Abstract
Cancer cells with BRCA1/2 deficiencies are sensitive to poly (ADP-ribose) polymerase (PARP) inhibitors. We evaluated the efficacy of talazoparib in DNA-Damage Repair (DDR)-altered patients. In this phase II trial, patients were enrolled onto one of four cohorts based on molecular alterations: (1) somatic BRCA1/2, (2) other homologous recombination repair pathway, (3) PTEN and (4) germline BRCA1/2. The primary endpoint was a clinical benefit rate (CBR): complete response, partial response or stable disease ≥24 weeks. 79 patients with a median of 4 lines of therapy were enrolled. CBR for cohorts 1–4 were: 32.5%, 19.7%, 9.4% and 30.6%, respectively. PTEN mutations correlated with reduced survival and a trend towards shorter time to progression.Talazoparib demonstrated clinical benefit in selected DDR-altered patients. PTEN mutations/loss patients derived limited clinical benefit. Further study is needed to determine whether PTEN is prognostic or predictive of response to PARP inhibitors.
Subject terms: Targeted therapies, Targeted therapies
Introduction
Cancer cells frequently have defects in deoxyribose nucleic acid (DNA) repair pathways1,2. Cells deficient in breast cancer gene 1 or 2 (BRCA1 or BRCA2) function have a high level of chromosomal instability3–5. Poly (ADP-ribose) polymerase (PARP) inhibitors have been shown to be selectively lethal to cells lacking functional BRCA1 or BRCA2 with minimal toxicity to normal cells6. Further, tumors with germline BRCA mutations demonstrated defects in homologous recombination (HR) and have been shown in several trials to respond to PARP inhibitors7,8.
Preclinical studies suggest that cancer cells with alterations in other homologous recombination (HR) repair pathway genes (e.g., ataxia telangiectasia mutated (ATM) and partner and localizer of BRCA2 (PALB2) may also be sensitive to PARP inhibition9–12. ATM is recruited and activated by DNA double-strand breaks where it phosphorylates several proteins that are important in activating the DNA damage checkpoint leading to cell cycle arrest, DNA repair or apoptosis. Heterozygous germline mutation of ATM is a moderate-risk factor for developing breast cancer11. Montani and colleagues found that ATM-depletion can sensitize breast cancer cells to PARP inhibition thus PARP inhibition could be of benefit in patients with low ATM protein expression/activity such as would be seen in heterozygous carriers of germline ATM mutation11. Also of interest, the PALB2 gene encodes for a protein that functions in repair of double stranded DNA breaks. Patients with variants of the PALB2 gene have an increased risk of developing breast cancer10. Further, cells deficient in PALB2 have been found to be sensitive to PARP inhibitors9.
Additional genes such as Fanconi Anemia (FA) genes were of interest for this study as identification of the FANCD1 gene as BRCA2 provided the first direct link between FA proteins and DNA repair13,14. Because BRCA2 deficient cells are sensitive to PARP inhibition, we wished to determine whether or not tumors that have deleterious alterations of the FA gene family would share this trait.
Phosphatase and tensin homolog (PTEN) acts as a tumor suppressor and mutations of this gene are a step in the development of many cancers. PTEN is important in the regulation of the cell cycle thus preventing the rapid growth and division of cells15. Studies have shown that PTEN deficiency caused a homologous repair deficiency, potentially due to downregulation of Rad51, a critical HR component16,17. Further, PTEN loss has been shown to be synthetic lethal with PARP, with PTEN loss sensitizing to PARP inhibitors in vitro and in vivo16. However, the role of PTEN remains controversial, as some data suggest that it may not regulate Rad51 expression or HR function. Additionally, PTEN inactivation has been found to render BRCA1/2 aberrant cells resistant to PARP18,19.
Talazoparib exerts its antineoplastic effects through the selective inhibition of PARP enzymes, which are pivotal in the single-strand DNA break repair mechanism20. The pharmacological blockade of PARP by talazoparib leads to an accumulation of DNA lesions, precipitating the formation of double-strand breaks. In neoplastic cells characterized by mutations in BRCA1/2, the intrinsic defect in the homologous recombination repair pathway renders these double-strand breaks irreparable. This culminates in heightened genomic instability and triggers apoptosis, a process termed synthetic lethality20,21. This mechanism is particularly effective against cancer cells harboring specific genetic aberrations in DNA repair pathways. Notably, talazoparib exhibits a distinct pharmacodynamic property of forming PARP-DNA complexes, further potentiating its cytotoxic profile22. This attribute of talazoparib underscores its clinical utility, leading to its approval by the U.S. Food and Drug Administration (FDA) in October 2018 for the management of HER2-negative advanced breast cancers with deleterious or suspected pathogenic mutations in the BRCA genes23. Furthermore, talazoparib is more potent at lower concentrations than earlier generation PARP1/2 inhibitors (such as olaparib, rucaparib and veliparib)24,25.
In recent years, mounting clinical evidence has substantiated the rationale for extending the application of talazoparib beyond the confines of BRCA1/2 mutation-positive malignancies, aligning with our trial’s molecularly driven strategy. This extension is predicated on the paradigm of ‘BRCAness’, where tumors share similar vulnerabilities to BRCA mutant cancers due to deficiencies in the DNA damage repair mechanisms20,26. For instance, a meta-analytic survey encompassing 21 diverse tumor types identified that a significant subset (17.4%) exhibits mutations in homologous recombination repair (HRR)-associated genes, thus providing compelling rationale for exploring talazoparib in these broader oncogenic contexts27. Notably, primary prostate cancers exhibit a HR deficiency mutation prevalence of approximately 20%, with higher incidence observed in metastatic lesions, ranging up to 23%28. These findings underscore the need to re-evaluate and expand current oncological paradigms for the clinical deployment of PARP inhibitors, reflecting our study’s commitment to a molecularly centered therapeutic approach22–25,29,30.
Based on this scientific background, we conducted a phase II study to formally evaluate whether talazoparib achieves clinical benefit (complete response (CR), partial response (PR) or stable disease (SD) ≥ 24 weeks) in metastatic or inoperable locally advanced or locally recurrent cancer patients who have germline mutations in BRCA1 or BRCA2 with cancers other than breast or ovarian cancer, PTEN mutation/deletion or loss by IHC and in patients with somatic mutations or homologous deletions in BRCA1 or BRCA2, or alterations in other HR repair pathway genes.
Results
Demographic and clinical characteristics
A total of 94 patients consented to participate in the study. However, 15 were excluded: 12 patients screen-failed and 3 withdrew their consent prior to enrollment. Seventy-nine patients with advanced, metastatic malignancies were enrolled and treated on study between December 2014 and July 2021 (Supplementary Fig. 1). Demographic and clinical characteristics are summarized in Table 1. The median age of patients was 61 (range, 22–84 years). The median number of prior systemic therapies was 4 (range, 1–13). No patients had received prior PARP inhibitor therapies. Fifty-seven patients (72.2%) had received prior platinum therapy. The most common cancer types enrolled were sarcoma followed by colorectal cancer, breast, and cholangiocarcinoma. The median number of cycles (cycle = 28 days) completed for all patients was 2 (range, 1–47 + ). Thirty-five patients (44%) received more than two cycles. For patients with SD or better, the median number of cycles completed was 6 (range, 2–47 + ). Pre-identified pathogenic or activating molecular aberration that were used for study enrollment are summarized in Table 1.
Table 1.
Baseline demographics and clinical characteristics
| Characteristic | Somatic BRCA1/2 | Other HR pathways | PTEN mutation/loss | Germline BRCA1/2 | All |
|---|---|---|---|---|---|
| N = 18 | N = 31 | N = 14 | N = 16 | N = 79 | |
| Gender, n (%) | |||||
| Male | 5 (28) | 15 (48) | 3 (21) | 5 (31) | 28 (35) |
| Female | 13 (72) | 16 (52) | 11 (79) | 11 (69) | 51 (65) |
| Median age | 59.5 (32–81) | 59 (32–75) | 63.5 (22–84) | 61 (53–72) | 61 (22–84) |
| ECOG | |||||
| 0 | 3 (17) | 6 (19) | 4 (29) | 2 (13) | 15 (19) |
| 1 | 15 (83) | 25 (81) | 10 (71) | 14 (88) | 64 (81) |
| Tumor Type | |||||
| Sarcomaa | 3 (16.7) | 5 (16.1) | 3 (21.4) | 0 | 11 (13.9) |
| Colorectal | 2 (11.1) | 4 (12.9) | 1 (7.1) | 3 (18.8) | 10 (12.7) |
| Breast | 3 (16.7) | 2 (6.5) | 2 (14.3) | 0 | 7 (8.9) |
| Cholangiocarcinoma | 1 (5.6) | 4 (12.9) | 1 (7.1) | 1 (6.3) | 7 (8.9) |
| Ovarian | 2 (11.1) | 3 (9.7) | 1 (7.1) | 0 | 6 (7.6) |
| Pancreas | 0 | 1 (3.2) | 1 (7.1) | 4 (25) | 6 (7.6) |
| Head and neck | 1 (5.6) | 2 (6.5) | 1 (7.1) | 1 (6.3) | 5 (6.3) |
| Ampulla of Vater | 0 | 2 (6.5) | 0 | 1 (6.3) | 3 (3.8) |
| Bladder | 0 | 1 (3.2) | 0 | 2 (12.5) | 3 (3.8) |
| Gastric | 1 (5.6) | 1 (3.2) | 0 | 1 (6.3) | 3 (3.8) |
| Renal | 1 (5.6) | 1 (3.2) | 0 | 1 (6.3) | 3 (3.8) |
| Endometrial | 0 | 1 (3.2) | 1 (7.1) | 0 | 2 (2.5) |
| Gall bladder | 0 | 1 (3.2) | 1 (7.1) | 0 | 2 (2.5) |
| Lung | 2 (11.1) | 0 | 0 | 0 | 2 (2.5) |
| Melanoma | 1 (5.6) | 0 | 0 | 0 | 1 (1.3) |
| Salivary gland | 1 (5.6) | 0 | 0 | 0 | 1 (1.3) |
| Other | 0 | 3 (9.7)b | 2 (14.3)c | 2 (12.5)d | 7 (8.9) |
| Number of prior systemic therapies | |||||
| Median | 4 (1–13) | 3 (1–13) | 5 (2–11) | 5 (2–11) | 4 (1–13) |
| Prior treatment | |||||
| Surgery | 16 (89) | 26 (84) | 12 (86) | 11 (69) | 65 (82.3) |
| Chemotherapy | 18 (100) | 31 (100) | 14 (100) | 16 (100) | 79 (100) |
| Radiation | 14 (78) | 19 (61) | 10 (71) | 7 (44) | 50 (63.3) |
| Prior Platinum | 13 (72) | 23 (74) | 9 (64) | 12 (75) | 57 (72.2) |
| Molecular Aberration for enrollment | |||||
| Somatic BRCA1 | 9 (50) | 0 | 0 | 0 | 9 (11.4) |
| Somatic BRCA2 | 9 (50) | 0 | 0 | 0 | 9 (11.4) |
| Germline BRCA1 | 0 | 0 | 0 | 8 (50) | 8 (10.1) |
| Germline BRCA2 | 0 | 0 | 0 | 8 (50) | 8 (10.1) |
| ATM | 0 | 10 (32) | 0 | 0 | 10 (12.7) |
| FA genes | 0 | 5 (16) | 0 | 0 | 5 (6.3) |
| ARID1A | 0 | 5 (16) | 0 | 0 | 5 (6.3) |
| PALB2 | 0 | 4 (13) | 0 | 0 | 4 (5.1) |
| ATR | 0 | 2 (6.5) | 0 | 0 | 2 (2.5) |
| PTEN mutation | 0 | 0 | 8 (57) | 0 | 8 (10.1) |
| PTEN gene loss | 2 (14) | 0 | 2 (2.5) | ||
| PTEN loss by IHC | 4 (29)f | 0 | 4 (5.1) | ||
| Othere | 0 | 5 (16) | 0 | 0 | 5 (6.3) |
ARID1A AT-Rich Interaction Domain 1A, ATM Ataxia-Telangiectasia Mutated, ATR Ataxia Telangiectasia and Rad3 related, BRCA Breast Cancer gene, FA Fanconi Anemia, PALB2 Partner and Localizer of BRCA2, PTEN Phosphatase and Tensin homolog, IHC immunohistochemistry, ECOG Eastern Cooperative Oncology Group.
