Abstract
Reverse transcriptase PCR (RT-PCR) consistently detected bovine leukemia virus transcripts in fresh cells, and competitive RT-PCR enumerated these transcripts. The detection of transcripts in limited numbers of tumor cells indicated that expression occurs in a minority of cells. The data suggest that individual cells contain hundreds of copies of the tax/rex transcript in vivo.
Bovine leukemia virus (BLV), a B-cell lymphotropic retrovirus of the BLV–human T-cell leukemia virus (HTLV) genus, causes a persistent B-lymphocytosis (PL) and/or a neoplastic proliferation of B lymphocytes in a minority of infected animals after prolonged infection (reviewed in reference 28). Virus expression in vivo is highly restricted throughout disease progression and in leukosis. This has made it difficult to identify a discrete mechanism by which BLV can mediate leukemogenesis. Amplification by reverse transcriptase PCR (RT-PCR) has detected BLV mRNAs in freshly isolated peripheral blood mononuclear cells (PBMC) and tumor cells of infected animals (1, 18, 19, 23, 38). Overt virus expression occurs upon cultivation of these cells in vitro.
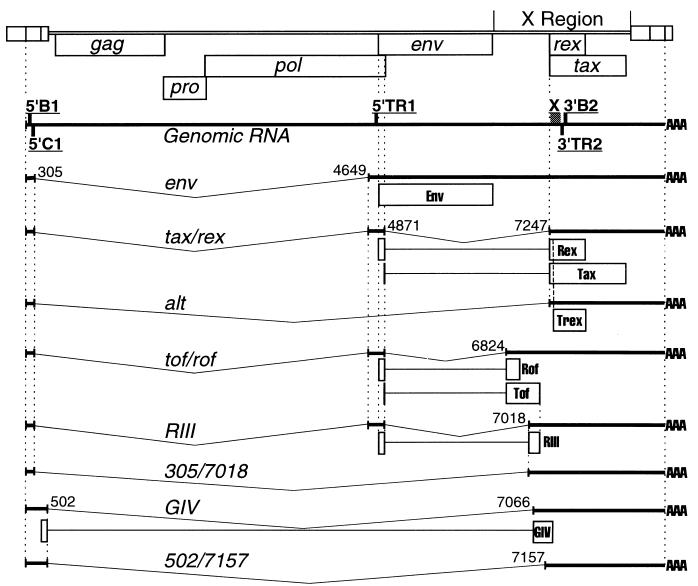
The genome of BLV, like that of HTLV type 1 (HTLV-1), contains a region, X, which encodes proteins that regulate virus expression, have the capacity to mediate host gene expression, and have been implicated in cellular transformation (11, 12, 26, 40, 49, 51). The expression of the open reading frames of the X region is dependent upon spliced transcripts (Fig. 1). pp18 Rex, a transregulator of mRNA processing, and p34 Tax, a transcriptional transactivator, are encoded by overlapping reading frames in the 3′ proximal portion of the X region from a doubly spliced transcript, tax/rex (39, 41, 55), which has been detected in vivo (1, 18, 19, 23, 38). A singly spliced version of the tax/rex transcript, alt, has the second exon excluded and has also been detected in vivo (19). This transcript is capable of encoding a truncated Rex protein (Trex; Fig. 1) which migrates at 14 to 16 kDa (1, 19, 42, 55). The alt transcript is similar to a singly spliced transcript of HTLV-1, which produces a truncated Rex protein, p21 Rex. This splicing pattern is common to all members of the BLV-HTLV genus (34). Several spliced transcripts with the capacity to express 5′ proximal portions of the X region have been identified (1, 7) (Fig. 1). These transcripts are present at low levels in productively infected cells. Their significance is indicated by their conservation in members of the BLV-HTLV genus and the reduction of in vivo replicative capacity of viruses with mutations in this region (7, 10, 27, 52, 54).
FIG. 1.
Representation of the BLV genome, open reading frames, transcripts, and the products of translation of spliced transcripts. The locations of primers are indicated on the genomic RNA, and their identifiers are underlined. Vertical dashed lines align common termini of transcripts, exons, and reading frames. Splice donor and acceptor sites are numbered according to the proviral sequence described by Sagata et al. (42). Names and splice sites of alternate transcripts and their predicted protein products are from references 19 (alt), 7 (tof/rof), and 1 (RIII, 305/7018, GIV, and 502/7157).
A sensitive quantitative competitive (QC) RT-PCR technique utilizes a size- and sequence-matched RNA competitor in order to allow high PCR cycle numbers without loss of quantitation (31). RT-PCR products of competitor constructs with a modified restriction endonuclease sequence are distinguished by restriction digestion. In addition to the quantitation of viral transcripts in a pool of cells, mRNAs can be detected by RT-PCR in limited numbers of cells to assess their distribution in the population (14). BLV expression has been detected, rarely, in individual PBMC by in situ hybridization (30) and RT-PCR on sorted cells (18) and in fresh lymphoid tissues and lymphomas by immunohistochemistry (20).
We sought to quantitate BLV X region transcripts in the PBMC of animals with PL (PL animals) and cells from malignant lymphomas (ML) and to assess the distribution of these transcripts in ML cells. QC RT-PCR was utilized to measure tax/rex and alt transcripts, and direct RT-PCR amplification of RNA from limited numbers of cells was used to enumerate cells with these transcripts.
Detection of BLV X region transcripts.
