Abstract
Autoimmune regulator (aire) is a transcription factor that controls the self-reactivity of the T cell repertoire. Although previous results indicate that it exerts this function in part by promoting ectopic expression of a battery of peripheral-tissue antigens in epithelial cells of the thymic medulla, recent data argue for additional roles in negative selection of thymocytes by medullary cells. As one approach to exploring such roles, we performed computational analyses of microarray data on medullary RNA transcripts from aire-deficient versus wild-type mice, focusing on the genomic localization of aire-controlled genes. Our results highlight this molecule's transcriptional activating and silencing roles and reveal a significant degree of clustering of its target genes. On a local scale, aire-regulated clusters appeared punctate, with aire-controlled and aire-independent genes often being interspersed. This pattern suggests that aire's action may not be a simple reflection of the wide action of a chromatin remodeling enzyme. Analysis of the identity of certain of the clustered genes was evocative of aire's potential roles in antigen presentation and the coordination of intrathymic cell migration: for example, major histocompatibility complex class I and class II gene products and certain chemokine genes are targets of aire-regulated transcription.
Keywords: autoimmunity, microarray, T cell tolerance, thymus, gene expression
In organisms that possess an adaptive immune system, the establishment of immunological tolerance to self-antigens is a vital, but complex, process. For T lymphocytes, there appears to be a bulwark of overlapping and complementary mechanisms that operate at different stages of the T cell differentiation process (reviewed in refs. 1 and 2). One such mechanism involves the exposure of immature thymocytes to a wide repertoire of antigens usually associated with peripheral tissues (reviewed in ref. 3), promoting the elimination of cells whose antigen-specific receptors (T cell receptors) are reactive to them. This ectopic gene expression is controlled, in part, by the AIRE/aire (autoimmune regulator, aire) gene. Loss-of-function mutations of AIRE in humans result in the multiorgan autoimmune disease autoimmune polyendocrine syndrome type 1 (APS-1, also called autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy or APECED); mutation of the aire homolog in mice results in multiorgan inflammatory infiltrates and autoantibodies of diverse specificities (4, 5). Exploration of these mutant animals revealed that aire exerts its antiautoimmunity function primarily in thymic medullary epithelial cells (MECs) and that it does so in part by controlling the expression of peripheral-tissue antigens (5).
Aire possesses several structural features indicative of a transcription factor: a SAND domain with sequence similarity to DNA-binding domains in nonmammalian species (although the domain in aire lacks certain amino acids required for DNA binding in other factors) (6, 7), a nuclear localization signal (8, 9), and three nuclear-receptor-binding LXXLL domains (10, 11). It exhibits significant transactivation potential in a classical GAL4 transactivation assay (8, 12) and can interact with the common transcriptional coactivator, CREB-binding protein (12). Aire also shows E3 ubiquitin-ligase activity (13) through which it could potentially modulate the activity of chromatin elements.
How aire controls ectopic expression of peripheral-tissue antigens remains an open question, in part because DNA-binding activity has proven difficult to evidence reproducibly (14). The genes whose transcription is activated by aire are numerous (estimated as 200-1,200 (5). They include a broad diversity of protein functional classes (hormones such as insulin, enzymes such as cytochrome p450 subunits, structural molecules such as keratins, and growth factors such as insulin-like growth factor 2) and tissue localizations (essentially every organ is represented). These genes are controlled in the relevant peripheral tissues by very specific, and different, transcriptional programs. One clue to aire's mechanism of action may lie in the observation that reproductive organs are the other locale where it is strongly expressed. This intriguing finding prompts one to ask whether aire activates transcription as a classic transcription factor by binding (directly or indirectly) to specific motifs in promoter/enhancer regions or whether it effects a broader program of chromatin remodeling, as encountered during genomic reprogramming in the germ line.
Herein, we have performed a bioinformatic analysis of the genomic localization of aire-controlled genes in hopes that the footprint of this molecule's action on the genome might provide new insights into its regulatory mechanism.
Materials and Methods
Microarray Data Preprocessing. Raw microarray data (.CEL files) from Affymetrix MgU74Av2 gene chips (three replicates each of WT and aire-deficient (KO) sorted thymic MECs; ref. 5) or MOE430A chips (two replicates each of WT and class II transactivator (CIITA) KO sorted dendritic cells, ref. 15) were processed by using the s+ array analyzer 1.1 (Insightful, Seattle) and the RMA probe-level normalization algorithm (16).
