Abstract
Several studies published so far used highly selective image datasets from unclear sources to train computer vision models and that may lead to overestimated results, while those studies conducted in real-life remain scarce. To avoid image selection bias, we stacked convolutional and recurrent neural networks (CNN-RNN) to analyze complete optical coherence tomography (OCT) cubes in a row and predict diabetic macular edema (DME), in a real-world diabetic retinopathy screening program. A retrospective cohort study was carried out. Throughout 4-years, 5314 OCT cubes from 4408 subjects who attended to the diabetic retinopathy (DR) screening program were included. We arranged twenty-two (22) pre-trained CNNs in parallel with a bidirectional RNN layer stacked at the bottom, allowing the model to make a prediction for the whole OCT cube. The staff of retina experts built a ground truth of DME later used to train a set of these CNN-RNN models with different configurations. For each trained CNN-RNN model, we performed threshold tuning to find the optimal cut-off point for binary classification of DME. Finally, the best models were selected according to sensitivity, specificity, and area under the receiver operating characteristics curve (AUROC) with their 95% confidence intervals (95%CI). An ensemble of the best models was also explored. 5188 cubes were non-DME and 126 were DME. Three models achieved an AUROC of 0.94. Among these, sensitivity, and specificity (95%CI) ranged from 84.1–90.5 and 89.7–93.3, respectively, at threshold 1, from 89.7–92.1 and 80–83.1 at threshold 2, and from 80.2–81 and 93.8–97, at threshold 3. The ensemble model improved these results, and lower specificity was observed among subjects with sight-threatening DR. Analysis by age, gender, or grade of DME did not vary the performance of the models. CNN-RNN models showed high diagnostic accuracy for detecting DME in a real-world setting. This engine allowed us to detect extra-foveal DMEs commonly overlooked in other studies, and showed potential for application as the first filter of non-referable patients in an outpatient center within a population-based DR screening program, otherwise ended up in specialized care.
Keywords: Deep learning, Diabetic Macular Edema, Diabetic Retinopathy, Optical Coherence Tomography, Screening, Telemedicine
Subject terms: Predictive medicine, Type 2 diabetes
Introduction
Diabetes mellitus (DM), specially type II, remains a challenge for public health as the incidence continues to rise1,2, and with it the burden of associated comorbidities as diabetic eye disease3. Among the latter, diabetic macular edema (DME) is the most prevalent sight-threatening (ST) condition although treatable while macular changes are not yet permanent4. Several countries have implemented successful diabetic retinopathy (DR) screening programs, mainly based in fundus retinography (FR), that lowered the overall incidence of blindness5. Nevertheless, recent reports advocated for the potential benefit of adding optical coherence tomography (OCT) to FR, as it may detect indirect early DME signs often missed by FR in a non-negligible proportion,6 improving the cost-effectiveness of screening models7. As a counterpart, the deployment of such program faces new resourcing challenges that could jeopardize the sustainability of any public health system; adding OCT to FR raises the instrumentation costs, and highly skilled specialists are needed to read the OCT cubes to achieve the best results6,8.
To address some of these challenges, artificial intelligence, especially deep convolutional neural networks (CNNs), have been widely tested for the automated classification of macular pathologies in FR and/or OCT scans, as well as to assist the retina specialists to speed-up their decision-making process9. In this sense, several studies published using CNNs for the classification of DME showed specialist-level or even outperformed the results achieved by retina specialists, although their generalizability raises important concerns10,11. In some studies, authors used 2D-OCT scans (B-scans) and that introduced a selection bias as training images must be previously selected from the OCT cube12,13. Moreover, the majority used central fovea B-scans that could potentially miss both non-foveal involving anatomical signs and DMEs10–13. Some authors relied in FR alone to detect DME, thus based on indirect signs that likely indicate progression which in turn may be easier to detect by the models10,14. Furthermore, in most of them the authors did not state the criteria followed to assess DME10. In consequence, most of the datasets used in these studies may not represent the actual distribution of the disease in the general population, so that may lead to overestimated performances and poor generalizability in turn.
Recurrent neural networks (RNN) are deep learning models suitable to analyze time-series data and could be stacked at the output of several CNNs to predict time-dependent outcomes in images15,16. Hence, such hybrid model (CNN-RNN) applied to OCT B-scans would allow to analyze the OCT cube in a row and sequentially in time.
Thus, the aim of the present study was to build hybrid deep learning models (CNN-RNN) with different settings and trained with an “ad hoc” ground truth of DME, then to test them for the detection of DME in an unselected dataset from a real-world DR screening program.
Methods
Model development and training with a ground truth of DME
A detailed description of all models settings, training and testing, as well as image processing was shown in Supplementary Material 1. Briefly, we pre-trained a backbone custom CNN using a publicly available dataset comprised of B-scans of DME, drusen, choroidal neovascularization, and normal macula (Supplementary Material 1.1–1.2)12. Then, twenty-two (22) of these pre-trained CNNs were stacked in parallel, initialized with the pre-training weights, and fed with 22 unselected B-scans extracted from the OCT cube. Researchers did not intervene in the selection of images for training or testing, so that avoided image selection bias. To this end, we divided each 128-slice cube into those that mainly comprised the foveal zone and those which mainly comprised the parafoveal zone. The foveal zone was captured between slices 60 to 85 from where we automatically extracted every two (12 B-scans), being the remaining the parafoveal zone from where we automatically extracted every ten (10 B-scans; Supplementary Material 1.3). Every pre-trained CNN outputs an embedding of image features from the flatten layer. Then, all were concatenated into a sequence which was forwarded to the bidirectional recurrent layer. The bidirectional wrapper moves a cell forward and another backwards along the sequence to learn dependencies between time-dependent features. Finally, the output from the RNN layer was fully-connected to a sigmoid layer to predict the probability of DME for the 22-slice OCT cube (Supplementary Fig. 1).
We used a ground truth of DME and normal macula, described elsewhere6 but enriched with additional OCT cubes from a second DR screening program (Hospital Clínic of Barcelona, Spain). Additional samples were graded following the same criteria6. All images were acquired using the Topcon 3D OCT-Maestro 1. The ground truth dataset was split into training, validation, and test, ensuring a similar proportion of DME between folds, and avoiding data leakage by creating splits of unique subjects. The binary cross-entropy was used as the loss function, although accuracy, the area under the receiver operating characteristic (ROC) curve (AUROC), and area under the precision-recall curve (AUPRC) were also computed and compared. From the pool of trained models, we selected those with the best generalizable metrics in the test set.