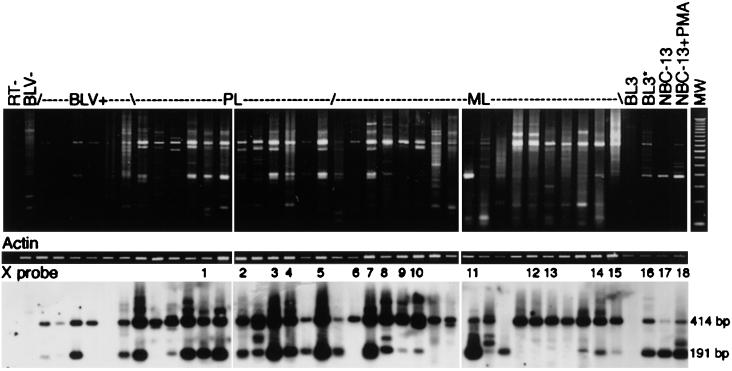
Primers 5′B1 and 3′B2 (Table 1; Fig. 1) are capable of amplifying all reported variations in splicing of BLV X region transcripts (1, 7, 19). Figure 2 shows the products of RT-PCR of 50 ng of RNA with this primer pair after 45 cycles of amplification with a 63°C annealing temperature. All RNAs were treated with DNase I, selected by chromatography on oligo(dT) cellulose, and quantitated by spectrophotometry. Southern blotted amplicons were hybridized with an end-labeled oligonucleotide probe, X (Table 1; Fig. 1), which is also common to known BLV transcripts and is internal to the primer sequences. PBMC and tumor cells were collected from naturally infected animals. Samples included RNAs from the PBMC of one BLV-negative animal, six BLV-positive, hematologically normal animals (non-PL animals), and 12 PL animals and ML cells from 18 animals. Five-nanogram quantities of RNAs from BLV-negative BL3 cells (ATTC CRL 8037), BLV-positive BL3* cells (18) and NBC-13 cells (16), and NBC-13 cells treated with 10 nM phorbol-12-myristate-13-acetate (PMA; Sigma) were also assayed. The BL3* and NBC-13 cell lines were previously single cell cloned by endpoint dilution. Mock cDNA preparations from NBC-13 RNA (RT−) served as a negative control.
TABLE 1.
Sequences and positions of oligonucleotide primers and probes
| Primer | Sequence (5′–3′) | Nucleotide position |
|---|---|---|
| 5′ACT1 | GAGAAGATCTGGCACCACAC | 19–38a |
| 3′ACT2 | AGCCATCTCCTGCTCGAAGT | 456–437a |
| 5′ACTN1 | CTGCGTGTGGCCCCCGAGGA | 52–71a |
| 3′ACTN2 | GGTGCGGCCAGAGGCGTACA | 216–197a |
| 5′B1 | GTGCTCAGCTCTCGGTCCTG | 237–256b |
| 3′B2 | TTCGAATTGGAGTCGCTCAT | 7368–7349b |
| 5′C1 | GAGCTCTCTTGCTCCCGAGA | 256–275b |
| 5′TR1 | CAAAACAATCGTCGGTGGCT | 4705–4724b |
| 3′TR2 | GATGGTGACATCATTGGACA | 7320–7301b |
| 3′TR2Hind | GGCATCGATGGTGACATCATTGGACAAAACCAGGGCCGGGCAAGCTTGTAGc | 7326–7276b |
| 5′ALT | TCCGGCAGCGGTCAG/CAAGTd | 291–305/7247–7251b |
| X probe | GTTGTTGGTTGGGGGCCCCACTCTCTACA | 7252–7280b |
| ACT probe | CCTCTGAACCCCAAGGCCAACCGTGAGAAGATGACCCAGATC | 97–138a |
FIG. 2.
Amplification of X region transcripts. Fifty-nanogram quantities of DNase I-treated, poly(A)+ RNA preparations were amplified by RT-PCR with BLV primers 5′B1 and 3′B2 (top panel) or with actin primers 5′ACT1 and 3′ACT2 (middle panel). The bottom panel shows the results of hybridization of BLV amplicons with X probe. Samples include PBMC from a BLV-seronegative animal (BLV−), PBMC from seropositive, non-PL animals (BLV+), PBMC from seropositive, PL animals (PL), and ML cells. Five-nanogram quantities of control poly(A)+ RNA preparations were used (BL3, BL3*, NBC-13, and NBC-13 plus PMA). The numbers over the bottom panel indicate samples which were subsequently subjected to QC RT-PCR. The molecular weight (MW) lane contained a 50-bp ladder. The amplified products of tax/rex and alt transcripts are 414 and 191 bp, respectively.
Products corresponding to the tax/rex and alt transcripts (414 and 191 bp, respectively) were detected in most, but not all, samples tested (Fig. 2). tax/rex was detected in five of six samples from non-PL animals, and alt was detected in four of six. In PL samples, tax/rex was detected in all samples, and alt was detected in 11 of 12. All of the tumor samples contained BLV transcripts, but one contained alt and not tax/rex, and 12 of 18 contained alt. Additional RT-PCR products, with sizes corresponding to previously detected BLV transcripts as well as to unidentified transcripts, were observed to varying degrees in PL and ML samples.
The detection of tax/rex and alt transcripts in fresh cells has been observed previously (1, 18, 19, 23, 38). One tumor sample was negative for tax/rex, and although it was positive for alt, this example further weakens the case for viral protein function in the maintenance of the transformed state. At this stage of disease the cell genome is marked by chromosomal abnormalities (43), which could account for this phenotype. BLV transcripts were not detected in cells from all BLV-positive, non-PL animals, but few of the PBMC from these animals are infected (9, 32); the inability to detect BLV transcripts in all samples at this stage of disease could be due to limitations of the assay.