aincludes angiosarcoma (2), spindle cell (1), leiomyosarcoma (4), pleomorphic liposarcoma (1), desmoplastic small round cell (1), soft tissue sarcoma (1) and synovial sarcoma (1).
bincludes urachal (1), appendiceal (1) and unknown primary (1).
c includes squamous cell carcinoma of skin (1) and cervical (1).
dincludes sebaceous adenocarcinoma on upper eyelid (1) and esophageal (1).
eincludes BRIP1, BRCA1-Interacting Protein 1 (1); EMSY, EMSY Transcriptional Repressor (1): RAD51, RAD51 recombinase (1); BARD1, BRCA1-associated ring domain protein 1 (1); and BAP1, BRCA 1-associated protein 1 (1).
fone patient had co-occurring PTEN_Q245*/ PTEN loss by IHC.
Efficacy
Table 2 summarizes mean clinical benefit rate (CBR) which is the primary end point of study. Among the 79 patients treated across the study, there was one (1.3%) CR, seven (8.9%) PRs and ten (12.7%) SD ≥ 24 weeks. A total of 18/79 (22.8%) patients derived clinical benefit (CR/PR/SD ≥ 24 weeks). The overall response rate (ORR) was 10.1% (8/79; 95% CI: 4.5%, 19.0%). Table 3 shows detail on molecular alterations, cohort assignment, duration of treatment and best response by RECIST v1.1 in patients that demonstrated clinical benefit. Supplementary Table 4 shows a breakdown of ORR and CBR by tumor type.
Table 2.
Treatment response per cohort as assessed by RECIST v1.1
| Response category | Somatic BRCA1/2 Mutation (N = 18) | Other HR repair pathway gene aberrationsb (N = 31) | PTEN Mutation or Loss by IHC (N = 14) | Germline BRCA1/2 Mutation (N = 16) | All (N = 79) |
|---|---|---|---|---|---|
| Mean CBRa - n (%) | 6 (32.5) (90% credible interval: 16.7, 50.3) | 6 (19.7) (90% credible interval: 9.6, 31.9) | 1 (9.4) (90% credible interval: 1.2, 23.3) | 5 (30.6) (90% credible interval: 14.5, 49.2) | 18 (22.8) |
| ORR - n (%) (95% CI) | 4 (22.2) (6.4, 47.6) | 3 (9.7) (2.0, 25.8) | 0 | 1 (6.3) (0.2, 30.2) | 8 (10.1) (4.5, 19.0) |
| CR - n (%) | 1 (5.6) | 0 | 0 | 0 | 1 (1.3) |
| PR - n (%) | 3 (16.7) | 3 (9.7) | 0 | 1 (6.3) | 7 (8.9) |
| SD ≥ 24 weeks - n (%) | 2 (11.1) | 3 (9.7) | 1 (7.1) | 4 (25.0) | 10 (12.7) |
| SD < 24 weeks - n (%) | 4 (22.2) | 5 (16.1) | 2 (14.3) | 6 (37.5) | 17 (21.5) |
| PD - n (%) | 8 (44.4) | 17 (54.8) | 10 (71.4) | 3 (18.8) | 38 (48.1) |
| NE - n (%) | 0 | 3 (9.7) | 1 (7.1) | 2 (12.5) | 6 (7.6) |
| Median PFS (weeks) (95% CI) | 11.5 (7.9, 23.3) | 7.9 (7.7, 21.1) | 7.7 (7.6, 9.9) | 23.6 (7.4, 33.9) | 9.6 (7.7, 15.9) |
| PFS at 4 weeks (%) (95% CI) | 94 (84, 100) | 90 (80, 100) | 93 (80, 100) | 88 (73, 100) | 91 (85, 98) |
| PFS at 8 weeks (%) (95% CI) | 61 (42, 88) | 43 (29, 65) | 29 (12, 65) | 69 (49, 96) | 50 (40, 62) |
| Median OS (months) (95% CI) | 14.6 (5.4, 36.4) | 8.9 (6.0, 11.8) | 8.5 (5.1, 102) | 12.3 (7.6, 21.1) | 10.3 (8.5, 12.9) |
| OS at 6 months (%) (95% CI) | 71 (52, 96) | 67 (52, 86) | 69 (48, 100) | 81 (64, 100) | 71 (62, 82) |
| OS at 12 months (%) (95% CI) | 65 (46, 92) | 32 (19, 54) | 23 (9, 62) | 50 (31, 82) | 42 (32, 55) |
RECIST v1.1 response evaluation criteria in solid tumors guideline version 1.1, HR Homologous Recombination, IHC immunohistochemistry, CBR clinical benefit rate, ORR objective response rate, PFS progression-free survival, OS overall survival, CR complete response, PR partial response, SD stable disease, PD progressive disease, NE non-evaluable, BRCA Breast Cancer gene, PTEN Phosphatase and Tensin homolog, CI confident interval, n number.
aThe mean clinical benefit rate in each cohort was assessed using posterior probabilities with corresponding credible interval.
bIncludes mutations, deletions and amplifications.
Table 3.
Patients with SD ≥ 24 weeks, CR or PR by RECIST v1.1
| Subject ID | Cancer Type | Cohort Assignment | Cohort Specific Molecular Alteration | Best Response by RECIST v1.1 | # of Prior Systemic Therapies | Duration of Treatment (weeks) | Response length (weeks) |
|---|---|---|---|---|---|---|---|
| 01-040 | Leiomyosarcoma of uterus | 1 | BRCA2 S2186fs*3 | −1% (SD) | 4 | 32 | 24.1 |
| 01-048 | Soft tissue sarcoma | 1 | BRCA1 V788fs*10 | −54% (PR) | 6 | 23 | 7.4 |
| 01-061 | Ovarian | 1 | BRCA2 R3052W | −100% (CR) | 3 | 182+ | 31+ |
| 01-073 | Breast | 1 | BRCA1 Q356* | −14% (SD) | 7 | 40 | 32.6 |
| 01-079 | Salivary Gland | 1 | BRCA1_M1I | −73% (PR) | 2 | 31 | 23.4 |
| 01-082 | Breast Cancer | 1 | BRCA1_Q380* | −81% (PR) | 3 | 112+ | 88.6+ |
| 02-054 | Appendiceal | 2 | ATM S1599* | −33% (PR) | 2 | 61 | 12 |
| 02-007 | Ovarian | 2 | BRIP1 (FANCJ) A402Vfs*21 | 3% (SD) | 4 | 34 | 26.1 |
| 02-032 | Cholangiocarcinoma | 2 | ATM_ G1676fs | −38% (PR) | 2 | 60 | 16 |
| 02-033 | Myoxid Spindle Cell sarcoma | 2 | FANCC Loss of exon 28 | −10% (SD) | 3 | 25 | 24.6 |
| 02-035 | Urachal adenocarcinoma | 2 | PALB2 R170fs14 | −35% (PR) | 3 | 25 | 17 |
| 02-062 | GE Junction | 2 | ATM deletion ATM E2052* | 8% (SD) | 3 | 31 | 24 |
| 03-008 | Gallbladder | 3 | PTEN L70F | 5% (SD) | 6 | 32 | 24 |
| 04-034 | Cholangiocarcinoma | 4 | BRCA2 2041insA | −14% (SD) | 6 | 34 | 25.7 |
| 04-029 | Ampulla of Vater | 4 | BRCA2_c.8633-1 G > C | −20% (SD) | 2 | 124 | 116.3 |
| 04-052 | Adenocarcinoma of cecum | 4 | BRCA2 p.K944* | −100% (PR)a | 2 | 62 | 25.1 |
| 04-065 | Gastric adenocarcinoma | 4 | BRCA2 c.7878 G > A p.Trp2628Ter | −11% (SD) | 4 | 87 | 81 |
| 04-075 | Pancreatic | 4 | BRCA 2 D252Vfs*24 | −11% (SD) | 4 | 44 | 36.4 |
ARID1A AT-Rich Interaction Domain 1A, ARFRP1 ADP Ribosylation factor related protein 1, ATM Ataxia-Telangiectasia Mutated, BRCA Breast Cancer gene, BRIP1 BRCA1-Interacting Protein 1, FANCC Fanconi Anemia Complementation group C, FANCJ Fanconi Anemia Complementation group J, PALB2 Partner and Localizer of BRCA2, Trp tryptophan, Ter termination, ID identifier, CR complete response, PR partial response, SD stable disease.
aSubject ID 04-052 was considered PR given persistence of ascites.
