Abstract
Thirteen point mutations targeting predicted domains conserved in homologous protein kinases were introduced into the UL97 coding region of the human cytomegalovirus. All mutagenized proteins were expressed in cells infected with recombinant vaccinia viruses (rVV). Several mutations drastically reduced ganciclovir (GCV) phosphorylation. Mutations at amino acids G340, A442, L446, and F523 resulted in a complete loss of pUL97 phosphorylation, which was strictly associated with a loss of GCV phosphorylation. Our results confirm that in rVV-infected cells pUL97 phosphorylation is due to autophosphorylation and show that several amino acids conserved within domains of protein kinases are essential for this pUL97 phosphorylation. GCV phosphorylation is dependent on pUL97 phosphorylation.
It has been shown that open reading frame (ORF) UL97 of human cytomegalovirus (HCMV) encodes a protein that has homologies with protein kinases (PK) but that also directs the essential phosphorylation of ganciclovir (GCV) and other antiviral nucleoside analogs to their monophosphates (2, 11, 19, 24). The UL97 protein (pUL97) is detectable in the nuclei of infected and transfected cells but is not localized in the nucleoli (15). Multiple clustered mutations in the coding region of UL97 have been described to induce phenotypical resistance to GCV in HCMV strains (1, 3, 7, 12, 19, 23). Until now, no clinical HCMV strains carrying extensive deletions or lacking pUL97 have been isolated.
Although investigated by different groups, the biological relevance of UL97 is still unknown. Very recently it has been published that a recombinant HCMV with a large deletion in UL97 has a severe replication deficiency (16a). pUL97 is present in the virus particles and is able to partially substitute in function for the herpes simplex virus UL13 protein (16, 21, 22). Evidence obtained from recombinant vaccinia viruses (rVV) indicated that pUL97 is not a nucleoside kinase (14). Furthermore, it was shown that, after expression in the baculovirus system, the protein autophosphorylates at serines and threonines (8). By investigating pUL97 derivatives carrying larger deletions in the vaccinia virus system, we could show that the N terminus contains a nuclear localization signal and that a central domain (amino acids [aa] 305 to 365) is essential for pUL97 phosphorylation as well as for GCV phosphorylation (15). Only one artificial amino acid exchange has been published so far, proving the importance of an invariant lysine at position 355 for autophosphorylation (8).
In PK from different species, several domains, I to XI, have been defined by sequence alignments (5, 6). Very recently the crystal structure of the type I transforming growth factor β receptor with an autophosphorylating serine/threonine kinase activity has also been reported to contain these regions (9). In pUL97, homologous sequences have been found for the PK domains (Fig. 1). However, an additional putative functional domain which is not detectable in PK seems to be located between domains VII and VIII. Since no other natural substrate has been identified so far, even today the assumption that pUL97 is a PK is based mainly on the above-mentioned partial sequence homologies and on the autophosphorylating capacity of the protein.
FIG. 1.
Alignment of some conserved motifs in different UL97 homologues. The numbers indicate amino acid positions of the UL97 protein sequence. Published mutations conferring GCV phenotypic resistance on HCMV strains are indicated by boldface, underlined, italic letters. Domain, conserved domains in PK as defined by Hanks et al. (5, 6); S/T-PK, amino acids conserved in serine/threonine PK; Herp, amino acids that are conserved in the UL97 homologues of herpes simplex virus, human herpesvirus 6, varicella-zoster virus, and Epstein-Barr virus according to the work of Chee et al. (2); RhCMV, rhesus cytomegalovirus; MCMV, murine cytomegalovirus; GCMV, guinea pig cytomegalovirus (4, 20); Cons, amino acids that are conserved among the cytomegalovirus proteins; MUTA, mutations introduced by site-directed mutagenesis.
Site-directed mutagenesis in the UL97 ORF.