BLV transcription commonly occurs upon in vitro cultivation of infected cells. To address the possible onset of transcription ex vivo, the following conditions were used during the collection of tumors: (i) tumors were excised within 10 min of the death of the animal; (ii) tumor masses were immediately frozen on dry ice or cut with scissors in phosphate-buffered saline on ice to release cells into suspension, and (iii) 500 μg of actinomycin D per ml was included in cell suspension buffers, in selected samples, to block transcription. No differences were observed for such preparations, in either the detection or relative levels of the BLV transcripts, compared to identical samples prepared at ambient temperatures and without actinomycin D (data not shown).
Figure 2 also shows the products of RT-PCR of the same poly(A)+ RNA preparations with primers 5′ACT1 and 3′ACT2 (Table 1) for 22 cycles in separate reactions. These primers yield a 437-bp amplicon from bovine β-actin mRNA. Amplification of serial twofold RNA dilutions showed that 22 cycles provided a linear response of amplicon quantity to input RNA in these reactions (data not shown). This allowed an evaluation of the RNAs, quantitated by spectrophotometry, in the context of RT-PCR.
QC RT-PCR of BLV transcripts.
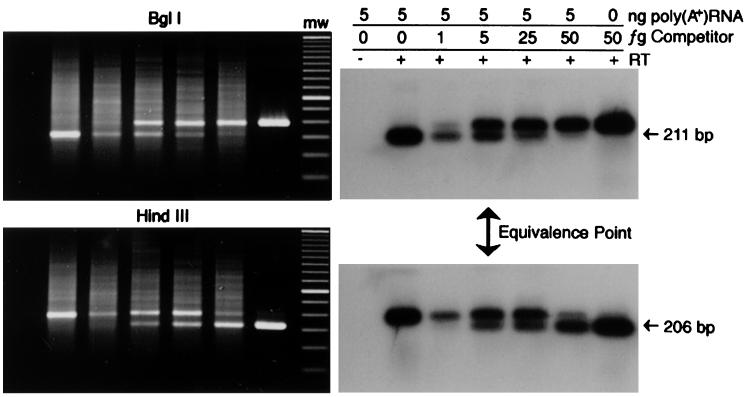
Analysis of the levels of viral gene transcription in vivo focused on the tax/rex and alt transcripts. Clones of these transcripts were prepared in pBluescript SK(−) (Stratagene) by RT-PCR of RNA from persistently infected fetal lamb kidney cells (FLK-BLV [48]) with primers 5′C1 and 3′TR2Hind (Table 1; Fig. 1). 3′TR2Hind introduces two point mutations, which alter a native BglI site to a HindIII site. Competitor RNAs transcribed from these clones were quantitated by spectrophotometry, and their sizes were confirmed by gel electrophoresis. Competitor dilutions were included in RT-PCRs of 5 ng of DNase I-treated, poly(A)+, cellular RNAs. Specific amplification of either the tax/rex or alt transcript was achieved with the common 3′ primer 3′TR2 and a 5′ primer in the second exon of tax/rex, 5′TR1, or a 5′ primer which crosses the alt splice junction, 5′ALT (Table 1; Fig. 1), and 40 cycles of amplification with 63°C annealing. Twenty-microliter aliquots of QC RT-PCR products were digested with 5 U of HindIII or BglI for 2 h at 37°C and subjected to electrophoresis. Southern blots were hybridized with the end-labeled oligonucleotide probe X. The quantity of native transcripts was determined to be equal to the quantity of competitor when the products of BglI and HindIII digestion, 211 and 206 bp, respectively, were equivalent.
Figure 3 shows an example of quantitation of 5 ng of NBC-13 poly(A)+ RNA, where the tax/rex transcript quantity was assessed at 5 fg of competitor. Samples were first tested against fivefold competitor dilutions in order to define a range of appropriate twofold dilutions. Results are presented in Table 2 as femtogram equivalents of competitor RNA per nanogram of cellular RNA and corresponding estimates of copies of mRNA per cell. These estimates were based upon an approximation of 500 fg of total mRNA per cell (from typical lymphocyte yields of poly[A]+ RNA). Molecules of individual transcripts or competitor RNAs per femtogram were calculated by their lengths (tax/rex mRNA, 1,800 bases; tax/rex competitor RNA, 398 bases; alt mRNA, 1,600 bases; alt competitor RNA, 176 bases).
FIG. 3.
Quantitation of tax/rex transcript in NBC-13 cells. BglI- or HindIII-digested products of QC RT-PCR were subjected to electrophoresis and Southern analysis with the X probe. The input of cellular RNA, in nanograms of poly(A)+ RNA, and competitor RNA, in femtograms of competitor, and the presence of RT in reaction mixtures are shown above the autoradiographs. An arrow indicates the relative equivalence point. The molecular weight (mw) lane contains a 50-bp ladder with an intense band at 350 bp. BglI digestion yields a 211-bp fragment from tax/rex transcript amplicons, and HindIII digestion yields a 206-bp band from tax/rex competitor RNA amplicons.
TABLE 2.