Random Number Generation and False Positive Rate (FPR) Derivation. Randomized data sets were constructed by random-number generation based on genewise expression means and intrareplicate coefficients of variation. FPRs were calculated by dividing subsets (selected on fold change) of the randomized data set with equivalent subsets from the experimental data set.
Annotation. Probesets were annotated by using the April 29, 2004, Affymetrix NetAffx annotation, based on National Center for Biotechnology Information mouse genome release 32. In cases where a single transcript was targeted by multiple probesets, the one with the highest Affymetrix sequence alignment score was selected. Fold-change was calculated as WT/KO, and each transcript was assigned a fold-change ranking, (for example, no. 1 corresponds to the transcript with the highest fold change).
Clustering Analysis. Algorithms were implemented on the s+ 6.x platform (Insightful) to calculate intergenic distances between genes significantly differentially regulated and to assess the number of such genes within 200 kb of each other. This procedure was applied to the experimental data set and 10,000 randomly generated data sets (see Fig. 3C). Alternatively, the number of genes within 200 kb of each other was assessed in a sliding window of 100 genes by fold-change rank (see Fig. 4).
Fig. 3.
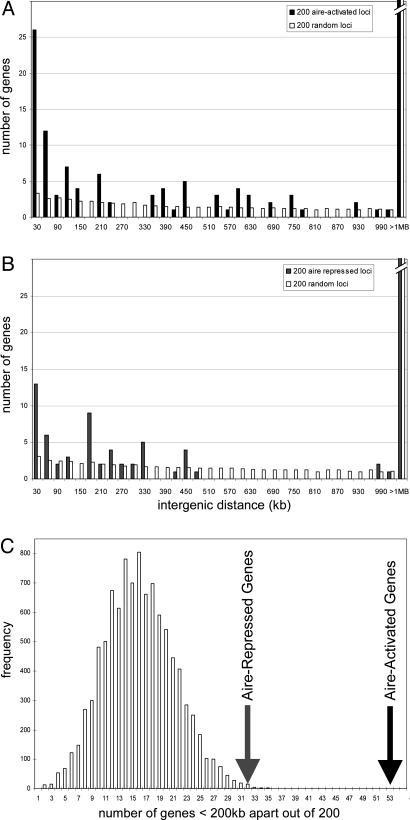
Aire-regulated genes are clustered. (A) Histogram of intergenic distances <1 Mb for the top 200 aire-activated genes (filled bars) compared with the mean of 1,000 randomly drawn sets of 200 genes. (B) Histogram for the top 200 aire-repressed genes. (C) Two hundred genes were randomly drawn 10,000 times, and the number of genes <200 kb apart were calculated.
Fig. 4.
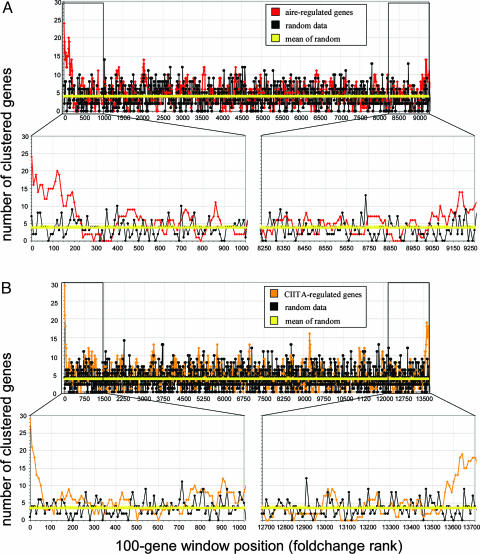
Clustering analysis in data sets from multiple transcription factor KO experiments. Genes were ranked by WT/KO fold-change (x axis), and the number of genes within 200 kb of each other was calculated within a sliding window of 100 genes (y axis). (A) Significant clustering among aire-activated genes, with slightly less pronounced clustering in aire-repressed genes. (B) A similar clustering pattern in CIITA-regulated genes.
To identify aire-regulated gene clusters, we designed an algorithm to identify all gene clusters in the Affymetrix genome, as defined by two or more genes within 200 kb of each other. Those gene clusters that contained one or more aire-regulated genes were flagged. All S+ algorithms are available upon request.