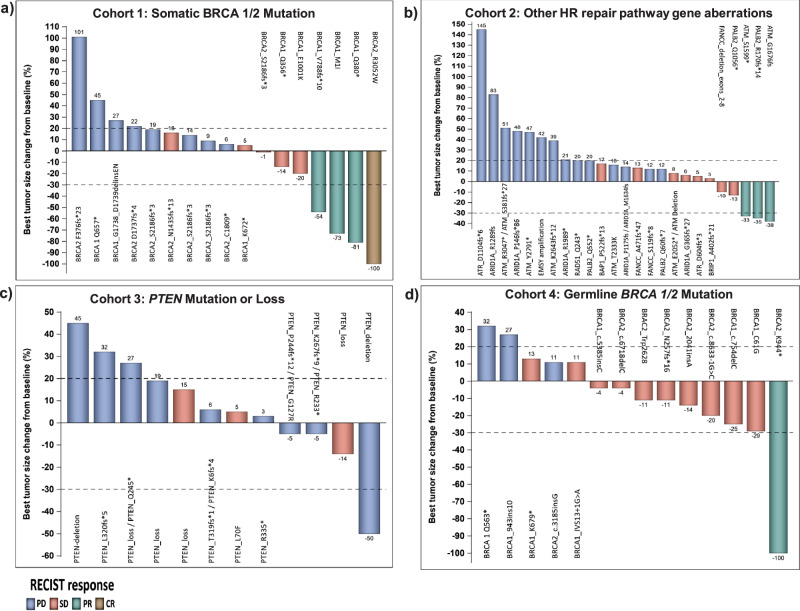
Of 79 patients treated on trial, 68/79 (86.1%) had disease that was measurable by RECISTv1.1 and had post-baseline radiographic imaging studies performed. However, 11/79 did not complete tumor re-assessment and were not included in the waterfall plots (Fig. 1, Supplementary Fig. 1). Evaluation of tumor response per RECIST v1.1 is shown in Fig. 1 and Table 2. In Fig. 1, waterfall plots depict the best RECIST response per patient broken down by cohort. In cohort 1 (Fig. 1a and Table 2), six of 18 patients had a mean CBR of 32.5% (90% credible interval: 16.7%, 50.3%), including one with CR, three with PR and two with prolonged SD. In cohort 2 (Fig. 1b and Table 2), six of 31 patients had a mean CBR of 19.7% (90% credible interval: 9.6%, 31.9%) including three patients with PR and three with prolonged SD. Limited CBR was observed in patients with PTEN mutation/loss in cohort 3 (Fig. 1c and Table 2). Among these patients, one with gallbladder cancer and somatic PTEN L70F mutation had prolonged SD (32 weeks). The mean CBR for cohort 3 was 9.4% (90% credible interval: 1.2%, 23.3%). In cohort 4 (Fig. 1d and Table 2), the mean CBR was 30.6% (90% credible interval: 14.5%, 49.2%) including one patient with PR and 4 with prolonged SD.
Fig. 1. Waterfall plots depicting best RECIST response by patient broken down by cohort.
Individual patients are represented by vertical bars on the X-axis. The best RECIST response (%) is depicted on the Y-axis. Of 79 patients treated on trial, 68/79 (86.1%) had disease that was measurable by RECISTv1.1 and had post-baseline radiographic imaging studies performed. Molecular aberrations corresponding to the enrolled cohorts are shown on the top of each waterfall plot. Patients with somatic BRCA1/2 mutations (cohort 1) are shown in Fig. 1a; patients with other HR repair pathway mutations (cohort 2) are shown in Fig. 1b; patient with PTEN mutation/loss (cohort 3) are shown in Fig. 1c; and patients with germline BRCA1/2 mutations (cohort 4) are shown in Fig. 1d.
The study had a median follow-up duration of 9.9 months, with data cut-off occurring on August 1, 2022. The median PFS on this trial was 9.6 weeks (95% CI: 7.7, 15.9 weeks). The median OS across all cohorts was 10.3 months (95% CI: 8.5, 12.9 months). The median DOR was 6.3 months (95% CI: 1.7, not reached) in the eight patients that were objective responders (CR or PR).
We found that patients with PTEN mutation exhibited a higher risk of death and disease progression compared to patients with PTEN wildtype (WT) status. Our prespecified cohort survival analyses revealed that the risk of death among the patients in cohort 3 was 2.93 (95% CI: 1.22, 7.02, p = 0.016) times the risk among that of cohort 1, 1.33 (95% CI: 0.68, 2.60, p = 0.407) times that of cohort 2, and 1.93 (95% CI: 0.88, 4.22, p = 0.101) times that of cohort 4 (Fig. 2a, c). Similarly, the risk of disease progression among the PTEN mutation/loss (cohort 3) was higher with a hazard ratio of 1.84 (95% CI: 0.86, 3.94, p = 0.115) times that of cohort 1, 1.51 (95% CI; 0.76, 2.99, p = 0.238) times that of cohort 2 and 2.26 (95% CI:0.97,5.25, p = 0.058) times that of cohort 4 (Fig. 2b, c).
Fig. 2. Overall survival and progression free survival analysis.
Patients were grouped by mutation status/cohort. a Kaplan–Meier plot of overall survival. b Kaplan–Meier plot of progression-free survival. c Table shows the hazard ratios and p-value from Cox proportional hazards regression model.
PTEN analysis
PTEN IHC was performed for 8 of the 14 patients enrolled to cohort 3 that had pre-treatment biopsy tissue available. PTEN IHC levels overall correlated with mutation and copy number status. All patients with deleterious PTEN mutations or deletion had either no or low PTEN expression (Fig. 3a).
Fig. 3. PTEN status and overall survival and progression free survival analysis.
a PTEN mutation status and PTEN-IHC results are shown in tile and horizontal bar plots. PTEN mutations are represented in the left tile plot with the allelic frequency (AlleleFreq) of somatic mutations represented by the fill and the amino acid change indicated by text. PTEN copy number deletions are indicated as H.DEL. The middle tile plot represents the percentage of cells with the respective PTEN-IHC staining level; score 0 = no staining, score 1 = low, score 2 = intermediate, and score 3 = high staining levels. The right horizontal bar plot represents the H-score calculated from the PTEN-IHC staining levels. Of note, patients with either PTEN truncating mutations or H.DEL had PTEN loss on IHC testing. Additionally, two of three patients with PTEN frameshift mutations also had PTEN loss by IHC. b Kaplan–Meier plot of overall survival. Sixty-eight patients were stratified by PTEN status (either wild-type, mutated or deletion/loss by IHC). Table shows the hazard ratios and p-value from Cox proportional hazards regression model. c Kaplan–Meier plot of progression-free survival. Sixty-eight patients were stratified by PTEN status (either wild-type, mutated or deletion/loss by IHC). Table shows the hazard ratios and p-value from Cox proportional hazards regression model.
To investigate the impact of PTEN status, at study enrollment, on the efficacy of talazoparib, we conducted a comparative analysis of patients with PTEN WT versus those with deleterious PTEN alterations including PTEN loss by IHC. The analysis encompassed 70 of the 79 patients whose PTEN status was known at the time of enrollment. Amongst these 70 patients, 50 (71.4%) exhibited PTEN WT, while 18 (25.7%) displayed deleterious PTEN alterations, which included 6 cases of PTEN deletion or loss by IHC and 12 cases of PTEN inactivating mutations. Two patients from cohort 2 harbored PTEN mutations of indeterminate significance and were excluded from the overall survival (OS) and progression-free survival (PFS) evaluations. This stratification allowed for a more nuanced understanding of the impact of specific PTEN deleterious alterations on patient outcomes when treated with talazoparib.
Our analysis revealed a correlation between PTEN inactivating alterations and diminished survival outcomes, including OS and PFS. We observed that the median OS varied across the groups, with PTEN WT patients showing a median OS of 11.7 months (95% CI: 7.8, 15.4). In contrast, patients with PTEN inactivating mutations had a median OS of 6.5 months (95% CI:4.4,14.6) and those with PTEN deletion or loss by IHC exhibited a median OS of 10.2 months (95% CI:6.3,16.3) (Fig. 3b).The median PFS in PTEN WT, mutated and PTEN deletion or loss were 3.1 months (95% CI:1.8,4.9), 1.8 months (95% CI:0.5,5.7) and 1.8 months (95% CI: 1.0, not reached), respectively. The risk of disease progression in patients with PTEN alterations was higher compared to those with PTEN WT though not statistically significant. The hazard ratios for disease progression in PTEN inactivating mutations and PTEN deletion or loss by IHC were 1.37 (95% CI: 0.71–2.64, p = 0.352) and 2.02 (95% CI: 0.75-5.24, p = 0.147) when compared with PTEN WT, respectively (Fig. 3c). Furthermore, the analysis revealed a trend towards an increase in risk of death, with a HR of 1.71 (95% CI, 0.87–3.37, p = 0.118) in PTEN mutated and HR of 1.38 (95% CI, 0.54–3.54, p = 0.505) in PTEN deletion or loss by IHC when compared to the PTEN WT group.