Quantitation of tax/rex and alt transcripts
| Sample
|
tax/rex
|
alt
|
|||
|---|---|---|---|---|---|
| No. | Typea | Ratio (fg/ngb) | Copy no.c | Ratio (fg/ng) | Copy no. |
| 1 | PL | 1 | 2 | 0.02 | 0.10 |
| 2 | PL | 0.5 | 1 | 0.008 | 0.04 |
| 3 | PL | 0.5 | 1 | 0.02 | 0.10 |
| 4 | PL | 0.5 | 1 | 0.004 | 0.02 |
| 5 | PL | 1 | 2 | 0.02 | 0.10 |
| 6 | ML | 0.1 | 0.2 | 0.00 | 0.00 |
| 7 | ML | 0.5 | 1 | 0.01 | 0.05 |
| 8 | ML | 1 | 2 | 0.02 | 0.10 |
| 9 | ML | 0.5 | 1 | 0.004 | 0.02 |
| 10 | ML | 0.1 | 0.2 | 0.004 | 0.02 |
| 11 | ML | 0.1 | 0.2 | 1 | 5 |
| 12 | ML | 0.5 | 1 | 0 | 0 |
| 13 | ML | 0.5 | 1 | 0 | 0 |
| 14 | ML | 2.5 | 5 | 0.01 | 0.05 |
| 15 | ML | 2 | 4 | 0.04 | 0.2 |
| 16 | BL3* | 50 | 100 | 50 | 250 |
| 17 | NBC-13 | 1 | 2 | 50 | 250 |
| 18 | NBC-13+PMA | 50 | 100 | 50 | 250 |
PL PBMC, ML, or BLV-infected cells, BL3*, NBC-13, and NBC-13 with PMA.
Femtograms of competitor RNA equivalent to 1 ng of input RNA determined from twofold dilutions.
Estimates of copy number per cell based on a theoretical yield of 500 fg of mRNA per cell.
A total of 18 samples, indicated by numbers in Fig. 2, were quantitated (Table 2). Samples included PBMC from five PL animals, 10 tumor samples, and the BLV-positive cell lines BL3*, NBC-13, and PMA-treated NBC-13. Quantities of the tax/rex transcript were consistent in PL PBMC at approximately one to two copies per cell. In tumor cells, tax/rex levels varied, from approximately 0.2 to 5 copies per cell. Levels of the alt transcript were significantly lower, with copy numbers in the range of 0.02 to 0.1 per cell. Most of the samples contained less than one copy of the alt transcript per cell, and three tumor samples were negative for alt, but one tumor contained approximately five copies per cell. Quantities of transcripts lower than a single copy per cell suggest that these transcripts are present only in a subpopulation of the cells.
Levels of the tax/rex transcript in the NBC-13 cell line were comparable to those observed in tumor cells. This cell line can be stimulated with PMA to induce BLV expression (24), and such stimulation, here, increased tax/rex levels in these cells approximately 50-fold to levels similar to those observed in productively infected BL3* cells. NBC-13 and BL3* cells had approximately 250 copies of the alt transcript per cell, and the levels of the alt transcript were not significantly enhanced by treatment of NBC-13 cells with PMA.
RT-PCR of limited numbers of cells.
The detection of transcripts in limited numbers of tumor cells in a direct lysis procedure allowed the evaluation of the distribution of the tax/rex and alt transcripts in this cell population. Selected tumors were 3 to 5 cm in diameter, and gross tissue was completely replaced by neoplastic cells. Suspensions of cells were derived from the interior of the tumor mass by cutting fresh, decapsulated tissues with scissors in phosphate-buffered saline supplemented with actinomycin D. Cells were diluted to appropriate concentrations to yield fixed cell numbers in 1-μl aliquots and distributed into wells of a 96-well plate on ice. Cell numbers were visually confirmed. The cells were lysed by the addition of 9 μl of 0.025% Nonidet P-40 (Sigma), 2 mM dithiothreitol, and 1 U of RNasin per μl, incubated on ice for 20 min, and frozen at −80°C. Lysates were thawed on ice, supplemented with 2 μl of 25 mM Tris-HCl (pH 8.3), 37.5 mM KCl, 1.5 mM MgCl2, and 1 U of DNase I per μl, and incubated for 15 min at room temperature. Three microliters of 1 mM EDTA was added, and the lysates were heated to 65°C for 10 min and chilled on ice. After the addition of 10 μl of buffer containing 125 mM Tris-HCl (pH 8.3), 187.5 mM KCl, 7.5 mM MgCl2, a 62.5 mM concentration of each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2.5 U of RNasin per μl, 5 μM primer 3′B2, 0.25 μM primer 3′ACT2 (Table 1; Fig. 1), and 10 U of RT (Superscript II; Life Technologies), samples were incubated for 1 h at 45°C followed by 10 min at 70°C. Negative controls received the same mix prior to the addition of RT. cDNA preparations were overlaid with a PCR mix containing 2 μM primer 5′B1 and 0.1 μM primer 5′ACT1 (Table 1) and amplified for 45 cycles with a 63°C annealing temperature. One microliter of a 1:100 dilution of the product of this PCR was reamplified with nested primer pairs for either tax/rex (5′TR1 and 3′TR2; 2 μM), alt (5′ALT and 3′TR2; 2 μM), or β-actin (5′ACTN1 and 3′ACTN2; 0.1 μM; Table 1) for 35 cycles, and 20 μl was subjected to electrophoresis and Southern analysis with the oligonucleotide probe X or ACT (Table 1).