Results
Both Positive and Negative Aire Influences on Gene Expression in the Thymus. To initiate a deeper exploration of transcriptional control by aire, we performed a reanalysis of published microarray data on transcripts from MECs isolated from WT versus KO mice. As detailed in ref. 5, the raw data sets originated from cytofluorimetrically sorted MECs, from which labeled probes were prepared after amplification; three independent replicates were hybridized to Affymetrix MgU74Av2 microarrays. In this reanalysis, preprocessing of the raw microarray data was performed by using the rma algorithm (16), a statistical method not available at the time of our initial report. This technique uses probe-level normalization, without noise from mismatched-probe data, to generate more reliable estimates of gene-expression values, particularly in the low end of the expression range, albeit at the cost of compressing apparent fold-change values. The greater reliability was important in the interpretation of aire control of transcription, because ectopic expression of a given gene in the thymus is considerably lower than its expression in the relevant peripheral tissue(s).
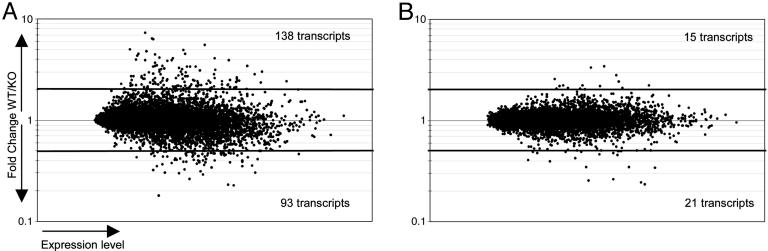
Fig. 1A shows that, in the absence of aire, large numbers of transcripts were up- or down-regulated. At a conservative fold-change value of 2, which corresponds to an FPR of 16%, 138 genes were activated by aire and 93 repressed. The former value is consistent with published results in ref. 5 and includes a preponderance of genes encoding peripheral-tissue antigens. However, we had previously underappreciated the number of aire-repressed genes.
Fig. 1.
Aire both activates and represses the transcription of a multitude of transcripts. (A) RNA from WT and aire KO thymi was hybridized to Affymetrix MgU74Av2 microarrays. Expression level vs. fold change (WT/KO) is plotted for all 12,000 probesets, showing that aire's absence affects the transcription of many genes. (B) Randomized data.
Both the list of repressed loci (Table 2, which is published as supporting information on the PNAS web site) and the subset of activated loci that encode molecules other than peripheral-tissue antigens are of particular interest in light of recent indications that aire not only regulates transcription of peripheral-tissue antigens in MECs, but also somehow controls their antigen-presentation capacity (E.S.V., unpublished data). Indeed, this more comprehensive list of aire-activated and repressed genes includes a number that might be expected to modify the ability of MECs to present antigens and thereby clonally delete differentiating thymocytes capable of recognizing the peripheral-tissue antigens they display (17, 18). For example, H2-DMβ and H2-DOβ are both involved in editing the repertoire of peptides bound to major histocompatibility complex (MHC) class II molecules, and their disruption could significantly influence the range of peptides presented by MECs (19). The chemokines CCL5, CCL9, and CCL25 could potentially influence the interaction of differentiating thymocytes with MECs, modifying their migration or the persistence of their contacts with deleting stromal cells (20).
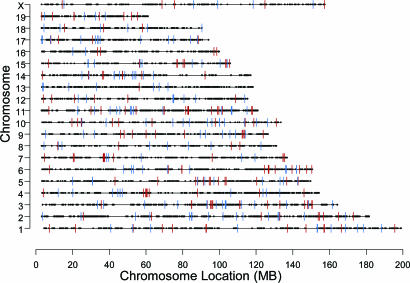
A Clustered Distribution of Aire-Controlled Genes. We then analyzed the chromosomal localization of genes whose expression in MECs was affected by aire deficiency, retrieving the positions of all genes represented on the MgU74Av2 chip (taking chromosomal positions from the April 2004 annotation of the MgU74Av2 chip, based on release 32 of the mouse genome sequence) and keeping only one example of those entities that corresponded to the same locus (filtering based on shared UnigeneID, retaining the best feature based on alignment score). On a broad scale, aire target loci were distributed throughout the genome, seeming to occur without chromosomal preference; all chromosomes included examples of aire-activated and -repressed genes (Fig. 2). On this scale, any appearance of clustering (e.g., ≈60 Mb on Chr4) was not significantly different from what was obtained through random sampling.