Very few amino acids conserved in PK have been proven experimentally to be indispensable for the functions of these proteins, an example being the GXGXXG motif involved in nucleotide binding in domain I or the invariant lysine in domain II (2, 6). In addition, by sequence alignments conserved amino acids or even sequence motifs implicating functional relevance and allowing for the definition of different conserved motifs have been detected. In order to get further evidence for the PK nature of pUL97, we wanted to experimentally prove the existence of assumed functional regions of the viral protein that are obviously involved in phosphorylation. Therefore, site-directed mutagenesis was performed at specific amino acids in different domains which are conserved among the pUL97 homologues of different cytomegaloviruses, human herpesviruses, and nonviral serine/threonine kinases, including the PK region of the transforming growth factor β receptor (9). Other mutations were introduced in the vicinities of putative functional amino acids in order to estimate the extension of the presumed domains (Fig. 1). None of the mutations have been found in phenotypically resistant HCMV. We also introduced a stop codon at aa Y617 to truncate the protein beyond this position. Therefore, from plasmid p214 containing the entire pUL97 coding region (14, 17) a BamHI/EcoRI fragment, aa 20 to 707, was extracted. After this fragment was introduced into plasmid pALTER (Promega), site-directed mutagenesis was performed by using the following oligonucleotides: 335Y>D, 5′-C ATG AGC GAC GAG AGC GAC CGC CTG-3′; 340G>V, 5′-GGC CAG GTC TCC TTC GGC-3′; 354V>L, 5′-C TAT CGC GTG CTC AAG GTG-3′; 359K>Q, 5′-G GCG CGT CAG CAC AGC GAG ACG-3′; 380E>V, 5′-GCT GGC GTG CAA CAG CAG C-3′; 442A>V, 5′-C ACG TTG GCC GAC GTT ATC AAA TT-3′; 446L>R, 5′-TTT CGC AAT CAC CAG TGT CG-3′; 523F>C, 5′-CAC CCT GCT TGC CGA CCC ATG CCG C-3′; 526M>I, 5′-GCT TTC CGA CCC ATC CCG CTG CAG AAG-3′; 574D>A, 5′-CGG CGC GGT CTG GCC GAG GTG CGC-3′; 579G>A, 5′-G CGC ATG GCC ACG GAG G-3′; 583L>F, 5′-G GAG GCG TTT CTC TTT AAG-3′; and 617Y>Stop, 5′-A ATG AGC TAA GGC GCC TGT CTC CTG-3′ (where underlining indicates the presumed functional amino acid and boldface indicates the mutation introduced).
Expression of the pUL97 derivatives after infection with rVV.
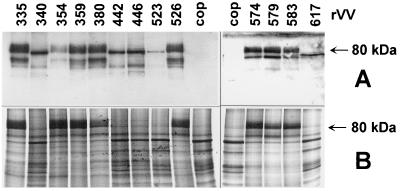
The mutagenized UL97 coding regions were introduced into vaccinia viruses in order to test functional properties, i.e., nuclear localization, GCV phosphorylation, and pUL97 phosphorylation. The expression of all mutagenized UL97 derivatives was confirmed by Western blot analysis (10) (Fig. 2A). None of the mutations disturbed the intracellular localization of the UL97 proteins, which were all found in the nuclei of infected cells (data not shown). However, the migrations and shapes of the different pUL97 signals in the Western blot were dependent on the ability to phosphorylate pUL97 itself (see Fig. 2B and below). Loss of this function resulted in a sharp band at 80 or 70 kDa for rVV617, whereas autophosphorylation caused an apparently more intensive signal. However, these signals were due mainly to several additional, slower-migrating bands, resulting in a smear between 80 and approximately 100 kDa. No signals were observed in cell extracts harvested after mock infection or after infection with the wild-type vaccinia virus strain Copenhagen carrying the thymidine kinase of vaccinia virus.
FIG. 2.
Expression and phosphorylation of newly mutagenized UL97 proteins in cells infected with rVV. Lanes labeled 335 to 617 indicate rVV carrying different point mutations in the UL97 ORF introduced by site-directed mutagenesis; cop, cells infected with the vaccinia virus strain Copenhagen (without the UL97 ORF). CV1 cells were infected at an MOI of 5 with the indicated rVV and were harvested 17 to 20 h postinfection for preparation of nuclear extracts. The extracts were used in parallel for both Western blot analyses and PK assays. (A) For Western blotting nuclear matrix extracts were separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis. Here pUL97 was visualized by chemiluminiscence with the pUL97 antiserum. (B) For determination of pUL97 phosphorylation, nuclear matrix fractions were tested for PK activity. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography.