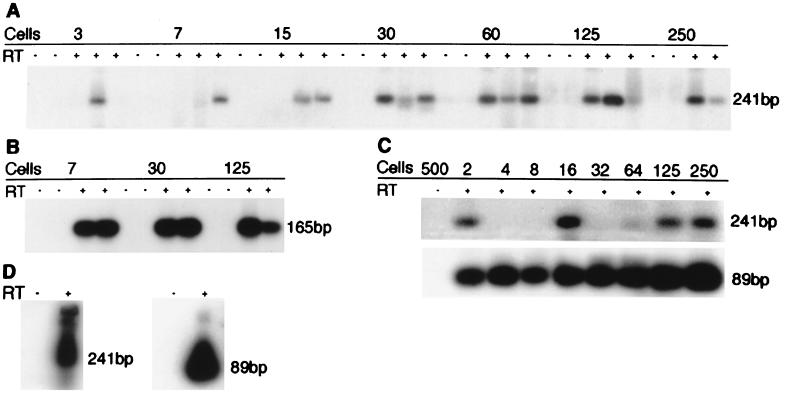
Figure 4 shows an example of the products from tax/rex amplification of serial twofold cell dilutions. Only three of five preparations of each dilution received RT. The number of cells required to yield a consistent PCR signal in three of three samples with RT was interpreted as containing, at least, a single cell with tax/rex transcripts. This cutoff provides a conservative estimation of the numbers of positive cells within the population. Signals obtained in fewer than three of three dilutions suggested that individual or few cells contained sufficient transcripts for detection. This assay detected β-actin in single tumor cells. Results for the five tumors tested ranged from 1 cell in 30 (3.3%; sample shown in Fig. 4) to 1 in 250 (0.4%), with an average of 1 in 125 cells or 0.8%. In similar assays the tax/rex transcript was limited to approximately 5% of NBC-13 cells and present in single BL3* cells (Fig. 4). The alt transcript was rarely detected in fewer than 100 tumor cells (not shown) but was readily detected in single NBC-13 or BL3* cells (Fig. 4).
FIG. 4.
RT-PCR of limited numbers of BLV-infected cells. Twofold serial dilutions of cells were lysed and subjected to RT-PCR for BLV or β-actin transcripts. The actual numbers of cells in dilutions under 50 were confirmed visually and varied by no more than three cells. (A) Five samples at each dilution of ML cells were tested for tax/rex transcripts, two without and three with RT enzyme. The products of nested primers, 5′TR1 and 3′TR2, are 241 bp. (B) Samples were the same as in panel A but were amplified with nested β-actin primers 5′ACTN1 and 3′ACTN2, which yield a 165-bp product. (C) RT-PCR of lysates from serial twofold dilutions of NBC-13 cells with tax/rex-specific primers 5′TR1 and 3′TR2 (241 bp) or alt specific primers 5′ALT and 3′TR2 (89 bp). (D) Amplification of two BL3* cells as described for panel C. Southern blots were hybridized with X probe or actin probe (Table 1).
The possibility that the positive cells present in the tumor mass do not belong to the neoplastic pool cannot be excluded. Such tumors do contain low levels of infiltrating T lymphocytes as determined by immunohistology (6), but these cells have not been found to be significant targets of BLV infection (32) and are unlikely to be responsible for the signal observed in the tumor cell preparations.
Further characterization of NBC-13 cells.
BLV expression in NBC-13 cells is restricted and can be induced with PMA (24). Analysis of NBC-13 DNA revealed the presence of a full-length provirus and one with a deletion of 4,620 bp between positions 2217 and 6837 (data not shown). The deletion precludes the expression of tax/rex, but not singly spliced alt, from this provirus.
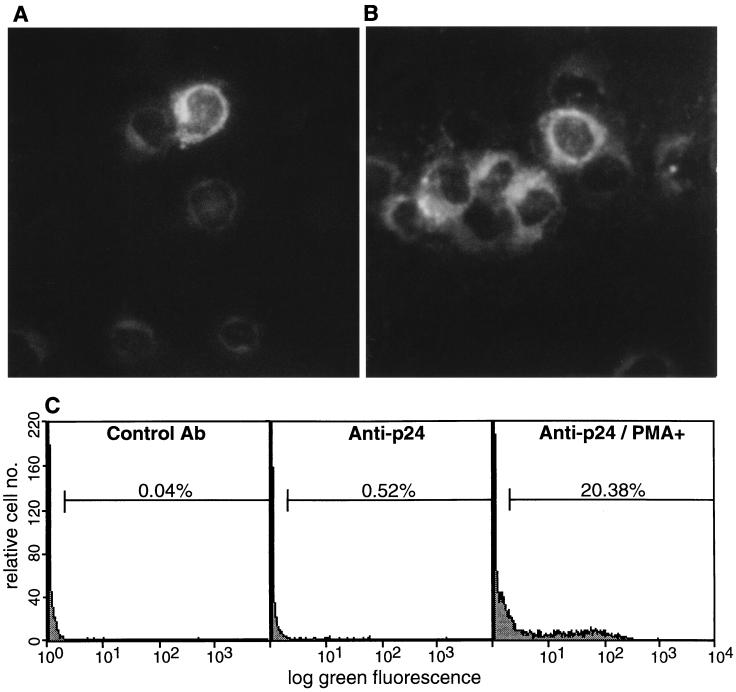
Immunofluorescence assays of acetone/methanol-fixed preparations of NBC-13 cells with monoclonal anti-BLV p24 (4′G9) demonstrated the presence of a small number of virus-expressing cells (Fig. 5A). Flow cytometric analysis of 10,000 methanol-fixed NBC-13 cells labelled with anti-p24 demonstrated that 0.5% of this clonal cell line expressed BLV antigen (Fig. 5C). Treatment of these cells with PMA stimulated the expression of p24 in 20% of cells after 24 h (Fig. 5B and C). Analysis regions included less than 0.04% of antibody controls.
FIG. 5.
Indirect immunofluorescence staining of NBC-13 cells. (A) Photomicrograph of a rare positive cell, stained with a monoclonal antibody for BLV p24 and goat anti-mouse immunoglobulin G-fluorescein isothiocyanate, prior to PMA treatment (magnification, ×600; Evans blue counterstain). (B) NBC-13 cells 24 h after treatment with 10 nM PMA (magnification, ×600). (C) Flow cytometric analysis of NBC-13 cell preparations as described for panels A and B stained with a control monoclonal antibody (Ab) or with anti-p24.
