Abstract
Neoadjuvant therapy (NAT) is an important treatment for patients with resectable locally advanced esophageal squamous cell carcinoma (ESCC), but neoadjuvant resistance affects the overall treatment outcome. Therefore, it is particularly important to accurately screen the population for NAT and explore the mechanism of resistance. Usually, different chemotherapy regimens cause different drug resistance mechanisms. Prior to combining immunotherapy with chemotherapy, extensive research has been conducted on previous drug resistance mechanisms. Currently, the mainstream NAT for ESCC involves chemotherapy combined with immunotherapy. We have witnessed the remarkable effect of this combination therapy; however, there are still a considerable number of patients whose tumor tissues show no change or even progress after NAT, and their drug resistance mechanisms remain unclear. Hence, we aim to identify relevant evidence that can distinguish and predict the effectiveness of NAT from a clinical perspective in order to provide a clinical basis for future screening of suitable populations for NAT and discovery of drug resistance mechanisms. This study is based in China's high incidence area of esophageal cancer, where enrolled patients all receive the current mainstream NAT regimen resulting in more reliable outcomes.
Keywords: Esophageal squamous cell carcinoma, Neoadjuvant therapy, Tumor regression grade, Clinical and genetic characteristics
Subject terms: Cancer genomics, Cancer therapy, Cancer, Genetics, Medical research, Oncology
Introduction
Esophageal cancer (EC) is one of the common malignant tumors in the digestive tract. Its incidence rate ranks eighth among all malignant tumors worldwide, and its mortality rate ranks sixth1. In China, squamous cell carcinoma of the esophagus (ESCC), accounting for over 90% of cases, poses a serious threat to human health due to its hidden onset, long duration, and regional characteristics of affected population2,3. Globally, new cases and deaths from EC continue to increase4,5. In 2020, there were approximately 604,000 new cases and 544,000 deaths from EC worldwide. In China alone, there were 324,000 new cases and 301,000 deaths6. Particularly in Linzhou City, China has the highest incidence and mortality rates globally7. Currently, surgical resection remains the primary treatment option for EC8, but a significant number of patients are diagnosed at intermediate or advanced stages where immediate surgery may not completely eradicate the cancerous focus9.
In recent years, with the maturity and promotion of preoperative neoadjuvant chemotherapy, especially the emergence of neoadjuvant chemotherapy combined with immunotherapy, an increasing number of patients with advanced EC have been given the opportunity to undergo surgery. This development greatly encourages doctors and patients10. Currently, as neoadjuvant regimens continue to improve and optimize, some patients' cancer foci even completely disappear (TRG 0) after NAT. This not only reduces the surgical difficulty but also significantly improves the complete resection rate of cancer foci. As a result, it further reduces the risk of postoperative recurrence and metastasis11,12.
Although neoadjuvant therapy (NAT) has benefited most patients, there is still a considerable number of patients who cannot meet the standard for surgical resectability after NAT. In some cases, cancer lesions may even progress during NAT, which not only delays treatment but also increases the difficulty of surgery and may ultimately result in abandoning surgical treatment altogether13. Therefore, more accurate population screening for NAT is necessary14. This retrospective study collected clinical data from esophageal cancer patients who received NAT and combined transcriptome sequencing data from pathological tissues before and after NAT to explore the clinical and genetic characteristics of TRG 0 and TRG III patients after NAT. The goal was to identify their clinical and genetic differences as a foundation for subsequent precise neoadjuvant population screening and drug resistance mechanism mining.
Object and method
Data and criteria of enrolled patients
Studying subjects and medication regimens
Patients with locally advanced ESCC who underwent neoadjuvant therapy (NAT) at the Department of Thoracic Surgery, First Affiliated Hospital of Zhengzhou University, from January 2022 to December 2023 were enrolled.
Both groups received preoperative NAT with the regimen of "cisplatin + paclitaxel (albumin-bound type) + carrelizumab." Cisplatin: 75 mg/m2, intravenous infusion on day 1 every 21 days for two cycles; paclitaxel (albumin-bound type): 125 mg/m2, intravenous infusion on day 1 and day 8, every 21 days for two cycles; carrelizumab: 200 mg, intravenous infusion on day 1 for each cycle, lasting no less than 20 min and no more than 60 min, every 21 days for two cycles.
Case inclusion and exclusion criteria
Inclusion criteria:
Age 45–90 years old, male or female;
ESCC confirmed by pathology;
Patients with locally advanced ESCC who have received two neoadjuvant therapies;
Surgery had been completed and tumor tissue available for transcriptome sequencing;
Surgical plan: radical resection of ESCC with esophagogastric reconstruction and at least 2-field lymph node dissection was performed.
The tumor regression grade was TRG 0 or TRGIII.
Exclusion criteria:
High ESCC (the distance between the upper end of the lesion and the incisor is less than 20 cm);
Multiple systemic lymph node metastasis (except para-cardia lymph node metastasis and left gastric lymph node metastasis);
Patients who did not undergo surgery after NAT;
Patients with missing clinical laboratory and examination data;
The grade of tumor regression was between TRG 0 and TRGIII.
Patients who died after surgery;
TRG classification criteria (NCCN)
TRG 0: no residual cancer cells;
TRG I: single cancer cell or cancer cell cluster;
TRG II: fibrotic response over residual cancer cells;
TRG III: almost no fibrosis, visible large residual cancer;
A total of 54 patients were enrolled in this study, including 33 patients in TRG 0 group and 21 patients in TRG III group. There was no significant difference in gender, age, body mass index, preoperative complications, adverse habits and other related data between the two groups (P > 0.05) (Table 1).
Table 1.
Basic information of enrolled patients.
| Project | Group | P | |
|---|---|---|---|
| TRGa 0 (n = 33) |
TRG III (n = 21) |
||
| Gender | |||
| Male | 23 (69.7%) | 15 (71.43%) | > 0.9999 |
| Female | 10 (30.3%) | 6 (28.57%) | |
| Age (years) | 64.55 ± 8.77 | 62.69 ± 6.67 | 0.4993 |
| BMIb (kg/cm2) | 24.61 ± 3.40 | 24.18 ± 3.12 | 0.6634 |
| Complication | |||
| Fitness | 20 (60.61%) | 16 (76.19%) | 0.3749 |
| Underlying diseasesc | 13 (39.39%) | 5 (23.81%) | |
| Bad hobbies (Male, smoking or/and alcohol) | |||
| Yes | 17 (73.91%) | 10 (66.67%) | 0.7219 |
| No | 6 (26.09%) | 5 (33.33%) | |
aTumor regression classification.
bBody Mass Index.
cHypertension, diabetes mellitus, Coronary heart disease.