Fig. 2.
Genomic locations of the top 200 aire-activated (red) and the top 200 aire-repressed (blue) loci. Aire-regulated genes appear to distribute without preference among the chromosomes.
To examine chromosomal localizations at a finer resolution, we implemented an algorithm that tests, for each aire-regulated gene, the distance to its closest neighbor also controlled by aire. These distances were then used to generate a histogram (Fig. 3A) of the intergenic distances between the 200 genes most activated by aire (this particular threshold was chosen initially as corresponding to an FPR of ≈0.1). A substantial proportion of genes, 53 of 200, had another aire-activated neighbor within 200 Kb or less (filled bars). For comparison, only 16 genes had neighbors within 200 Kb when 200 genes were drawn at random from the ≈9,000 unique genes on the chip (white bars). To estimate the significance of this distribution, we performed 10,000 draws of 200 random genes and counted the number of genes with neighbors within 200 Kb. As illustrated in Fig. 3C, this procedure resulted in a Gaussian distribution with a mean at ≈15 genes. Although this distribution of randomly picked loci was itself higher than would be expected from a truly random placement of genes across all chromosomes (reflecting the fact that genes are actually significantly clustered across the chromosomes), it never reached the 53 genes counted for aire-controlled loci (P < 0.001). The same operations were repeated with the top 200 genes whose expression in MECs was repressed by aire. In this instance, 32 of 200 genes were found to have an aire-repressed neighbor within 200 Kb (Fig. 3B), again outside of the range observed with randomly picked genes (Fig. 3C; P < 0.005). Thus, genes whose ectopic expression in MECs were controlled by aire clearly had a nonrandom placement in the genome and were often clustered within quite short genomic intervals.
Because our choice of the 200 genes most affected by aire was somewhat arbitrary, we repeated the analysis, this time sliding a window of 100 genes across the entire list of unique loci ranked according to fold change in aire-deficient versus WT MECs (Fig. 4A). Significant clustering was observed at the left end of the spectrum, corresponding to aire-activated genes, clearly departing from the profile generated by randomized loci (black line). Clustering, although to a lesser extent, was also evident at the right end of the spectrum, representing the genes most repressed by aire. Interestingly, the clustering of aire-activated genes converged to the random baseline (genome-wide average shown as a yellow line) when the sliding window was set at >200. This value is in line with our previous estimates of the number of genes activated by aire (5) and with values from FPR estimates (data not shown). Thus, in those cases where the effect of a mutation or other phenotypic alteration acts on chromosomally clustered genes, the clustering index can become a useful indication of the significant fold-change range. Such an independent indication would be unaffected by the multiple-sampling issues that often confound the interpretation of microarray data.
We then asked whether the genomic clustering detected with aire-activated genes also applied to loci controlled by other transcription factors. The same analytical process was applied to data sets from dendritic cells of mice WT or deficient in CIITA (15). CIITA is the “master” transcriptional regulator of MHC class II genes and ancillary loci that encode molecules involved in antigen processing and presentation (21, 22). For those genes in dendritic cells influenced by CIITA (Fig. 4B), the analysis showed clear evidence of genomic clustering at both the activated and repressed end of the spectrum. This clustering was even more marked than for aire-regulated loci, albeit impacting a smaller number of genes, with the clustering curve converging to the random baseline at ≈50 activated and 150 repressed loci. As was the case in the analysis of aire-regulated loci, genes activated by CIITA were relatively more clustered than those repressed by CIITA. Aire and CIITA thus demonstrate comparable chromosomal profiles of regulatory impact. However, no clustering was detected in microarray data sets when comparing B cells from mice WT or deficient in NF-κB1 (S., Moran, A. Caniapa, and S. Pillai, unpublished data), possibly because, in a control system such as NF-κB, with its partially redundant elements, only a fraction of the true target genes may be evident in a microarray analysis based on fold change.
Aire-Regulated Genomic Regions. As aire-regulated genes appeared clustered, we then asked whether, on a local scale, all of them were similarly affected: control mechanisms involving general derepression of a gene block within a chromatin loop might be expected to influence entire blocks of neighboring loci. To this end, we implemented a computer algorithm that lists all gene clusters in the genome, clusters being defined as 2 or more genes within 200 Kb of each other. From this list, those clusters that contained one or more aire-regulated loci were identified (by using the top 200 each of the aire-activated and aire-repressed distributions).
