Abstract
The expression of clock genes has been observed to be impaired in biopsies from patients with inflammatory bowel disease (IBD). Disruption of circadian rhythms, which occurs in shift workers, has been linked to an increased risk of gastrointestinal diseases, including IBD. The peripheral circadian clock in intestinal epithelial cells (IECs) was previously shown to balance gastrointestinal homeostasis by regulating the microbiome. Here, we demonstrated that the intestinal clock is disrupted in an IBD-relevant mouse model (IL-10−/−). A lack of the intestinal clock gene (Bmal1) in intestinal epithelial cells (IECs) in a chemically and a novel genetically induced colitis model (DSS, Bmal1IEC−/−xIL-10−/−) promoted colitis and dramatically reduced survival rates. Germ-free Bmal1IEC−/− mice colonized with disease-associated microbiota from IL-10−/− mice exhibited increased inflammatory responses, highlighting the importance of the local intestinal clock for microbiota-induced IBD development. Targeting the intestinal clock directly by timed restricted feeding (RF) in IL-10−/− mice restored intestinal clock functions, including immune cell recruitment and microbial rhythmicity; improved inflammatory responses; dramatically enhanced survival rates and rescued the histopathological phenotype. In contrast, RF failed to improve IBD symptoms in Bmal1IEC−/−xIL-10−/− mice, demonstrating the significance of the intestinal clock in determining the beneficial effect of RF. Overall, we provide evidence that intestinal clock dysfunction triggers host immune imbalance and promotes the development and progression of IBD-like colitis. Enhancing intestinal clock function by RF modulates the pathogenesis of IBD and thus could become a novel strategy to ameliorate symptoms in IBD patients.
Keywords: Intestinal circadian clock, Gastrointestinal inflammation, IBD, Restricted feeding, Microbiota
Subject terms: Mucosal immunology, Mechanisms of disease
Introduction
The circadian (lat. circa = approximately, dies = day) system consists of the central pacemaker in the suprachiasmatic nucleus (SCN), which drives behavioral rhythms and orchestrates peripheral clocks [1]. Disruption of the circadian system, for example, as experienced during jet lag [2], is related to immune deficits and pathologies, such as cancer and infections [3, 4], and has been associated with a wide range of diseases, including inflammatory bowel disease (IBD) [5].
As previously shown, tissue-specific functions are controlled by local peripheral clocks [2, 6]. Importantly, circadian rhythms, including those related to mucosal immunity and the microbiome, have been identified in the intestine [7–9]. Disturbance of the gastrointestinal (GI) physiology results in changes in the microbiota composition, which plays a role in the development of GI diseases, including IBD [10]. Our recent work highlights the importance of functional intestinal clocks for host–microbe crosstalk to balance GI homeostasis [11]. Hence, a local intestinal oscillator may impact the development of GI disorders. Indeed, epidemiological studies have reported suppressed clock gene expression in biopsies from IBD patients [12], indicating a link between the intestinal clock and disease occurrence. However, the role of the intestinal clock in GI diseases remains to be addressed. Here, we hypothesized that the intestinal clock may constitute a mechanism by which mucosal immune functions and the microbiome promotes GI inflammation.
Recently, time-restricted eating/feeding (RF) has become increasingly popular due to its positive effects on metabolic health and inflammation and its impact on the gut microbiome [13]. Preclinical evidence showed that RF improves systemic inflammation [14]. RF is also a powerful tool for regulating peripheral clocks [15], including in the intestinal tissues [9], and microbial rhythmicity [8]. Consequently, we hypothesize that RF may influence the development and progression of IBD by targeting intestinal clock functions.
In this study, we demonstrated the causal relationship between intestinal clock dysfunction and the severity of GI inflammation in chemically and genetically induced colitis models (IL-10−/−, Bmal1IEC−/−xIL-10−/−, dextran sulfate sodium (DSS)) and microbiota transfer experiments. Potential mechanisms include regulation of key genes relevant for inflammatory and bacterial responses by the intestinal clock, which were identified by RNA-sequencing analysis. Importantly, for the first time, we demonstrated the beneficial effects of nighttime-restricted feeding (RF) on circadian colon functions, the microbiome, IBD progression and survival of IL-10−/− mice, a well-described mouse model of colitis [16]. Remarkably, RF failed to ameliorate colitis symptoms and improve survival in intestinal clock-deficient Bmal1IEC−/−xIL-10−/− mice, demonstrating the relevance of a functional intestinal clock in mediating the positive effect of RF; thus, RF might be a novel target for future IBD therapies.
Results
Intestinal circadian disruption in an IBD-relevant mouse model
The development of various GI diseases, including IBD [5], has been linked to circadian disruption, such as that occurring during shift work and frequent travel across time zones [2]. IL-10-deficient mice on a Sv129 background (IL-10−/− Sv129) exhibit a normal circadian behavioral phenotype (Fig. 1A, Supplementary Fig. 1A). Central circadian clock functions, such as rhythmic wheel-running activity and food intake during a light–dark (LD) cycle or constant darkness (DD), were indistinguishable from those of the controls, although total activity was slightly reduced in the IL-10−/− Sv129 mice (Fig. 1A, Supplementary Fig. 1B–D).
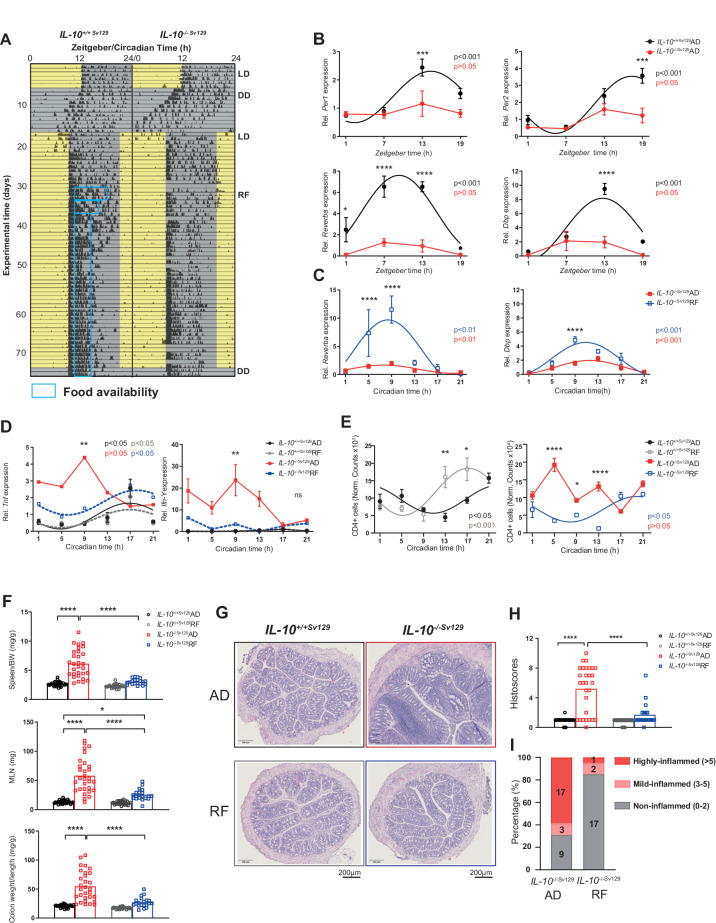
Fig. 1.