Collection and analysis of clinical laboratory and examination data during NAT
Clinical laboratory data included white blood cell count and tumor marker levels during NAT. Clinical examination data included CT and endoscopic ultrasound data before and after NAT to collect information on tumor location, size, invasion level, and lymph node-related imaging. Pathological data mainly focused on the immunohistochemical indicators of tumor tissues before NAT.
Collection and analysis of surgical data and postoperative hospital stay
The surgical data mainly included the visual description of tumors and lymph node status in the surgical field, as well as the operation time, intraoperative blood loss, and duration of postoperative drainage tube indwelling. The overall postoperative hospital stay was defined as the time from completion of the operation to recovery in the hospital after discharge.
Collection and analysis of postoperative pathological data
The postoperative pathological data should include the number of lymph node dissections and the number of patients with lymph node metastasis, as well as information on vascular invasion, nerve invasion, and resection margin invasion.
Transcriptome sequencing of paired pathological tissues before and after NAT
The pathological tissues of 7 patients before and after NAT were randomly selected from each TRG 0 group and TRG III group for transcriptome sequencing to explore the genetic differences between the two groups. In this study, the transcription levels of pathological tissues were detected using the Illumina Novaseq 6000 sequencing platform. Paired tumor tissue RNA was extracted and isolated with TRIzol reagent (MJZol total RNA extraction kit). The concentration and purity of total RNA were immediately detected after extraction from the tissue samples. The integrity of the RNA was assessed by agarose gel electrophoresis, followed by measurement of RIN value. Then mRNA was isolated from total RNA and fragmented into small fragments approximately 300 bp in size. mRNA served as a template for reverse synthesis of cDNA, which was then used for PCR amplification. The purified products were used to construct the final library. Bridge PCR amplification was performed on the Illumina Novaseq 6000 sequencing platform to generate clusters. Significantly differentially expressed genes were selected based on default criteria: FDR < 0.05 & |log2FC|≥ 1. GO database and KEGG database were respectively utilized for gene function enrichment analysis using GO terms and pathway enrichment analysis using KEGG pathways.
Statistical methods
The statistical data from this study were analyzed using SPSS 21.0 software. Univariate analysis was conducted using Chi-square, T test, or Fisher's exact test, while multivariate analysis was performed using a logistic regression model. Unpaired T test was used for analyzing unpaired sample data. A significance level of P < 0.05 was considered statistically significant.
Statement
This project has been approved by the Ethics Committee of Scientific Research and Clinical Experiment at the First Affiliated Hospital of Zhengzhou University, confirming that all methods were performed in accordance with relevant guidelines and regulations. All enrolled patients signed informed consent to ensure their right to make an informed decision.
Retrospective registration
The study has been registered inthe Ethics Committee of Scientific Research and Clinical Experiment of the First Affiliated Hospital of Zhengzhou University, Ethics code: 2022-KY-0175-002.
Ethical approval and consent to participate
This project has been approved by the Ethics Committee of Scientific Research and Clinical Experiment of the First Affiliated Hospital of Zhengzhou University and confirmed that all methods were performed in accordance with the relevant guidelines and regulations. All enrolled patients signed informed consent to ensure their right to informed consent.
Consent for publication
All authors approve the manuscript for publication.
Results
Based on clinical data
Clinical test results during NAT have been obtained
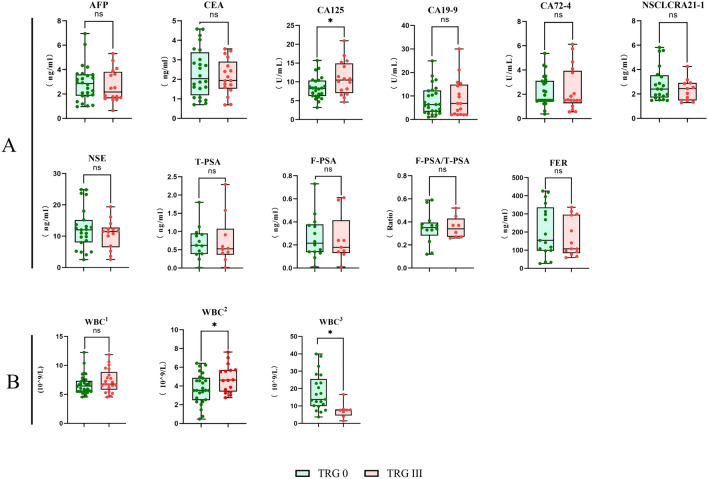
The level of CA125 in the TRG 0 group was significantly lower than that in the TRG III group before neoadjuvant therapy (*P < 0.05). Analysis of white blood cell levels during neoadjuvant therapy showed that the white blood cell level in the TRG 0 group was lower than that in the TRG III group, and this difference was statistically significant (*P < 0.05). After using drugs to increase white blood cell count, the white blood cell level in the TRG 0 group became higher than that in the TRG III group, and this difference was also statistically significant (*P < 0.05) (Fig. 1).
Figure 1.
Clinical blood test results of the enrolled patients. (A) Tumor marker data from patients prior to NAT. (B) Information on the white blood cell counts in enrolled patients during various time periods:1The initial white blood cell count in enrolled patients; 2The toxicity reaction of white blood cells towards chemotherapy drugs; 3The sensitivity of white blood cells towards leukocyte-raising drugs.
Original lesion information of the enrolled patients
The data of endoscopic ultrasound and CT scans were collected for the enrolled patients before NAT. The results showed no significant difference in the location, length, maximum thickness, and depth of invasion of the primary tumor between the TRG 0 group and TRG III group (P > 0.05). There was also no significant difference in neck, chest, abdominal lymph nodes, and periesophageal lymph nodes between the two groups (P > 0.05). Additionally, there was no significant difference in cTNM stage between the two groups (P > 0.05) (Table 2).
Table 2.