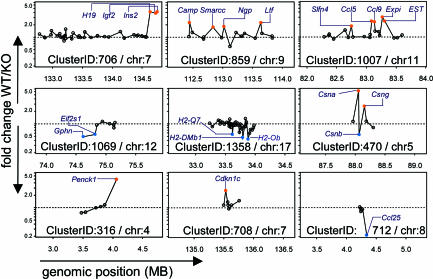
The software identified 1,538 gene clusters (Table 3, which is published as supporting information on the PNAS web site) within the genome defined by the coverage of the MgU74Av2 chip. Of these clusters, 251 (16%) contained an aire-regulated target gene, and 54 contained two or more target loci (Table 1). Yet, these clusters were far from being uniformly affected by aire, as illustrated by the examples shown in Fig. 5. In many, only one gene was affected. When two or more genes in a cluster were regulated by aire, no particular pattern was apparent: aire-controlled loci were scattered and separated by one or more noncontrolled genes in some clusters (e.g., ClusterIDs 859 and 1007) or were grouped together in others (e.g., ClusterID 1069); some clusters contained both aire-activated and -repressed loci (e.g., ClusterID 470). These results suggest that the genomic regions that aire interacts with retain locus-specific control over the expression of individual genes.
Table 1. Summary of clusters containing aire-regulated genes.
| No. (%) | |
|---|---|
| Total no. of clusters in the Affymetrix genome | 1,538 |
| Clusters with one or more aire-regulated genes | 251 (16) |
| Clusters with only activated genes | 93 |
| Clusters with only repressed genes | 104 |
| Clusters with two or more aire-regulated genes | 54 (4) |
| Clusters with only activated genes | 24 |
| Clusters with only repressed genes | 16 |
| Clusters with both activated and repressed genes | 14 |
Clusters are two or more genes within 200 kb of each other.
Fig. 5.
Aire-regulated gene clusters. Each cluster is plotted as genomic position (in Mb) vs. fold change (WT/KO). Clusters contain aire-regulated genes that are either directly adjacent, separated by nonregulated intervening genes, or even divergently regulated; this heterogeneity in expression profiles suggests that aire does not control the expression of all genes in a region. The names of aire-regulated genes are indicated.
The identities of certain of the gene clusters nicely reflected aire's proposed roles in negative selection and hint at additional mechanistic details. Several clusters included loci encoding peripheral-tissue antigens: for example, ClusterID 407 contained casein genes, which are found in mammary tissue, and ClusterID 646 encompassed genes encoding kallikrein proteases, many of which are thyroid-specific. The MHC, encoding a multitude of genes involved in antigen presentation, housed clusters, including MHC class I and class II genes (Cluster ID 1358 and 1359 (Table 4, which is published as supporting information on the PNAS web site). As discussed above, certain chemokine genes were controlled by aire, and there is an impressive group of these genes in ClusterID 1007. Perhaps giving a clue to aire's molecular mechanism, one of the most highly differentially regulated clusters was ClusterID 706, which contains the imprinted genes H19, insulin II, and igf2. Another interesting cluster was ClusterID 269, which has been previously detected and termed the epidermal differentiation complex in studies characterizing the differentiation program in epidermal tissue (23, 24); the existence of this cluster perhaps bears some relation to the fact that APS-1 patients usually present first with mucocutaneous candidiasis. A number of aire-regulated gene clusters also contained loci encoding keratin products (ClusterIDs 1019, 1020, and 1287; Table 2). Different keratins have been used as markers to define subsets of medullary and cortical epithelia within the thymus and may be important in structuring the stromal network that nurtures thymocyte maturation (25).
Discussion
Aire as a Positive and Negative Transcriptional Regulator. Previous analyses of the genes controlled by aire emphasized its role as a transcriptional activator. In this reanalysis, however, we noted that a substantial number of transcripts were up-regulated in the absence of aire, highlighting a role in transcriptional repression. As with its role in activation, aire may act directly on the genes whose transcription it represses or may function, indirectly, by promoting the expression of downstream factors that inhibit transcription. In any event, these in vivo results are consistent with recent findings from transfection experiments showing a negative impact by aire on transcription of the MHC-II and IL-1ra genes (26); our data corroborate these two specific findings (see Table 2), and suggest a broader, genome-disseminated role for aire in transcriptional repression. Competition for the transcription factor CREB-binding protein has been proposed as a potential mechanism for a silencing effect of aire (26), and this hypothesis has also been advanced to explain the emerging role for CIITA in transcriptional repression (27). Interestingly, there was some overlap in the sets of genes repressed by aire and CIITA (e.g., IL-4, see Table 2).