Restricted feeding restores the disrupted colonic clock in IL-10−/−Sv129 mice and ameliorates colitis. A Representative actogram of wild type (IL-10+/+Sv129) and IL-10−/−Sv129 mice exposed to a light (yellow)-dark (gray) cycle (LD), constant darkness (DD), and nighttime-restricted feeding (RF, blue boxes). Tick marks represent running wheel activity. B Clock gene expression in colonic tissues from IL-10−/−Sv129 mice (Control AD and RF: n = 24; IL-10−/−Sv129AD: n = 24; IL-10−/−Sv129RF: n = 19; n = 3-4 mice/time points for each group) C Clock gene and D inflammatory genes expression in colonic tissues. E Quantification of the amount of CD4 + T cells in the colon LP from wild type and IL-10−/−Sv129 mice under AD and RF conditions. F Organ weights, G representative colonic cross section stained with H&E and H histopathological scores of colonic tissues from wild-type and IL-10−/−Sv129 mice under AD and RF conditions (top) and I quantification of individual mice with different degrees of inflammation (bottom). Tissues with scores between 0 and 2 were classified as noninflamed, those with scores between 3 and 5 were classified as mildly inflamed, and those with scores >5 were classified as highly inflamed. The data points represent individual mice (n = 3–4/time point for each group). Significant rhythms are illustrated with fitted cosine-wave regression using a line (significance: cos-fit p value ≤ 0.05). Two-way ANOVA with Tukey’s correction was also used. The data are presented as the means ± SEMs. Asterisks indicate significant differences; *p < 0.05, ***p < 0.001, ****p < 0.0001. Details on the number of mice per time point are summarized in the Supplementary Table 6
Recently, we demonstrated an imbalance in GI immune homeostasis in mice lacking a functional intestinal clock [11]. This prompted us to characterize the molecular intestinal clock in inflamed IL-10−/− Sv129 mice. Robust circadian rhythms were found in the expression of the clock genes Per1/2 (Period 1/2), Rev-erbα (NR1D1) and Dbp in the colonic epithelial cells of wild-type mice. This rhythmicity was lost, and gene expressions were strongly suppressed in IL-10−/− Sv129 mice (Fig. 1B), indicating a dysfunctional colonic clock. The circadian origin of these results was validated in colonic tissues collected at 6 different circadian times from IL-10−/− Sv129 mice kept in constant darkness and in cultured colonic organoids (Supplementary Fig. 1E, F). This effect was restricted to the colon since the major clock genes remained rhythmic in the liver, cecum and cultured jejunal organoids (Supplementary Fig. 2). Loss of rhythmicity in IL-10−/− Sv129 mice was also observed for inflammatory genes, e.g., Tnf, Ifnγ, Cxcr4, and Vcam-1, and T-cell recruitment to the lamina propria (LP) (Fig. 1D, E, Supplementary Fig. 1G, H). Furthermore, the peak expression of most clock genes was strongly correlated with the colonic histological score; the frequency of immune cells in the lamina propria, including macrophages, neutrophils, CD4+ cells and dendritic cells; and the expression of genes involved in barrier function (Supplementary Fig. 1I, J). These results indicate a causal relationship between colon clock dysfunction in IL-10−/− Sv129 mice and their inflammatory phenotype.
Nighttime-restricted feeding restored the colonic clock and ameliorated pathological changes in IL-10−/− mice
Nighttime-restricted feeding (RF) is a widely used approach in mice to influence the rhythmicity of peripheral clocks [15]. We tested whether RF in IL-10−/− Sv129 mice restores the colonic clock and impacts their Crohn’s disease-like phenotype. Therefore, RF was gradually introduced to IL-10−/− Sv129 mice and controls, which were euthanized after 4 weeks of RF on 6 different circadian times (Fig. 1A, Supplementary Fig. 1A). Food intake and nighttime activity levels were comparable between genotypes and feeding conditions (Supplementary Fig. 1B–D). Importantly, RF substantially improved the circadian rhythmicity of clock gene expression in IL-10−/− Sv129 colonic tissue to similar levels as those observed in control mice under ad libitum (AD) conditions (Fig. 1C, Supplementary Table 1). Moreover, a restored rhythmicity of Tnf and reduced levels of Tnf and Ifn-y as well as a restored rhythmicity in the number of CD3+CD4+ cells isolated from the colonic LP and an overall reduced number of CD4 + T lymphocytes were observed in IL-10−/− Sv129 mice during RF in comparison to those in AD-fed IL-10−/− Sv129 mice (Fig. 1D, E, right). In contrast, CD3+CD8+ cell recruitment remained arrhythmic (Supplementary Fig. 1H). Consistently, the weights of the spleen and mesenteric lymph nodes (MLNs) and the colon density, which were much heavier in the AD-fed IL-10−/− Sv129 mice than in the control mice, were significantly lower during RF and were undistinguishable from those of the controls (Fig. 1F). Histological analysis of colon cross-sections further revealed that more than 58% of AD-fed IL-10−/−Sv129 mice were highly inflamed (Histoscore>5), which was dramatically reduced to 5% by RF (Fig. 1G–I). These data demonstrate that RF restores the disruption of colon clock function and completely ameliorates the immune phenotype in IL-10−/−Sv129 mice.
Loss of microbial rhythmicity during colonic inflammation in IL-10−/− mice is restored by RF
Gut microbiota dysbiosis has been associated with the development of IBD in humans and mouse models [10]. Previously, we identified a novel link between the intestinal clock, microbiome rhythms and GI homeostasis [11]. Consistently, the microbiota composition differed significantly between genotypes despite the feeding conditions (Fig. 2A). Circadian rhythmicity in community diversity (normalized species richness), the abundance of both major phyla, Bacteroidota and Firmicutes, and highly abundant families, including Lachnospiraceae and Oscillospiraceae, as well as zOTUs, followed circadian oscillation in the wild type, whereas these rhythms were abolished in AD-fed IL-10−/−Sv129 mice (Fig. 2B–E). In accordance with the results obtained from IBD patients [17], alterations in the abundance of Lachnospiraceae, Oscillospiraceae and Erysipelotrichaceae were observed in IL-10−/− mice (Supplementary Fig. 3A). The abundances of the genera Oscillibacter, Eubacterium, Clostridium and Pseudoflavonifractor significantly differed among the genotypes (Supplementary Fig. 3Bleft, Supplementary Table 2). Moreover, we detected significant correlations between disease markers, such as histological score, inflammatory marker gene expression and zOTUs belonging to Lachnospiraceae and Oscillospiraceae (Supplementary Fig. 3Bright), which is consistent with our recent findings showing a correlation between Lachnospiraceae and active disease in IBD patients [18]. Importantly, RF not only restored the differences in abundance between genotypes (Supplementary Fig. 3B, Supplementary Table 3) but also restored microbial rhythmicity in IL-10−/−Sv129, as illustrated by the community diversity and at the levels of the major phyla and families (Fig. 2B–E). Enhanced microbial rhythmicity was also found among the 237 identified zOTUs visualized by heatmaps (Fig. 2D, Supplementary Fig. 3C). More than 50% of the zOTUs that gained rhythmicity during RF in IL-10−/− mice belonged to the Lachnospiraceae family (Supplementary Table 2), which includes bacterial taxa important for SCFA production and secondary bile acid (BA) conversion and thus plays crucial roles in IBD progression [19, 20].