The alt transcript is persistently expressed in NBC-13 cells. The Rex amino terminus, deleted with the second exon in the singly spliced alt transcript, includes an arginine-rich region implicated in RNA binding and nuclear localization. The HTLV-1 protein homolog p21X has been identified in tumor cells and transformed cell lines (3, 35). p21X is localized in the cytoplasm, and its HTLV-2 homolog has been demonstrated to act as a dominant-negative inhibitor of Rex function (8, 29). Considering that the Rex proteins of BLV and HTLV-1 are functionally interchangeable (15), the BLV truncated Rex protein could also act as an inhibitor of Rex function. Such a mechanism could explain the restricted BLV expression observed in NBC-13 cells, and the ability to manipulate this expression experimentally offers an in vitro model for the control of BLV expression.
The work presented here addresses the long-standing paradox of pathogenesis in BLV and HTLV-1: the paucity of expression of virus. While much effort has been expended to define the oncogenicity of proteins encoded by these viruses, evidence for significant levels of virus expression during disease progression is lacking. While it remains clear that BLV expression in vivo is limited, the results demonstrate the continued presence of X region transcripts in significant numbers of cells. The tax/rex transcript was present at levels approximating 0.2 to 5 copies per cell in PL PBMC and ML cells, and detection of this transcript in limited numbers of tumor cells indicated expression in a minimum of 0.8% of cells. In light of the estimates of copy numbers, the results suggest that these cells actually contained hundreds of copies of the tax/rex transcript and could have contained functional levels of tax/rex protein products. Limited virus expression was also observed in NBC-13 cells, and these results demonstrate that variation in BLV expression exists in cloned cell populations. This selective viral gene expression was further demonstrated in NBC-13 cells by immunofluorescence. The mechanism whereby viral gene transcription is restricted to a subpopulation of clonal cells is unknown. A possible distinction of these cells is their position in the cell cycle. BLV expression has been shown to be cell cycle dependent in FLK-BLV cells (45) and associated with the induction of cell division by PMA in NBC-13 cells (24). BLV transcription could also be associated with apoptosis.
A product of translation of the tax/rex transcript, p34 Tax, has the demonstrated capacity to affect cell function. It facilitates the activation of cyclic AMP response element-binding protein (CREB) to transactivate viral gene transcription (4, 13, 26, 51, 53). Tax has immortalization potential in cell culture (50, 51) and activates the transcription of cellular genes via CREB (26). The HTLV-1 Tax protein, in addition to the transactivation of viral gene transcription via CREB, can activate or repress the transcription of cellular genes involved in the control of cell proliferation (2, 17, 21, 46), DNA repair (22), tumor suppression (5), and apoptosis (47), induce cell division by interacting directly with proteins, such as p53 (36, 37), p16INK4a (44), and cyclin D (33), and disrupt the mitotic checkpoint by binding to human mitotic arrest-defective protein 1 (25). The loss of this checkpoint allows the premature onset of anaphase and the development of karyotypic abnormalities similar to those observed in malignancy. If the Tax protein of BLV is capable of functions similar to that of HTLV-1, then it has the capacity to promote proliferation and perturb host chromosomal integrity. The evidence presented here shows that the tax/rex gene of BLV is transcribed throughout infection in significant quantities in a minority of infected cells. The interrelationship between selective viral gene expression and BLV disease progression remains enigmatic.
Acknowledgments
This work was supported in part by USDA grant 93-37204-9213. J.R. was supported by USDA National Needs Graduate Fellowship 92-38420-7366.
We thank Richard A. Reyes, Gary L. Cockerell, and Thomas J. Divers for the generous provision of samples and Craig Schultz and the USDA staff at the Taylor Packing Co. for support in the rapid collection of tumor material. NBC-13 cells were provided by Jorge F. Ferrer, and 4′G9 anti-p24 monoclonal antibody was provided by Daniel Portetelle.
REFERENCES
- 1.Alexandersen S, Carpenter S, Christensen J, Storgaard T, Viuff B, Wannemuehler Y, Belousov J, Roth J A. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J Virol. 1993;67:39–52. doi: 10.1128/jvi.67.1.39-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Béraud C, Sun S-C, Ganchi P, Ballard D W, Greene W C. Human T-cell leukemia virus type I Tax associates with and is negatively regulated by the NF-κB2 p100 gene product: implications for viral latency. Mol Cell Biol. 1994;14:1374–1382. doi: 10.1128/mcb.14.2.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat N K, Adachi Y, Samuel K P, Derse D. HTLV-1 gene expression by defective proviruses in an infected T-cell line. Virology. 1993;196:15–24. doi: 10.1006/viro.1993.1450. [DOI] [PubMed] [Google Scholar]
- 4.Boros I M, Tie F, Giam C-Z. Interaction of bovine leukemia virus transactivator Tax with bZip proteins. Virology. 1995;214:207–214. doi: 10.1006/viro.1995.9939. [DOI] [PubMed] [Google Scholar]
