Abstract
The cellular localization of the Epstein-Barr virus-encoded RK-BARF0 protein was analyzed by fluorescence microscopy and immunoblotting. The recombinant RK-BARF0 protein was found to be tightly bound to nuclear structures, whereas 16- to 20-kDa RK-BARF0 derivatives, generated by differential splicing of the RK-BARF0 transcript, were present throughout the cell. Moreover, a previously generated anti-RK-BARF0 rabbit serum was found to cross-react with cellular proteins, showing that the previously identified 30- to 35-kDa membrane-associated proteins do not represent RK-BARF0.
Epstein-Barr virus (EBV) is associated with three lymphoproliferative diseases of B-cell origin: acute infectious mononucleosis, endemic Burkitt’s lymphoma (BL), and lymphoma occurring in immunocompromised individuals. In addition, it has been shown that there is a likely etiologic role for EBV in the development of undifferentiated nasopharyngeal carcinoma (NPC) (14). Studies have shown the presence of rightward transcripts from the BamHI A region of EBV in NPC tumor tissue. Sequence analyses of cDNA clones from a transcription library of a tumor xenograft (C15) suggested that primary transcripts were subjected to differential splicing, giving rise to the existence of a family of related transcripts in NPC tissues (4, 5, 7, 9, 18). All NPC tissues tested contained either four or five rightward transcripts of BamHI A which have a common 3′ terminal open reading frame (ORF), termed BARF0. PCR analyses and in situ hybridization, using a riboprobe specific for the BARF0 ORF, also detected BamHI A transcripts in EBV-positive BL cell lines as well as in all EBV-transformed lymphoblastoid cell lines (LCLs) studied (4).
In a recent study (15), cDNA cloning, reverse transcriptase-PCR (RT-PCR), and Northern blotting were used to further define the structures of the BamHI A rightward transcripts. Three BamHI A cDNAs were isolated and found to be previously unidentified mRNAs that contained the BARF0 ORF and additional ORFs encoded by multiple exons, including one, termed RK-BARF0, which extended the size of the BARF0 ORF from 174 to 279 amino acids (aa). Fries et al. (6) raised a rabbit antiserum to a synthetic peptide containing an amino acid sequence encoded within the BARF0 ORF. This antiserum detected a glutathione S-transferase–BARF0 fusion protein and both BARF0 and RK-BARF0 proteins expressed from transfected constructs in H1299 epithelial cells. The serum also immunoprecipitated the 20-kDa protein BARF0 and the 30-kDa protein RK-BARF0 translated in vitro and identified a membrane-associated doublet of 30- and 35-kDa proteins in all of the EBV-infected cell lines tested.
In further work the BARF0 ORF was shown to encode an HLA class I-restricted cytotoxic T lymphocyte (CTL) epitope which was recognized by CTLs from EBV-seropositive, but not -seronegative, individuals (11). In EBV-positive cells this CTL epitope was found to be effectively removed by differential splicing of the BARF0 ORF. These splicing events also resulted in the generation of novel RK-BARF0 protein derivatives of 16 to 20 kDa (10).
Cellular localization of RK-BARF0.