Study cohort and data collection
A retrospective cohort study nested in a teleophthalmology real-world DR screening program was conducted. From November 2015 to March 2019, we included all diabetic patients (either type), of any gender, and aged ≥ 18 years. We included one eye per subject, which was the affected in case of unilateral DME, or a random sample if both eyes had the same diagnosis (DME or non-DME).
The characteristics of the screening program were described elsewhere6. In short, screening visits were conducted by a technician in an outpatient center and collected health data of interest, measured the best-corrected visual acuity (BCVA), and acquired a 3-field FR (first centered on the macula, second on the disc, and third supero-temporal)17, and a 6 × 6 mm OCT macular cube scan. Patients were re-imaged under pupil dilation in case of low quality of images. Then, the technician forwarded all the abovementioned data, also including the ETDRS average thicknesses, the ETDRS topographic map, and the macular volume, to a retina specialist in the Hospital who acts as the gatekeeper to specialized care. There, the retina specialist makes the initial diagnosis, and decides whether to refer the patient. The retina specialist assessed DME based on the presence of macular thickening ≥ 300 µm with anatomical signs of DME (cysts, microaneurysms, exudates, neurosensory detachment, and hyperreflective dots), without signs of another macular disease in the FR18,19. The severity of DME was also assessed depending on the distance to central fovea as proposed by the International Clinical Diabetic Retinopathy and Diabetic Macular Edema Disease Severity Scales (ICDRSS), into mild, moderate and severe20. Subjects whose OCT and FR were missing or ungradable were excluded from the study cohort.
Model evaluation in the study cohort
All 22-slice OCT cubes from the study cohort were extracted and pre-processed, then fed to the CNN-RNN models to predict the probability of DME. Model predictions were evaluated against the diagnosis made by the retina specialist in the screening program, as described above. ROC curves, AUROC, and the partial AUROC (pAUROC) at a range of false positive rate (FPR) < 0.05 and < 0.1, along with their 95% confidence intervals (95%CI) were computed. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and the cumulative incidence of DME were calculated at a range of 1000 prospectively tuned thresholds set to predict the best parameters for screening: (1) Youden index (threshold 1), (2) the highest sensitivity (threshold 2), and (3) the highest specificity (threshold 3). For the latter two, we established a baseline specificity and sensitivity of at least 0.8 or greater. Only the models that yielded the best AUROC with a pAUROC over 0.80 were considered onwards. The selected models were ensembled to build a voting classifier based on the mode of class predictions obtained at each model threshold.
Additionally, the characteristics of false positives and false negatives obtained were reviewed by a third retina specialist.
Finally, as an exploratory analysis, we tested the potential generalizability of the models for classification of referable DR. In this case, we graded DR as mild non-proliferative, moderate non-proliferative, and referable DR, which included severe non-proliferative and proliferative plus moderate and severe DME20.
Statistical analysis
Baseline characteristics of the population were expressed as median and interquartile range (IQR) for quantitative variables, and as frequencies and percentages for qualitative variables. Differences between two medians were tested using the Mann–Whitney’s U, and the test on the equality of proportions was used to compare two qualitative variables. A p-value < 0.05 was set as statistically significant.
ROC curves and AUROC (95%CI) were computed to test the model performance as it is independent of the prevalence of the disease. Partial AUROC (pAUROC; 95%CI) was also computed to obtain more nuanced models21. Youden (J) index was computed to represent the maximum sensitivity and specificity given for a single point on the ROC curve. Diagnostic accuracy for binary outcomes was assessed using the sensitivity, specificity, PPV, and NPV, after probability thresholding. Diagnostic accuracy was also stratified by gender, age, laterality, BCVA, grade of DR, OCT quality of image, and grade of DME. For the latter, due to low numbers, we collapsed moderate and severe. Incidence of DME predicted by the models was calculated as the number of true positives divided by the total number of subjects. Intervals at 95% of confidence (95%CI) were calculated using the standard normal distribution or the binomial distribution for proportions22.
To test the assumption of no missing cases due to lower resolution of images (22-slice cubes) we carried out a sensitivity analysis by severity of DME, as well as by other covariates as grade of DR, BCVA, quality of image, among others.
Models were developed in GPU-enabled Tensorflow v.2.4, and diagnostic accuracy was computed with Scikit-learn v. 1.2.2, for Python. The remaining analyses were run with STATA/MP v.17 (Stata Corp LLC, College Station, TX, USA).
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the University Hospital “Príncipe de Asturias” on March 2, 2020. The need for informed consent was waived due to the retrospective nature of the study . This study complied with the provisions of Spanish and European laws on personal data as well as with the Declaration of Helsinki (Fortaleza 2013).
Results
Baseline and clinical characteristics of the study cohort
The characteristics of the study population were detailed in Table 1. We included 5314 screens from 4408 subjects. Of them, 126 (2.37%) were DMEs, and 5188 (97.6%) had no record of DME throughout the study period. The majority of DMEs were mild (77; 61.1%), while moderate and severe contributed equally (23; 18.3%, each). Compared to subjects without, those with DME had fewer screening visits, were older, predominantly males, with longer duration of DM, and had a greater prevalence of acute myocardial infarction (Table 1). Regarding vision, subjects with DME had lower BCVA, greater prevalence of moderate and severe non-proliferative DR, as well as proliferative DR, and had greater proportion of epiretinal membranes, pupil dilation or previous macular scars. In OCT, subjects with DME showed greater average macular thicknesses and volume across the ETDRS grid (Table 1).
Table 1.