The initial lesion data of the patients included.
| Project | Group | P | |
|---|---|---|---|
| TRG 0 (n = 33) |
TRG III (n = 21) |
||
| Tumor location (cm) | |||
| Upper enda | 27.13 ± 3.04 | 28.07 ± 2.84 | 0.3602 |
| Lower endb | 31.96 ± 3.45 | 32.73 ± 3.32 | 0.5037 |
| Tumor length (cm) | 5.25 ± 1.70 | 5.10 ± 1.58 | 0.7560 |
| Tumor thickness (mm) | 16.02 ± 6.02 | 15.67 ± 4.80 | 0.8722 |
| Degree of infiltration (Ultrasound gastroscopy) | |||
| Muscularis propria | 2 (6.06%) | 2 (9.52%) | |
| Full-thickness | 17 (51.52%) | 5 (23.81%) | 0.1300 |
| Outer mold | 14 (42.42%) | 14 (66.67%) | |
| Lymph nodes located in the cervical, thoracic, and abdominal region (CTc) | |||
| Cervical | |||
| Swollen | 1 (3.03%) | 1 (4.76%) | 0.6814 |
| Normal | 32 (96.97%) | 20 (95.24%) | |
| Thoracic | |||
| Swollen | 14 (42.42%) | 10 (47.62%) | 0.7827 |
| Normal | 19 (57.58%) | 11 (52.38) | |
| Aabdominal regions | |||
| Swollen | 7 (21.21%) | 5 (23.81%) | > 0.9999 |
| Normal | 26 (78.79%) | 16 (76.19%) | |
| Peri-esophageal lymph nodes (EUSd) | |||
| Swollen | 17 (51.52%) | 10 (47.62%) | > 0.9999 |
| Normal | 16 (48.48%) | 11 (52.38%) | |
| TNMe staging (AJCCf 8th edition) | |||
| II | 5 (15.15%) | 3 (14.28%) | |
| III | 22 (66.67%) | 14 (66.67%) | 0.9942 |
| IV | 6 (18.18%) | 4 (19.05) | |
aDistance between upper end and incisor.
bDistance between lower end and incisor.
cComputed Tomography.
dEndoscopic Ultrasonography.
eClassification of malignant tumors.
fAmerican Joint Committee on Cancer.
The analysis of immunohistochemical results from pathological tissue revealed no significant difference in AE1/AE3, CK5/6, P40, P63, CK7, CK8/18 , SYN , CD56 , CgA , Ki-67 (%) , and CPS between the TRG 0 group and TRG III group (P > 0.05) (Table 3).
Table 3.
Immunohistochemical data of the original tumor tissue in enrolled patients (Ratio of patients with positive indicators to all patients tested).
| Project | Group | P | |
|---|---|---|---|
| TRG 0 | TRG III | ||
| AE1/AE3 | |||
| + | 7 (87.5%) | 4 (100%) | > 0.9999 |
| − | 1 (12.5) | 0 (0) | |
| CK5/6 | |||
| + | 9 (81.9%) | 4 (80%) | > 0.9999 |
| − | 2 (18.1%) | 1 (20%) | |
| P40 | |||
| + | 8 (88.9%) | 5 (83.3%) | > 0.9999 |
| − | 1 (11.1%) | 1 (16.7%) | |
| P63 | |||
| + | 5 (83.3%) | 4 (100%) | > 0.9999 |
| − | 1 (16.7%) | 0 (0) | |
| CK7 | |||
| + | 2 (33.3%) | 2 (50%) | > 0.9999 |
| − | 4 (66.7%) | 2 (50%) | |
| CK8/18 | |||
| + | 4 (57.1%) | 3 (100%) | 0.4750 |
| − | 3 (42.9%) | 0 (0) | |
| SYN | |||
| + | 0 (0) | 1 (25%) | > 0.9999 |
| − | 3 (100%) | 3 (75%) | |
| CD56 | |||
| + | 0 (0) | 2 (50%) | 0.4667 |
| − | 2 (100%) | 2 (50%) | |
| CgA | |||
| + | 1 (25%) | 1 (25%) | > 0.9999 |
| − | 3 (75%) | 3 (75%) | |
| Ki-67(%) | 50.2 ± 30.1 | 50.7 ± 24.8 | > 0.9999 |
| CPSa | 8.26 ± 11.42 | 11.7 ± 11.28 | 0.4234 |
aCombined Positive Score.
Surgical and postoperative pathological data of (the enrolled patients
Based on the surgical records of the enrolled patients, the results showed that there was no significant difference in tumor invasion within the surgical field, cervical and abdominal lymph node enlargement, number of lymph nodes removed, positive surgical margins, operation time, intraoperative blood loss, and postoperative hospital stay between the TRG 0 group and the TRG III group (P > 0.05). However, there were significant differences in chest lymph node enlargement within the surgical field, number of patients with lymph node metastasis, vascular invasion), perineural invasion and postoperative drainage tube indwelling time between the two groups (*P < 0.05) (Table 4).
Table 4.
Surgical and postoperative pathological data of enrolled patients.