Molecular analysis
To date, we have analyzed the pre-treatment biopsies collected on patients in cohort 3. Among 14 patients enrolled into this cohort, samples for 12 patients underwent whole exome sequencing (WES) and analysis. Our focus was on confirming molecular alterations in PTEN along with identifying alterations in DNA Damage Repair (DDR) genes including ATM, BRCA2, BRCA1, ARID1A, PALB2, ATR, CDK12, BARD1, CHEK1, EMSY, BRIP1, FANCC, CHEK2, RAD51D and RAD51. The WES results were then compared to the patient’s historical molecular report used for enrollment purposes alongside their best RECIST response, as illustrated in Fig. 4.
Fig. 4. Overview of mutational landscape and treatment response of patients in cohort 3 (PTEN alterations and PTEN loss by IHC).
The heatmap illustrates alterations detected in PTEN and in the DNA Damage Repair (DDR) genes (including ATM, BRCA2, BRCA1, ARID1A, PALB2, ATR, CDK12, BARD1, CHEK1, EMSY, BRIP1, FANCC, CHEK2, RAD51D and RAD51) with corresponding treatment response. The heatmap analysis presents results from the whole exome sequencing (WES) analysis using pre-treatment tumor biopsy samples, alongside corresponding pre-identified molecular alterations allowing for enrollment onto study (identified as CLIA alterations). Patient with clinical progression as assessed by the treating physician and no post-baseline scans was assigned a value of +21% (*).
Our analysis confirmed the pre-identified molecular aberration used for study enrollment in 7 of 9 patients’ (78%) pre-treatment tumor biopsy samples (3 patients enrolled for PTEN loss by IHC were excluded including patients 005, 021 and 030).
Adverse events
Talazoparib therapy was generally well tolerated with most AEs being grade 1–2. In total, treatment-related AEs occurred in 57/79 (72.2%) patients with the most common being thrombocytopenia (37.9%), anemia (32.9%) fatigue (24.1%) and leukopenia (24.1%) (Supplementary Table 1). Treatment-related AEs that were grade 3–4 occurred in 31/79 (39.2%) patients. Grade 3–4 thrombocytopenia was observed in 17/79 (21.5%) of patients. Fifteen patients (18.9%) experienced grade 3–4 anemia and 12 patients (15.2%) experienced grade 3–4 neutropenia (Supplementary Table 2). All patients who experienced grade 3–4 anemia were transfused with packed red blood cells (PRBCs). Thrombocytopenia and neutropenia were transient and resolved with treatment break. Supplementary Table 3 outlines AEs resulting in dose reduction.
Apart from hematological toxicities, we also encountered one patient with grade 3 fatigue (1.3%), 1 patient with grade 3 diarrhea (1.3%) and 1 patient with grade 3 proteinuria (1.3%). No other grade 3–4 laboratory AEs were encountered during this study. As of August 1, 2022, a total of 40 SAEs were reported during this study, 3 of which were attributed to talazoparib including one patient who died during treatment. This patient was a 67-year-old female with metastatic urothelial cancer who completed only 1 cycle of treatment. She developed grade 3 thrombocytopenia that was attributed to talazoparib, but her death was of unknown cause. The second treatment related SAE was reported for a 51-year-old male with metastatic appendiceal adenocarcinoma who developed grade 4 anemia following 49 days of study treatment. The anemia resolved to baseline after patient received PRBC transfusion. The last treatment related SAE reported was grade 3 diarrhea, which led to discontinuation from study treatment for a 45-year-old male with primary papillary renal cell carcinoma.
Treatment related AEs leading to talazoparib dose modification occurred in 19% of patients. One patient had dose modification due to grade 2 fatigue while the remaining 14 participants were due to hematologic toxicities (Supplementary Table 3).
Discussion
Recent evidence underscores the versatility of talazoparib, advocating for its use in a variety of contexts, across a spectrum of molecular backgrounds and tumor types. In the era of personalized cancer therapy, where patients are often molecularly profiled at or shortly after the time of cancer relapse, we need to have a better understanding of the biologic and clinical significance of the genomic testing results. We attempted to ascertain the clinical actionability of somatic alterations of BRCA1/2, mutations/deletions/amplification in other HR repair pathway genes, PTEN genomic alterations or PTEN loss, and germline BRCA1/2 mutations beyond breast or ovarian cancer, as it pertains to sensitivity to the PARP inhibitor talazoparib.
Patients with deleterious germline BRCA1/2 mutations have tumors that have demonstrated defects in HR and have been shown in several trials to have response to PARP inhibitors7,8. Currently trials have focused on patients with breast and ovarian cancer. We were interested to see if the treatment benefit translated to other tumor types. This phase II study demonstrated encouraging activity of talazoparib in cohorts with somatic and germline BRCA1/2 mutations, where mean clinical benefit rates (CBRs) were 32.5% and 30.6%, respectively. However, patients with PTEN alterations showed limited benefit with mean CBR of 9.4%. Analysis of treatment response by tumor type revealed lower ORR (14.3%) and CBR (28.6%) in our patients compared to previous studies, where ORRs ranging from 31%–33% and CBRs from 48–50% in breast cancers31. Despite attempts to stratify by tumor type, our study did not show improvements in ORR and CBR. This outcome could stem from our study’s design, which focused on a range of molecular alterations and deliberately excluded established “responder” disease types such as ovarian and breast cancers in the germline cohort. Our tissue agnostic basket study resulted in varied sample sizes among different tumor types. Additionally, our patient population did not include individuals with known germline PALB2 mutations at the time of enrollment, a molecular aberration known to have sensitivity to PARP inhibition32. Finally, the lower response rate in our study may also be attributable to the extensive prior lines of therapy that our patients had received. This is in line with a previous study of twenty-six breast cancer patients who had undergone up to seven lines of prior chemotherapy, where no objective responses were observed in patients with either BRCA mutant or wild-type tumors33.
The tolerability of talazoparib in our study aligns with findings from de Bono’s earlier phase I study34. Both our study and his predominantly reported anemia, thrombocytopenia, and neutropenia as the most common grade 3 or 4 adverse events (AEs)34. Yet, our study observed a higher incidence of thrombocytopenia (Grade 1–2: 37.9% vs. 21%; Grade 3–4: 21.5% vs. 18%), potentially due to our patients undergoing more extensive prior treatments, with a median of 4 systemic therapies compared to 2.5 in de Bono’s study. Contrary to de Bono’s findings, our study reported a lower incidence of alopecia34.
In our study, cohort one, focusing on patients with deleterious somatic BRCA1/2 mutations, showed a mean CBR of 32.5%. Notable responses included a sustained complete response in an ovarian cancer patient with a BRCA2 R3052W mutation, partial responses in a soft tissue sarcoma patient (BRCA1 V788fs*10 mutation) and a salivary gland cancer patient (BRCA1 M1I mutation), and prolonged partial response in a breast cancer patient (BRCA1 Q380* mutation). Additionally, two patients exhibited stable disease for over 24 weeks. Cohort two, which included patients with alterations in other homologous recombination repair pathway genes, had a CBR of 19.7%. This cohort observed partial responses in patients with appendiceal carcinoma (ATM S1599* mutation), cholangiocarcinoma (ATM G1676fs mutation), and urachal adenocarcinoma (PALB2 R170fs14 mutation), along with three cases of prolonged stable disease in patients with mutations in Fanconi Anemia genes and ATM mutations. These findings are consistent with previous studies that have demonstrated anti-tumor efficacy with PARP inhibition for patients with HRD gene alterations. For example, a patient with advanced pancreatic cancer that responded to talazoparib in the first-in-human phase I study was found to harbor a PALB2 mutation34. Further, in a phase 2 trial of olaparib in patients with advanced, castration-resistant prostate cancer, responses were observed in 4 patients with ATM aberrations35.
Studies have shown that some cancers with PTEN loss are sensitive to PARP inhibitors, and this may be due to down regulation of Rad51, a critical HR component16,17. This concept has been supported by Shen and colleagues who found that in mouse embryonic fibroblast, inactivation of PTEN induced chromosomal instability through defective Rad51-mediated HR DNA repair36. Additionally, Minami and colleagues found that in PTEN-deficient lung cancer cells and xenograft tumor models, combination therapy with cisplatin and olaparib proved to be synergistic37. This effect was not seen when PTEN was restored in the lung cancer cells or when tested on PTEN+ xenograft models37. Minami and colleagues also found that PTEN deficiency led to reductions in nuclear RAD51, RPA focus formation and phosphorylated Chk1 and Mre1137. Thus, PTEN inactivation leads to suppression of DNA repair37. However, conflicting data challenges the notion of a direct causal link between PTEN and RAD51 expression, these studies suggest that PTEN may not regulate Rad51 and that PTEN inactivation renders BRCA 1/2 aberrant cells resistant to PARP18,19.
Fourteen patients with PTEN mutation/deletion or PTEN loss by IHC were enrolled onto cohort three. The CBR when the cohort enrollment was halted early due to lack of efficacy was 9.4%. We performed a subgroup analysis within the PTEN-altered cohort, differentiating between PTEN deletion or loss by IHC and PTEN inactivating mutations. This stratification yielded a trend towards diminished OS and PFS in both PTEN altered subgroups when compared to PTEN wildtype and contrasts with previous findings showing that cancers with PTEN loss exhibited increased sensitivity to PARP inhibitors16,17.