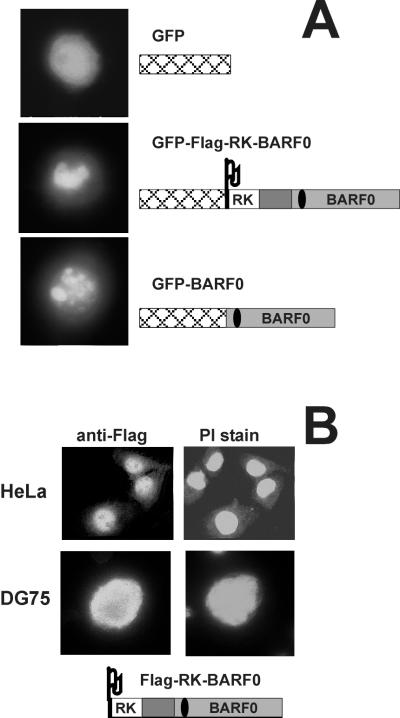
In order to further characterize the biological functions of RK-BARF0 and BARF0 the cellular localization of these proteins was analyzed. The Flag-RK-BARF0 and BARF0 cDNAs (6) were cloned into the KpnI-BamHI and BamHI sites, respectively, of the vector pEGFP-C1 (Clontech) and were transiently expressed in the EBV-negative BL cell line DG75 (3). Localization of the green fluorescent protein (GFP) fusion proteins (GFP-Flag-RK-BARF0 and GFP-BARF0) and GFP alone was determined by fluorescence microscopy on living cells (Fig. 1A). As expected, GFP alone, being present in both the nucleus and cytoplasm, was distributed throughout the cell. In contrast, both the GFP-Flag-RK-BARF0 and GFP-BARF0 cell transfectants showed prominent nuclear staining that formed bright condensed structures. This result was surprising, as RK-BARF0 was thought to be membrane associated (6). As the cytoplasm of DG75 cells is fairly small it was difficult to determine the proportions of GFP-Flag-RK-BARF0 and GFP-BARF0 that were present in the cytoplasm. To confirm that RK-BARF0 was indeed associated with the nucleus, Flag-tagged RK-BARF0 cDNA, cloned into the expression vector EBO-pLPP (10), was transiently introduced into HeLa cells or stably expressed in DG75 cells. The transfected cell lines were treated with propidium iodide (2 μg per ml) to stain nucleic acid and with anti-Flag monoclonal antibody (MAb) (diluted 1:60 in phosphate-buffered saline) (Eastman Kodak) to identify Flag-tagged proteins. Indirect immunofluorescence showed that the Flag-tagged proteins were present throughout the cell but were predominantly localized in the parts of the nucleus other than the nucleoli (Fig. 1B). These results confirmed the data for localization of the GFP fusion proteins and indicated that a sequence within the BARF0 ORF was sufficient for targeting the proteins to the nucleus.
FIG. 1.
Fluorescence microscopy in transfected EBV-negative cells. The tagged RK-BARF0 and BARF0 gene constructs are illustrated and include the RK exon (open box), the BARF0 ORF (grey box), and the in-frame sequence (dark grey box) located 5′ of the BARF0 ORF (according to references 6 and 15) as well as the Flag epitope (Flag) and GFP (cross-hatched box). The location of the 20-mer peptide used to raise the polyclonal anti-RK-BARF0 rabbit serum (6) is shown by a black oval. (A) DG75 cells were transiently transfected with plasmids encoding GFP, GFP-Flag-RK-BARF0, or GFP-BARF0, and GFP fluorescence was analyzed in living cells. (B) The Flag-RK-BARF0 gene was either transiently expressed in HeLa cells or stably expressed in DG75 cells. Cells were stained with an anti-Flag MAb (anti-Flag) and propidium iodide (PI stain) and analyzed by indirect immunofluorescence of fixed cells.
Fractionation of cells expressing Flag-RK-BARF0 and its derivatives.
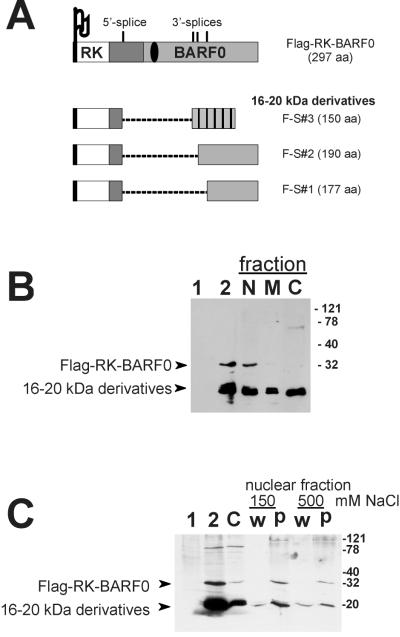
We have previously shown that the RK-BARF0 transcript undergoes differential splicing utilizing an identical 5′ splice site and one of three different 3′ splice sites. This results in the generation of three RK-BARF0 derivatives of 190 or 177 aa and a frameshifted chimeric protein product (which is totally unrelated to RK-BARF0 in its C-terminal half) of 150 aa (10) (Fig. 2A). To confirm the fluorescence study results and to determine whether the RK-BARF0 16- to 20-kDa protein derivatives were also localized to the nucleus, DG75 cells stably expressing Flag-RK-BARF0 were fractionated into nuclear, membrane, and cytoplasmic fractions as described recently (6). Protein was detected by immunoblotting by using an anti-Flag antibody, diluted 1:300 (12). The total lysate of transfected cells showed low amounts of full-length RK-BARF0 protein but high levels of 16- to 20-kDa Flag-tagged RK-BARF0 derivative proteins (Fig. 2B). The 32-kDa Flag-RK-BARF0 protein was found to be almost exclusively associated with the nuclear fraction, while the 16- to 20-kDa derivatives were present in all cellular fractions. These results indicated that the sequence responsible for the nuclear association of RK-BARF0 was encoded within the region which was removed by splicing of the RK-BARF0 transcript. Interestingly, the region removed was arginine rich and had been suggested to be involved in binding of nucleic acids (15).