Baseline characteristics of the study cohort.
| Non-DME (N = 5188) | DME (N = 126) | p-value | |
|---|---|---|---|
| Age, years, median (IQR) | 65 (56–72) | 68 (58–75) | 0.01 |
| Gender, females, n (%) | 2129 (41.0) | 41 (32.5) | 0.06 |
| Right eye, n (%) | 2512 (48.4) | 61 (48.4) | 1 |
| CV risk factors, n (%) | |||
| Current smoker | 960 (18.5) | 17 (13.5) | 0.15 |
| Hypertension | 3305 (63.7) | 88 (69.8) | 0.16 |
| Dyslipidemia | 3517 (67.8) | 88 (69.8) | 0.63 |
| Acute myocardial infarction | 616 (11.9) | 26 (20.6) | 0.003 |
| Stroke | 31 (0.60) | 1 (0.79) | 0.78 |
| Type of diabetes, n (%): | |||
| 1 | 443 (8.57) | 6 (4.76) | 0.13 |
| 2 | 4728 (91.4) | 120 (95.2) | |
| Duration of diabetes, n (%): | |||
| ≤ 15 years | 3710 (72.7) | 67 (53.6) | < 0.001 |
| > 15 years | 1392 (27.3) | 58 (46.4) | |
| BCVA, Snellen decimal, n (%): | |||
| ≤ 0.5 | 790 (15.2) | 46 (36.5) | < 0.001 |
| > 0.5 | 4398 (84.8) | 80 (63.5) | |
| Diabetic retinopathy, n (%) | |||
| No abnormalities | 4147 (80.0) | 12 (9.52) | < 0.001 |
| Mild non-proliferative | 593 (11.4) | 19 (15.1) | |
| Moderate non-proliferative | 340 (6.56) | 63 (50.0) | |
| Severe non-proliferative | 18 (0.35) | 19 (15.1) | |
| Proliferative* | 26 (0.50) | 12 (9.52) | |
| Ungradable | 64 (1.23) | 1 (0.79) | |
| Other retinal pathologies, n (%): | |||
| Age-related macular degeneration | 175 (3.37) | 1 (0.79) | 0.13 |
| Severe myopia | 33 (0.64) | – | – |
| Epiretinal membranes | 50 (0.96) | 7 (5.56) | < 0.001 |
| Grade of DME, n (%): | |||
| Mild | – | 77 (61.1) | |
| Moderate | – | 23 (18.3) | |
| Severe | – | 23 (18.3) | |
| Not recorded | – | 3 (2.38) | - |
| Pupil dilation, n (%) | 662 (12.8) | 29 (23.0) | 0.001 |
| Macular scars, n (%) | 51 (0.98) | 5 (3.97) | 0.01 |
| Corneal and vitreal opacities, n (%) | 28 (0.54) | – | – |
| Scans with artifacts, n (%): | 39 (0.75) | 1 (0.79) | 0.62 |
| Screening visits, n (%): | |||
| 1 | 3553 (68.5) | 120 (95.2) | < 0.001 |
| 2 | 1298 (25.0) | 6 (4.76) | < 0.001 |
| 3 | 309 (5.96) | – | – |
| 4 | 28 (0.54) | – | – |
| OCT measurements, median (IQR) | |||
| Quality of image, TopQ | 48.3 (42.0–53.8) | 43.8 (37.9–49.9) | < 0.001 |
| Macular volume, mm3 | 7.45 (7.18–7.71) | 7.89 (7.57–8.39) | < 0.001 |
| CST, µm | 201.0 (184.0–224.0) | 262.5 (218.0–316.0) | < 0.001 |
| Center macula, µm | 237.8 (222.4–254.0) | 284.8 (254.9–329.3) | < 0.001 |
| Temporal inner macula, µm | 285.0 (273.5–295.9) | 305.5 (285.3–331.6) | < 0.001 |
| Superior inner macula, µm | 296.6 (284.9–307.9) | 311.2 (295.5–334.4) | < 0.001 |
| Nasal inner macula, µm | 298.9 (286.7–310.2) | 315.9 (301.0–336.2) | < 0.001 |
| Inferior inner macula, µm | 292.3 (280.3–304.4) | 309.9 (291.4–337.1) | < 0.001 |
| Temporal outer macula, µm | 244.2 (234.0–254.1) | 259.2 (244.2–277.9) | < 0.001 |
| Superior outer macula, µm | 254.9 (244.2–264.7) | 266.0 (252.8–284.6) | < 0.001 |
| Nasal outer macula, µm | 271.9 (260.5–282.6) | 280.4 (270.1 –302.8) | < 0.001 |
| Inferior outer macula, µm | 252.7 (242.1–263.5) | 265.7 (252.8–286.5) | < 0.001 |
DME diabetic macular edema, IQR interquartile range, CV cardiovascular, BCVA best corrected visual acuity, OCT optical coherence tomography, CST central subfield thickness.
*Includes proliferative and photocoagulated proliferative DR.
Training of hybrid models with a ground truth dataset of DME
The ground truth dataset comprised 650 OCT cubes from unique subjects, being 433 (66.6%) of them in the training set (127 -29.3%- DMEs, and 306 -70.7%- non-DMEs), 100 (15.4%) in the validation set (20 -20%- DMEs, and 80 -80%- non-DMEs), and 117 (18%) in the test set (40 -34.2%- DMEs, and 77 -65.8%- non-DMEs). Seven (7) different hybrid models yielded the best predictive and generalizable results in the test set (Supplementary material 1.4–1.6).
Model selection and diagnostic accuracy in the study cohort
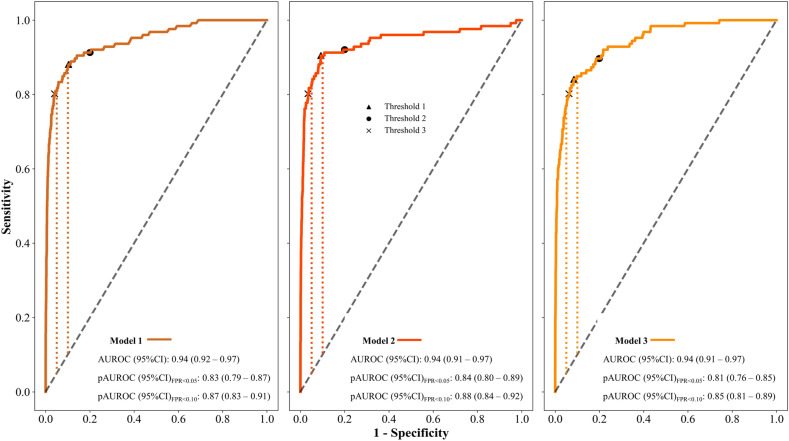
The abovementioned models were then tested in the study cohort and three showed the best performance. Their architecture characteristics and results were detailed in Supplementary material 1.5–1.6. AUROC (95%CI) was 0.94 (0.92–0.97) for model 1, 0.94 (0.91–0.97) for model 2, and 0.94 (0.91–0.97) for model 3. For the region at FPR < 0.05, pAUROC (95%CI) was 0.83 (0.79–0.87), 0.84 (0.80–0.89), and 0.81 (0.76–0.85) for models 1, 2, and 3, respectively, while at FPR < 0.10, pAUROC (95%CI) was 0.87 (0.83–0.91), 0.88 (0.84–0.92), and 0.85 (0.81–0.89) for models 1, 2, and 3, respectively (Fig. 1).
