 nm. A positive low-energy exciton band such as that observed in this spectrum is expected for exciton coupling of a dinucleotide containing identical bases in the right-handed helical conformation of B-form DNA (18, 19).
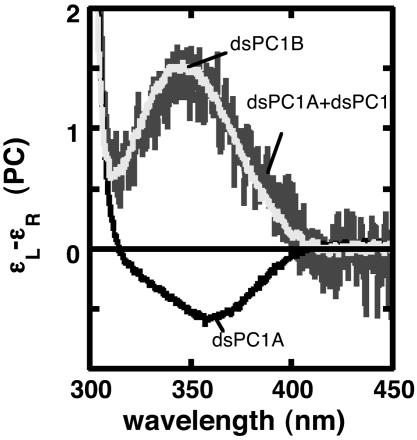
dsPC1A and dsPC1B contain two PC residues separated by three canonical bases (Fig. 1b). The CD spectrum of the ds oligonucleotide dsPC1A shows a trough with Δε = -0.55 [liters (mol PC)-1·cm-1] at 360 nm, whereas dsPC1B shows a peak with Δε = 1.5 [liters (mol PC)-1·cm-1] at 345 nm (Fig. 4). These different spectra appear to be an effect of sequence context on the CD of the PC residues. In the oligonucleotide dsPC1A, both PC residues are in C(PC)A sequences. Hence, the negative trace in Fig. 4 can be considered to represent the molar CD of PC in this sequence. Likewise, the positive CD of dsPC1 (black trace in Fig. 3a) would be the molar CD of PC in an A(PC)C sequence. dsPC1B contains two PC residues, one in a C(PC)A sequence and the other in the A(PC)C sequence, and its CD spectrum equals the sum of the dsPC1 and dsPC1A CD spectra (Fig. 4). These results show that the low-energy CD spectra produced by isolated PC residues depend on the nearest neighbor sequence and are additive.
Fig. 4.
Low-energy CD spectra of 36 μM dsPC1A and 24 μM dsPC1B, containing two PC residues separated by 3 nt. The dark gray line is the sum of the dsPC1A and dsPC1 (Fig. 3a) spectra. Concentration and CD units are as in Fig. 3.
Discussion
Low-Energy CD of PC. The most important result of this study is the demonstration that the low-energy CD of PC can be used to investigate the conformation of DNA at selected cytosine residues. Low-energy CD spectra reflect local base stacking at PC residues during the transition from ss to ds oligonucleotide (Fig. 3). In particular, the low-energy CD spectrum of dsPC2 exhibits exciton coupling characterized by two bands of opposite sign centered on the low-energy UV absorption maximum of PC. A CD spectra of this shape indicates base stacking between adjacent PC residues in a right helical conformation (18, 19). In contrast, the maximum intensity of the low-energy CD spectrum of ssPC1 occurred at the absorption maximum. Formation of dsPC1 increased the CD, and this spectroscopic change reflects the chiral environment of nucleotides in the DNA sequence near the PC residue.
A CD signal characteristic of exciton coupling has also been reported for adjacent 2-AP residues in dsDNA. A low-energy peak is observed at 326 nm, and its intensity is three times larger than for dsPC2 (1). It is worth emphasizing that the low-energy CD signal of PC is well separated from the CD spectra of DNA and protein. Hence, the exciton couplet with equal intensities and opposite signs that is expected from the conservative nature of exciton coupling (18, 19) is more clearly visible for dsPC2 (Fig. 3) than for 2-AP dimers in dsDNA (1). The wavelength where the CD intensity is zero corresponds exactly to the absorption maximum of PC. CD spectra of this quality open the possibility for theoretical calculations of the relation between structure and CD for a single base pair within a particular polynucleotide. Finally, because their CD signals above 300 nm are well separated, it may be possible to use 2-AP and PC together in low-energy CD experiments and simultaneously measure local conformations at two different sites within a given nucleic acid construct.