Whatever its mechanism, the silencing role of aire is likely to have significant consequences: for example, the expression of several genes encoding molecules involved in antigen presentation (e.g., H2-DMβ and H2-DOβ) was altered in the absence of aire, which may contribute to the ineffectiveness of aire-deficient thymi displaying self-antigens to elicit proper clonal deletion of thymocytes that recognize them (ref. 18; E.S.V., unpublished data) whether through direct presentation by MECs or through cross-presentation by dendritic cells (17).
Chromosomal Clustering and Implications for Aire's Effects on Transcription. The clustering of coregulated genes is a quite frequent phenomenon in eukaryotic genomes (28). In the simplest of cases, the clustered genes are a product of comparatively recent duplication events, where evolutionary divergence has yet to generate different expression patterns. In other cases, genes whose products participate in the same pathways are often colocalized, somewhat reminiscent of a prokaryotic operon (29, 30). There are regulatory advantages to such an arrangement: the same enhancers or locus-control regions are shared and ensure coordinated expression in single cells. There may be evolutionary advantages as well, such as coevolution of genes in tight linkage disequilibrium that contribute to a function as a haplotype (reviewed in ref. 28). The notion of chromosomal domains of gene expression has been recently validated in whole-genome microarray analyses (31, 32). Here, we observed clustering of the genes transcribed in the thymus under the control of aire, which was perhaps less intuitive, because many of these genes are expressed in the periphery in a variety of tissues and cells according to a diversity of developmental and environmental dictates.
To a significant extent, the influence on neighboring loci corresponded to an effect on duplicated or closely homologous genes, but colocalized family members were not the only instances of the clustered targets of aire (Fig. 5). Additionally, rather than the consistent effect one might have expected from a broad activity on chromatin conformation, the impact of aire was much more punctate within the chromosomal stretches, often affecting only two or three genes while leaving neighboring and interspersed genes unaffected. In several cases, such as the casein gene cluster (Fig. 5; ClusterID 470), aire even had opposing effects on directly adjacent neighboring genes. Thus, it does not seem likely that this protein's influence can be explained simply in terms of chromatin remodeling: either a more complex mechanism involving gene-specific binding of additional positive and negative factors may come into play, or aire behaves like a conventional transcription factor, binding itself to cis-acting control elements within individual genes. Either way, it is worth keeping in mind that many transcription factors and enhancer elements have long been known to have dual activating/silencing capabilities (33).
It may be worthwhile to highlight the relationship between our study and that of Gotter et al. (34). The latter provided evidence that genes expressed differentially between human medullary and cortical epithelial cells were chromosomally clustered. As was the case in our study, many of these loci did encode peripheral-tissue antigens, and certain of the same gene clusters (e.g., the epidermal differentiation complex) were noted. However, our data set differs in focusing on aire-regulated loci and in not filtering out genes similarly expressed in cortical and medullary epithelial cells. Thus, it will include genes encoding peripheral-tissue antigens that are expressed at about the same levels in the two epithelial cell types (e.g., proteoplipid protein or myelin basic protein; ref. 34) but will not include peripheral-tissue antigens whose MEC expression is not controlled by aire (e.g., C-reactive protein or GAD67; ref. 5). In short, our data set is focused on a particular function of MECs, i.e., aire-promoted tolerance induction, rather than on their developmental/functional distinction from another stromal cell type.
Chromosomal Clustering as an Internal Validation in Microarray Experiments. Statistical validation of microarray data is a thorny issue, facing an extreme version of the classic “many variables/few observations” problem, when true differences dangerously approach experimental noise. Even with techniques for estimation of FPR, the investigator is faced with the issue of setting thresholds for inclusion, usually arbitrarily. The representation of Fig. 4 suggests that information derived from genomic clustering, when applicable, may be quite valuable: first, at the level of individual genes, because it would be very unlikely that experimental noise would similarly affect two neighboring genes (the actual FPR could be estimated as the product of the two individual FPRs); second, by providing an independent estimate of the range of the fold-change curve that includes significant alterations. Even if not all genes whose expression is affected in a particular condition are clustered, the clustering signal highlights quite clearly the areas where they are found and, thus, gives greater confidence in these segments of the distribution. This finding was the case with CIITA in this study: the very clear genomic clustering of genes whose expression is down-regulated by CIITA provided independent evidence for the initially underappreciated role of this factor in transcriptional repression (27, 35).