Figure 1.
AUROC and pAUROC (95%CI) for the classification of DME, with the best three models. ROC: receiver operating characteristics; AUROC: area under the ROC curve; DME: diabetic macular edema. Vertical dotted and colored lines represented pAUROC at FPR (1-specificity) < 0.05 and FPR < 0.10.
Sensitivity and specificity were calculated at the three thresholds. Model 2 showed the most balanced results at threshold 1: sensitivity (95%CI) was 90.5 (84.0–95.0), and specificity (95%CI) was 90.8 (90.0–91.6). Similar results but with higher specificity was reached by the ensemble model: sensitivity (95%CI) was 88.9 (82.1–93.8), and specificity (95%CI) was 93.3 (92.6–94.0). The ensemble model showed the best results regarding sensitivity and specificity at the remaining thresholds: sensitivity (95%CI) was 92.1 (82.1–93.8), and specificity (95%CI) was 83.1 (82.1–84.1) at threshold 2, while sensitivity (95%CI) was 81.0 (73.0–87.4), and specificity (95%CI) was 97.0 (96.5–97.4) at threshold 3 (Table 2).
Table 2.
Diagnostic accuracy of the three best models after threshold tuning.
| Model | Sensitivity % (95% CI) | Specificity % (95% CI) | NPV % (95% CI) | PPV % (95% CI) | Cumulative incidence % (95% CI) | |
|---|---|---|---|---|---|---|
| Threshold* 1 |
1 2 3 Ensemble |
88.1 (81.1–93.2) 90.5 (84.0–95.0) 84.1 (76.6–90.0) 88.9 (82.1–93.8) |
89.7 (88.8–90.5) 90.8 (90.0–91.6) 91.6 (90.8–92.3) 93.3 (92.6–94.0) |
99.7 (99.5–99.8) 99.8 (99.5–99.9) 99.6 (99.4–99.8) 99.7 (99.5–99.8) |
17.2 (14.3–20.3) 19.3 (16.2–22.7) 19.5 (16.3–23.1) 24.5 (20.6–28.7) |
2.09 (1.72–2.51) 2.15 (1.77–2.57) 1.99 (1.64–2.41) 2.11 (1.74–2.53) |
| Threshold† 2 |
1 2 3 Ensemble |
91.3 (82.1–93.8) 92.1 (82.1–93.8) 89.7 (82.1–93.8) 92.1 (82.1–93.8) |
80.0 (78.9–81.1) 80.0 (78.9–81.1) 80.1 (78.9–81.1) 83.1 (82.1–84.1) |
99.7 (99.5–99.8) 99.8 (99.5–99.9) 99.7 (99.5–99.8) 99.8 (99.5–99.9) |
10.0 (8.32–11.9) 10.1 (8.39–12.0) 9.84 (8.18–11.7) 11.7 (9.77 – 13.9) |
2.16 (1.79–2.59) 2.18 (1.81–2.61) 2.13 (1.76–2.55) 2.18 (1.81–2.61) |
| Threshold‡ 3 |
1 2 3 Ensemble |
80.2 (72.1–86.7) 80.2 (72.1–86.7) 80.2 (72.1–86.7) 81.0 (73.0–87.4) |
96.0 (95.4–96.5) 96.6 (96.0–97.0) 93.8 (93.1–94.4) 97.0 (96.5–97.4) |
99.5 (99.3–99.7) 99.5 (99.3–99.7) 99.5 (99.3–99.7) 99.5 (99.3–99.7) |
32.8 (27.6–38.3) 36.1 (30.4–42.0) 23.9 (19.9–28.2) 39.5 (33.5–45.8) |
1.90 (1.55–2.30) 1.90 (1.55–2.30) 1.90 (1.55–2.30) 1.92 (1.57–2.33) |
NDME: 126; Nnon-DME: 5188.
DME diabetic macular edema, CI confidence interval, PPV positive predictive value, NPV negative predictive value.
*Threshold 1: Youden index.
†Threshold 2: highest sensitivity.
‡Threshold 3: highest specificity.
Negative predictive values (NPV; 95%CI) were high and similar in all scenarios (above 99.5). As expected, positive predictive values (PPV; 95%CI) were low (as it drastically depends on the prevalence of the disease but increases with higher specificity); with the ensemble model at threshold 3, PPV was 39.5 (33.5–45.8). Cumulative incidence of DME for the study period was similar across all thresholds and for all models (Table 2).
Diagnostic accuracy of the ensemble model by different covariates
The stratified analysis showed a similar diagnostic accuracy across all strata, excepting for moderate and severe DME which resulted in higher sensitivity and specificity compared to mild DME and overall; the highest sensitivity (95%CI) was 100 (92.9–100) at threshold 2, and the highest specificity was reached at threshold 3 (specificity; 95%CI = 97.0; 96.5–97.4). In contrast, the specificity observed among subjects with ST-DR (including severe non-proliferative and proliferative) was lower as compared to overall; at threshold 3 it was 62.9 (44.9–78.5), and even lower at thresholds 1 and 2 (Table 3).
Table 3.
Sensitivity and specificity of the ensemble model at all thresholds, by different covariates.