Effect of Nearest Neighbor Bases on Low-Energy CD of Isolated PC Residues. We observed an unexpected effect of sequence context on the low-energy CD spectra of isolated PC residues in dsDNA oligonucleotides. The CD of PC in an A(PC)C sequence is positive (black trace in Fig. 3a), whereas the CD of PC in C(PC)A is negative (black trace in Fig. 4). These signals arise from the sequence context of the PC residue and are additive, as shown by the molar CD of dsPC1B, which contains two PC residues, one in each sequence (Fig. 4).
The origin of these different signals is uncertain. Geometric factors involved in determining the total chirality of adjacent bases are modified when the polarity of the trinucleotide sequence is reversed and could change the sign of the CD spectrum. In addition, the PC residue may have a different range of conformations in the two sequences. An unusually large flexibility of the CA dinucleotide step in dsDNA has been reported from gel mobility assays (22), from fluctuations in protein-DNA structures (23), and from EPR experiments (24); averaging over a larger range of structures due to the greater flexibility of a CA dinucleotide base pair could contribute to the negative CD band of C(PC)A. Although further studies will be necessary to understand this phenomenon, the low-energy CD of an isolated PC residue appears to be sensitive to nearest neighbor bases and may give information about local conformation and mobility at GC sequences.
The local behavior of RNA and DNA at the nucleotide and dinucleotide level has many biological consequences. For example, local conformations are thought to contribute to protein-nucleic acid recognition primarily through indirect readout in which specific binding occurs because the protein recognizes sequence-dependent conformational features of DNA or RNA (25, 26). Conceptually, local deformations could produce a fraction of the duplex population in the product conformation; specific protein binding would then “trap” this conformation and displace the equilibrium toward the bound complex. In this case, free oligonucleotide would be expected to have structural characteristics similar to the specifically bound oligonucleotide (25, 27). Experiments with techniques such as gel mobility assays, NMR spectroscopy, and x-ray crystallography have been carried out for a number of years to study specific recognition of DNA sequences by proteins. Observation of local, sequence-dependent deformation of duplex DNA or RNA by low-energy CD, such as demonstrated in this paper, appears to be a useful complementary method to study this and related phenomena.
In conclusion, we have demonstrated the feasibility of using low-energy CD to determine local conformations of nucleic acids at AT sequences (1) and GC sequences (this work). The structural basis of exciton coupling of 2-AP or PC dinucleotides is well understood (18, 19), and these signals can be used to measure local base stacking. Isolated 2-AP or PC mononucleotides also display chirality that reflects their asymmetric environment and is sensitive to sequence context. Local DNA conformation and mobility play a role in much nucleic acid biochemistry, and low-energy CD should be a useful technique to study such processes.
Acknowledgments
We thank John Schellman, Monique Erard, and members of our laboratories for many helpful discussions and John Randolf for his insightful comments. This work was supported in part by National Institutes of Health Grants GM-15792 and GM-29158 (to P.H.v.H.) and the Centre National de la Recherche Scientifique (N.P.J.). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Author contributions: N.P.J. and P.H.v.H. designed research; N.P.J. performed research; N.P.J. wrote the first draft of paper.; W.A.B. performed and analyzed most of the CD experiments; P.H.v.H. and N.P.J. revised the paper; and P.H.v.H. provided the materials and laboratory facilities.
Abbreviations: 2-AP, 2-aminopurine; ds, double-stranded; PC, pyrrolo-cytosine (as the base) or pyrrolo-cytidine (as the nucleoside or nucleotide); ss, single-stranded.