| Project | Group | P | |
|---|---|---|---|
| TRG 0(n = 33) | TRG III (n = 21) | ||
| Assessment of tumor infiltration during surgery | |||
| No visible infiltration | 27 (81.82%) | 13 (61.91%) | 0.1036 |
| Infiltration | 6 (18.18%) | 8 (38.09%) | |
| Intraoperative assessment of lymph node status | |||
| Cervical | |||
| Swollen | 7 (21.21%) | 2 (9.52%) | 0.4538 |
| Normal | 26 (78.79%) | 19 (90.48%) | |
| Thoracic | |||
| Swollen | 20 (60.61%) | 19 (90.48%) | 0.0378 |
| Normal | 13 (39.39%) | 2 (9.52%) | |
| Aabdominal regions | |||
| Swollen | 21 (63.64%) | 17 (80.95%) | 0.2924 |
| Normal | 12 (36.36%) | 4 (19.05%) | |
| Number of lymph nodes removed | 35.32 ± 9.89 | 34.13 ± 10.06 | 0.7318 |
| Number of patients with lymph node metastasis | |||
| Yes | 8 (24.24%) | 11 (52.38%) | 0.0448 |
| No | 25 (75.76%) | 10 (47.62%) | |
| Vascular invasion | |||
| Yes | 0 (0) | 7 (33.33%) | 0.0007 |
| No | 33 (100%) | 14 (66.67%) | |
| Perineural invasion | |||
| Yes | 2 (6.06%) | 11 (52.38%) | 0.0004 |
| No | 31 (93.94%) | 10 (47.62%) | |
| Positive surgical margins | |||
| Yes | 0 (0) | 2 (9.52%) | 0.1468 |
| No | 33 (100%) | 19 (90.48%) | |
| Duration of surgery (min) | 286.5 ± 61.5 | 222 ± 60.8 | 0.1741 |
| Hemorrhage during surgical procedures (ml) | 150 ± 50 | 134.5 ± 68.5 | 0.7694 |
| Postoperative drainage tube duration (d) | 7.65 ± 2.21 | 9.56 ± 3.99 | 0.0348 |
| Postoperative hospital stay (d) | 9.5 (3 ± 2.89 | 10.79 ± 3.82 | 0.1988 |
Based on the transcriptome data )
Sensitive gene characteristics of tumor tissues before and after NAT in TRG 0 group
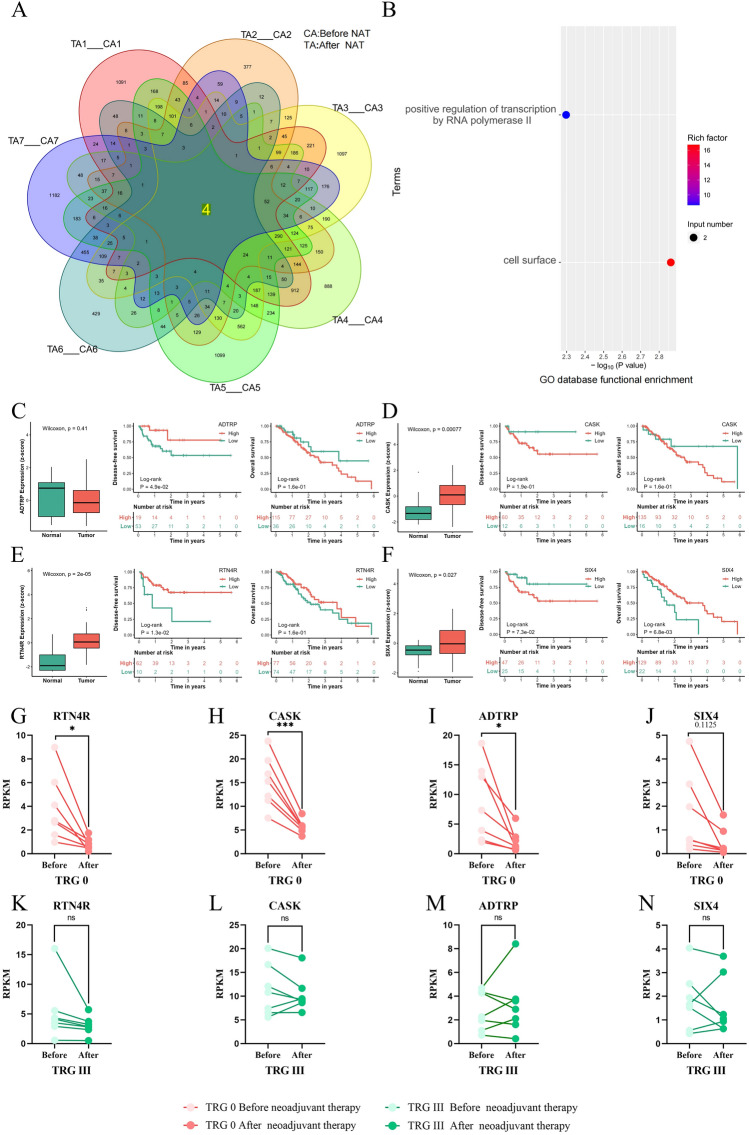
In the paired detection analysis of clinical samples from TRG 0 patients before and after NAT, there were four genes (RTN4R, ADTRP, CASK and SIX4)whose expression levels were significantly decreased after NAT. These genes were common to all patients in this group. The expression levels of RTN4R, ADTRP, and CASK showed statistical significance (*P < 0.05) (Fig. 2G–I). They play important roles in mediating axon growth, cell migration, protein scaffold formation, and neural cell differentiation (Fig. 2A). Functional enrichment analysis using the GO database revealed that CASK and SIX4 is associated with the function of RNA polymerase II while RTN4R and ADTRP are associated with cell surface composition (Fig. 2B). Analysis of The Cancer Genome Atlas (TCGA) database demonstrated high expression levels of CASK (Fig. 2D), RTN4R (Fig. 2E) in EC tissues with statistically significant differences observed (*P < 0.05). Patients with high RTN4R expression had longer disease-free survival compared to those with low RTN4R expression; this difference was statistically significant as well (*P < 0.05). Similarly, patients with high SIX4expression had longer overall survival than those with low SIX4 expression,and the difference was also statistically significant (*P < 0.05). Transcriptome cross-validation indicated no significant difference in the expression levels of selected genes before and after NAT in the TRG III group (P > 0 0.05 ) (Fig. 2K–N).
Figure 2.
Gene characteristics of TRG 0 patients, their expression in cancer and adjacent tissues, and their correlation with prognosis.