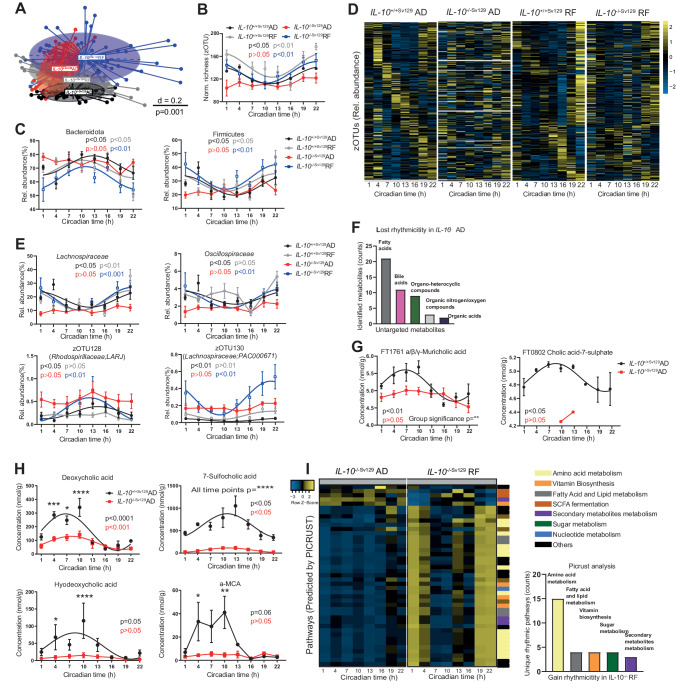
Fig. 2.
Impaired microbiome rhythms in IL-10−/−SV129 mice are restored after RF. A Beta-diversity illustrated by MDS plots of fecal microbiota based on generalized UniFrac distances (GUniFrac) in wild type and IL-10−/−Sv129 mice under AD and RF conditions. Comparison of genotype and feeding effects was performed by PERMANOVA (adj.p = 0.001). Data points represent individual sample collected from different mice at different time points (n > 5 mice/time points for each group). B Circadian profile of normalized richness and C relative abundance of phyla in wild type and IL-10−/−Sv129 mice under AD and RF conditions. D Heatmap depicting the relative abundance of zOTUs (mean relative abundance>0.1%; prevalence > 10%). Data from IL-10−/−Sv129 mice under AD or RF conditions are normalized to the peak of each zOTU and ordered by the peak phase in wild type mice under AD or RF conditions, respectively. Yellow and blue indicate high and low abundance, respectively. Each column represents the group average for each time point. E Relative abundance of highly abundant families and representative zOTUs from wild type and IL-10−/−Sv129 mice under AD and RF conditions. F Classification of the top 5 fully annotated super classes of metabolites that lost rhythmicity in IL-10−/−Sv129 mice under AD. Circadian profiles of bile acids (BAs) G measured by untargeted or H targeted metabolomics of wild type and IL-10−/−Sv129 mice under AD conditions. At some time points (CT 1, 4, 7, 16, 19, and 22) the levels for the IL-10−/− group were under the detection limit and thus not illustrated in the graph. I Heatmap of pathways (calculated by PICRUST 2.0) with restored rhythms in IL-10−/−Sv129 mice under RF (left) and the quantification of the superclass (right). Rhythmicity identified by JTK_Cycle (Bonferroni adj. p value ≤ 0.05). Significant rhythms are illustrated with fitted cosine-regression; data points connected by straight lines indicate no significant cosine fit curves (p > 0.05) and thus no rhythmicity. Comparisons of groups were performed by two-way ANOVA followed by Sidak correction. The results are summarized in Supplementary Table 3. Data are represented as the mean ± SEM. Details on mouse numbers per time point are summarized in Supplementary Table 6
To address the physiological relevance of disturbed microbial oscillations in IL-10−/−Sv129 mice, (un)targeted metabolite analysis was performed on feces. Indeed, 55 fully annotated metabolites were clustered according to genotype (Supplementary Fig. 3D) in IL-10−/−Sv129 mice, most of which were lipids and lipid-like metabolites, including fatty acids and BAs (Fig. 2F). For example, the rhythmicity of bile acids, including muricholic acid and 7-sulfocholic acid, as well as the long-chain fatty acid oleic acid and linoleic acid, was lost and suppressed (Fig. 2G, Supplementary Fig. 3E). Targeted metabolomics further validated these genotype differences (Fig. 2H, Supplementary Fig. 3F). For example, in IL-10−/−sv129 mice, a highly suppressed amplitude or lost time difference was observed for primary and secondary bile acids (BAs), such as deoxycholic acid (DCA), hyodeoxycholic acid, 7-sulfocholic acid (7-sulfo-CA) and α-muricholic acid (Fig. 2H). Most of these secondary BAs are key mediators involved in gut dysbiosis, especially in IBD [21]. Additionally, reduced SCFA levels, such as those of acetate, butyrate and propionate, were found in IL-10−/−sv129 mice (Supplementary Fig. 3F), similar to observations in IBD patients and related mouse models [19]. The concentrations of valeric acid and desaminotyrosin were also suppressed in IL-10−/−Sv129 mice (Supplementary Fig. 3F). Importantly, PICRUSt2 analysis of samples obtained from IL-10−/−Sv129 mice under RF conditions revealed restored rhythmicity of the assigned pathways involved in fatty acid synthesis and SCFA fermentation (Fig. 2I). Taken together, these results indicate that the changes in microbial rhythmicity and composition observed in IL-10−/−Sv129 mice can be reversed by RF; thus, microbial rhythmicity might be involved in the beneficial effect of RF on colonic inflammation.
Germ-free intestinal clock-deficient mice develop an increased inflammatory response following the transfer of IL-10−/−-associated microbiota
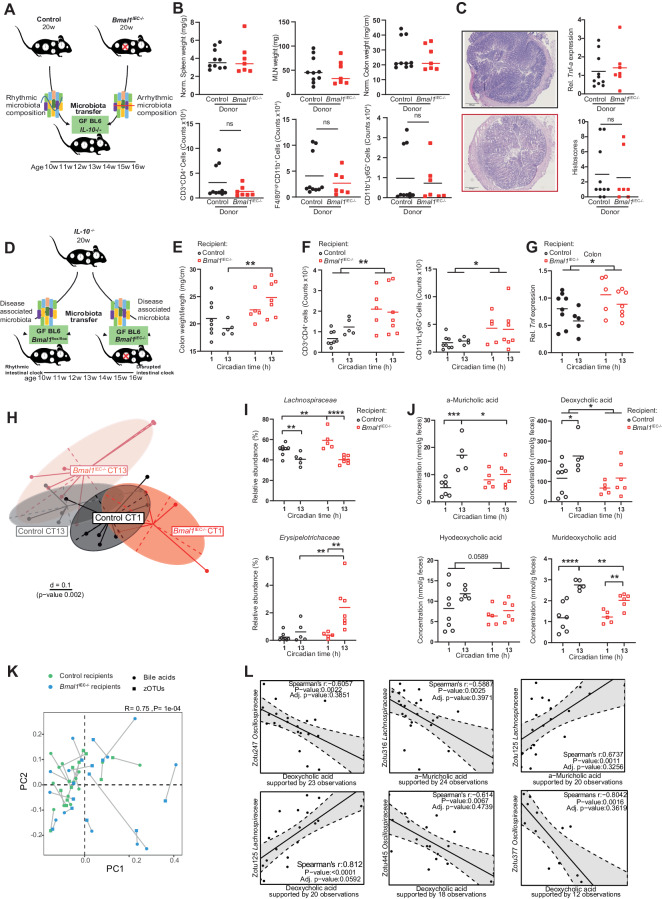
Recently, we demonstrated that a functional intestinal clock is required to maintain GI homeostasis by driving the microbiome, including Lachnospiraceae and microbiota-derived DCA and 7-sulfo-CA [11]. To determine whether a dysfunctional intestinal clock in IL-10−/−Sv129 mice or the arrhythmicity of the microbiome directly affects the severity of inflammation, we performed two distinct cecal microbiota transfer experiments. First, colonialization of cecal content from intestine-specific clock-deficient (Bmal1IEC−/−) and littermate (Bmal1flox/flox) donors was performed to transfer rhythmic and arrhythmic microbiota into germ-free (GF) IL-10−/−BL6 recipients (Fig. 3A). Surprisingly, the weight of the immune organs and the amount of immune cell infiltration to the LP did not differ among the recipients (Fig. 3B). Consistently, histological staining, scoring, and inflammatory marker gene expression revealed no pathological differences between recipients (Fig. 3C), indicating that microbial rhythmicity might not directly induce intestinal inflammation. However, when GF Bmal1IEC−/− mice received disease-associated microbiota from inflamed IL-10−/−BL6 mice, colon weight, immune cell recruitment to the colonic LP and Tnf gene expression were severely elevated in Bmal1IEC−/− recipients compared to controls, although the histological score obtained from a single colon cross-section showed no clear difference between the recipients (Fig. 3D–G, Supplementary Fig, 4A–C). Interestingly, clustering between circadian time points and recipients was found for beta diversity in cecal samples (Fig. 3H). Alterations in the abundance of Lachnospiraceae and Erysipelotrichaceae in Bmal1IEC−/− recipients were similar to those in IL-10−/−Sv129 mice (Fig. 3I, Supplementary Fig. 3A). Accordingly, the cecal concentrations of α-MCA, DCA, HDCA and MDCA were lower than those in control recipients (Fig. 3J, Fig. 2H). Procrustes analysis (PA) revealed an association between zOTUs and BA production; in particular, Lachnospiraceae and Oscillospiraceae were correlated with DCA and aMCA concentrations (Fig. 3K, L). Taken together, these results reflect an early stage of inflammation following colonization with disease-associated microbiota when recipients lack a functional intestinal clock. Consequently, the intestinal clock represents a functional link between the microbiome and GI inflammation.