GCV phosphorylation and phosphorylation of pUL97 in cells infected with rVV.
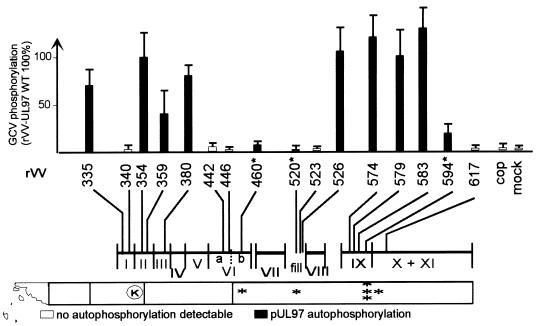
Two advantages of the pUL97 vaccinia virus system are the possibilities of quite easily generating recombinants (13) and of investigating mutated UL97 proteins that might cause a nongrowing or slow-growing HCMV phenotype. Since purification of the foreign protein, biochemical quantification, and in vitro enzymology have not been possible so far, we have performed extensive experiments to correlate the multiplicities of the infecting vaccinia viruses with pUL97 expression, quantitative GCV phosphorylation, and pUL97 phosphorylation (18). Quantitative determination of GCV phosphorylation with a standard deviation of 20% was possible when a multiplicity of infection (MOI) between 10 and 40 was used (18). Hence, for all GCV phosphorylation experiments in this study 143B (tk−) cells were infected with vaccinia virus recombinants at an MOI of 20. High-pressure liquid chromatography analysis was performed as described previously (14, 24). For fractionation of nuclei and cytoplasms, CV1 cells were infected with rVV at an MOI of 5 and harvested 20 h postinfection as described previously (14, 24). Phosphorylation of pUL97 was determined according to the method of He et al. (8). The phosphorylating capacities of the different UL97 mutants in the rVV-infected cells are summarized in Fig. 3, whereas the data showing the phosphorylation of pUL97 itself are shown in Fig. 2B. Two point mutations at D574 and G579, amino acids conserved in UL97 homologues of other herpesviruses, as well as the nearby mutation at L583 (all domain IX [Fig. 1]) in the N-terminal part of the region, where most of the natural mutations that confer GCV resistance are clustered (aa 591, 592, 594, 595, 596, 598, 603, and 607) (1, 3, 7, 12, 18, 19, 23), did not interfere with GCV or pUL97 phosphorylation. However, the stop codon at the tyrosine at position 617 caused a complete loss of GCV phosphorylation and of pUL97 phosphorylation as well, suggesting that the region beyond the stop codon is also involved in protein function.
FIG. 3.
GCV phosphorylation in rVV-infected 143B (thymidine kinase-deficient) cells. The numbers 335 to 617 indicate the different rVV; cop, vaccinia virus strain Copenhagen. The bars represent mean values and standard errors of the results of 6 to 10 independent experiments. The Roman numerals denote conserved domains in PK (5). Point mutations which were found in phenotypically resistant HCMV at positions 460, 520, and 594 have been previously described (14) and are indicated by asterisks. The invariant lysine (K) at position 355 reported by He et al. (8) is indicated.
The substitution of the second glycine residue at position 340 of the highly conserved GXGXXG motif which is part of domain I of PK and involved in nucleotide binding resulted in the complete loss of GCV phosphorylation and pUL97 phosphorylation. Interestingly, the alteration of the Y amino acid at position 335, located only 5 aa upstream of this crucial position, only slightly influenced GCV phosphorylation and did not inhibit pUL97 phosphorylation. The substitution of the lysine at position 359, which we had suspected to be the invariant lysine of PK (15), did not lead to the expected loss of GCV or pUL97 phosphorylation, although a decline of approximately 50% was measured in GCV phosphorylating activity. The mutation of the adjacent V354 did not influence either phosphorylating activity. This result is in accordance with the assumption that it is in fact the lysine located at a typical distance of 14 aa from the GXGXXG domain that is essential for phosphate binding.