Despite our observations of poorer outcomes in patients with PTEN-altered tumors, a 61-year-old breast cancer patient treated in cohort 1 with co-occurring deleterious BRCA1 and PTEN mutations experienced a prolonged PR. It is unclear if the patient’s anti-tumor efficacy is due to the deleterious BRCA1 mutation and response to PARP inhibition and if this was enough to overcome the trend towards poorer OS and PFS we saw in patients with PTEN mutations This case illustrates the complexity and personalized nature of cancer treatment responses and the need for more in depth analysis. Clearly, further studies are needed to determine if PTEN alterations are simply a marker of poor prognosis in the spectrum of cancers assessed or if they are true negative predictors for PARP inhibitor therapy benefit. It would also be of interest to determine if the lack-of single agent efficacy could be overcome with combination therapy targeting the PI3K/AKT/mTOR pathway given the PTEN mutation status. Interestingly, despite the limitation of a small sample size, Westin and colleagues demonstrated encouraging clinical outcomes in patients with PTEN mutation treated with a combination of the PARP and AKT inhibitors, olaparib and capivasertib38. Indeed there are emerging preclinical data demonstrating enhanced antitumor activity with the combination of PARP and PI3K pathway inhibitors and PI3K inhibitors were found to have in vivo synergy with PARP inhibitors for the treatment of an endogenous mouse model for BRCA1-related breast cancers and patient derived xenograft models of TNBC without BRCA mutations39,40. Further, there is emerging clinical data suggesting clinical benefit of combinations with PARP and PI3K pathway inhibitors in patients with germline BRCA mutations as well as wildtype patients38,40,41.
Cohort four, which consisted of diverse tumor types beyond the traditional scope of breast and ovarian cancers, yielded a mean CBR of 30.6%. This expansion of PARP inhibitor use is compelling given the partial response observed in a colorectal cancer patient with a germline BRCA2 p.K944*. Additionally, 4 patients attained SD ≥ 24 weeks including two patients with ampullary and gastric cancers, carrying germline BRCA2_c.8633-1 G > C and BRCA2_Trp2628Ter, respectively, who exhibited sustained response over a year. These findings echo the successes observed with olaparib in germline BRCA-mutated metastatic pancreatic cancer26. These findings underscore the therapeutic potential of PARP inhibitors across a range of tumors.
Analyzing the outcomes across various cohorts treated with talazoparib, we observed a disparity in the trend of OS and PFS emerging when comparing hazard ratios between germline BRCA 1/2 mutated patients (cohort 4) and those with somatic BRCA 1/2 mutations (cohort 1). The hazard ratio for OS in cohort 4 versus cohort 1 was 1.54 (95% CI: 0.72, 3.27), contrasting with the PFS hazard ratio of 0.91 (95% CI: 0.45, 1.83). This observed discrepancy could be explained by the distinct patient characteristics in cohort 4. Notably, 25% of patients in cohort 4 were diagnosed with pancreatic cancer, known for its aggressive behavior and challenging prognosis. Additionally, there were differences in the clinical characteristics between cohort 1 and 4. Patients in cohort 4 had a median age of 61 years (range 53–72 years), whereas the median age in cohort 1 was 59.5 years (range 32–81 years). Importantly, after discontinuing talazoparib, the median number of treatments received by patients in cohort 1 was 1 (range 0–4), compared to 0.5 (range 0–5) in cohort 4. This suggests that patients in cohort 1 had access to more treatment options post-talazoparib, potentially impacting their survival outcomes.
Our study has several limitations. Firstly, we were limited by the small numbers of patients in each cohort, which may impact the statistical power and generalizability of our findings. Additionally, the patients included in our study were heavily pretreated, having undergone a median of 4 prior systemic therapies. Furthermore, our patient population was heterogenous consisting of various tumor types. The pretreatment history and heterogeneity could have influenced their response to the intervention tested in our study. Another important limitation to consider is that the enrollment criteria were contingent upon molecular aberrations identified in pre-existing molecular reports. This process, while allowing for expedited study enrollment, raises concerns regarding the fidelity of tumor characterization. Moreover, the study did not define a standardized temporal window for molecular testing, which could potentially lead to the inclusion of patients with outdated molecular data.
To address these limitations, it would be beneficial to incorporate real-time comprehensive molecular testing prior to study commencement. This would help ensure a more accurate understanding of the determinates of treatment response and facilitate the identification of reliable biomarkers. Additionally, real-time molecular testing results would help us better match patients to targeted treatments potentially improving the efficacy and precision of our intervention.
In summary, talazoparib was well tolerated by patients and showed promising clinical benefit in a subset of patients with germline as well as somatic alterations in HR repair pathway genes. Further study is needed to determine the efficacy of talazoparib in larger cohorts, and to identify predictive markers of response and resistance that can refine patient selection.
Methods
Study design and dosing
This was a prospective study conducted as a single-center, investigator-initiated non-randomized, multi-cohort, phase II study to determine the response of patients with advanced solid tumors who had no curative therapeutic options and pre-identified molecular aberrations appropriate for enrollment to the study. Written Informed consent was obtained and patients were treated in accordance with University of Texas, MD Anderson Cancer Center Institutional Review Board (IRB) guidelines. Treatment with the PARP inhibitor, talazoparib, was administered on an outpatient basis at MD Anderson. Biomarin (and later Pfizer) supplied talazoparib tosylate in 250 µg capsules and subjects in all cohorts were instructed to take 1 mg daily. Compliance was determined through pill diaries and the return of unused drugs at the end of every cycle. Potential toxicities and patient safety were evaluated at baseline, weekly during cycle 1 and before clearance to begin every new 28-day cycle. Toxicities were assessed by adverse events monitoring, physical exams and laboratory tests. Complete blood counts (CBCs) continued to be performed weekly beyond cycle 1 to ensure adequate bone marrow function. Adverse events (AEs) were graded according to the National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events (CTCAE) Version 4.03. Patients who experienced Grade 3 or 4 AEs, resumed drug at 25% dose reduction after recovery with a maximum of 2 dose reductions allowed per patient. Clinical trial information: NCT02286687.
Ethics approval and consent to participate
All participants signed the informed consent form prior to study enrollment and study related procedures. The study was approved by MD Anderson Institutional Review Board (IRB) in accordance with the Declaration of Helsinki, Good Clinical Practice, and all federal, state and local regulatory guidelines.
Cohort assignment
Subjects were assigned to one of four cohorts based on pre-identified molecular aberrations determined through a Clinical Laboratory Improvement Amendments (CLIA) validated approach. To be eligible for enrollment, the molecular aberrations had to be predicted to be deleterious, or loss of PTEN expression by gene or immunohistochemistry (IHC). The cohorts included: (1) somatic mutations or deletions of BRCA1/2; (2) mutations, homozygous deletions or amplification in other HR repair pathway genes (e.g., ATM, PALB2, Fanconi Anemia genes, ARID1A, MRE11, RAD50, NBS1, ATR or EMSY amplification); (3) mutations or homozygous deletions in PTEN and/or PTEN loss by IHC; and (4) germline BRCA1/2 mutations (not breast or ovarian cancer). Patients with breast or ovarian cancer were eligible if they did not have germline BRCA1/2 mutations but had another alteration qualifying for cohorts 1–3.
Germline testing was done locally as part of standard-of-care for patients on the germline cohort (cohort 4). All other cohorts had germline sequencing in the research environment after informed consent. Patients with somatic mutations in BRCA1/2 were initiated on treatment prior to germline testing, but germline testing was performed to confirm cohort stratification (i.e., to determine if the BRCA1/2 mutation was somatic or germline). In Cohort 2 of our study, eligibility for patient enrollment was contingent upon the identification of either germline or somatic pathogenic variants in specific homologous recombination (HR) DNA repair pathway genes, excluding BRCA1/2. The genes targeted for Cohort 2 enrollment included PALB2, EMSY, ATR, ATM, RAD51, BAP1, BARD1, and various Fanconi Anemia genes and others. Based on the specific pathogenic variant detected. Patients with somatic as well as deleterious germline alterations in PTEN were eligible for cohort 3. For patients with more than one alteration, they were assigned as follows:
Patients with germline BRCA1/2 alterations (not breast or ovarian cancer) were assigned to cohort 4, regardless of other alterations.
Patients with somatic BRCA1/2 mutations or deletions were assigned to cohort 1, regardless of other somatic alterations.
Patients with PTEN mutations or deletions or loss of PTEN expression were assigned to cohort 3, in the absence of BRCA alterations, and regardless of other HR repair pathway alterations.
Patients with mutations or deletions in ATM; PALB2; Fanconi Anemia genes; ARID1A; MRE11; RAD50; NBS1; and ATR; or amplification of EMSY were assigned to cohort 2, in the absence of other BRCA alterations.
Eligibility criteria
Key inclusion criteria for patients meeting the above-outlined requirements included disease that is biopsiable and measurable by RECIST v1.1, Eastern Cooperative Oncology Group (ECOG) performance status (PS) of 0–1, at least 4 weeks beyond last treatment and adequate organ function defined as absolute neutrophil count (ANC) ≥ 1500/mL, platelet count ≥100,000/mL, hemoglobin 9 ≥ g/dL, serum creatinine ≤ 1.5X the upper limit of normal (ULN) (or glomerular filtration rate (GFR) ≥ 60 mL/min), serum total bilirubin ≤ 1.5 x ULN (or direct bilirubin ≤ ULN), or alanine aminotransferase (ALT) and aspartate aminotransferase (AST) ≤ 2.5 x ULN or ≤ 5 x ULN with liver metastases. Key exclusion criteria were prior treatment with a PARP inhibitor, known hepatitis B, hepatitis C or human immunodeficiency virus (HIV) infection, active central nervous system (CNS) metastases and/or carcinomatous meningitis, additional malignancy that is progressing or requires active treatment, clinically significant cardiovascular disease, active infection requiring IV antibiotics or other uncontrolled intercurrent illness requiring hospitalization, inability to swallow and pregnancy/breastfeeding.
Genomic eligibility
All genomic alterations in eligible genes were reviewed by the MD Anderson Precision Oncology Decision Support (PODS) prior to patient enrolment. Alterations were researched within the published literature for any known effect on function, stability, expression, or therapeutic sensitivity. Alterations were then classified for their functional significance and variant-level actionability, as previously described42. Those with a known loss-of-function (Inactivating) or where a loss-of-function can be predicted due to a premature truncation or frameshift prior to well-characterized essential domains (Inactivating: Inferred) were considered eligible for the trial. Comprehensive details regarding the type of sample utilized for molecular testing, sample collection date, molecular report date, study entry date, specific commercial platform used in molecular testing (including panel version), and lines of systematic treatment administered after molecular testing (used for study enrollment) and preceding the initiation of talazoparib treatment are summarized in Supplementary Table 5.