Fig. 3.
Dysfunction of the host intestinal clock promotes microbiota-induced colonic inflammation. A Schematic illustration of the experimental design. Cecal contents from control (rhythmic) and Bmal1IEC−/− (arrhythmic) donors were transferred to germ-free IL-10−/−BL6 mice. B Organ weights (top) and number of immune cells recruited into the colonic LP (bottom) of the recipients. C Representative H&E staining of colonic cross sections from the recipients (left) and Tnf gene expression, as well as histopathological scores (right). D Schematic illustration of the experimental design. Disease-associated microbiota from IL-10−/− donors was transferred to germ-free control (Bmal1flox/flox) and Bmal1IEC−/− recipients. E Colon weight, F number of immune cells recruited into the colonic lamina propria and G Tnf expression in colon tissues from recipients. H Beta diversity illustrated by MDS plots of cecal microbiota based on generalized UniFrac distances (GUniFrac) in recipients at circadian times (CTs) 1 and 13. I Relative abundance of families and J amounts of α-muricholic acid (α-MCA), deoxycholic acid (DCA), hyodeoxycholic acid (HDCA) and murideoxycholic acid (MDCA) in the cecal contents of control and Bmal1IEC−/− recipients. K Procrustes analysis (PA) of the cecal microbiota and bile acid (BA) levels. The length of the line is proportional to the divergence between the data from the same mouse. L Representative correlation plot between zOTUs and BAs. The data points represent individual mice (n > 5/time points for each genotype). Mann–Whitney U tests and two-way ANOVA followed by Benjamini–Hochberg correction were used. Asterisks indicate significant differences; *p < 0.05, **p < 0.01, ****p < 0.0001. The data are presented as the mean ± SEMs. Details on the number of mice per time point are summarized in Supplementary Table 6
Genetic intestinal clock dysfunction alters colonic immune functions
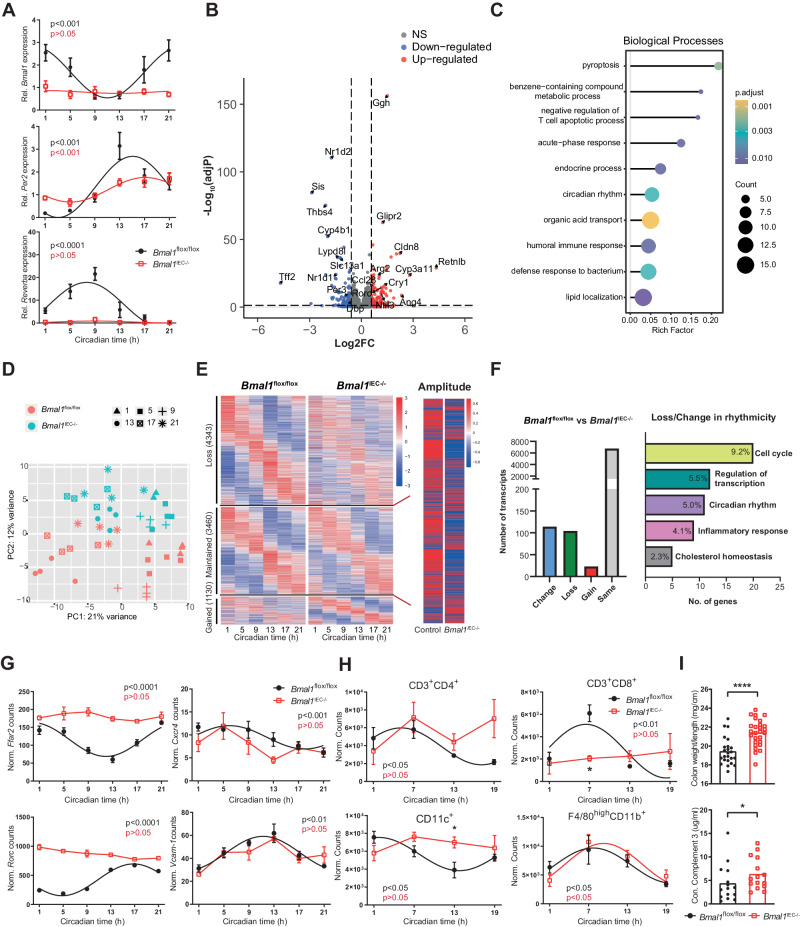
To determine whether intestinal clock dysfunction influences the colonic immune response, we used Bmal1IEC−/− mice. Colonic IEC clock dysfunction was validated by arrhythmic and suppressed clock gene expression (Fig. 4A). Furthermore, bulk RNA-sequencing analysis of colonic tissue samples obtained during the circadian day revealed differential expression between genotypes (fold change>1.5, adj. p < 0.05), such as Retnlb (Resistin-like beta) and Tff2 (Trefoil factor 2) (Fig. 4B), which are involved in colitis development [22, 23]. Consistently, gene ontology enrichment analysis highlighted pathways relevant for ‘humoral immune response,’ ‘defense response to bacterium’ and ‘T-cell apoptotic process’ (Fig. 4C). Clustering according to the genotype was observed by principal component analysis (PCA), but clustering according to the collection time point was less pronounced in Bmal1IEC−/− (Fig. 4D). Consistently, 55% of rhythmic transcripts in the controls were arrhythmic in Bmal1IEC−/− mice (Fig. 4Eleft, Supplementary Table 4). The majority of the resilient rhythmic transcripts showed a highly suppressed amplitude in Bmal1IEC−/− mice (Fig. 4E, right). DODR analysis further confirmed that >200 genes either lost or changed rhythmicity (114/104, respectively) in Bmal1IEC−/− mice and were enriched in pathways related to “circadian rhythm” and “inflammatory response” (Fig. 4F). In particular, genes involved in the inflammatory and immune responses [24–26], including Ffar2, Hc, Abcc2 and Rorc, goblet cell function [27] and immune cell migration [28, 29], such as Cxcr4 and Vcam-1, lost rhythmicity in Bmal1IEC−/− mice (Fig. 4G, Supplementary Fig. 5A, B, Supplementary Table 4). Accordingly, immune cell frequency (CD4+ and CD8+ T cells and dendritic cells) lost rhythmicity in the colonic LP of Bmal1IEC−/− mice (Fig. 4H), similar to the results obtained for IL-10−/−Sv129 mice (Fig. 1E). Although a lack of the intestinal clock did not result in a pathophysiological phenotype (Supplementary Fig. 5C–E), neither the colon weight nor the concentration of complement 3 in the stools of Bmal1IEC−/− mice was significantly increased (Fig. 4I). These findings are consistent with the results obtained from IL-10−/− mice (Fig. 1F, Supplementary Fig. 5F), suggesting that intestinal clock dysfunction might indeed magnify inflammatory processes in IL-10−/− mice.