- 5.Brauweiler A, Garrus J E, Reed J C, Nyborg J K. Repression of bax gene expression by the HTLV-I Tax protein: implications for suppression of apoptosis in virally infected cells. Virology. 1997;231:135–140. doi: 10.1006/viro.1997.8509. [DOI] [PubMed] [Google Scholar]
- 6.Chiba T, Ajito T, Okada K. Phenotype analysis of lymphocytes present in different stages of neoplasia induced by bovine leukemia virus. Leukemia. 1994;8:S206–S210. [PubMed] [Google Scholar]
- 7.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciminale V, Zotti L, D’Agostino D M, Chieco-Bianchi L. Inhibition of human T-cell leukemia virus type 2 Rex function by truncated forms of Rex encoded in alternatively spliced mRNAs. J Virol. 1997;71:2810–2818. doi: 10.1128/jvi.71.4.2810-2818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cockerell G L, Rovnak J. The correlation between the direct and indirect detection of bovine leukemia virus infection in cattle. Leuk Res. 1988;12:465–469. doi: 10.1016/0145-2126(88)90112-9. [DOI] [PubMed] [Google Scholar]
- 10.Cockerell G L, Rovnak J, Green P L, Chen I S Y. A deletion in the proximal untranslated pX region of human T-cell leukemia virus type II decreases viral replication but not infectivity in vivo. Blood. 1996;87:1030–1035. [PubMed] [Google Scholar]
- 11.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derse D. trans-acting regulation of bovine leukemia virus mRNA processing. J Virol. 1988;62:1115–1119. doi: 10.1128/jvi.62.4.1115-1119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse D, Casey J W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986;231:1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- 14.Edmands S, Kirk J, Lee A, Radich J. Rapid RT-PCR amplification from limited cell numbers. PCR Methods Appl. 1994;3:317–319. doi: 10.1101/gr.3.6.317. [DOI] [PubMed] [Google Scholar]
- 15.Felber B K, Derse D, Athanassopoulos A, Campbell M, Pavlakis G N. Cross-activation of the Rex proteins of HTLV-I and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol. 1989;1:318–328. [PubMed] [Google Scholar]
- 16.Ferrer J F, Stock N D, Lin P. Detection of replicating C-type viruses in continuous cell cultures established from cows with leukemia: effect of the culture medium. J Natl Cancer Inst. 1971;47:613–621. [PubMed] [Google Scholar]
- 17.Fujii M, Sassone-Corsi P, Verma I M. c-fos promoter trans-activation by the tax1 protein of human T-cell leukemia virus type 1. Proc Natl Acad Sci USA. 1988;85:8526–8530. doi: 10.1073/pnas.85.22.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaynor E M, Mirsky M L, Lewin H A. Use of flow cytometry and RT-PCR for detecting gene expression by single cells. BioTechniques. 1996;21:286–291. doi: 10.2144/96212rr02. [DOI] [PubMed] [Google Scholar]
- 19.Haas L, Divers T, Casey J W. Bovine leukemia virus gene expression in vivo. J Virol. 1992;66:6223–6225. doi: 10.1128/jvi.66.10.6223-6225.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heeny J L, Valli P J S, Jacobs R M, Valli V E O. Evidence for bovine leukemia virus infection of peripheral blood monocytes and limited antigen expression in bovine lymphoid tissue. Lab Investig. 1992;66:608–617. [PubMed] [Google Scholar]
- 21.Hirai H, Suzuki T, Fujisawa J, Inoue J, Yoshida M. Tax protein of human T-cell leukemia virus type I binds to the ankyrin motifs of inhibitory factor kappa B and induces nuclear translocation of transcription factor NF-kappaB proteins for transcriptional activation. Proc Natl Acad Sci USA. 1994;91:3584–3588. doi: 10.1073/pnas.91.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeang K-T, Widen S G, Semmes O J, Wilson S H. HTLV-I transactivator protein, Tax, is a trans-repressor of the human beta-polymerase gene. Science. 1990;247:1082–1084. doi: 10.1126/science.2309119. [DOI] [PubMed] [Google Scholar]
- 23.Jensen W A, Rovnak J, Cockerell G L. In vivo transcription of the bovine leukemia virus tax/rex region in normal and neoplastic lymphocytes of cattle and sheep. J Virol. 1991;65:2484–2490. doi: 10.1128/jvi.65.5.2484-2490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen W A, Wicks-Beard B J, Cockerell G L. Inhibition of protein kinase C results in decreased expression of bovine leukemia virus. J Virol. 1992;66:4427–4433. doi: 10.1128/jvi.66.7.4427-4433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin D-Y, Spencer F, Jeang K-T. Human T-cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 26.Katoh I, Yoshinaka Y, Ikawa Y. Bovine leukemia virus trans-activator p34Tax activates heterologous promoters with a common sequence known as a cAMP-responsive element or the binding site of a cellular transcription factor ATF. EMBO J. 1989;8:497–503. doi: 10.1002/j.1460-2075.1989.tb03403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkhofs P, Heremans H, Burny A, Kettman R, Willems L. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J Virol. 1998;72:2554–2559. doi: 10.1128/jvi.72.3.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammericks M, Willems L, Portetelle D. Bovine leukemia virus. Vol. 3. New York, N.Y: Plenum Press; 1994. [Google Scholar]
- 29.Kubota S, Hatanaka M, Pomerantz R J. Nucleo-cytoplasmic redistribution of the HTLV-I rex protein: alterations by coexpression of the HTLV-I p21x protein. Virology. 1996;220:502–507. doi: 10.1006/viro.1996.0339. [DOI] [PubMed] [Google Scholar]
- 30.Lagarias D M, Radke K. Transcriptional activation of bovine leukemia virus in blood cells from experimentally infected, asymptomatic sheep with latent infections. J Virol. 1989;63:2099–2107. doi: 10.1128/jvi.63.5.2099-2107.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCulloch R K, Choong C S, Hurley D M. An evaluation of competitor type and size for use in the determination of mRNA by competitive PCR. PCR Methods Appl. 1995;4:219–226. doi: 10.1101/gr.4.4.219. [DOI] [PubMed] [Google Scholar]