Assessment of tumor response
Baseline radiographic imaging (e.g., computed tomography (CT) scan or magnetic resonance imaging (MRI)) was performed within four weeks of the start of treatment. Tumor measurements were performed on patients at baseline and at the end of every two cycles (three cycles after 24 weeks). Measurable target lesions were evaluated for response using the Response Evaluation Criteria in Solid Tumors (RECIST v1.1). For purposes of this report, prolonged stable disease (SD) was defined as SD lasting ≥ 24 weeks. Clinical benefit was defined as CR, PR, or SD ≥ 24 weeks.
PTEN IHC testing
For cohort 3, patients could enroll with PTEN loss on any CLIA test on archival samples. For patients treated on cohort 3, central immunohistochemistry for PTEN was performed on pre-treatment formalin fixed and paraffin embedded (FFPE) core needle biopsies. IHC was performed as previously described43,44. Briefly, the staining was performed on 4-micron sections using a monoclonal mouse anti-Human PTEN antibody, Clone 6H2.1 (DAKO, Santa Clara, CA, USA) with a 1:100 dilution. Control tissue was stained with the same run. All stains were reviewed by a qualified pathologist blinded to clinical data.
Both intensity and percent positivity of cytoplasmic staining was assessed on the biopsies. Stromal cells and/or endothelial cells with PTEN expression were used as internal positive controls. Any signal arising within the nucleus or from the cell surface membrane was documented but not used for scoring purpose. PTEN expression was semi-quantitatively scored as 0 (no staining), 1 (light staining), 2 (moderate staining) and 3 (strong staining). If there was no staining of tumor cells and no PTEN expression in stroma or endothelial cells the staining was considered as inconclusive, and the result was not considered for analysis.
Histologic score (H-Score) for cytoplasmic staining was calculated by using the following previously published criteria45. H-score ranged from 0 (no staining) to 300 (maximum staining) and was calculated by using the formula: H Score = 1x (% light staining) + 2x (% moderate staining) + 3x (% strong staining).
Whole exome sequencing of tumor and analysis
Pre-treatment tumor biopsies were obtained from 12 patients in cohort 3 (PTEN alterations and PTEN loss by IHC), from which DNA was extracted and purified. Whole-exome libraries were constructed using biotin-labeled probes from the SeqCap EZ Exome V3 (Roche, Pleasanton, CA, USA) and subsequently sequenced on an Illumina HiSeq4000 sequencer (Illumina Inc., San Diego, CA, USA). Sequence reads were aligned to the human reference genome Hg19 using BWA, with somatic variants identified using MuTect46, germline mutations called using Platypus47, and indels detected using Pindel48. Alignment of FASTQ files to the reference genome utilized specific parameters with BWA49, and preprocessing steps including duplicate marking, realignment, and recalibration were carried out using Picard and GATK50. DNA copy number analysis employed HMMcopy for whole genome sequencing data and an in-house application, ExomeLyzer51, for whole exon sequencing data, with segmentation performed using CBS52. WES variant calls list is provided in Supplementary Note 1.
Statistical considerations and analysis
The study employed Simon’s Two-stage design53, which incorporates early stopping rules for futility. Initially, a minimum of 10 evaluable patients were enrolled onto each cohort and assessed for clinical benefit during the first stage. If a cohort met the predefined criteria for >25% clinical benefit rate, the study progressed to the second stage, wherein additional 20 patients were enrolled. The maximum sample size to each cohort was set at 30, except for cohort 2 (other HR pathways, which permitted enrollment up to 30 patients for each sub-cohort). The PTEN cohort was terminated early due lack of response.
The primary endpoint of this study was clinical benefit rate (CBR) in each cohort. Clinical benefit (CB) is defined as any of the following, CR, PR, or SD for ≥ 24 weeks. A Bayesian design was used to calculate efficacy (targeting a clinical benefit rate of 25%, 90% credible interval:14–38%) and monitor toxicity54. If the lower bound of the 90% posterior credible interval is ≥14%, the drug will be recommended for a subsequent independent confirmatory study.
CBR was assessed using posterior probabilities along with 90% credible interval. Objective response rate (ORR) with 95% confidence interval (CI) was calculated using exact binomial method. Secondary endpoints included progression-free survival (PFS), overall survival (OS), duration of response (DOR) and toxicities. PFS was defined as from treatment start to progression or death, whichever occurred first, or last follow up. OS was defined as from treatment start to death or last follow up. DOR was defined as time from response to progression, death or last follow up among the objective responders (CR or PR). Toxicities were summarized by frequency. Distributions of PFS and OS were estimated using the Kaplan Meier method55. The differences in PFS or OS were estimated using the Cox proportional hazards regression model.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We sincerely thank Wen Xiong for her support in patient recruitment and data collection. Special appreciation goes to Xueyao Fu, Meng Gao, William Brugmann, Rosa Mostorino and Binoj Nair for data collection and patient coordination efforts. Our gratitude also extends to Mary Nowak for administrative support and Argun Akcakanat and Kurt W Evans for their support in DNA analysis and sample management. Lastly, we acknowledge Amber Johnson and the support from MD Anderson Precision Oncology Decision Support Services (PODSS). This study was supported in part by the Once Upon a Time Foundation, The Cancer Prevention and Research Institute of Texas (RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy, NCATS Grant UL1 TR000371 (Center for Clinical and Translational Sciences), The Bosarge Family Foundation, and the MD Anderson Cancer Center Support Grant (P30 CA016672). The authors are fully responsible for the content of this manuscript, and the views and opinions described in the publication reflect only those of the authors.
Author contributions
All authors conceived and/or designed the work that led to the submission, acquired data and/or played an important role in interpreting the results; drafted or revised the manuscript; approved the final version and agreed to be accountable for all aspects of the work. The manuscript has been reviewed and approved by Pfizer.
Data availability
The whole-exome sequencing data generated and analyzed in this study have been deposited at MD Anderson OpenWorks repository, accessible via 10.52519/00133, under controlled access as per MD Anderson compliance guideline56. These data may be subject to patient confidentiality and may require a material transfer agreement. The de-identified participant data and dataset generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The R code used to generate heatmap is provided in Supplementary Note 2.
Competing interests
SPP reports clinical trial research funding/grant support through the institution from AbbVie, Inc.; ABM Therapeutics, Inc.; Acepodia, Inc; Alkermes; Aminex Therapeutics; BioMarin Pharmaceutical, Inc; Boehringer Ingelheim; Bristol Myers Squib; Cerulean Pharma, Inc.; Chugai Pharmaceutical Co., Ltd; Curis, Inc.; Cyclacel Pharmaceuticals; Daiichi Sankyo; Eli Lilly; ENB Therapeutics; Epigenetix Inc.; Five Prime Therapeutics; F-Star Beta Limited; F-Star Therapeutics; Gene Quantum; Genmab A/S; Gilead Sciences, Inc.; GlaxoSmithKline; Helix BioPharma Corp.; Hengrui Pharmaceuticals, Co., Ltd.; HiberCell, Inc.; Immorna Biotherapeutics, Inc.; Immunomedics, Inc.; Incyte Corp.; Jacobio Pharmaceuticals Co., Ltd.; Jiangsu Simcere Pharmaceutical Co., Ltd.; Loxo Oncology, Inc.; Lytix Biopharma AS; Medimmune, LLC.; Medivation, Inc.; Merck Sharp and Dohme Corp.; Nectin Therapeutics, Ltd.; Novartis Pharmaceuticals; Nurix; Pieris Pharmaceuticals, Inc.; Pfizer; Phanes Therapeutics; Principia Biopharma, Inc.; Puma Biotechnology, Inc.; Purinomia Biotech, Inc.; Rapt Therapeutics, Inc.; Replimune; Roche/Blueprint; Seattle Genetics; Silverback Therapeutics; Shasqi, Inc.; Synlogic Therapeutics; Taiho Oncology; Tesaro, Inc.; Theradex Oncology; Toragen Therapeutics, Inc.; TransThera Bio; Xencor, Inc; ZielBio, Inc.;NCI/NIH; P30CA016672 – Core Grant (CCSG Shared Resources). CHK reports research funding from NIH/NCI P30CA016672 and Biostatistics Resource Group. YY served as a statistical consultant to AbbVie, Amgen, Bexion, BeyondSpring, Boehringer Ingelheim, Bristol Myers Squibb, Century, Enliven, GT Medical, NeoImmueTech, Merck, NGM, Repare, Servier, Starpax, Transthera, Xinthera, and Vertex. DDK reports research funding/grant support for clinical trials from Pfizer, Phoplatin Therapeutic, Symphogen, NIH clinical translation science award, clinical translation NCI grant and consultancy/advisory board participation for Black Beret Life Science, Affigen and Phosplatin. VS reports research funding/grant support for clinical trials from AbbVie, Agensys, Inc., Alfasigma, Altum, Amgen, Bayer, BERG