Fig. 4.
The intestinal clock regulates transcripts involved in intestinal immune responses. A Clock gene expression and B volcano plot of transcriptional changes (upregulated: adjusted p < 0.05, FC > 1.5, red; downregulated: adjusted p < 0.05, FC > 1.5, blue) in colon tissues from control and Bmal1IEC−/− mice. C Gene Ontology enrichment analysis of differentially expressed genes. D Principal component analysis of 46 colon samples obtained from control (red) and Bmal1IEC−/− (blue) mice. Different shapes indicate different circadian time (CT) points. E Heatmap of transcripts that lost, maintained or gained rhythmicity in colon tissues from Bmal1IEC−/− mice (left) and amplitude comparison of the transcripts that maintained rhythmicity (right) (JTK_cycle adj.p < 0.05 as rhythmic). Transcripts are ordered according to the peak phase of the control groups. Each column represents the group average for each time point. F Number of transcripts whose rhythmic changes were lost/changed according to comparison (left) and GO analysis (top 5, right). Circadian profiles of G gene expression and immune cell frequency H in the colonic LP of control and Bmal1IEC−/− mice. I Colon weight (top) and complement 3 levels in the feces of control and Bmal1IEC−/− mice. The data points represent individual mice (n = 4/time points for each genotype). Mann–Whitney U test. Asterisks indicate significant differences; *p < 0.05, ****p < 0.0001. Significant rhythms are illustrated with fitted cosine-wave regression using a line (significance: cos-fit p value ≤ 0.05). Details on the number of mice per time point are summarized in Supplementary Table 6
Bmal1IEC−/− mice are susceptible to acute and chronic colon inflammation
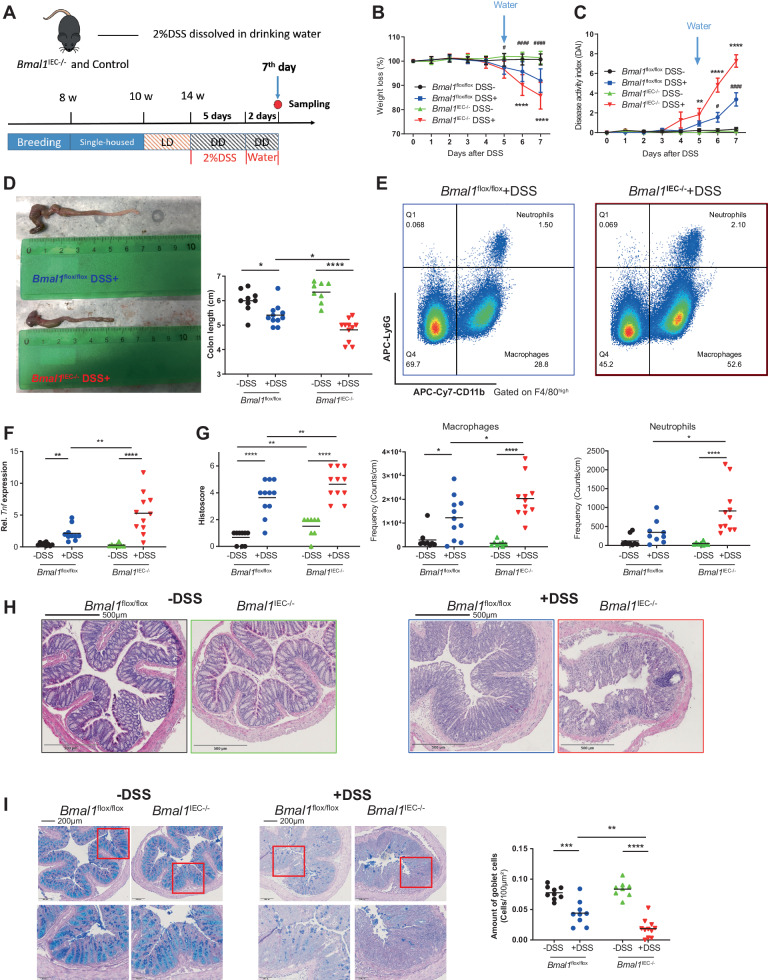
To directly determine the cause‒effect relationship between intestinal clock dysfunction and GI inflammation, Bmal1IEC−/−mice and controls were released into DD condition and received drinking water supplemented with 2% DSS (Fig. 5A) to induce tissue damage and create a model of acute colitis [30]. DSS treatment of Bmal1IEC−/− mice caused a significant reduction in body weight, an increase in the disease activity index (DAI) [31], and a decrease in colon length compared to DSS treated controls (Fig. 5B–D). Accordingly, the expression of inflammatory markers, such as Tnf, and neutrophil and macrophage recruitment to the colonic LP, were strongly enhanced in Bmal1IEC−/− mice (Fig. 5E, F). Consistently, in the same mice, histopathological evaluation of colon cross-sections revealed significant differences between genotypes after DSS treatment, which was reflected by increased histological scores in Bmal1IEC−/−mice (Fig. 5G, H). Additionally, intestinal clock dysfunction caused the loss of goblet cells following DSS treatment (Fig. 5I), which represents a critical factor during DSS-induced colitis [31]. These results demonstrated increased sensitivity to DSS-induced colitis in Bmal1IEC−/− mice.
Fig. 5.