The glutamic acid of PK in domain III can be detected only in some of the viral pUL97 homologues. However, it is found in the protein derived from human herpesvirus 6 and in pUL97, where it is supposed to be located at position 380. The substitution of this amino acid caused only a small decrease in GCV phosphorylation and no alteration in pUL97 phosphorylation.
Contrary to this small decrease, dramatic effects were observed with the substitutions A442V, L446R, and F523C. The first and second were located in domain VIa, and the last was located in the putative specific domain unique for UL97 homologues of herpesviruses. All three point mutations conferred a complete loss of GCV phosphorylation down to levels observed in mock-infected cells and also a loss of pUL97 phosphorylation. It should be mentioned that both of the two mutations (M460V and H520O) from naturally resistant HCMV strains found in this regions drastically reduced GCV phosphorylation but not pUL97 phosphorylation (15). Until now only reductions in GCV phosphorylation have been observed for GCV-resistant clinical isolates. It is even more significant that all point mutations or small deletions found in pUL97s of HCMV isolates gave rise to proteins which were still able to autophosphorylate when they were investigated with our vaccinia virus system (14, 15, 18). Together with the known fact that phosphorylation is a widespread mechanism of activation or inactivation in biological systems, our results strengthen the view that autophosphorylation of the protein may well be associated with its natural, biological function. Additionally, we could demonstrate for the first time that autophosphorylation seems to be a prerequisite for GCV phosphorylation, since mutations abrogating autophosphorylation concomitantly led to a complete loss of GCV phosphorylation.
Phosphorylation of pUL97 expressed by rVV is also due to autophosphorylation.
In conclusion, rVV-expressed pUL97 is biologically active as determined by GCV phosphorylation. Under the conditions used (1 M NaCl and pH 9.0), an 80-kDa protein was the major labeled species in the nuclear matrix fractions (Fig. 2B). These labeled proteins comigrated with pUL97 as detected by parallel Western blot analysis performed with identical samples (Fig. 2A). However, the phosphorylated protein at 80 kDa was not observed with rVV340, rVV442, rVV446, rVV523, or rVV617, whereas in the Western blot analysis the 80-kDa proteins were present in all samples tested except for rVV617, which produced a smaller band since the protein was C-terminally truncated. Virtually no comparable labeling at 80 kDa was observed when nuclear matrix extracts of mock-infected 143B (thymidine kinase-deficient) cells or 143B cells infected with the wild-type vaccinia virus strain Copenhagen were incubated with [γ-33P]ATP.
To further examine whether pUL97 phosphorylation in the vaccinia virus system is indeed due to autophosphorylation, protein phosphorylation was investigated under higher-concentration salt conditions (1.5 M NaCl), with a shorter incubation time (20 min), and in the presence of heparin (50 μg/ml). pUL97 phosphorylation was still observed under these conditions (data not shown). Finally, we performed a time kinetic analysis of pUL97 phosphorylation, which showed that after 30 s of incubation, almost 50% of the protein was already phosphorylated and that after 10 min, the reaction reached its maximum (data not shown). In conclusion, these findings further support the presence of an intrinsic PK activity in pUL97 as well as in the vaccinia virus system, and the activity seems not to be derived from contaminations in the nuclear extracts.
Here, we could show that several amino acids which are conserved among serine/threonine kinases and which can also be found in pUL97 and its homologues in herpesviruses have a functional relevance for pUL97. The region present in the herpesvirus proteins but which is not found in the S/T PK might be connected with the substrate and the function of the protein. We could show for the first time that phosphorylation of pUL97 is essential for GCV phosphorylation. The existence of amino acids indispensable for protein function again indicate that pUL97 may be an interesting direct target for antiviral therapy.
Acknowledgments
The skillful technical assistance of Anke Lüske and the valuable help of Albert Zimmermann in the performance of the high-pressure liquid chromatography analyses and the kinase assays are greatly appreciated.
P.S. is a student of the DFG-Graduiertenkolleg Biomolekulare Medizin; D.M. was supported by a grant of the BMBF (01KI9603). The work was supported in part by a grant from the European community (PL960471).