Non-sensitive gene characteristics of tumor tissues were analyzed before and after NAT in TRG III group
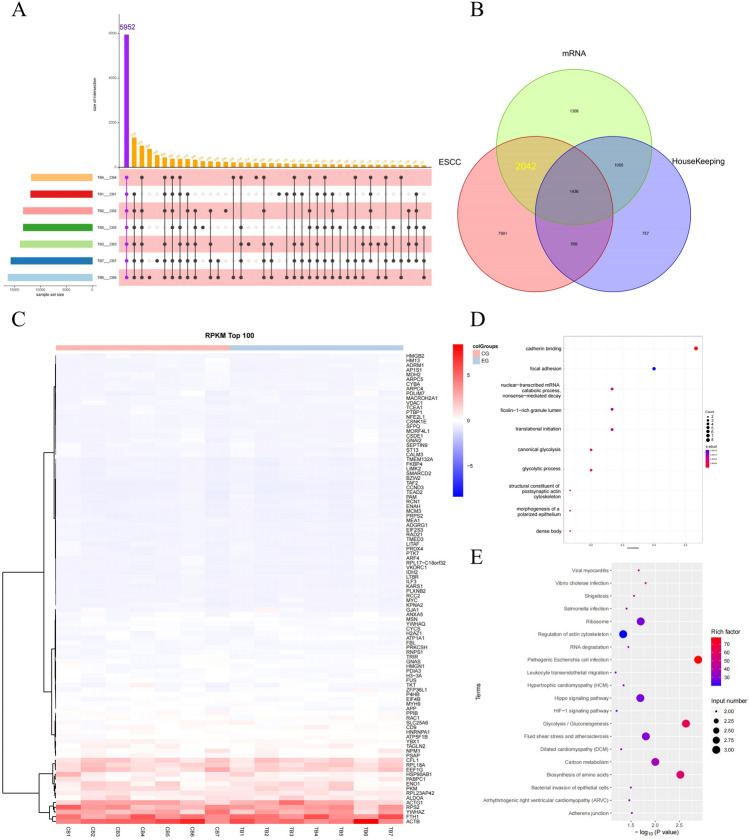
In the paired analysis of clinical samples from TRG III patients before and after NAT, a total of 5952 genes whose expression levels did not change significantly before and after NAT were common to all patients in the TRG III group (Fig. 3A). A total of 12,024 EC-related genes and 3804 housekeeping genes were detected from the TCGA database. The sequencing gene set was overlapped with the EC gene set to obtain the EC-related genes in these sequencing results, and then the housekeeping genes were removed. Finally, 2042 NAT-insensitive genes were obtained (Fig. 3B). The genes were ranked according to their level of gene expression, and the top 100 genes with RPKM values were selected for cluster analysis. The results showed that ACTB, FTH1, YWHAZ, RPS2, ACTG1, ALDOA, RPL23AP42, PKM, ENO1, PABPC1 HSP90AB1 EEF1G RPL18A CFL1 had the highest gene expression levels among them (Fig. 3C). Functional enrichment analysis using GO database on this TOP14 gene set revealed that these genes mainly functioned in cadherin binding nuclear transcription mRNA catabolic process glycolysis (Fig. 3D). KEGG pathway enrichment analysis also indicated that these TOP14 genes played important roles in glycolysis glucose metabolism synthesis amino acid biosynthesis carbon metabolism signaling pathways (Fig. 3E).
Figure 3.
Gene characteristics, GO function enrichment, and KEGG pathway enrichment information are provided for TRG III patients.
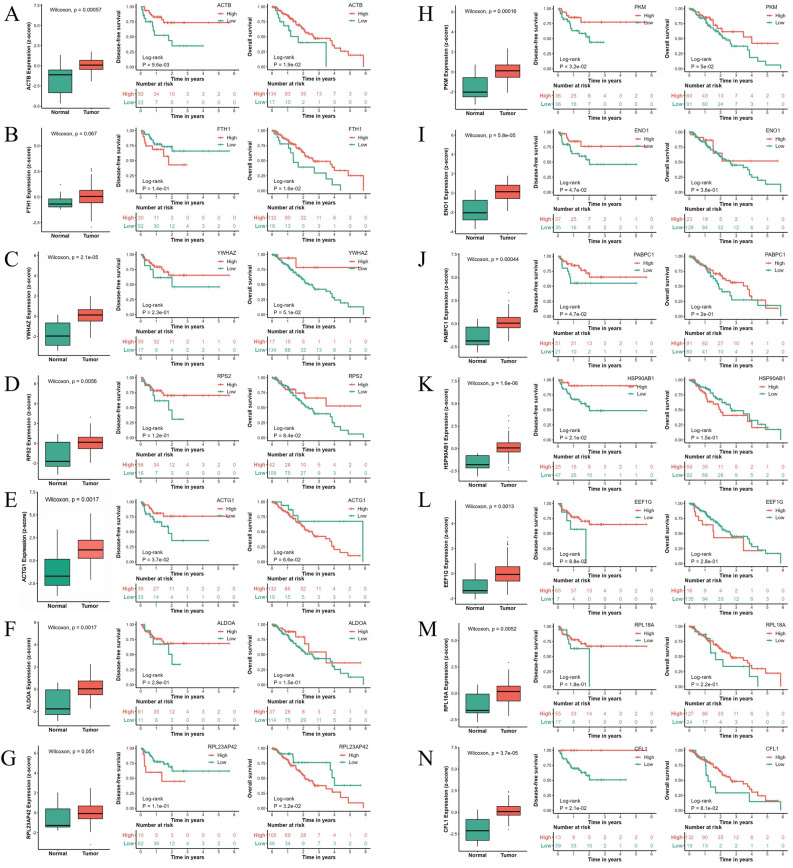
The TCGA database showed that in the expression analysis of EC and adjacent tissues, ACTB (Fig. 4A), YWHAZ (Fig. 4C), RPS2 (Fig. 4D), ACTG1 (Fig. 4E), ALDOA (Fig. 4F), PKM (Fig. 4H), ENO1 (Fig. 4I), PABPC1 (Fig. 4J), HSP90AB1 (Fig. 4K), EEF1G (Fig. 4L), RPL18A (Fig. 4M) and CFL1 (Fig. 4N) were highly expressed in EC tissues, and the difference was statistically significant (*P < 0.05). In the analysis of DFS in patients with EC, patients with high expression of ACTB (Fig. 4A), ACTG1 (Fig. 4E), PKM (Fig. 4H), ENO1 (Fig. 4I), PABPC1 (Fig. 4J) , HSP90ABl (Fig. 4K) , and CFLl (Fig. 4N) in cancer tissues had longer DFS than those with low expression.The difference was statistically significant (*P < 0.05). In the OS analysis of patients with EC, patients with high expression of ACTB (Fig. 4A), FTHl (Fig. 4B), and PKM (Fig. 4H) in cancer tissues had longer OS than those with low expression,and the difference was statistically significant (*P < 0.05). Patients with low expression of RPL23AP42 in cancer tissues had longer OS than those with high expression (Fig. 4G) and the difference was statistically significant (*P < 0.05). This gene may have potential value as a drug target.
Figure 4.
The expression of drug-insensitive genes in cancer and paracancerous tissues and the relationship between their content and prognosis in TRG III patients.