| NDME (%) | Threshold 1 | Threshold 2 | Threshold 3 | ||||
|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity 95% CI) | ||
| Gender | |||||||
| Males | 85 (67.5) | 89.4 (80.9–95.0) | 93.4 (92.5–94.3) | 94.1 (86.8–98.1) | 84.4 (83.1–85.7) | 78.8 (68.6–86.9) | 97.0 (96.3–97.5) |
| Females | 41 (32.5) | 87.8 (73.8–95.9) | 93.2 (92.1–94.3) | 87.8 (73.8–95.9) | 81.3 (79.5–82.9) | 85.4 (70.8–94.4) | 97.0 (96.2–97.7) |
| Age | |||||||
| ≤ 65 | 56 (44.4) | 94.6 (85.1–98.9) | 95.8 (95.0–96.5) | 94.6 (85.1–98.9) | 88.4 (87.1–89.6) | 91.1 (80.4–97.0) | 98.0 (97.4–98.5) |
| > 65 | 70 (55.6) | 84.3 (73.6–91.9) | 90.7 (89.6–91.8) | 90.0 (80.5–95.9) | 77.5 (75.8–79.1) | 73.0 (61.4–82.6) | 96.0 (95.2–96.7) |
| Laterality | |||||||
| Left eye | 61 (48.4) | 89.2 (79.1–95.6) | 94.2 (93.3–95.1) | 90.8 (81.0–96.5) | 85.5 (84.1–86.8) | 78.5 (66.5–87.7) | 97.1 (96.3–97.7) |
| Right eye | 65 (51.6) | 88.5 (77.8–95.3) | 92.4 (91.3–93.4) | 93.4 (84.1–98.2) | 80.7 (79.1–82.2) | 83.6 (71.9–91.8) | 96.9 (96.2–97.6) |
| BCVA, Snellen decimal: | |||||||
| ≤ 0.5 | 46 (36.5) | 95.7 (85.2–99.5) | 81.8 (78.9–84.4) | 97.8 (88.5–99.9) | 63.4 (60.0–66.8) | 82.6 (68.6–92.2) | 91.5 (89.4–93.4) |
| > 0.5 | 76 (63.5) | 84.2 (74.0–91.6) | 95.5 (94.8–96.1) | 88.2 (78.7–94.4) | 86.9 (85.9–87.9) | 80.3 (69.5–88.5) | 98.0 (97.6–98.4) |
| Type of diabetes | |||||||
| 1 | 6 (4.76) | 83.3 (35.9–99.6) | 95.0 (92.6–96.9) | 83.3 (35.9–99.6) | 88.7 (85.4–91.5) | 33.3 (4.33–77.7) | 97.3 (95.3–98.6) |
| 2 | 120 (95.2) | 89.2 (82.2–94.1) | 93.2 (92.4–93.9) | 92.5 (86.2–96.5) | 82.6 (81.5–83.7) | 83.3 (75.4–89.5) | 97.0 (96.5–97.5) |
| Duration of diabetes | |||||||
| ≤ 15 years | 67 (53.2) | 95.5 (87.5–99.1) | 94.7 (93.9–95.4) | 97.0 (89.6–99.6) | 84.6 (83.4–85.8) | 85.1 (74.3–92.6) | 97.8 (97.3–98.3) |
| > 15 years | 58 (46.8) | 81.0 (68.6–90.1) | 89.6 (87.9–91.1) | 86.2 (74.6–93.9) | 79.2 (76.9–81.3) | 75.9 (62.8–86.1) | 94.7 (93.4–95.8) |
| Grade of DME | |||||||
| Mild | 77 (61.1) | 84.4 (74.4–91.7) | 93.3 (92.6–94.0) | 88.3 (79.0–94.5) | 83.1 (82.1–84.1) | 74.0 (62.8–83.4) | 97.0 (96.5–97.4) |
| Moderate and severe | 46 (38.9) | 97.8 (88.5–99.9) | 93.3 (92.6–94.0) | 100 (92.9–100) | 83.1 (82.1–84.1) | 93.5 (82.1–98.6) | 97.0 (96.5–97.4) |
| Diabetic retinopathy | |||||||
| Non-ST | 97 (77.0) | 86.6 (78.2–92.7) | 93.7 (93.0–94.4) | 90.7 (83.1–95.7) | 83.5 (82.5–84.5) | 78.4 (68.8–86.1) | 97.3 (96.8–97.7) |
| ST-DR* | 28 (23.0) | 96.4 (81.7–99.9) | 51.4 (34.0–68.6) | 96.4 (81.7–99.9) | 40.0 (23.9–57.9) | 89.3 (71.8–97.7) | 62.9 (44.9–78.5) |
| OCT quality of image (TopQ) | |||||||
| ≤ 40 | 43 (34.1) | 90.7 (77.9–97.4) | 80.8 (78.2–83.2) | 95.4 (84.2–99.4) | 61.0 (57.8–64.1) | 83.7 (69.3–93.2) | 92.3 (90.4–93.9) |
| > 40 | 83 (65.9) | 88.0 (79.0–94.1) | 96.2 (95.6–96.8) | 90.4 (81.9–95.8) | 88.3 (87.2–89.2) | 79.5 (69.2–87.6) | 98.1 (97.6–98.5) |
NDME: 126; Nnon-DME: 5188.
DME diabetic macular edema, CI confidence interval, BCVA best corrected visual acuity, ST sight-threatening, OCT optical coherence tomography.
*ST-DR included severe non-proliferative and proliferative DR.
Characteristics of false positives and false negatives
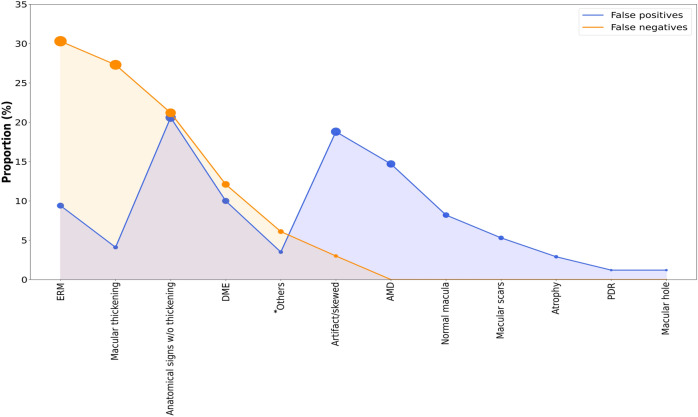
The characteristics of false positives and negatives obtained with the ensemble model at threshold 3 were explored. In this configuration, we obtained 24 false negatives and 156 false positives. According to the review made by the third retina specialist, 87.8% of all false negatives predicted by the model were correctly classified indeed. They were mainly characterized by epiretinal membranes, macular thickening without signs of DME, and anatomical signs of DME without macular thickening, and just 4 DMEs (12.1%) were missed by the model. On the other hand, anatomical signs of DME without macular thickening, images with artifacts (eye blinking, cropped images) or skewed, and age-related macular degeneration accounted for 54% of false positives, and 17 (10%) DMEs were misdiagnosed by the retina specialist, thus correctly classified by the model (Fig. 2).
Figure 2.