FIG. 2.
Fractionation of EBV-negative DG75 cells expressing Flag-RK-BARF0. (A) Summary of the differential splicing of RK-BARF0 as reported recently (10). The locations of the splice sites are shown, and the sizes (in amino acids) of the full-length Flag-tagged RK-BARF0 and its 16- to 20-kDa protein derivatives are given. For graphic details see the legend of Fig. 1. Note that the products F-S#1 and F-S#2 maintain the BARF0 frame, whereas F-S#3 undergoes a frameshift in its C terminus (cross-hatched). (B) Cells stably expressing Flag-RK-BARF0 were separated into nuclear (N), membrane (M), and cytoplasmic (C) fractions. For controls, total extracts of cells expressing either the control vector (lane 1) or Flag-RK-BARF0 (lane 2) were used. Total and fractionated proteins were separated by SDS–12% polyacrylamide gel electrophoresis, and their expression was analyzed by immunoblotting by using an anti-Flag MAb. Molecular size markers (expressed in kilodaltons) are shown, and the positions of Flag-RK-BARF0 and its 16- to 20-kDa derivatives are indicated by arrows. (C) Cells stably expressing Flag-RK-BARF0 were separated into cytoplasmic (C) and nuclear fractions. Samples of the nuclear extract were then incubated with buffers containing either low (150 mM) or high (500 mM) NaCl concentration, and the corresponding nuclear supernatant (w) and pellet (p) were collected and analyzed by immunoblotting.
To determine whether full-length RK-BARF0 was free in the nucleoplasm or was associated with structures within the nucleus, nuclei were prepared from DG75 cells stably expressing Flag-RK-BARF0 and were extracted with buffers containing different concentrations of NaCl according to the method of Sambrook et al. (16). Flag-RK-BARF0 was detected by immunoblotting by using the anti-Flag MAb and was not removed from the nucleus by washing with 150 mM NaCl, and the majority of the protein remained associated with the nuclear pellet even after washing with 500 mM NaCl (Fig. 2C). Taken together these data clearly demonstrated that the RK-BARF0 protein was not membrane associated, as previously thought (6), but was instead localized to structures within the nucleus.
Specificity of the polyclonal rabbit RK-BARF0 antiserum.
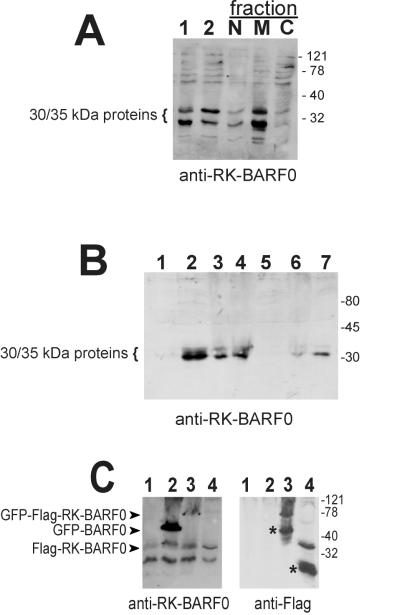
The study generating the original data on the membrane localization of RK-BARF0 utilized cellular fractionation of an EBV-positive LCL and a polyclonal rabbit anti-RK-BARF0 serum (6). This anti-RK-BARF0 serum (diluted 1:1,000) was therefore utilized to probe the Flag-RK-BARF0-expressing DG75 cell fractions. As shown in Fig. 3A, the 30- and 35-kDa proteins, previously identified by Fries et al. (6), were as expected present primarily in the membrane fraction, suggesting that they did not represent RK-BARF0. More importantly, these proteins were also detected in the EBV-negative DG75 cells (Fig. 3C, lane 1), indicating that the anti-RK-BARF0 serum cross-reacted with cellular proteins.