Health, Blueprint Medicines Corporation, Boston Biomedical, Inc., Boston Pharmaceuticals, Celgene Corporation, D3 Bio, Inc., Dragonfly Therapeutics, Inc., Exelixis, Fujifilm, GlaxoSmithKline, Idera Pharmaceuticals, Inc., Incyte Corporation, Inhibrx, Loxo Oncology, MedImmune, MultiVir, Inc., NanoCarrier, Co., National Comprehensive Cancer Network, NCI-CTEP, Northwest Biotherapeutics, Novartis, PharmaMar, Pfizer, Relay Therapeutics, Roche/Genentech, Takeda, Turning Point Therapeutics, UT MD Anderson Cancer Center, and Vegenics Pty Ltd.; travel support from ASCO, ESMO, Helsinn Healthcare, Incyte Corporation, Novartis, and PharmaMar; consultancy/advisory board participation for Helsinn Healthcare, Jazz Pharmaceuticals,Incyte Corporation, Loxo Oncology/Eli Lilly, MedImmune, Novartis, QED Therapeutics, Relay Therapeutics, Daiichi-Sankyo, and R-Pharm US; and other relationship with Medscape. DH reports research funding/grant support for clinical trials from AbbVie, Adaptimmune, Adlai-Nortye, Amgen, Astra-Zeneca, Bayer, Biomea, Bristol-Myers Squibb, Daiichi-Sankyo, Deciphera, Eisai, Eli Lilly, Endeavor, Erasca, F. Hoffmann-LaRoche, Fate Therapeutics, Genentech, Genmab, Immunogenesis, Infinity, Kyowa Kirin, Merck, Mirati, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Pyramid Bio, Revolution Medicine, SeaGen, STCube, Takeda, TCR2, Turning Point Therapeutics, VM Oncology. Also received travel, Accommodations, Expenses from AACR, ASCO, CLCC, Bayer, Genmab, SITC, Telperian and serves as consulting, Speaker, or Advisory Role for 28Bio, Abbvie, Acuta, Adaptimmune, Alkermes, Alpha Insights, Amgen, Affini-T, Astellas, Aumbiosciences, Axiom, Baxter, Bayer, Boxer Capital, BridgeBio, CARSgen, CLCC, COG, COR2ed, Cowen, Ecor1, EDDC, Erasca, Exelixis, Fate Therapeutics, F.Hoffmann-La Roche, Genentech, Gennao Bio, Gilead, GLG, Group H, Guidepoint, HCW Precision Oncology, Immunogenesis, Incyte Inc, Inhibrix Inc, InduPro, Janssen, Jounce Therapeutics Inc, Liberium, MedaCorp, Medscape, Novartis, Numab, Oncologia Brasil, ORI Capital, Pfizer, Pharma Intelligence, POET Congress, Prime Oncology, Projects in Knowledge, Quanta, RAIN, Ridgeline, SeaGen, Stanford, STCube, Takeda, Tavistock, Trieza Therapeutics, Turning Point Therapeutics, WebMD, YingLing Pharma, Ziopharm. DH also reports Other ownership interests: Molecular Match (Advisor), OncoResponse (Founder, Advisor), Telperian (Founder, Advisor). SQF reports research funding/grant support for clinical trials from NIH/NCI P30CA016672 – Core Grant (CCSG Shared Resources); Abbisko; BeiGene; BioAtla, LLC.; Boehringer Ingelheim; CUE Biopharma, Inc.; Eli Lilly & Co.; Exelixis; Greenfire Bio, Inc.; Hookipa Biotech; IMV, Inc.; Innovent Biologics, Co., Ltd.; K-Group Beta; Lyvgen Biopharm, Co., Ltd.; MacroGenics; MediLink Therapeutics, Co. Ltd.; Millennium Pharmaceuticals, Inc.; Nerviano Medical Sciences; NeuPharma, Inc.; NextCure, Inc.; Ningbo NewBay Technology Development Co., Ltd.; Novartis; NovoCure; Nykode Therapeutics AS.; Parexel International, LLC; Pionyr Immunotherapeutics, Inc.; PureTech Health, LLC; Sellas Life Sciences Group; Soricimed Biopharma, Inc.; SQZ Biotechnologies; Sumitomo Dainippon; Taiho Oncology and NCCN; Treadwell Therapeutics; Turnstone Biologics; Tyligand Bioscience, Ltd.; Virogin Biotech, Ltd. AN reports research funding/grant support for clinical trials from NCI, EMD Serono, MedImmune, Healios Onc. Nutrition, Atterocor/Millendo, Amplimmune, ARMO BioSciences, Karyopharm Therapeutics, Incyte, Novartis, Regeneron, Merck, Bristol-Myers Squibb, Pfizer, CytomX Therapeutics, Neon Therapeutics, Calithera Biosciences, TopAlliance Biosciences, Eli Lilly, Kymab, PsiOxus, Arcus Biosciences, NeoImmuneTech, Immune-Onc Therapeutics, Surface Oncology, Monopteros Therapeutics, BioNTech SE, Seven & Eight Biopharma, and SOTIO Biotech AG. Consulting fees from CTI, Deka Biosciences, Janssen Biotech, NGM Bio, PsiOxus Therapeutics, Immune-Onc Therapeutics, STCube Pharmaceuticals, OncoSec KEYNOTE-695, Genome & Company, CytomX Therapeutics, Nouscom, Merck Sharp & Dohme Corp, Servier, Lynx Health, AbbVie, PsiOxus. AN received travel and accommodation expense from ARMO BioSciences, NeoImmuneTech, NGM Biopharmaceuticals and honoraria for speaking engagement from AKH Inc, The Lynx Group, Society for Immunotherapy of Cancer (SITC), Korean Society of Medical Oncology (KSMO), Scripps Cancer Care Symposium, ASCO Direct Oncology Highlights, European Society for Medical Oncology (ESMO), CME Outfitters. JR reports non-financial support and reasonable reimbursement for travel from European Society for Medical Oncology; receiving consulting and travel fees from Ellipses Pharma and IONCTURA (including serving on the scientific advisory board); Consulting fees from Aadi Bioscience, Clarion Healthcare, Debiopharm, Monte Rosa Therapeutics, Cullgen. Pfizer, Merus N.V., Macorgenics, Oncology One, Envision Pharma, Columbus Venture Partners, Sardona Therapeutics, Avoro Capital Advisors, Vall d’Hebron Institute of Oncology/Ministero De Empleo Y Seguridad Social, Chinese University of Hong Kong, Boxer Capital, LLC, Tang Advisors, LLC, Incyte, Alnylam Pharmaceuticals ; receiving research funding from Blueprint Medicines, Black Diamond Therapeutics, Merck Sharp & Dohme, Hummingbird, Yingli and Vall d’Hebron Institute of Oncology/Cancer Core Europe; and serving as investigator in clinical trials with Novartis, Spectrum Pharmaceuticals, Symphogen, BioAlta, Pfizer, GenMab, CytomX, Kelun-Biotech, Takeda-Millenium, GalxoSmithKline, Taiho, Roche Pharmaceuticals, Hummingbird, Yingli, Bycicle Therapeutics, Merus, Curis, Bayer, AadiBioscience, Nuvation, ForeBio, BioMed Valley Discoveries, Loxo Oncology, Hutchinson MediPharma, Cellestia, Deciphera, Ideaya, Amgen, Tango Therapeutics, Mirati, Linnaeus Therapeutics, and Cancer Core Europe. MJ reports grants from QED, personal fees from Incyte, grants from Taiho, other from Novartis, personal fees from Oncosil, grants and personal fees from MERCK, grants from EMD SERONO, personal fees and non-financial support from MECLUN, outside the submitted work. JA serves as ad hoc paid advisor to: BMS, Merck, Astellas, Daiichi, Zymeworks, Jazz, Novartis, Gilead, Taiho, Amgen. KR reports research funding/grant support from AbbVie, AstraZeneca, Bayer, Daiichi Sankyo, Eisai, Genentech, Guardant Health, HiberCell, Innovent, Janssen, Merck, Seattle Genetics, UCB Biosciences, Xencor and serves as consultant for AstraZeneca, Bayer, Daiichi Sankyo, Eisai, Genentech, Seattle Genetics. NS reports current clinical trial research funding/grant support through the institution from AstraZeneca, Innovent, Ascentage, Deciphera, Boehringer Ingelheim, and Advisory Board consultant for Aadi Biosciences, Bayer, and Boehringer Ingelheim. GBM reports SAB/Consultant: Amphista, Astex, AstraZeneca, BlueDot, Chrysallis Biotechnology, Ellipses Pharma, GSK, ImmunoMET, Infinity, Ionis, Leapfrog Bio, Lilly, Medacorp, Nanostring, Nuvectis, PDX Pharmaceuticals, Qureator, Roche, Signalchem Lifesciences, Tarveda, Turbine, Zentalis Pharmaceuticals; Stock/Options/Financial: Bluedot, Catena Pharmaceuticals, ImmunoMet, Nuvectis, SignalChem, Tarveda, Turbine; Licensed Technology: HRD assay to Myriad Genetics, DSP patents with Nanostring; Sponsored research: AstraZeneca. AT reports research funding/grant support for clinical trials from OBI Pharma USA Inc., Baxalta, Bayer, Boston Biomedical, Cancer Prevention & Research Institute of Texas (CPRIT), EMD Serono, IMMATICS, Parker Institute for Cancer Immunotherapy, Placon Therapeutics, Agenus, Tachyon, Tempus Labs, Tvardi Therapeutics, Karus Therapeutics; Novocure Ltd., Orionis, NIH/NCI P30CA016672 – Core Grant (CCSG Shared Resources). AT served as consulting or advisory role for Vincerx, BrYet, and Diaccurate, NEX-I, Macrogenics, Avstera. FM-B reports research funding/grant support for clinical trials from Aileron Therapeutics, AstraZeneca, Bayer Healthcare Pharmaceutical, Calithera Biosciences, Curis, CytomX Therapeutics, Daiichi Sankyo, Debiopharm International, eFFECTOR Therapeutics, Genentech, Guardant Health, Klus Pharma, Takeda Pharmaceutical, Novartis, Puma Biotechnology, Taiho Pharmaceutical. FM-B served as advisory committee for Black Diamond, Biovica, Eisai, FogPharma, Immunomedics, Inflection Biosciences, Karyopharm Therapeutics, Loxo Oncology, Mersana Therapeutics, OnCusp Therapeutics, Puma Biotechnology Inc., Seattle Genetics, Sanofi, Silverback Therapeutics, Spectrum Pharmaceuticals, Zentalis, personal fees for consulting/travel related from AbbVie, Aduro BioTech, Alkermes, AstraZeneca, Daiichi Sankyo, DebioPharm, Ecor1 Capital, eFFECTOR Therapeutics, F. Hoffman-La Roche, GT Apeiron, Genentech, Harbinger Health, IBM Watson, Infinity Pharmaceuticals, Jackson Laboratory, Kolon Life Science, Lengo Therapeutics, Menarini Group, OrigiMed, PACT Pharma, Parexel International, Pfizer, Protai Bio, Samsung Bioepis, Seattle Genetics, Tallac Therapeutics, Tyra Biosciences, Xencor, Zymeworks, European Organization for Research and Treatment of Cancer (EORTC), European Society for Medical Oncology (ESMO), personal fees for honoraria from Chugai Biopharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00634-6.