Conclusion
This analysis of chromosomal clustering offers a perspective on the action of the aire transcription factor. Aire not only promotes the ectopic thymic transcription of genes encoding peripheral-tissue antigens, but also affects, either positively or negatively, the expression of a number of other genes. Some of these alterations may be important in eliciting the deletion of thymocytes reactive to the presentation of peripheral-tissue antigens, and many are localized to previously uncharacterized gene clusters, the precise control of which remains to be elucidated. It appears that these transcriptional influences may not be explained by a simple influence of aire on chromatin remodeling.
Supplementary Material
Acknowledgments
We thank C. John Luckey, Reinhard Obst, Vincent Butty, and other members of the Mathis/Benoist laboratory for inspiration and helpful discussion; Leslie Jerkins for assistance in preparing the manuscript; and Stuart Moran, Annaiah Caniapa, and Shiv Pillai for sharing unpublished data. This work was supported by National Institutes of Health Grant RO1 DK60027-03 and Young Chair funds (to D.J.M. and C.O.B.), Joslin's National Institute of Diabetes and Digestive and Kidney Diseases-funded Diabetes and Endocrinology Research Center core facilities, Harvard College Research Program Fund (to J.B.J.), and National Institutes of Health Grant 2T32 DK07260-26 (to E.S.V.).
Abbreviations: aire, autoimmune regulator; CIITA, class II transactivator; FPR, false positive rate; KO, knockout; MEC, medullary epithelial cell; MHC, major histocompatibility complex.
References
- 1.Ohashi, P. S. (2003) Curr. Opin. Immunol. 15, 668-676. [DOI] [PubMed] [Google Scholar]
- 2.Walker, L. S. & Abbas, A. K. (2002) Nat. Rev. Immunol. 2, 11-19. [DOI] [PubMed] [Google Scholar]
- 3.Kyewski, B., Derbinski, J., Gotter, J. & Klein, L. (2002) Trends Immunol. 23, 364-371. [DOI] [PubMed] [Google Scholar]
- 4.Ramsey, C., Winqvist, O., Puhakka, L., Halonen, M., Moro, A., Kampe, O., Eskelin, P., Pelto-Huikko, M. & Peltonen, L. (2002) Hum. Mol. Genet. 11, 397-409. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, M. S., Venanzi, E. S., Klein, L., Chen, Z., Berzins, S., Turley, S. J., von Boehmer, H., Bronson, R., Dierich, A., Benoist, C., et al. (2002) Science 298, 1395-1401. [DOI] [PubMed] [Google Scholar]
- 6.Gibson, T. J., Ramu, C., Gemund, C. & Aasland, R. (1998) Trends Biochem. Sci. 23, 242-244. [DOI] [PubMed] [Google Scholar]
- 7.Bottomley, M. J., Collard, M. W., Huggenvik, J. I., Liu, Z., Gibson, T. J. & Sattler, M. (2001) Nat. Struct. Biol. 8, 626-633. [DOI] [PubMed] [Google Scholar]
- 8.Bjorses, P., Halonen, M., Palvimo, J. J., Kolmer, M., Aaltonen, J., Ellonen, P., Perheentupa, J., Ulmanen, I. & Peltonen, L. (2000) Am. J. Hum. Genet. 66, 378-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyoshi, H., Hatakeyama, S., Pitkanen, J., Mouri, Y., Doucas, V., Kudoh, J., Tsurugaya, K., Uchida, D., Matsushima, A., Oshikawa, K., et al. (2004) J. Biol. Chem. 279, 33984-33991. [DOI] [PubMed] [Google Scholar]
- 10.Nagamine, K., Peterson, P., Scott, H. S., Kudoh, J., Minoshima, S., Heino, M., Krohn, K. J., Lalioti, M. D., Mullis, P. E., Antonarakis, S. E., et al. (1997) Nat. Genet. 17, 393-398. [DOI] [PubMed] [Google Scholar]
- 11.Pitkanen, J., Vahamurto, P., Krohn, K. & Peterson, P. (2001) J. Biol. Chem. 276, 19597-19602. [DOI] [PubMed] [Google Scholar]