Characteristics of false positives and negatives obtained with the ensemble model at threshold 3. ERM: epiretinal membranes; DME: diabetic macular edema; AMD: age-related macular degeneration; PDR: proliferative diabetic retinopathy. *Includes ischemia and thinning, asteroid hyalosis, central serous choroidopathy, lamellar macular hole, vascular tortuosity, and epithelial pigment detachment.
Generalizability of models for classification of referable DR
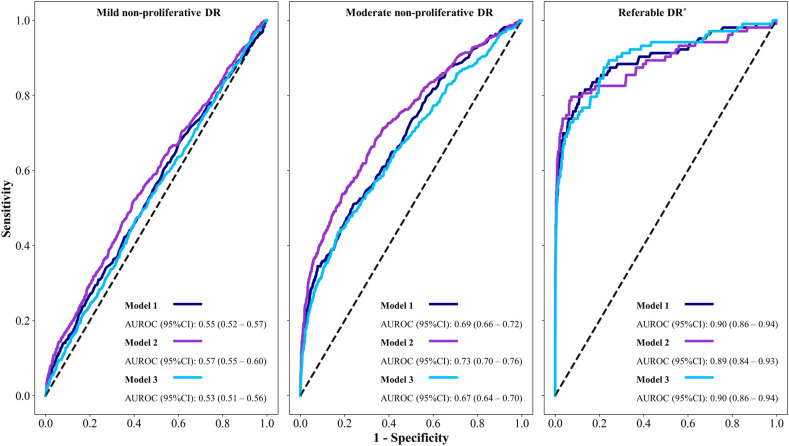
The three selected models yielded almost chance prediction in the classification of mild non-proliferative DR. Conversely, an increasing trend to a high performance was observed with severity. Overall, AUROC for referable DR was equal or above 0.89 (Fig. 3).
Figure 3.
ROC and AUROC (95%CI) for different grades of diabetic retinopathy with the best three models. ROC: receiver operating characteristics; AUROC: area under the ROC curve; DR: diabetic retinopathy; DME: diabetic macular edema. *Referable DR included severe non-proliferative DR, proliferative DR, moderate DME, and severe DM.
Discussion
Results from the present study showed that our hybrid (CNN-RNN) deep learning models achieved high diagnostic accuracy in the identification of DME, overall and by severity, in a dataset of OCT cubes generated in a real-world DR screening program as a product of routine clinical care.
Computer vision models based on CNNs have been widely tested for the detection of DR or DME so far23. Nevertheless, very few have been designed for direct clinical application in real-life, with prospectively collected datasets, and well-defined diseases10,13,14,24. CNNs require significant amounts of manually expert-labeled data for training and validation. At present, some publicly available datasets comprised of OCT B-scans or FR have been used in numerous studies, however, using flat images as those implies selective sampling10,12. The screening of DR in FR alone leads to low sensitivity in the diagnosis of DME, as the criteria to assess DME is based on the presence of indirect signs that are indeed markers of progression, and a non-negligible proportion of false positives and negatives (almost 50% of DME without indirect signs are missed)6 Likewise, selective sampling also occurs in B-scans as they must be previously selected or, in other cases, only central fovea B-scans were extracted from the OCT cube10,12. For these reasons, DMEs with clear signs of the disease are overrepresented in existing datasets, and may introduce a significant bias with misleadingly outstanding results10. Otherwise, when transported to real-world datasets, much more realistic results have been reported13,14,24–26.
To our knowledge, this is the first study that applied hybrid (CNN-RNN) models to analyze OCT cubes with the aim to detect DMEs of either grade. Hybrid models have been proposed to predict time-dependent outcomes as treatment response in colorectal and lung cancer15,16, although we took advantage of this feature to analyze OCT cubes with just one expert annotation per volume, and also to avoid misalignments when re-assembling the cube if 3D-CNNs were used11. Furthermore, another potential benefit of our approach is the management of time, making our models transferable to predict time-to-event outcomes as treatment response (e.g. vascular endothelial growth factor inhibitors) or risk of progression. At present, other authors have also developed a model for segmentation and identification of several referable retinal diseases using OCT cubes, so that precluded the comparability with our results27.
Regarding the screening of DME, Liu et al.13 carried out a study nested in a real-word DR screening program and reported a sensitivity, specificity, and AUROC of 91.3, 97.5, and 0.944, respectively, using a combination of FR and OCT. Taking the Youden index as reference, our ensemble model yielded a sensitivity (95%CI) and specificity (95CI%) of 88.9 (82.1–93.8) and 93.3 (92.6–94.0), while AUROC for the best three models was 0.94. Our results were slightly lower, but some considerations should be made to adequately compare the results: Liu et al. used B-scans and FR but without assessment by grade of DME13. Of note, using a modest population of 600 diabetics, they reported a PPV of 75.4% strongly influenced by a high prevalence of DME of almost fourfold the observed in our population. In practical terms, since PPV depends on prevalence, with the sensitivity and specificity reported by them we would barely reach a PPV of 50% in our population, and suggesting PPV alone as inadequate metric for screening in imbalanced datasets. Moreover, baseline and clinical characteristics of the study population were not reported, so that prevented an extensive interpretation of results13.
This pioneer study evaluated the ability of the models to detect different grades of DME, and showed no differences in performance to detect mild DMEs, but outperformed the overall results in moderate and severe DMEs, so this trend may support, in part, our concerns about the misleading results observed in studies using selected datasets10,28. It is important to stress that, in our population, the majority of DMEs were mild (61.1%) as a consequence of a well-established screening program where patients are detected earlier6. Nevertheless, is also relevant to detect early DMEs to personalize screening visits, adjust antidiabetic treatments, and manage other cardiovascular risk factors, to improve their visual prognosis29,30.
The design of the best strategy for DME screening in the community must consider multiple dimensions, as well as the impact of potential misdiagnoses, the number of false positives that specialized care could undertake or the cost-effectiveness, among others. Population-based screening of a disease with such low prevalence would benefit most from higher specificity as increases PPV to a greater extent than higher sensitivity31, and would rule out most subjects screened in a first step, reducing drastically the workload of the specialist. Based on our results, that would cost 3–6% (156–311 subjects) false positives, and almost 20% false negatives (24–25 DMEs missed), throughout the study period. Conversely, screening based on sensitivity would result more efficient although highly challenging in turn since advanced stages of DR share indirect markers with DME and both entities often concur. In this scenario, we would miss 7.9% (10) DMEs at the cost of 16.9% (877) false positives, at best. Alternatively, the Youden index would return more balanced metrics overall; 7–9% false positives, and 9–11% false negatives. Of note, after exploring the characteristics of false negatives, only 4 DMEs were incorrectly predicted by the model, so re-calculated sensitivity would rise to 94% with specificity unchanged. On the other hand, the characteristics of false positives revealed that the models may learn anatomical signs of DME that could be shared with ST-DR, explaining the low specificity of the models among these subjects. Regarding the latter, our models showed potential transferability to referable DR, although specific training would be needed specially for mild and moderate stages32.