FIG. 3.
Specificity of the RK-BARF0 antiserum. (A) DG75 cells stably expressing Flag-RK-BARF0 were separated into nuclear (N), membrane (M), and cytoplasmic (C) fractions and subjected to SDS–12% polyacrylamide gel electrophoresis. For controls, total extracts of cells expressing either the control vector (lane 1) or Flag-RK-BARF0 (lane 2) were used. (B) Total cell extracts of the EBV-negative B-cell lymphoma line BJAB (lane 1), of the EBV-positive LCL IS (lane 2), and of the EBV-negative BL cell lines BL41 (lane 3), BL30K (lane 4), Ramos (lane 5), DG75 (lane 6), and BL30A (lane 7) were separated by SDS–12% polyacrylamide gel electrophoresis. (C) Total cell extracts of DG75 cells transfected with plasmids encoding GFP (lane 1), GFP-BARF0 (lane 2), GFP-Flag-RK-BARF0 (lane 3), and Flag-RK-BARF0 (lane 4) were separated by SDS–10% polyacrylamide gel electrophoresis. Protein expression was analyzed by immunoblotting by using either the polyclonal anti-RK-BARF0 serum or an anti-Flag MAb (indicated at the bottom). Molecular size markers (expressed in kilodaltons) are shown, and the positions of the cross-reactive 30- to 35-kDa proteins and the tagged RK-BARF0 and BARF0 proteins are indicated at the left. The asterisks mark proteins which were detected by the anti-Flag MAb but not by the anti-RK-BARF0 serum.
To confirm that the 30- and 35-kDa proteins detected with the rabbit antiserum were of cellular origin, a panel of EBV-negative B-cell lines was examined by immunoblotting by using this antiserum (Fig. 3B). The results were in agreement with the data presented by Fries et al. (6) in that neither BJAB nor Ramos cell lines (1) expressed the 30- and 35-kDa proteins. However, the 30- and 35-kDa proteins were identified in the EBV-negative BL cell lines BL41, BL30K, DG75, and BL30A (13) and in an EBV-positive control LCL. The BL41, BL30, and DG75 cell lines were confirmed to be EBV negative by DNA PCR, RT-PCR, and immunoblotting (data not shown). To ensure that the sample of rabbit serum being used still retained its reactivity to the epitope within the BARF0 ORF, against which it was raised, immunoblot analyses of DG75 cells expressing either GFP-Flag-RK-BARF0, GFP-BARF0, or Flag-RK-BARF0 were performed. By using the rabbit antiserum, GFP-Flag-RK-BARF0 and GFP-BARF0 were detected on the immunoblots, demonstrating that the rabbit serum indeed contained antibodies specific for BARF0 (Fig. 3C, lanes 2 and 3). Detection of Flag-RK-BARF0 was difficult not only because this protein has the same molecular weight as the 35-kDa cellular cross-reactive protein but also because the differential splicing of the RK-BARF0 message largely reduced the amount of full-length protein (lanes 1 and 4). By using the anti-Flag MAb both the GFP-Flag-RK-BARF0 and the Flag-RK-BARF0 proteins were detected. The anti-Flag MAb also reacted with smaller Flag-tagged protein products, of approximately 60 kDa and 16 to 20 kDa in size, which originated from differential splicing of the GFP-RK-BARF0 and RK-BARF0 messages, respectively (Fig. 3C, lanes 3 and 4). Consequently, these proteins were not detected by the rabbit anti-RK-BARF0 serum, as the differential splicing caused the loss of the B-cell epitope region used to raise the rabbit antiserum. These data demonstrated that the membrane-associated 30- and 35-kDa proteins, detected by the rabbit anti-RK-BARF0 serum, did not represent RK-BARF0 and were cellular in origin. The presence of these cellular proteins would have masked any virus-encoded RK-BARF0 protein expressed in the EBV-positive cell lines.