REFERENCES
- 1.Baldanti F, Silini E, Sarasini A, Talarico C L, Stanat S C, Biron K K, Furione M, Bono F, Palu G, Gerna G. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J Virol. 1995;69:796–800. doi: 10.1128/jvi.69.2.796-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee M S, Lawrence G L, Barrell B G. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 3.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 4.Fox D S, Schleiss M R. Sequence and transcriptional analysis of the guinea pig cytomegalovirus UL97 homolog. Virus Genes. 1997;15:255–264. doi: 10.1023/a:1007988705909. [DOI] [PubMed] [Google Scholar]
- 5.Hanks S, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–81. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 6.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 7.Hanson M, Preheim L C, Chou S, Talarico C L, Biron K K, Erice A. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob Agents Chemother. 1995;39:1204–1205. doi: 10.1128/aac.39.5.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z, He Y, Kim Y, Chu L, Ohmstede C, Biron K K, Coen D M. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse M, Chen Y-G, Massagué J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGFβ receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:681–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Littler E, Stuart A D, Chee M S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analog ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 12.Lurain N S, Spafford L E, Thompson K D. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger C, Michel D, Schneider K, Lüske A, Schlicht H-J, Mertens T. Human cytomegalovirus UL97 kinase confers ganciclovir susceptibility to recombinant vaccinia virus. J Virol. 1994;68:8423–8427. doi: 10.1128/jvi.68.12.8423-8427.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michel D, Pavic I, Zimmermann A, Haupt E, Wunderlich K, Heuschmid M, Mertens T. The UL97 gene product of the human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J Virol. 1996;70:6340–6347. doi: 10.1128/jvi.70.9.6340-6346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel D, Schaarschmidt P, Wunderlich K, Heuschmid M, Simoncini L, Mühlberger D, Zimmermann A, Pavic I, Mertens T. Functional regions of the human cytomegalovirus protein pUL97 involved in nuclear localization and phosphorylation of ganciclovir and pUL97 itself. J Gen Virol. 1998;79:2105–2112. doi: 10.1099/0022-1317-79-9-2105. [DOI] [PubMed] [Google Scholar]
- 16.Ng T I, Talarico C, Burnette T C, Biron K, Roizman B. Partial substitution of the functions of the herpes simplex virus 1 UL13 gene by the human cytomegalovirus UL97 gene. Virology. 1996;225:347–358. doi: 10.1006/viro.1996.0609. [DOI] [PubMed] [Google Scholar]
- 16a.Prichard M N, Gao N, Jairath S, Mulamba G, Krosky P, Coen D M, Parker B O, Pari G S. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Simoncini, L., F. Baldanti, D. Michel, M. Heuschmid, A. Zimmermann, P. Schaarschmidt, G. Gerna, and T. Mertens. Mutations in the UL97 ORF found in resistant clinical human cytomegaloviruses differentially reduce ganciclovir (GCV) phosphorylation as determined by vaccinia virus recombinants. Submitted for publication. [DOI] [PubMed]
- 19.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 20.Swanson R, Bergquam E, Wong S W. Characterization of rhesus cytomegalovirus genes associated with anti-viral susceptibility. Virology. 1998;240:338–348. doi: 10.1006/viro.1997.8935. [DOI] [PubMed] [Google Scholar]
- 21.Van Zeijl M, Fairhurst J, Baum E Z, Sun L, Jones T R. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology. 1997;231:72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- 22.Wolf D G, Honigman A, Lazarovits J, Tavor E, Panet A. Characterization of the human cytomegalovirus UL97 gene product as a virion-associated protein kinase. Arch Virol. 1998;143:1223–1232. doi: 10.1007/s007050050370. [DOI] [PubMed] [Google Scholar]
- 23.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J Clin Investig. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann A, Michel D, Pavic I, Hampl W, Lüske A, Neyts J, De Clercq E, Mertens T. Phosphorylation of aciclovir, ganciclovir, penciclovir and S2242 by the cytomegalovirus UL97 protein: a quantitative analysis using recombinant vaccinia viruses. Antivir Res. 1997;36:35–42. doi: 10.1016/s0166-3542(97)00034-x. [DOI] [PubMed] [Google Scholar]