Our study has strengths and limitations that need to be discussed. We trained our models based on a ground truth of DME constructed by retinal experts with high expertise and low inter-individual variability, and then evaluated the models within a well-defined population extracted from the target population where these models could be further applied. In addition, our models were able to analyze OCT cubes, avoiding selective sampling and allowing to detect extra-foveal DMEs, which in turn were the majority in our population and often missed due to subclinical presentations. Finally, the ensemble model improved the results, but its complexity must be limited to be deployed effectively within a teleophthalmology screening program. By contrast, an external validation of the models, using datasets from other imaging sources and populations would be necessary. Despite we achieved good results indeed, we did not analyze the 128 B-scans from the OCT cube, so further approaches to build more complete samples would be needed. In this line, the combination of image-related data as thicknesses from the ETDRS grid may reduce the number of cases where anatomical signs were present but without reaching 300 µm. Finally, artificial intelligence is usually considered as black box models due to their complexity and lack of interpretability, and that reduces the trust of specialists in their predictions33. For this reason, medical researchers in this field must make efforts towards explainable artificial intelligence that support retina specialists to make meaningful clinical decisions.
Conclusions
Our model was able to detect DME in complete OCT cubes from a real-world screening program, making unnecessary to select training images and avoiding selection bias. This approach showed potential applicability to discard in a first step the majority of non-DMEs otherwise referred to specialized care for re-evaluation, then reducing the workload and needs of retina specialists, then speeding-up the access to the teleophthalmology screening program as a consequence.
Supplementary Information
Acknowledgements
The authors would also like to acknowledge Lucía Ocaña Rodríguez, Víctor Márquez Quesada, Montserrat Álvarez Beleño, Rafael Calvo Tello, and Guillermo González Sánchez, from the University of Alcalá, for their contribution in the seminal development of the custom convolutional neural network, although for other research purposes.
Abbreviations
- AUPRC
Area under the precision-recall curve
- AUROC
Area under the receiver operating characteristics curve
- BCVA
Best-corrected visual acuity
- CNN
Convolutional neural networks
- CI
Confidence interval
- DM
Diabetes mellitus
- DME
Diabetic macular edema
- DR
Diabetic retinopathy
- FPR
False positive rate
- FR
Fundus retinography
- ICDRSS
International Clinical Diabetic Retinopathy and Diabetic Macular Edema Disease Severity Scales
- IQR
Interquartile range
- NPV
Negative predictive value
- OCT
Optical coherence tomography
- pAUROC
Partial area under the receiver operating characteristics curve
- PPV
Positive predictive value
- RNN
Recurrent neural networks
- ROC
Receiver operating characteristic
- ST
Sight-threatening
- ST-DR
Sight-threatening diabetic retinopathy
Author contributions
ARM: Conceptualization, Methodology, Software, Formal Analysis, Investigation, Resources, Data Curation, Writing-Original Draft, Visualization, Supervision, Project Administration, Funding Acquisition; CA: Conceptualization, Validation, Resources, Data Curation, Writing-Review & Editing, Supervision, Project Administration, Funding Acquisition; GA: Validation, Data Curation, Writing-Review & Editing; MO: Writing-Review & Editing; JZV: Writing-Review & Editing; MAT: Resources, Writing-Review & Editing.
Funding
Antonio Rodríguez-Miguel and Carolina Arruabarrena received a research grant (FIB-PI19-01) from the Biomedical Research Foundation of the University Hospital “Príncipe de Asturias” (Alcalá de Henares, Madrid, Spain). The sponsor had no role in the conceptualization, design, data collection, analysis, decision to publish or preparation of the manuscript.
Data availability
In line with Spanish and European laws, data share must be authorized by the Ethics Committee of the University Hospital “Príncipe de Asturias”, so authors are not allowed to openly make them available. However, data could be available by reasonable request from any organization or researcher to the corresponding author, provided that the Ethics Committee authorize specifically the data transfer.
Competing interests
The authors declare no competing interests. Javier Zarranz-Ventura receives consultant fees from Topcon, although for other research purposes.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68489-2.
References
- 1.Lovic, D. et al. The growing epidemic of diabetes mellitus. Curr. Vasc. Pharmacol.18, 104–109 (2020). 10.2174/1570161117666190405165911 [DOI] [PubMed] [Google Scholar]
- 2.International Diabetes Federation, 2021. IDF Diabetes, 10th edition. Brussels: Atlas Press. https://diabetesatlas.org/. Accessed (28 06 2023).