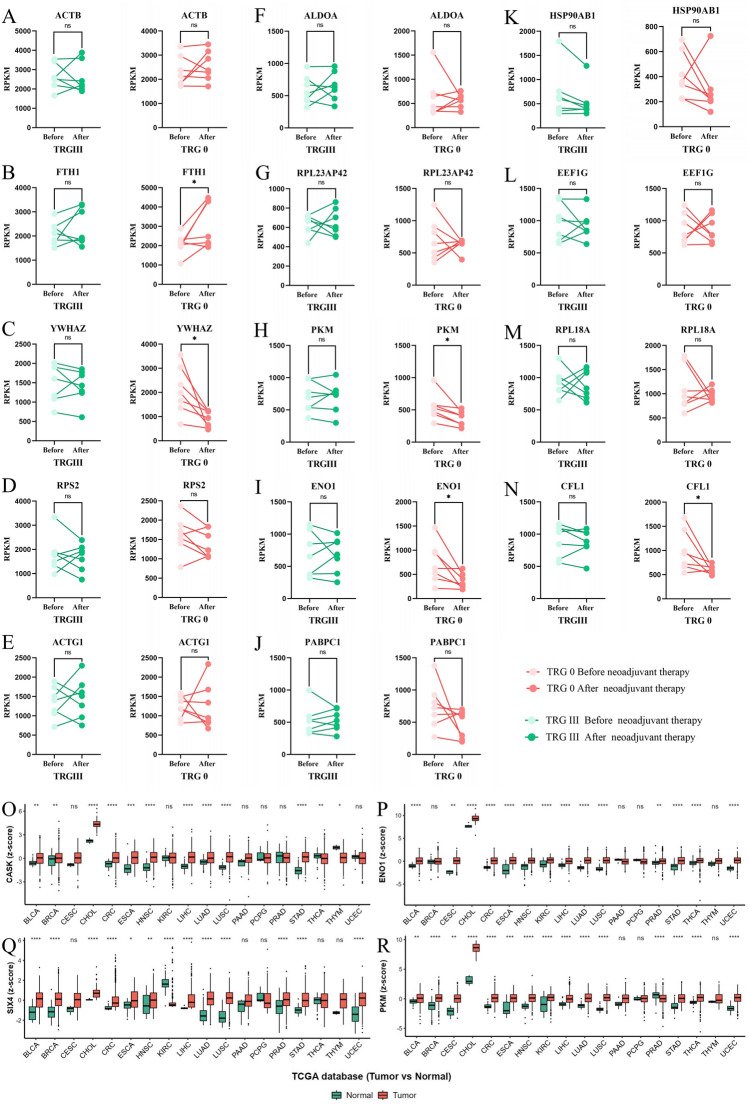
Transcriptome cross-validation showed that in the TRG 0 group, there was no significant difference before and after NAT (P > 0.05) for screening genes ACTB (Fig. 5A), RPS2 (Fig. 5C), ACTG1 (Fig. 5E), ALDOA (Fig. 5F), RPL23AP42 (Fig. 5G), PABPC1 (Fig. 5J), HSP90AB1 (Fig. 5K), EEF1G (Fig. 5L), and RPL18A (Fig. 5M). The expression levels of screened genes YWHAZ (Fig. 5C), PKM (Fig. 5H), ENO1 (Fig. 5I), and CFL1 (Fig. 5N) decreased significantly after NAT (*P < 0.05). The expression level of the screening gene FTH1 increased significantly after NAT (*P < 0.05). TCGA database showed that genes CASK, SIX4, ENO1 and PKM are highly expressed in a variety of cancer tissues (Fig. 5O–R).
Figure 5.
Transcriptome cross-validation of drug-insensitive genes in patients with TRG 0 is being conducted (A-N); Expression of CASK, SIX4, ENO1 and PKM in multiple cancer types (O-R).
Discussion
In this study, we found that the existing tests and examinations still cannot effectively distinguish between sensitive patients and non-genetic patients before NAT. Preoperative NAT has become an important means for the radical cure of EC in the current guidelines for diagnosis and treatment15. However, accurately screening the appropriate population for NAT still faces great challenges16. Although many studies have attempted to reveal the correlation between its clinical characteristics and therapeutic effect, its specific clinical characteristics have not been clearly established17–19.
This study demonstrated that the change in white blood cell levels during NAT can predict the effectiveness of NAT to a certain extent. Neoadjuvant sensitive patients appear to be more susceptible to the effects of chemotherapy drugs on white blood cell counts. In this study, the white blood cell levels of TRG 0 patients were significantly lower than those of TRG III patients after NAT. This can be explained by the homogeneity of patient cells; although cancer cells undergo heterogeneous transformation, all cells in the body still contain the same gene sequence20. Additionally, TRG 0 patients are more likely to recover from myelosuppression after neoadjuvant chemotherapy with leukemotropic drugs. The sensitivity of the body to leukemotropic drugs is closely related to the type and degree of bone marrow suppression caused by chemotherapy drugs, as well as the patient's overall physical condition21. Under identical neoadjuvant regimens, differences in patient constitution become a primary factor. However, this phenomenon also objectively predicts the therapeutic effect of NAT.
Neoadjuvant non-responsive patients appear to predict worse surgical and pathological outcomes. In this study, the proportion of patients with lymph node metastasis was higher in the TRG III group compared to the TRG 0 group. Postoperative pathological results revealed more severe vascular invasion and perineural invasion in tumor tissue from the TRG III group. The relationship between neoadjuvant resistance and tumor progression has not been fully established because it is uncertain whether there is heterogeneous transformation in tumor tissue during this period, wherein cells expressing chemotherapy-sensitive genes are eliminated and replaced by other non-sensitive cells; however, regardless of the mechanism, it leads to adverse clinical outcomes. This also underscores the necessity of combination therapy22.
The duration of drainage tube placement is shorter in the TRG 0 group compared to the TRG III group. Although there are various factors influencing the duration of drainage tube placement, postoperative complications like persistent pleural effusion, surgical site hemorrhage, chylothorax, and decreased albumin levels23 play a significant role. These factors greatly contribute to extended drainage tube placement after surgery. Therefore, it can be deduced that patients who have a complete response to neoadjuvant therapy have a lower occurrence rate of postoperative complications compared to non-responding patients.