Analyses of BARF0 transcripts in EBV-infected cells.
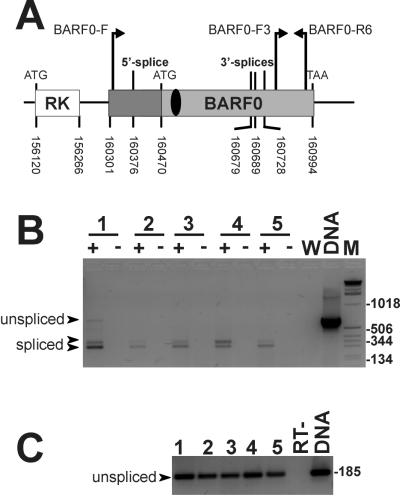
In EBV-positive BL cells, LCLs, and NPC cells, transcripts encoding the BARF0 ORF were recently shown to utilize a 5′ splice site and one of three different 3′ splice sites (10). The expression pattern of the BamHI A rightward messages is very complex, and so far no major transcript has been described. To obtain a semiqualitative estimation of the frequency of the differentially spliced BARF0 transcripts, expressed relative to the amounts of all of the BamHI A transcripts, RT-PCR was performed as outlined recently (10). Briefly, total RNA from a panel of EBV-positive cells was reverse transcribed with oligo-dT primers. For negative controls, water and the RT sample missing the RT enzyme were used. A positive control consisted of plasmid DNA of Flag-RK-BARF0. The primers BARF0-F3 (5′-TCTGCCGTGAAGGGTTG; position 160789 within the B95.8 EBV genome [2]) and BARF0-R6 (5′-GTGTTTTATTGCATGTCTCACACC; position 160973 within the same genome) were specific for the 185-bp-long 3′ end of the BARF0 ORF and should represent all BamHI A transcripts. The primers BARF0-F (5′-GCCCGAGGAGCTGTAGACC; position 160308 within the B95.8 EBV genome) and BARF0-R6 covered the complete BARF0 ORF and amplified full-length (666 bp) and spliced forms (314, 353, and 363 bp) (Fig. 4A). Comparison of the PCR-amplified products revealed that the differentially spliced BARF0 products (Fig. 4B) composed a small but significant proportion of the total viral BamHI A transcripts which were unspliced in their 3′-terminal ends (Fig. 4C).
FIG. 4.
RT-PCR of EBV-positive cells. (A) Schematic diagram of the genomic RK-BARF0 organization in EBV and the locations of the primers used. The numbering refers to the EBV sequence of strain B95.8 (2) and indicates start, stop, and splice sites as reported recently (10, 15). For graphical details see the legend of Fig. 1. (B) Total RNAs from the EBV-positive cell lines SB (an LCL) (lane 1), MutuI-BL (lane 2), and MutuIII-BL (lane 3), from the tumor xenograft C15-NPC (lane 4), and from EBV-negative DG75 cells stably expressing Flag-RK-BARF0 (lane 5) were analyzed by RT-PCR. The positive (+) and negative (−) RT samples, the water control (W), and the positive control DNA of the plasmid encoding Flag-RK-BARF0 (DNA) were amplified by using the primers BARF0-F and BARF0-R6. The PCR products were separated and visualized by electrophoresis on an ethidium bromide-containing 2.5% agarose gel. Some markers of the 1-kbp DNA ladder (M) are shown on the right, and the positions of the unspliced and spliced cDNAs are indicated by arrowheads. (C) RT-PCR performed by using the primers BARF0-F3 and BARF0-R6. Lanes 1 through 5 correspond to the positive RT samples used in the RT-PCR shown in panel B, and one negative RT sample is shown as a control. The molecular size of the unspliced cDNA is given.