Bmal1IEC−/− mice are more susceptible to DSS-induced colitis. A Schematic illustration of the DSS treatment procedure. Body weight changes B and disease activity indices C of control and Bmal1IEC−/− mice during DSS/water administration. D Representative images of the colon (left) and colon length measurements (right). E Gating strategy for macrophages and neutrophils in the colon LP and quantification. Tnf expression in colon tissues F, histopathological scores G, representative H&E staining H and PAS-AB staining quantification of the number of goblet cells based on a minimum of 20 crypts for each section I of colon sections from control and Bmal1IEC−/− mice with/without DSS treatment. The data points represent individual mice (n > 8 for each group). Two-way ANOVA and Benjamini‒Hochberg correction were used. Asterisks indicate significant differences between genotypes after DSS treatment; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Octothorpes indicate significant differences between Bmal1flox/flox mice after treatment (#p < 0.05, ##p < 0.01, ###p < 0.001). Details on the number of mice per time point are summarized in Supplementary Table 6
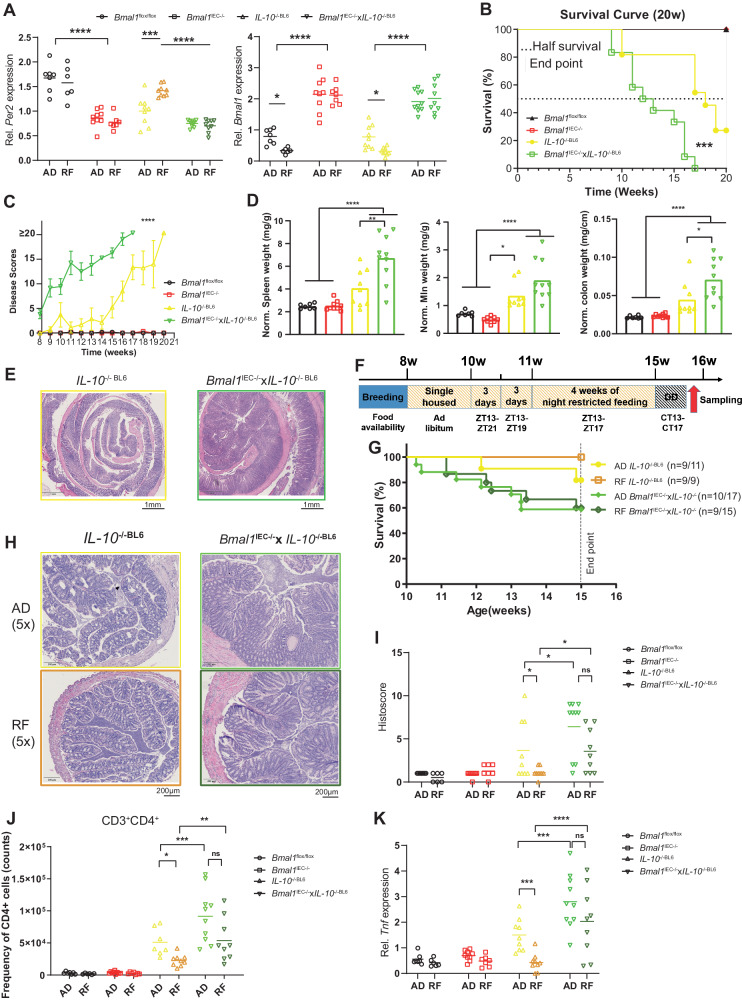
To further assess the impact of the intestinal clock on IBD development, we evaluated IBD pathology in a newly generated genetic mouse model prone to developing chronic colitis combined with a dysfunctional intestinal clock (Bmal1IEC−/−xIL-10−/−BL6). IL-10 deficiency on the BL6 background causes less severe colitis symptoms than on the SV129 background [16], which is likely a result of less severe alterations in clock gene expression (Fig. 6A, Fig. 1B, Supplementary Fig. 6F). The additional loss of the intestinal clock dramatically reduced the survival rates of Bmal1IEC−/−xIL-10−/−BL6 mice (Fig. 6B). In particular, all Bmal1IEC−/−x IL-10−/−BL6 mice were euthanized before the end of the experiment due to their severe disease burden, while approximately 30% of IL-10−/−BL6 mice survived. More severe tissue inflammation was observed in Bmal1IEC−/−xIL-10−/−BL6 mice, which was reflected by increased spleen, colon and MLN weights and increased disease scores (Fig. 6C–E). Similar to our previous results obtained from Bmal1IEC−/− mice [11], the microbiota diversity differed between genotypes, and circadian rhythmicity in terms of species richness and the abundance of the major phyla was dramatically disrupted in Bmal1IEC−/−xIL-10−/−BL6 mice, whereas only mild circadian changes were observed in IL-10−/−BL6 mice (Supplementary Fig. 6A–C). Moreover, a heatmap showing the peak relative abundances of zOTUs and their quantification confirmed the disruption of microbial rhythmicity in Bmal1IEC−/−xIL-10−/−BL6 mice, whereas intermediate phenotypes were found in IL-10−/−BL6 mice (Supplementary Fig. 6D, E). Taken together, these data demonstrate that a lack of the intestinal clock promotes sensitivity to acute and chronic colon inflammation.
Fig. 6.
Restricted feeding ameliorates the IBD-like symptoms in mice by targeting the intestinal clock. Colon clock genes expression A and survival B in control, Bmal1IEC−/−, IL-10−/−BL6 and Bmal1IEC−/−xIL-10−/−BL6 mice under AD and RF conditions. Disease scores C, organ weights D, and representative H&E staining images of colon sections E from IL-10−/−BL6 and Bmal1IEC−/−xIL-10−/−BL6 mice under AD conditions. F Schematic illustration of the experimental RF design. Survival analysis G, representative H&E staining of colon sections H, histopathological scores I, immune cell frequency in the colonic LP J and Tnf expression K in IL-10−/−BL6 and Bmal1IEC−/−xIL-10−/−BL6 mice under AD and RF conditions. The data points represent individual mice (n > 6/time points for each group). One-way ANOVA, two-way ANOVA following Benjamini‒Hochberg correction, and three-way ANOVA followed by Benjamini–Krieger-–Yekutieli correction were used. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data are presented as the mean ± SEM. Details on the number of mice per time point are summarized in Supplementary Table 6
Restricted feeding requires a functional intestinal clock to ameliorate colitis symptoms
To evaluate whether the beneficial effect of RF on the severity of GI inflammation observed in IL-10−/−Sv129 mice is caused directly by restoration of the gut clock function, Bmal1IEC−/−xIL-10−/−BL6 mice, IL-10−/−BL6 mice and controls were exposed to RF conditions (Fig. 6F). RF in IL-10−/−BL6 mice restored clock gene expression to control levels at CT13, whereas its expression was not restored in Bmal1IEC−/− mice or Bmal1IEC−/−xIL-10−/−BL6 mice due to a lack of intestinal Bmal1 (Fig. 6A, Supplementary Fig. 6F). Assessment of the humane endpoint of the experiment revealed that RF failed to improve the survival rate of Bmal1IEC−/−xIL-10−/−BL6 mice (9/15, 60%), but RF significantly improved the survival rate of IL-10−/−BL6 mice (9/9, 100%) (Fig. 6G). Consistently, according to the pathohistological evaluation, CD4+ T-cell recruitment and colonic Tnf expression were greater than the control levels in IL-10−/−BL6 mice under RF conditions (histology: p = 0.0138, CD4 cell recruitment: p = 0.0069, TNF expression: p = 0.0404) but remained indistinguishable between AD and RF conditions in Bmal1IEC−/−xIL-10−/−BL6 mice (histology: p = 0.7718, CD4 + T-cell recruitment: P = 0.6826, TNF expression: P = 0.7619; Fig. 6H–K), suggesting that the intestinal clock gates the beneficial effect of RF. Notably, RF in Bmal1IEC−/−xIL-10−/−BL6 mice enhanced species richness to control levels, whereas the microbiota remained arrhythmic (Supplementary Fig. 6G), indicating that a loss of microbial rhythms might be involved in the disease phenotype rather than the overall amount of species. Indeed, we identified 40 zOTUs that gained rhythmicity during RF in IL-10−/−BL6 mice but remained arrhythmic in Bmal1IEC−/−xIL-10−/−BL6 mice. The majority of these zOTUs belonged to the intestinal clock-controlled [11] families Lachnospiraceae, Muribaculaceae and Oscillospiraceae (Supplementary Fig. 6H, Supplementary Table 5).