Transcriptome sequencing results of enrolled patients showed that drug sensitivity genes CASK and SIX4 in TRG 0 patients may regulate the growth, proliferation, and differentiation of cancer cells and other advanced life processes by affecting the positive regulation of RNA polymerase II transcription24,25. This may be related to the mechanism of action of neoadjuvant drugs. For example, platinum drugs mainly bind to DNA and inhibit cell division and proliferation to achieve the effect of cancer treatment26. Paclitaxel can inhibit cell mitosis by enhancing and stabilizing tubulin polymerization while preventing microtubule depolymerization, thus achieving an anti-cancer effect27. Camrelizumab binds to the PD-1 receptor and blocks the PD-1/PD-L1 pathway, thereby blocking immune suppression mediated by the PD-1 pathway, especially immune suppression caused by tumor cells28. The premise for anti-tumor drugs is that their theoretical targets are not mutated; however, continuous heterogeneous changes in tumor cells are an unchangeable fact. In cross-validation of transcriptome data from TRG III patients, the expression levels of CASK and SIX4 remained relatively unchanged before and after NAT. We hypothesize that there might be a resistance regulatory mechanism in TRG III patients inhibiting down-regulation of CASK and SIX4 juvant resistance.
In this study, neoadjuvant drug non-sensitive genes ENO1 and PKM mainly focus on the basic life processes of cells such as amino acid biosynthesis, glycolysis, and glucose metabolism synthesis in TRG III patients. These two genes play a crucial role in the fundamental cellular processes29,30. The TCGA database showed a significant increase in the expression of ENO1 and PKM in EC tissues, which may be closely related to the rapid growth and invasion of tumor cells31. However, during cross-validation of transcriptome data from the TRG 0 group, there was a significant decrease in the expression levels of ENO1 and PKM after NAT. We hypothesize that there may be some resistance regulatory mechanism present in the tumor tissues of TRG III patients that maintains normal expression levels of ENO1 and PKM, thereby ensuring that cancer cell's normal anabolic process is not affected.
TCGA database showed that genes CASK, SIX4, ENO1 and PKM are highly expressed in a variety of cancer tissues and are important anticancer drug targets. Therefore, in-depth exploration of their abnormal regulatory mechanisms in NAT has important clinical value for increasing the drug sensitivity of neoadjuvant patients.
Conclusion
The change in white blood cell count during NAT for ESCC can be used as a clinical indication to predict the effect of NAT to some extent. Neoadjuvant resistance may indicate a worse pTNM stage. Transcriptome data suggest that there may be resistant regulatory mechanisms in the tumor tissues of TRG III patients, allowing normal growth and anabolic processes of tumor cells.
Acknowledgements
This work was supported by the key project of Henan Province Medical Science and Technology Research Plan in 2021 (No. SBGJ202102077) from the Health Commission of Henan Province.
Abbreviations
- NAT
Neoadjuvant therapy
- TRG
Tumor regression grade
- EC
Esophageal cancer
- ESCC
Esophageal squamous cell carcinoma
- NCCN
National Comprehensive Cancer Network
- BMI
Body mass index
- CT
Computed tomography
- EUS
Endoscopic ultrasonography
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- CPS
Cell positive expression rate
- TNM
Classification of malignant tumors
- AJCC
American Joint Committee on Cancer
- TCGA
The Cancer Genome Atlas
- OS
Overall survival
- DFS
Disease free survival
Author contributions
The study was designed by Professor Q.C.K. and Professor D.L.L. Y.N.S. was responsible for patient screening, while R.M.D., T.T.L., and L.Y. were responsible for clinical data collection and analysis. F.L. conducted clinical specimen collection, transcriptome sequencing, and follow-up work. This manuscript was written by Y.N.S. with editing contributions from Y.Q. and D.L.L. The manuscript underwent review by D.L.L.
Data availability
All data generated or analysed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally to this work: Yanan Song, Yu Qi and Feng Li.
Contributor Information
Duolu Li, Email: dorali1979@126.com.
Quancheng Kan, Email: kanqc@zzu.edu.cn.
References
- 1.Hassanipour, S., Mohammadian-Hafshejani, A., Ghoncheh, M. & Salehiniya, H. The incidence and mortality of esophageal cancer and its relationship with development in the world. Biomed Res Ther.4(9), 1607–1623 (2017). 10.15419/bmrat.v4i9.368 [DOI] [Google Scholar]
- 2.Sharma, T. et al. Cross-talk between the microbiome and chronic inflammation in esophageal cancer: potential driver of oncogenesis. Cancer Metastasis Rev.41(2), 281–299 (2022). 10.1007/s10555-022-10026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, S. et al. Mafb promotes the malignant phenotypes by IGFBP6 in esophageal squamous cell carcinomas. Exp. Cell Res.416(1), 113158. 10.1016/j.yexcr.2022.113158 (2022). 10.1016/j.yexcr.2022.113158 [DOI] [PubMed] [Google Scholar]
- 4.Tang, W. R., Fang, J. Y., Wu, K. S., Shi, X. J. & Lin, K. Epidemiological characteristics and prediction of esophageal cancer mortality in china from 1991 to 2012. Asian Pac. J. Cancer Prev.15(16), 6929–6934 (2014). 10.7314/APJCP.2014.15.16.6929 [DOI] [PubMed] [Google Scholar]
- 5.Yuan, L. et al. Analysis of living habit risk factors for esophageal cancer in central China: A bi-center case-control study. Front Oncol.13, 1077598. 10.3389/fonc.2023.1077598 (2023). 10.3389/fonc.2023.1077598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia, C. et al. Cancer statistics in china and united states profiles, trends, and determinants. Chin Med. J.135(5), 584–590 (2022). 10.1097/CM9.0000000000002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Q. et al. Effectiveness evaluation of organized screening for esophageal cancer: A case-control study in Linzhou city. China Sci. Rep.6, 35707. 10.1038/srep35707 (2016). 10.1038/srep35707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii, Y. et al. Non-curative resection for surgical t4b esophageal cancer: Esophagectomy or non-esophagectomy?. Langenbecks Arch. Surg.408(1), 201–206 (2023). 10.1007/s00423-023-02940-2 [DOI] [PubMed] [Google Scholar]