Analysis of RK-BARF0 expression in EBV-infected cells.
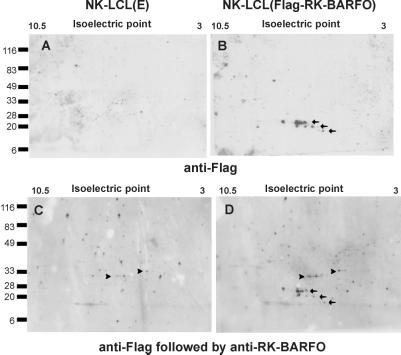
To further characterize the recombinant 16- to 20-kDa RK-BARF0 protein derivatives, two-dimensional (2D) gel electrophoresis was employed. Protein extracts of the EBV-positive cells of the NK LCL, which stably expressed either a control vector or Flag-RK-BARF0 (10), were subjected to 2D gel electrophoresis by using the Immobline Drystrip (pH range, 3 to 10.5) and an ExelGel sodium dodecyl sulfate (SDS) gradient (8 to 18%) according to the instructions of the manufacturer (Pharmacia), followed by immunoblotting by using an anti-Flag MAb. The anti-Flag MAb detected three sets of proteins, ranging from 16 to 20 kDa and with isoelectric point (pI) values ranging between 5 and 8, in the Flag-RK-BARF0-transfected cells but not in the control cells (Fig. 5A and B). These values correlated well with the predicted sizes (Fig. 2A) and pI values of the splice variants generated from the Flag-RK-BARF0 message. Each set of Flag-tagged proteins consisted of proteins with similar molecular weights but different pI values, suggesting that these proteins were posttranslationally modified. Since the pattern of differential splicing within the BARF0 mRNA also occurs in EBV-infected cells, as determined by RT-PCR, it is tempting to speculate that these truncated protein variants may be expressed in vivo and have some biological activity. Indeed, differential splicing is well known to regulate gene expression and to create protein isoforms with different functions (reviewed in reference 17).
FIG. 5.
2D gel electrophoresis. Total cell extracts of NK LCLs stably expressing a control vector (left) or Flag-RK-BARF0 (right) were separated according to their isoelectric points (first dimension) and molecular weights (second dimension). Protein expression was analyzed by immunoblotting by using an anti-Flag MAb (upper and lower) followed by the anti-RK-BARF0 serum (lower). Molecular size markers (expressed in kilodaltons) and the isoelectric point range are given. Arrows and arrowheads indicate the positions of proteins specifically detected by the anti-Flag MAb and by the anti-RK-BARF0 serum, respectively.
In order to analyze RK-BARF0 expression in vivo, the membranes were reprobed with the anti-RK-BARF0 serum, and the analysis revealed two sets of protein spots with pI values of approximately 5 to 7 and molecular masses of 30 to 35 kDa (Fig. 5C and D). As these proteins were present in both control transfectants and cell transfectants expressing Flag-RK-BARF0, they probably corresponded to the 30- to 35-kDa cross-reactive cellular proteins. Full-length RK-BARF0 was not detected in any of the transfected LCLs, and this was probably due to the small amounts of unspliced message expressed in the LCLs (Fig. 4, and see reference 10) and the high theoretically predicted pI values (10.2 and 10.9, respectively) (which were at the edge of the resolution of the 2D gel) for the Flag-tagged RK-BARF0 and viral RK-BARF0.
In summary, this study, in contrast to a recent report (6), demonstrates that the 30- to 35-kDa membrane-associated proteins are not encoded by the viral RK-BARF0 transcripts but instead are cellular proteins. This was clearly shown by immunoblot analyses identifying the 30- to 35-kDa membrane-associated proteins in a panel of EBV-negative BL cell lines. In addition 2D gel electrophoresis revealed that the apparent pIs of the 30- to 35-kDa membrane-associated proteins did not match the theoretical pI of the RK-BARF0 sequence. Support also came from fluorescence and fractionation assays using Flag-tagged RK-BARF0 protein, which revealed that, rather than being membrane associated, the recombinant Flag-RK-BARF0 protein was localized predominantly in the nucleus.