In summary, these results clearly demonstrated that intestinal clock dysfunction significantly contributes to colitis severity and that RF improved GI inflammation by targeting intestinal clock functions. Moreover, we provide the first evidence that the intestinal clock gates the inflammatory response and can be directly targeted by RF to mediate the severity and progression of IBD symptoms.
Discussion
Alterations in clock genes were found in colonic biopsy samples from IBD patients [12]. In this study on IL-10−/− mice, a previously described IBD-relevant mouse model of colitis [16], we provide evidence that a disrupted colonic circadian clock is a cause rather than a consequence of GI inflammation in IBD. We demonstrated that specific loss of the circadian function of the IEC clock enhances the severity and prevalence of DSS-induced colitis symptoms in mice. This finding is in accordance with previous literature showing that a lack of colonic Bmal1 worsened DSS-induced colitis [30].
In a novel genetic mouse model of colitis that also lacks a functional intestinal clock, we provide the first evidence for a cause‒effect relationship between intestinal clock function and colitis. These Bmal1IEC−/−xIL-10−/−BL6 mice develop a severely enhanced immune phenotype and experience dramatically reduced survival rates.
Potential mechanisms by which a disrupted colon clock in, e.g., IL-10−/− mice, might affect GI inflammation include genes involved in local immune functions and epithelial–microbe interactions. The intestinal clock regulates important inflammatory marker genes, such as Tnf, which are arrhythmic and enhanced in IL-10−/− mice and have previously been linked to the development of IBD [32].
Intestinal clock-controlled genes are also involved in immune cell recruitment, such as Ffar2, which plays an important role in intestinal homeostasis by regulating immune cell abundance in colonic lymphoid tissues and the secretion of mucus-associated proteins and antimicrobial peptides [24], as well as Abcc2, which actively regulates mucosal inflammation [26]. Both genes lost rhythmicity in Bmal1IEC−/− mice. Moreover, Rorc, which encodes the IBD risk factor RORγ [25], was elevated and arrhythmic in mice lacking an intestinal clock. These results further suggest the importance of the intestinal clock in modulating the IBD-like phenotype.
In addition, the expression of the promigratory genes Vcam-1 and Cxcr4 undergoes diurnal oscillation in multiple lymphoid and nonlymphoid tissues [28, 29]. Here, we revealed that the circadian rhythm of the expression of these two genes in the colon requires a functional intestinal clock. For example, the expression of Vcam-1 and Cxcr4 was arrhythmic in colonic tissues from mice harboring a dysfunctional intestinal clock (IL-10−/−Sv129 and Bmal1IEC−/−). Furthermore, Cxcr4 mediates diurnal oscillations in T-cell distribution and responses [28], and Vcam-1 is involved in the rhythmic pattern of leukocyte migration [29]. Therefore, the arrhythmicity of these genes likely contributes to immune cell recruitment to the colon LP and thus impacts the local immune response.
Here, we provide the first evidence for a functional role of the intestinal clock in regulating rhythmic immune and inflammatory processes, including immune cell recruitment and the microbiota composition, which are both key elements in IBD development and progression [32]. Dysfunctional colon clocks in IL-10−/− and Bmal1IEC−/− mice caused a loss of rhythmicity in the recruitment of T cells to the colonic lamina propria. Similarly, rhythmic leukocyte trafficking into the bloodstream and infiltration to the lymph nodes, spleen and bone marrow have previously been described for T cells and monocytes and depend on cell- and tissue-specific clocks [33].
The physiological importance of diurnal immune cell recruitment was demonstrated in lymphoid organs, which improved the immune response to antigens and bacterial infection [28]. Specific clustering of intestinal CD4+ cells induced colitis in an adoptive transfer mouse model [34]. Consequently, arrhythmic leukocyte recruitment to the colon might augment the inflammatory response in mice with a disrupted colon circadian clock.
The microbiota composition and function are associated with the development of GI inflammation [35], and the arrhythmicity of the microbiome is linked to the development of obesity and type 2 diabetes [36]. Interestingly, microbiome rhythmicity was disrupted in IL-10−/−Sv129 mice. This was likely caused by intestinal clock dysfunction in these mice because the intestinal clock was recently identified as a major driver of microbiota rhythmicity [11]. Indeed, Bmal1IEC−/−xIL-10−/−BL6 mice with a genetically dysfunctional intestinal clock exhibit a more pronounced loss of microbial rhythmicity than IL-10−/−BL6 mice. Similar to previous results obtained from Bmal1IEC−/− mice [11], intestinal clock-controlled taxa involved in SCFA fermentation and secondary bile acid formation lost rhythmicity in IL-10−/−Sv129 mice and thereby likely caused arrhythmicity in the levels of specific BAs, such as DCA, 7-sulfo CA, HDCA and α-MCA, which have frequently been linked to intestinal inflammation [37]. Notably, reduced levels of secondary BAs, e.g., DCAs and HDCAs, identified in Bmal1IEC−/− mice receiving disease-associated microbiota were also found in patients with UC, and supplementation, particularly with DCA, ameliorated experimental colitis in mice [20]. Therefore, the generation of arrhythmic microbial-derived products due to the composition of the arrhythmic microbiota in IL-10−/− mice might promote their inflammatory immune response.
Indeed, our recent transfer experiments provided evidence that the arrhythmic microbiota can alter GI immune homeostasis [11]. However, the transfer of the arrhythmic microbiota from Bmal1IEC−/− did not increase the severity of inflammation in germ-free IL-10−/− recipients (although 15% more IL-10−/− mice developed inflammation), suggesting that intestinal clock dysfunction rather than the loss of microbial rhythmicity induces colonic inflammation. Indeed, the transfer of IBD-associated microbiota provides direct evidence for the physiological relevance of the intestinal circadian clock in the microbiota-induced inflammatory response. In particular, the findings in Bmal1IEC−/− recipients reflect the immune response and altered microbiome observed in IL-10−/− donors and thus indicate an activated, early inflammatory response, although in colon cross-sections, no pathohistological tissue changes could be observed between genotypes. This might be due to patchy colonic inflammation associated with the Crohn’s disease-like phenotype reported in IL-10−/− mice [38]. Future transfer experiments (long-term and short-term) in IL-10−/− mice using “Swiss rolls” to investigate inflammation in the entire intestine might clarify whether the loss of microbial rhythmicity per se rather than the IBD-associated microbiota can promote gastrointestinal inflammation.
Taken together, these results highlight the relevance of intestinal clock function for GI inflammatory processes involved in microbiota-induced IBD development. Interestingly, a functional intestinal clock in control recipients was capable of restoring the circadian time differences of zOTUs. These rhythmic hosts showed reduced immune cell recruitment into the LP and inflammatory marker gene expression, indicating that the restoration of intestinal clock functions might reduce GI inflammation.