- 12.Pitkanen, J., Doucas, V., Sternsdorf, T., Nakajima, T., Aratani, S., Jensen, K., Will, H., Vahamurto, P., Ollila, J., Vihinen, M., et al. (2000) J. Biol. Chem. 275, 16802-16809. [DOI] [PubMed] [Google Scholar]
- 13.Uchida, D., Hatakeyama, S., Matsushima, A., Han, H., Ishido, S., Hotta, H., Kudoh, J., Shimizu, N., Doucas, V., Nakayama, K. I., et al. (2004) J. Exp. Med. 199, 167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson, P., Pitkanen, J., Sillanpaa, N. & Krohn, K. (2004) Clin. Exp. Immunol. 135, 348-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong, A. W., Brickey, W. J., Taxman, D. J., van Deventer, H. W., Reed, W., Gao, J. X., Zheng, P., Liu, Y., Li, P., Blum, J. S., et al. (2003) Nat. Immunol. 4, 891-898. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B. & Speed, T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallegos, A. M. & Bevan, M. J. (2004) J. Exp. Med. 200, 1039-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liston, A., Gray, D. H., Lesage, S., Fletcher, A. L., Wilson, J., Webster, K. E., Scott, H. S., Boyd, R. L., Peltonen, L. & Goodnow, C. C. (2004) J. Exp. Med. 200, 1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfonso, C. & Karlsson, L. (2000) Annu. Rev. Immunol. 18, 113-142. [DOI] [PubMed] [Google Scholar]
- 20.Petrie, H. T. (2003) Nat. Rev. Immunol. 3, 859-866. [DOI] [PubMed] [Google Scholar]
- 21.LeibundGut-Landmann, S., Waldburger, J. M., Krawczyk, M., Otten, L. A., Suter, T., Fontana, A., Acha-Orbea, H. & Reith, W. (2004) Eur. J. Immunol. 34, 1513-1525. [DOI] [PubMed] [Google Scholar]
- 22.Ting, J. P. & Trowsdale, J. (2002) Cell 109, S21-S33. [DOI] [PubMed] [Google Scholar]
- 23.Volz, A., Korge, B. P., Compton, J. G., Ziegler, A., Steinert, P. M. & Mischke, D. (1993) Genomics 18, 92-99. [DOI] [PubMed] [Google Scholar]
- 24.Salomon, B., Rhee, L., Bour-Jordan, H., Hsin, H., Montag, A., Soliven, B., Arcella, J., Girvin, A. M., Padilla, J., Miller, S. D., et al. (2001) J. Exp. Med. 194, 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klug, D. B., Carter, C., Crouch, E., Roop, D., Conti, C. J. & Richie, E. R. (1998) Proc. Natl. Acad. Sci. USA 95, 11822-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato, K., Sato, U., Tateishi, S., Kubo, K., Horikawa, R., Mimura, T., Yamamoto, K. & Kanda, H. (2004) Biochem. Biophys. Res. Commun. 318, 935-940. [DOI] [PubMed] [Google Scholar]
- 27.Zhu, X. S. & Ting, J. P. (2001) Mol. Cell. Biol. 21, 7078-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst, L. D., Pal, C. & Lercher, M. J. (2004) Nat. Rev. Genet. 5, 299-310. [DOI] [PubMed] [Google Scholar]
- 29.Boutanaev, A. M., Kalmykova, A. I., Shevelyov, Y. Y. & Nurminsky, D. I. (2002) Nature 420, 666-669. [DOI] [PubMed] [Google Scholar]
- 30.Spellman, P. T. & Rubin, G. M. (2002) J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen, B. A., Mitra, R. D., Hughes, J. D. & Church, G. M. (2000) Nat. Genet. 26, 183-186. [DOI] [PubMed] [Google Scholar]
- 32.Su, A. I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K. A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabellini, D., Tupler, R. & Green, M. R. (2003) Curr. Opin. Genet. Dev. 13, 239-245. [DOI] [PubMed] [Google Scholar]
- 34.Gotter, J., Brors, B., Hergenhahn, M. & Kyewski, B. (2004) J. Exp. Med. 199, 155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gourley, T. S., Patel, D. R., Nickerson, K., Hong, S. C. & Chang, C. H. (2002) J. Immunol. 168, 4414-4419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.