While these data demonstrate that the 30- to 35-kDa membrane-associated proteins do not represent RK-BARF0, they do not exclude the existence of the RK-BARF0 gene product in EBV-positive cells. Thus far the best evidence that RK-BARF0 and/or the BARF0 proteins are expressed in vivo comes from the observations that RNA transcripts capable of encoding these proteins exist in EBV-infected cells (4, 15, 18), that antisera from NPC patients reacted with in vitro-translated BARF0 protein (8), and that the BARF0 ORF encodes an HLA class I-restricted T-cell epitope which was recognized by CTLs from EBV-seropositive, but not -seronegative, individuals (11). The latter study and a subsequent report (10) indicated that the BARF0 antigen levels were low in EBV-infected cells, as LCLs and BL cell lines were poorly recognized by BARF0-specific CTLs due to differential splicing of the BARF0 ORF. It is apparent that final confirmation of the expression of the RK-BARF0 protein and of its 16- and 20-kDa derivatives in EBV-infected cells will require the generation of better antiserum. Since the 3′ region of the BARF0 ORF did not appear to undergo any additional splicing events, it should encode an amino acid sequence which is present in most BARF0-derived proteins, and thus the sequence could be used as a source of synthetic peptides for future antibody production.
Acknowledgments
We are grateful for the support of the members of the EBV unit at QIMR, particularly for technical help provided by L. Morrison and L. Poulsen, and are indebted to the generosity of P. Busson (Institute Gustave Roussy, Villejuif, France) and N. Raab-Traub (University of North Carolina, Chapel Hill) for providing NPC samples and plasmids.
This work was supported by grants from the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund (QCF), and the University of Queensland, Australia.
REFERENCES
- 1.Andersson M, Lindahl T. Epstein-Barr virus DNA in human lymphoid cell lines: in vitro conversion. Virology. 1976;73:96–105. doi: 10.1016/0042-6822(76)90064-7. [DOI] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Ben Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 4.Brooks L A, Lear A L, Young L S, Rickinson A B. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol. 1993;67:3182–3190. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H L, Lung M M, Sham J S, Choy D T, Griffin B E, Ng M H. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology. 1992;191:193–201. doi: 10.1016/0042-6822(92)90181-n. [DOI] [PubMed] [Google Scholar]
- 6.Fries K L, Sculley T B, Webster Cyriaque J, Rajadurai P, Sadler R H, Raab Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilligan K, Sato H, Rajadurai P, Busson P, Young L, Rickinson A, Tursz T, Raab-Traub N. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J Virol. 1990;64:4948–4956. doi: 10.1128/jvi.64.10.4948-4956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilligan K J, Rajadurai P, Lin J-C, Busson P, Abdel-Hamid M, Prasad U, Tursz T, Raab-Traub N. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kienzle N, Sculley T B, Greco S, Khanna R. Cutting edge: silencing virus-specific cytotoxic T cell-mediated immune recognition by differential splicing: a novel implication of RNA processing for antigen presentation. J Immunol. 1999;162:6963–6966. [PubMed] [Google Scholar]
- 11.Kienzle N, Sculley T B, Poulsen L, Buck M, Cross S, Raab-Traub N, Khanna R. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J Virol. 1998;72:6614–6620. doi: 10.1128/jvi.72.8.6614-6620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kienzle N, Young D B, Liaskou D, Buck M, Greco S, Sculley T B. Intron retention may regulate expression of Epstein-Barr virus nuclear antigen 3 family genes. J Virol. 1999;73:1195–1204. doi: 10.1128/jvi.73.2.1195-1204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenoir G M, Vuillaume M, Bonnardel C. The use of lymphomatous and lymphoblastoid cell lines in the study of Burkitt’s lymphoma. Int Agency Res Cancer Sci Publ. 1985;60:309–318. [PubMed] [Google Scholar]
- 14.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 15.Sadler R H, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Smith C W, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 18.Smith P R, Gao Y, Karran L, Michael D J, Snudden D, Griffin B E. Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors. J Virol. 1993;67:3217–3225. doi: 10.1128/jvi.67.6.3217-3225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]