- 32.Mirsky M L, Olmstead C A, Da Y, Lewin H A. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J Virol. 1996;70:2178–2183. doi: 10.1128/jvi.70.4.2178-2183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuveut C, Low K G, Maldarelli F, Schmitt I, Majone F, Grassmann R, Jeang K-T. Human T-cell leukemia virus type 1 Tax and cell cycle progression: role of cyclin D-cdk and p110Rb. Mol Cell Biol. 1998;18:3620–3632. doi: 10.1128/mcb.18.6.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orita O, Sato S, Aono Y, Minoura N, Yamashita T, Hinuma Y, Igarashi H. Identification of novel singly spliced pX mRNA transcripts common to all human T-cell leukemia virus type 1-related retroviruses. Virus Genes. 1993;7:197–204. doi: 10.1007/BF01702399. [DOI] [PubMed] [Google Scholar]
- 35.Orita S, Kobayashi H, Aono Y, Saiga A, Maeda M, Igarashi H. p21X mRNA is expressed as a singly spliced transcript from defective provirus genomes having a partial deletion of the pol-env region in human T-cell leukemia virus type 1-infected cells. Nucleic Acids Res. 1993;21:3799–3807. doi: 10.1093/nar/21.16.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pise-Masison C A, Radonovich M, Sakaguchi K, Appella E, Brady J N. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J Virol. 1998;72:6348–6355. doi: 10.1128/jvi.72.8.6348-6355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid R L, Lindholm P F, Mireskandari A, Dittmer J, Brady J N. Stabilization of wildtype p53 in human T lymphocytes transformed by HTLV-1. Oncogene. 1993;8:3029–3036. [PubMed] [Google Scholar]
- 38.Reyes R A, Cockerell G L. Unintegrated bovine leukemia virus DNA: association with viral expression. J Virol. 1996;70:4961–4965. doi: 10.1128/jvi.70.8.4961-4965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice N R, Stephens R M, Couez D, Deschamps J, Kettmann R, Burny A, Gilden R. The nucleotide sequence of the env and post-env region of bovine leukemia virus. Virology. 1984;138:82–93. doi: 10.1016/0042-6822(84)90149-1. [DOI] [PubMed] [Google Scholar]
- 40.Rosen C A, Sodroski J, Kettmann R, Burny A, Haseltine W. Trans activation of the bovine leukemia virus long terminal repeat in BLV-infected cells. Science. 1984;227:320–322. doi: 10.1126/science.2981432. [DOI] [PubMed] [Google Scholar]
- 41.Sagata N, Tsuzuku-Kawamura J, Nagayoshi-Aida M, Shimizu F, Imagawa K I, Ikawa Y. Identification of some biochemical properties of the major XBL gene product of bovine leukemia virus. Proc Natl Acad Sci USA. 1985;82:7879–7883. doi: 10.1073/pnas.82.23.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnurr M W, Carter R F, Dube I D, Valli V E, Jacobs R M. Nonrandom chromosomal abnormalities in bovine lymphoma. Leuk Res. 1994;18:91–99. doi: 10.1016/0145-2126(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Kitao S, Matsushime H, Yoshida M. HTLV-1 tax protein interacts with cyclin-dependent kinase inhibitor p16INK4A and counteracts its inhibitory activity towards CDK4. EMBO J. 1996;15:1607–1614. [PMC free article] [PubMed] [Google Scholar]
- 45.Takashima I, Olson C. Effect of mitogens and anti-bovine leukosis serums on DNA synthesis of lymphocytes from cattle. Eur J Cancer. 1980;16:639–645. doi: 10.1016/0014-2964(80)90204-2. [DOI] [PubMed] [Google Scholar]
- 46.Uittenbogaard M, Armstrong A P, Chiaramello A, Nyborg J K. HTLV-I Tax protein represses gene expression through the bHLH family of transcription factors. J Biol Chem. 1994;269:22466–22469. [PubMed] [Google Scholar]
- 47.Uittenbogaard M, Giebler H A, Reisman D, Nyborg J K. Transcriptional repression of p53 by human T-cell leukemia virus type 1 tax protein. J Biol Chem. 1995;270:28503–28506. doi: 10.1074/jbc.270.48.28503. [DOI] [PubMed] [Google Scholar]
- 48.Van Der Maaten M J, Miller J M. Replication of bovine leukemia virus in monolayer cell cultures. Bibl Haematol. 1976;43:360–362. doi: 10.1159/000399166. [DOI] [PubMed] [Google Scholar]
- 49.Willems L, Gegonne A, Chen G, Burny A, Kettmann R, Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987;6:3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willems L, Grimonpont C, Heremans H, Rebeyrotte N, Chen G, Portetelle D, Burny A, Kettmann R. Mutations in the bovine leukemia virus tax protein can abrogate the long terminal repeat-directed transactivating activity without concomitant loss of transforming potential. Proc Natl Acad Sci USA. 1992;89:3957–3961. doi: 10.1073/pnas.89.9.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems L, Kerkhofs P, Dequiedt F, Portetelle D, Mammerickx M, Burny A, Kettmann R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc Natl Acad Sci USA. 1994;91:11532–11536. doi: 10.1073/pnas.91.24.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willems L, Kettmann R, Chen G, Portetelle D, Burny A, Derse D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J Virol. 1992;66:766–772. doi: 10.1128/jvi.66.2.766-772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willems L, Kettmann R, Dequiedt F, Portetelle D, Vonèche V, Cornil I, Kerkhofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshinaka Y, Oroszian S. Bovine leukemia virus post-envelope gene coded protein: evidence for expression in natural infection. Biochem Biophys Res Commun. 1985;131:347–354. doi: 10.1016/0006-291x(85)91809-1. [DOI] [PubMed] [Google Scholar]