References
- 1.Birkelbach, M. et al. Detection of impaired homologous recombination repair in NSCLC cells and tissues. J. Thorac. Oncol.8, 279–286 (2013). 10.1097/JTO.0b013e31827ecf83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennessy, B. T. et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol.28, 3570–3576 (2010). 10.1200/JCO.2009.27.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George, J. et al. Nonequivalent gene expression and copy number alterations in high-grade serous ovarian cancers with BRCA1 and BRCA2 mutations. Clin. Cancer Res.19, 3474-3484 (2013). [DOI] [PubMed]
- 4.Gorringe, K. L. et al. Copy number analysis identifies novel interactions between genomic loci in ovarian cancer. PLoS One5, e11408 (2010). [DOI] [PMC free article] [PubMed]
- 5.Konishi, H. et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. USA108, 17773–17778 (2011). 10.1073/pnas.1110969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med.361, 123–134 (2009). 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]
- 7.Kummar, S. et al. A phase I study of veliparib in combination with metronomic cyclophosphamide in adults with refractory solid tumors and lymphomas. Clin. Cancer Res.18, 1726–1734 (2012). 10.1158/1078-0432.CCR-11-2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson, O. A. et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res.11, R47 (2009). 10.1186/bcr2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buisson, R. et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol.17, 1247–1254 (2010). 10.1038/nsmb.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P. et al. Association of common PALB2 polymorphisms with breast cancer risk: a case-control study. Clin. Cancer Res.14, 5931–5937 (2008). 10.1158/1078-0432.CCR-08-0429 [DOI] [PubMed] [Google Scholar]
- 11.Gilardini Montani, M. S. et al. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J. Exp. Clin. Cancer Res.32, 95 (2013). 10.1186/1756-9966-32-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, K. W. et al. A population of heterogeneous breast cancer patient-derived xenografts demonstrate broad activity of PARP inhibitor in BRCA1/2 wild-type tumors. Clin. Cancer Res.23, 6468–6477 (2017). 10.1158/1078-0432.CCR-17-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlett, N. G. et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science297, 606–609 (2002). 10.1126/science.1073834 [DOI] [PubMed] [Google Scholar]
- 14.Mathew, C. G. Fanconi anaemia genes and susceptibility to cancer. Oncogene25, 5875–5884 (2006). 10.1038/sj.onc.1209878 [DOI] [PubMed] [Google Scholar]
- 15.Chu, E. C. & Tarnawski, A. S. PTEN regulatory functions in tumor suppression and cell biology. Med Sci. Monit.10, RA235–RA241 (2004). [PubMed] [Google Scholar]
- 16.Mendes-Pereira, A. M. et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med.1, 315–322 (2009). 10.1002/emmm.200900041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEllin, B. et al. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res.70, 5457–5464 (2010). 10.1158/0008-5472.CAN-09-4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta, A. et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle8, 2198–2210 (2009). 10.4161/cc.8.14.8947 [DOI] [PubMed] [Google Scholar]
- 19.Peng, G. et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun.5, 3361 (2014). 10.1038/ncomms4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lord, C. J. & Ashworth, A. BRCAness revisited. Nat. Rev. Cancer16, 110–120 (2016). 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- 21.Pilie, P. G. et al. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin. Cancer Res.25, 3759–3771 (2019). 10.1158/1078-0432.CCR-18-0968 [DOI] [PubMed] [Google Scholar]
- 22.Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res.72, 5588–5599 (2012). 10.1158/0008-5472.CAN-12-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoy, S. M. Talazoparib: first global approval. Drugs78, 1939–1946 (2018). 10.1007/s40265-018-1026-z [DOI] [PubMed] [Google Scholar]
- 24.Shen, Y. et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res.19, 5003–5013 (2013). 10.1158/1078-0432.CCR-13-1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murai, J. et al. Stereospecific PARP trapping by BMN 673 and comparison with Olaparib and Rucaparib. Mol. Cancer Ther.13, 433–443 (2014). 10.1158/1535-7163.MCT-13-0803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golan, T. et al. Maintenance Olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med.381, 317–327 (2019). 10.1056/NEJMoa1903387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heeke, A. L., et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis. Oncol.201810.1200/PO.17.00286 (2018). [DOI] [PMC free article] [PubMed]
- 28.Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell162, 454 (2015). 10.1016/j.cell.2015.06.053 [DOI] [PubMed] [Google Scholar]
- 29.Agarwal, N. et al. Plain language summary of the design of the TALAPRO-2 study comparing talazoparib and enzalutamide versus enzalutamide and placebo in men with metastatic castration-resistant prostate cancer. Future Oncol.18, 2979–2986 (2022). 10.2217/fon-2022-0389 [DOI] [Google Scholar]
- 30.Agarwal, N. et al. Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial. Lancet402, 291–303 (2023). 10.1016/S0140-6736(23)01055-3 [DOI] [PubMed] [Google Scholar]
- 31.Tung, N. M. et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J. Clin. Oncol.38, 4274–4282 (2020). 10.1200/JCO.20.02151 [DOI] [PubMed] [Google Scholar]
- 32.Gruber, J. J. et al. A phase II study of talazoparib monotherapy in patients with wild-type BRCA1 and BRCA2 with a mutation in other homologous recombination genes. Nat. Cancer3, 1181–1191 (2022). 10.1038/s43018-022-00439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelmon, K. A. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol.12, 852–861 (2011). 10.1016/S1470-2045(11)70214-5 [DOI] [PubMed] [Google Scholar]
- 34.de Bono, J. et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov.7, 620–629 (2017). 10.1158/2159-8290.CD-16-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med.373, 1697–1708 (2015). 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen, W. H. et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell128, 157–170 (2007). 10.1016/j.cell.2006.11.042 [DOI] [PubMed] [Google Scholar]
- 37.Minami, D. et al. Synergistic effect of olaparib with combination of cisplatin on PTEN-deficient lung cancer cells. Mol. Cancer Res.11, 140–148 (2013). 10.1158/1541-7786.MCR-12-0401 [DOI] [PubMed] [Google Scholar]
- 38.Westin, S. N. et al. Phase Ib dose expansion and translational analyses of olaparib in combination with capivasertib in recurrent endometrial, triple-negative breast, and ovarian cancer. Clin. Cancer Res.27, 6354–6365 (2021). 10.1158/1078-0432.CCR-21-1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bian, X., et al. PTEN deficiency sensitizes endometrioid endometrial cancer to compound PARP-PI3K inhibition but not PARP inhibition as monotherapy. Oncogene (2017). [DOI] [PMC free article] [PubMed]
- 40.Juvekar, A. et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov.2, 1048–1063 (2012). 10.1158/2159-8290.CD-11-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matulonis, U. A. & Monk, B. J. PARP inhibitor and chemotherapy combination trials for the treatment of advanced malignancies: does a development pathway forward exist? Ann. Oncol.28, 443–447 (2017). 10.1093/annonc/mdw697 [DOI] [PubMed] [Google Scholar]
- 42.Johnson, A. et al. The right drugs at the right time for the right patient: the MD Anderson Precision Oncology Decision Support Platform. Drug Discov. Today20, 1433–1438 (2015). 10.1016/j.drudis.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djordjevic, B. et al. Clinical assessment of PTEN loss in endometrial carcinoma: immunohistochemistry outperforms gene sequencing. Mod. Pathol.25, 699–708 (2012). 10.1038/modpathol.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meric-Bernstam, F. et al. Influence of biospecimen variables on proteomic biomarkers in breast cancer. Clin. Cancer Res.20, 3870–3883 (2014). 10.1158/1078-0432.CCR-13-1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Detre, S., Saclani Jotti, G. & Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol.48, 876–878 (1995). 10.1136/jcp.48.9.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol.31, 213–219 (2013). 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet46, 912–918 (2014). 10.1038/ng.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, K. et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics25, 2865–2871 (2009). 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet43, 491–498 (2011). 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science346, 256–259 (2014). 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olshen, A. B. et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics5, 557–572 (2004). 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- 53.Simon, R. Optimal two-stage designs for phase II clinical trials. Control Clin. Trials10, 1–10 (1989). 10.1016/0197-2456(89)90015-9 [DOI] [PubMed] [Google Scholar]
- 54.Thall, P. F., Simon, R. M. & Estey, E. H. Bayesian sequential monitoring designs for single-arm clinical trials with multiple outcomes. Stat. Med14, 357–379 (1995). 10.1002/sim.4780140404 [DOI] [PubMed] [Google Scholar]
- 55.Kaplan, E. L. & Meier Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc.53, 457–481 (1958). 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 56.Piha-Paul, S. A., et al. Supplemental Data for A Phase II Study of Talazoparib Tosylate in Advanced Cancer Patients with Somatic and Germline (Not Breast or Ovarian Cancer) Alterations of BRCA1/2, Mutations/Deletions/Amplification in Other Homologous Recombination Repair Pathway Genes and PTEN or PTEN loss. 10.52519/00133.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-exome sequencing data generated and analyzed in this study have been deposited at MD Anderson OpenWorks repository, accessible via 10.52519/00133, under controlled access as per MD Anderson compliance guideline56. These data may be subject to patient confidentiality and may require a material transfer agreement. The de-identified participant data and dataset generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The R code used to generate heatmap is provided in Supplementary Note 2.