- 3.Teo, Z. L. et al. Global prevalence of diabetic retinopathy and projection of burden through 2045: Systematic review and meta-analysis. Ophthalmology128, 1580–1591 (2021). 10.1016/j.ophtha.2021.04.027 [DOI] [PubMed] [Google Scholar]
- 4.Schmidt-Erfurth, U. et al. Three-year outcomes of individualized ranibizumab treatment in patients with diabetic macular edema The restore extension study. Ophthalmology10.1016/j.ophtha.2013.11.041 (2014). 10.1016/j.ophtha.2013.11.041 [DOI] [PubMed] [Google Scholar]
- 5.Fenner, B. J. et al. Advances in retinal imaging and applications in diabetic retinopathy screening: A review. Ophthalmol. Ther.7, 333–346 (2018). 10.1007/s40123-018-0153-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arruabarrena, C. et al. Evaluation of the inclusion of spectral domain optical coherence tomography in a telemedicine diabetic retinopathy screening program: a real clinical practice. Retina43(8), 1308–1316 (2023). 10.1097/IAE.0000000000003832 [DOI] [PubMed] [Google Scholar]
- 7.Wong, I. Y. H., Wong, R. L. M., Chan, J. C. H., Kawasaki, R. & Chong, V. Incorporating Optical Coherence Tomography Macula Scans Enhances Cost-effectiveness of Fundus Photography-Based Screening for Diabetic Macular Edema. Diabetes Care43(12), 2959–2966 (2020). 10.2337/dc17-2612 [DOI] [PubMed] [Google Scholar]
- 8.Virgili, G. et al. Optical coherence tomography (OCT) for detection of macular oedema in patients with diabetic retinopathy. Cochrane Database Syst. Rev.10.1002/14651858.CD008081.pub3 (2015). 10.1002/14651858.CD008081.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, C. et al. The impact of artificial intelligence on retinal disease management: Vision Academy retinal expert consensus. Curr. Opin. Ophthalmol.34(5), 396–402 (2023). 10.1097/ICU.0000000000000980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, C. Y., Tang, F., Ting, D. S. W., Tan, G. S. W. & Wong, T. Y. Artificial intelligence in diabetic eye disease screening. Asia Pac J. Ophthalmol.10.22608/APO.201976 (2019). 10.22608/APO.201976 [DOI] [PubMed] [Google Scholar]
- 11.Yanagihara, R. T., Lee, C. S., Ting, D. S. W. & Lee, A. Y. Methodological Challenges of Deep Learning in Optical Coherence Tomography for Retinal Diseases: A Review. Transl. Vis. Sci. Technol.9(2), 11 (2020). 10.1167/tvst.9.2.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kermany, D. et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell.172(5), 1122-1131.e9 (2018). 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 13.Liu, R. et al. Application of artificial intelligence-based dual-modality analysis combining fundus photography and optical coherence tomography in diabetic retinopathy screening in a community hospital. Biomed. Eng. Online21(1), 47 (2022). 10.1186/s12938-022-01018-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raumviboonsuk, P. et al. Deep learning versus human graders for classifying diabetic retinopathy severity in a nationwide screening program. NPJ Digit Med.2, 25 (2019). 10.1038/s41746-019-0099-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, L., Dercle, L., Zhao, B. & Schwartz, L. H. Deep learning for the prediction of early on-treatment response in metastatic colorectal cancer from serial medical imaging. Nat. Commun.12(1), 6654 (2021). 10.1038/s41467-021-26990-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu, Y. et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res.25(11), 3266–3275 (2019). 10.1158/1078-0432.CCR-18-2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vujosevic, S. et al. Screening for diabetic retinopathy: 1 and 3 nonmydriatic 45-degree digital fundus photographs vs 7 standard early treatment diabetic retinopathy study fields. Am. J. Ophthalmol.148, 111–118 (2009). 10.1016/j.ajo.2009.02.031 [DOI] [PubMed] [Google Scholar]
- 18.Panozzo, G. et al. Diabetic macular edema: an OCT-based classification. Semin Ophthalmol.19(1–2), 13–20 (2004). 10.1080/08820530490519934 [DOI] [PubMed] [Google Scholar]
- 19.Ruia, S., Saxena, S., Gemmy Cheung, C. M., Gilhotra, J. S. & Lai, T. Y. Spectral Domain Optical Coherence Tomography Features and Classification Systems for Diabetic Macular Edema: A Review. Asia Pac J. Ophthalmol. (Phila)5(5), 360–367 (2016). 10.1097/APO.0000000000000218 [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson, C. P. et al. Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology110, 1667–1682 (2003). 10.1016/S0161-6420(03)00475-5 [DOI] [PubMed] [Google Scholar]
- 21.Kumar, R. & Indrayan, A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr.48(4), 277–287 (2011). 10.1007/s13312-011-0055-4 [DOI] [PubMed] [Google Scholar]
- 22.Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology143(1), 29–36 (1982). 10.1148/radiology.143.1.7063747 [DOI] [PubMed] [Google Scholar]
- 23.Shahriari, M. H., Sabbaghi, H., Asadi, F., Hosseini, A. & Khorrami, Z. Artificial intelligence in screening, diagnosis, and classification of diabetic macular edema: A systematic review. Surv. Ophthalmol.68(1), 42–53 (2023). 10.1016/j.survophthal.2022.08.004 [DOI] [PubMed] [Google Scholar]
- 24.Lin, D. et al. Application of Comprehensive Artificial intelligence Retinal Expert (CARE) system: a national real-world evidence study. Lancet Digit Health3(8), e486–e495 (2021). 10.1016/S2589-7500(21)00086-8 [DOI] [PubMed] [Google Scholar]
- 25.Beede E, Baylor E, Hersch F, et al. A human-centered evaluation of a deep learning system deployed in clinics for the detection of diabetic retinopathy. Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems. Association for Computer Machinery; Honolulu, (2020).
- 26.Abràmoff, M. D., Lavin, P. T., Birch, M., Shah, N. & Folk, J. C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med.1, 39 (2018). 10.1038/s41746-018-0040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Fauw, J. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med.24, 1342–1350 (2018). 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 28.Li, Z. et al. An automated grading system for detection of vision-threatening referable diabetic retinopathy on the basis of color fundus photographs. Diabetes Care41, 2509–2516 (2018). 10.2337/dc18-0147 [DOI] [PubMed] [Google Scholar]
- 29.Klein, R., Lee, K. E., Knudtson, M. D., Gangnon, R. E. & Klein, B. E. Changes in visual impairment prevalence by period of diagnosis of diabetes: the Wisconsin Epidemiologic Study of Diabetic Retinopathy. Ophthalmology116(10), 1937–1942 (2009). 10.1016/j.ophtha.2009.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UKPDS Group. Intensive blood-glucose control with sulphonylurea or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet1352, 837–853 (1998). [PubMed] [Google Scholar]
- 31.Westreich, D. Epidemiology by design. A causal approach to the health sciences (Oxford University Press, 2020). [Google Scholar]
- 32.Gargeya, R. & Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology124, 962–969 (2017). 10.1016/j.ophtha.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 33.Ting, D. S. W., Carin, L. & Abramoff, M. D. Observations and Lessons Learned From the Artificial Intelligence Studies for Diabetic Retinopathy Screening. JAMA Ophthalmol.137(9), 994–995 (2019). 10.1001/jamaophthalmol.2019.1997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In line with Spanish and European laws, data share must be authorized by the Ethics Committee of the University Hospital “Príncipe de Asturias”, so authors are not allowed to openly make them available. However, data could be available by reasonable request from any organization or researcher to the corresponding author, provided that the Ethics Committee authorize specifically the data transfer.