References
- 1.Johnson, N. P., Baase, W. A. & von Hippel, P. H. (2004) Proc. Natl. Acad. Sci. USA 101, 3426-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward, D. C., Reich, E. & Stryer, L. (1969) J. Biol. Chem. 244, 1228-1237. [PubMed] [Google Scholar]
- 3.Smagowicz, J. & Wierzchowski, K. L. (1974) J. Luminescence 8, 210-232. [Google Scholar]
- 4.Eritja, R., Kaplan, B. E., Mhaskar, D., Sowers, L. C., Petruska, J. & Goodman, M. F. (1986) Nucleic Acids Res. 14, 5869-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sowers, L. C., Fazakerley, G. V., Eritja, R., Kaplan, B. E. & Goodman, M. F. (1986) Proc. Natl. Acad. Sci. USA 83, 5434-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman, M. F., Hopkins, R. & Gore, W. C. (1977) Proc. Natl. Acad. Sci. USA 74, 4806-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, M. L., Stellwagen, R. H. & Goodman, M. F. (1981) J. Biol. Chem. 256, 7097-7100. [PubMed] [Google Scholar]
- 8.Frey, M. W., Sowers, L. C., Millar, D. P. & Benkovic, S. J. (1995) Biochemistry 34, 9185-9192. [DOI] [PubMed] [Google Scholar]
- 9.Fidalgo da Silva, E., Mandal, S. S. & Reha-Krantz, L. J. (2002) J. Biol. Chem. 277, 40640-40649. [DOI] [PubMed] [Google Scholar]
- 10.Mandal, S. S., Fidalgo da Silva, E. & Reha-Krantz, L. J. (2002) Biochemistry 41, 4399-4406. [DOI] [PubMed] [Google Scholar]
- 11.Purohit, V., Grindley, N. D. F. & Joyce, C. M. (2003) Biochemistry 42, 10200-10211. [DOI] [PubMed] [Google Scholar]
- 12.Rist, M., Wagenknecht, H. A. & Fiebig, T. (2002) Chemphyschem 3, 704-707. [DOI] [PubMed] [Google Scholar]
- 13.Woo, J., Meyer, R. B. & Gamper, H. B. (1996) Nucleic Acids Res. 24, 2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, C. & Martin, C. T. (2001) J. Mol. Biol. 308, 465-475. [DOI] [PubMed] [Google Scholar]
- 15.Liu, C. & Martin, C. T. (2002) J. Biol. Chem. 277, 2725-2731. [DOI] [PubMed] [Google Scholar]
- 16.Berry, D. A., Jung, K.-Y., Wise, D. S., Sercel, A. D., Pearson, W. H., Mackie, H., Randolf, J. B. & Somers, R. L. (2004) Tetrahedron Lett. 45, 2457-2461. [Google Scholar]
- 17.Dash, C., Rausch, J. W. & Le Grice, S. F. J. (2004) Nucleic Acids Res. 32, 1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, W. C., Jr., & Tinoco, I., Jr. (1969) Biopolymers 8, 715-731. [Google Scholar]
- 19.Cantor, C. R. & Schimmel, P. R. (1980) Biophysical Chemistry (Freeman, San Francisco).
- 20.Cavaluzzi, M. J. & Borer, P. N. (2004) Nucleic Acids Res. 32, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, R., Gaffney, B. L., Wang, C., Jones, R. A. & Breslauer, K. J. (1992) Proc. Natl. Acad. Sci. USA 89, 8832-8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington, R. E. & Winicov, I. (1994) Prog. Nucleic Acid Res. Mol. Biol. 47, 197-270. [DOI] [PubMed] [Google Scholar]
- 23.Olson, W. K., Gorin, A. A., Lu, X. J., Hock, L. M. & Zhurkin, V. B. (1998) Proc. Natl. Acad. Sci. USA 95, 11163-11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okonogi, T. M., Alley, S. C., Reese, A. W., Hopkins, B. H. & Robinson, B. H. (2002) Biophys. J. 83, 3446-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crothers, D. M. (1998) Proc. Natl. Acad. Sci. USA 95, 15163-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varani, G. (1997) Acc. Chem. Res. 30, 189-195. [Google Scholar]
- 27.Hizver, J., Rozenberg, H., Frolow, F., Rabinovich, D. & Shakked, Z. (2001) Proc. Natl. Acad. Sci. USA 98, 8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]