- 9.Shimada, A. et al. Utility of concurrent surgical treatment strategy with thoracoscopic esophagectomy for patients with synchronous esophageal and head and neck cancer. J. Laparoendosc. Adv. Surg. Tech. A. Part A32(5), 550–555 (2022). 10.1089/lap.2021.0441 [DOI] [PubMed] [Google Scholar]
- 10.Tian, Y. et al. A comparison of clinicopathologic outcomes and patterns of lymphatic spread across neoadjuvant chemotherapy, neoadjuvant chemoradiotherapy, and neoadjuvant immunochemotherapy in locally advanced esophageal squamous cell carcinoma. Ann. Surg. Oncol.31(2), 860–871 (2023). 10.1245/s10434-023-14534-9 [DOI] [PubMed] [Google Scholar]
- 11.Huang, R. et al. Neoadjuvant therapy for locally advanced esophageal cancers. Front. Oncol.12, 734581. 10.3389/fonc.2022.734581 (2022). 10.3389/fonc.2022.734581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hipp, J., Thomaschewski, M., Hummel, R. & Hoeppner, J. Complete response after neoadjuvant therapy for esophageal cancer: Implications for surgery. Der Chirurg. Ir. J. Med. Sci.93(2), 132–137 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Takeno, S. et al. Immunohistochemical and clinicopathologic analysis of response to neoadjuvant therapy for esophageal squamous cell carcinoma. Dis. Esophagus14(2), 149–154 (2001). 10.1046/j.1442-2050.2001.00174.x [DOI] [PubMed] [Google Scholar]
- 14.Khaitan, P. G. et al. Can clinical response predict pathologic response following neoadjuvant chemoradiation for esophageal cancer?. J. Gastrointest. Surg.26(7), 1345–1351 (2022). 10.1007/s11605-022-05315-y [DOI] [PubMed] [Google Scholar]
- 15.Yang, Y. et al. Safety and efficacy of neoadjuvant treatment with immune checkpoint inhibitors in esophageal cancer: Real-world multicenter retrospective study in China. Dis. Esophagus11, 1–8 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Feng, J., Wang, L., Yang, X., Chen, Q. & Cheng, X. Aso author reflections: A novel, sensitive, and effective index in predicting therapeutic response of neoadjuvant immunochemotherapy for esophageal squamous cell carcinoma. Ann. Surg. Oncol.31(1), 241–242 (2023). 10.1245/s10434-023-14464-6 [DOI] [PubMed] [Google Scholar]
- 17.Liu, Y. et al. Clinical value of hematologic test in predicting tumor response to neoadjuvant chemotherapy with esophageal squamous cell carcinoma. World J. Surg. Oncol.12, 43–48 (2014). 10.1186/1477-7819-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S. L. et al. Is there a correlation between clinical complete response and pathological complete response after neoadjuvant chemoradiotherapy for esophageal squamous cell cancer?. Ann. Surg. Oncol.23(1), 1–9 (2016). 10.1245/s10434-015-4764-0 [DOI] [PubMed] [Google Scholar]
- 19.Sato, H., Tsubosa, Y. & Kawano, T. Correlation between the pretherapeutic neutrophil to lymphocyte ratio and the pathologic response to neoadjuvant chemotherapy in patients with advanced esophageal cancer. World J. Surg.36(3), 617–622 (2012). 10.1007/s00268-011-1411-1 [DOI] [PubMed] [Google Scholar]
- 20.Ponder, B. A. J. Looking for genomic changes in cancer cells. Nature293(5828), 98–99 (1981). 10.1038/293098a0 [DOI] [PubMed] [Google Scholar]
- 21.Wang, C. et al. Safety and efficacy of pegylated recombinant human granulocyte colony-stimulating factor during concurrent chemoradiotherapy for small-cell lung cancer: A retrospective, cohort-controlled trial. BMC Cancer22(1), 1–10 (2022). 10.1186/s12885-023-11764-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong, L. Y. et al. The impact of neoadjuvant immunotherapy on perioperative outcomes and survival after esophagectomy for esophageal cancer. JTCVS Open14, 547–560 (2023). 10.1016/j.xjon.2023.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada, J. et al. Management of Pleural Effusion After Mediastinoscopic Radical Esophagectomy. Anticancer Res.38(12), 6919–6925 (2018). 10.21873/anticanres.13069 [DOI] [PubMed] [Google Scholar]
- 24.Moniaux, N. et al. The human rna polymerase ii-associated factor 1 (hpaf1): a new regulator of cell-cycle progression. PLoS ONE4(9), e70774. 10.1371/journal.pone.0007077 (2009). 10.1371/journal.pone.0007077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, L. et al. Polo-like kinase 1 inhibits the activity of positive transcription elongation factor of RNA Pol II b (P-TEFb). PLoS ONE8(8), e72289. 10.1371/journal.pone.0072289 (2013). 10.1371/journal.pone.0072289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajj, C., Becker-Flegler, K. A. & Haimovitz-Friedman, A. Novel mechanisms of action of classical chemotherapeutic agents on sphingolipid pathways. J. Biol. Chem.396(6/7), 669–679 (2015). 10.1515/hsz-2014-0302 [DOI] [PubMed] [Google Scholar]
- 27.Kase, A. M., Azzouqa, A.-G., Kochuveettil, S. & Colon-Otero, G. Efficacy of gemcitabine in combination with nanoparticle albumin-bound paclitaxel in the treatment of recurrent ovarian cancer: a retrospective single institution review. Cancer Med.12(8), 9434–9438 (2023). 10.1002/cam4.5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prodeus, A. & Gariepy, J. Abstract b032: targeting the pd-1/pd-l1 immune evasion axis with DNA aptamers. Cancer Immunol Res.4(1), B032–B032 (2016). 10.1158/2326-6074.CRICIMTEATIAACR15-B032 [DOI] [Google Scholar]
- 29.He, Y. et al. Linc-UROD stabilizes ENO1 and PKM to strengthen glycolysis, proliferation and migration of pancreatic cancer cells. Transl. Oncol.27, 101583. 10.1016/j.tranon.2022.101583 (2023). 10.1016/j.tranon.2022.101583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Z. H. et al. SIRT 1 binding with PKM and NSE and modulate their acetylation and activities. Biochim. Biophys. Acta Proteins Proteom.1867(9), 794–801 (2019). 10.1016/j.bbapap.2019.06.003 [DOI] [PubMed] [Google Scholar]
- 31.Tian, M., Zhu, R., Ding, F. & Liu, Z. Ubiquitin-specific peptidase 46 promotes tumor metastasis through stabilizing ENO1 in human esophageal squamous cell carcinoma. Exp. Cell Res.395(1), 112188. 10.1016/j.yexcr.2020.112188 (2020). 10.1016/j.yexcr.2020.112188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.