To determine whether intestinal clock function can be targeted by meal timing to influence the immune response to GI inflammation, IL-10−/− mice were exposed to nighttime-restricted feeding (RF). Here, we demonstrated that rhythmicity and the amplitude of the colonic circadian clock can indeed be restored by RF in a diseased mouse model (IL-10−/−) with disrupted colonic clock function. Moreover, targeting the colon clock by RF further led to the restoration of rhythmic colon clock functions, such as CD4+ T-cell recruitment into the colonic LP and microbiome oscillations. This finding is in line with the literature showing that RF in rodents influences various peripheral clocks [9, 15] and genes involved in inflammatory signaling, such as NF-kB, TLR and IL-17, in many tissues, including the intestine [39]. Consistent with our hypothesis, RF further ameliorated the IBD-like colitis phenotype in IL-10−/− mice and substantially enhanced their survival rate. This finding is in accordance with results obtained in mice showing that RF protects against DSS-induced colitis by affecting intestinal functions [40]. Interestingly, meal timing in humans has been tested for the treatment of a variety of diseases, including diabetes, cardiovascular diseases and cancer [13], and inconsistent meal times could be correlated with IBD symptoms [41], indicating that RF might become a strategy for the treatment of IBD patients. However, further experiments are required to validate the long-term effect of RF.
Importantly, we provide the first direct mechanistic evidence that the effect of RF on the severity of IBD is directly gated by the intestinal clock because RF fails to reduce the inflammatory phenotype (immune cell infiltration, inflammatory marker gene expression, histopathological scores) when the intestinal clock is genetically dysfunctional in Bmal1IEC−/−xIL-10−/− mice. Our results clearly demonstrated that intestinal clock dysfunction significantly contributes to colitis severity and that RF improved GI inflammation by targeting intestinal clock functions. Notably, we observed improved microbiota richness following RF in Bmal1IEC−/−xIL-10−/− mice (Supplementary Fig. 6B, G), which has been associated with improved IBD conditions [42]. Thus, we cannot exclude the possibility that other factors, such as the microbiome, which is known to respond to RF [8] independent of the intestinal clock, might also contribute to improved disease outcomes. Nevertheless, no significant improvements in tissue pathology, the inflammatory response or overall survival following RF were observed in Bmal1IEC−/−xIL-10−/− mice. Thus, these results demonstrate that the intestinal clock represents a target for the treatment of GI inflammation. Whether the beneficial effect of RF is solely due to intestinal clock functions or can be affected by additional factors remains to be addressed in future experiments. For example, the role of the intestinal clock in mediating the effect of RF could be further validated by subjecting intestinal clock-deficient mice to DSS treatment and placing them under RF or AD-fed conditions.
Taken together, our results demonstrate that a functional intestinal clock is essential for maintaining GI homeostasis and is a major player in IBD progression. In addition, our findings suggest that enhancing intestinal clock function by meal timing might become the next step in developing novel strategies for circadian therapies for IBD and potentially other metabolic diseases in humans.
Methods
Ethics statement and mouse models
For details, see the supplementary material.
Behavior analysis
Handling and activity measurements during the experiments were performed as previously described [43]. Wheel-running activity was analyzed using ClockLab software (Actimetrics). The last 10–14 days of each condition were used to determine the period (tau, calculated using an X2 periodogram and confirmed by fitting a line to the onset of activity), the duration of the active period (alpha), the amount of activity, and the subjective day/night activity ratio (where the subjective day under DD conditions is the inactive period between the cessation of activity and the onset of activity and the subjective night is the active period between the onset of activity and the cessation of activity). The average daily food intake was measured in the second week under different light conditions and ad libitum feeding conditions. For the restricted feeding condition, the average daily food intake was measured manually over 3 consecutive days in the last week of the restricted feeding period.
Light–dark (LD) and constant darkness (DD) conditions
Male IL-10−/−Sv129 mice and their wild-type littermates were housed individually at 8 weeks of age under an LD cycle for 2 weeks (aged 8–10 weeks), switched to a DD cycle for 2 more weeks (aged 10–12 weeks) and subsequently returned to the LD phase.
Nighttime-restricted feeding
Fourteen-week-old IL-10−/−Sv129 mice and wild-type littermates were gradually introduced to the restricted feeding (RF) protocol starting with 8 h of food availability (ZT13-ZT21) for the first 3 days, followed by 6 h of food availability (ZT13-ZT19) for the following 3 days and finally 4 h of food availability (ZT13-ZT17) for four weeks. The same RF regimen was also applied to Bmal1flox/flox, Bmal1IEC−/−, IL-10−/−BL6 and IL-10−/−BL6 x Bmal1IEC−/− mice, with an earlier start point at the age of 10 weeks. Tissues were harvested at the end of the 4 week RF period on the 2nd day in constant darkness.
Tissue collection
For details, see the supplementary material.
Organoids
For details, see the supplementary material.
Gene expression analysis
For details, see the supplementary material.
RNA sequencing
RNA quality was verified using an Agilent2100 Bioanalyzer with RNA 6000Nano Reagents. Library preparation and rRNA depletion were performed using the TruSeq Stranded mRNA Library Prep Kit. After the final QC step, the libraries were sequenced in a paired-end mode (2 × 150 bases) on a Novaseq6000 sequencer (Illumina) with a depth of ≥ 12 million paired reads per sample.
Pre-processing
The quality of the next-generation sequencing data was assessed with FastQC v0.11.5 (RRID:SCR_014583, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality samples were filtered out. Adapter content and low-quality reads were removed using Trimmomatic v0.39 [44].
Trimmed FASTQ files were then mapped against the mouse mm10 genome with the STAR v2.7.5c aligner [45]. Format conversions were performed using SAMtools v1.3.1 [46]. The featureCounts program v1.4.6 [47] was used to count reads located within an exon that did not overlap with multiple features, with a threshold of MAPQ ≥ 4, and that were not chimeric.
Normalization and differentially expressed gene analysis
DESeq2 version 1.22.0 (RRID:SCR_015687 [48]) was used to normalize the read count matrix and perform differential expression analysis. The Bioconductor package “biomaRt” version 2.38 (RRID:SCR_002987 [49]) was used to map the MGI symbols to Ensembl gene IDs. To identify general DEGs between WT and KO mice, an initial model (Genotype + Time) with a filtering FC > 1.5 and Benjamini‒Hochberg-Adj.p < 0.05 was used to identify the differentially expressed genes (DEGs). Then, a multimodel (Genotype + Period + Genotype: Period) was used to compare day vs. night differences.
Gene Ontology biological process enrichment was performed using clusterProfiler (RRID: SCR_016884) [50], and GO terms were considered to be significantly enriched when the q value was <0.05. All genes expressed with a minimum of 1 count in any of the samples were used as the background. Redundant terms were removed manually.
Circadian analysis
The rhythmicity of the oscillating transcripts was measured by the JTK cycle [51] through the MetaCycle R package [52]. With the settings of Period=24 h and adj.p < 0.05, filtered genes were then defined as rhythmic. The package compareRhythms [53] was modified to compare rhythmicity differences in transcripts between control and Bmal1IEC−/− mice as previously described [11].
The associated RNA-seq data are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE230078.
High-throughput 16S ribosomal RNA (rRNA) gene amplicon sequencing analysis
For details, see the supplementary material.
Targeted and untargeted metabolite analyses
For details, see the supplementary material. The associated untargeted metabolomics data are available at MSV000091092.
Dextran sulfate sodium (DSS)-induced acute experimental colitis
Fourteen-week-old male Bmal1IEC−/− mice and controls were maintained in constant darkness after the experimental protocol started. Briefly, Bmal1IEC−/− and control mice were administered 2% DSS dissolved in drinking water for 5 days, followed by normal water for 2 days. The onset of disease in each mouse was calculated separately, and the mice were sacrificed at circadian time (CT) 7 on day 7, when most of the immune cells peaked in the colon lamina propria.
Transfer experiment
For details, see the supplementary material.
Immune cell isolation from the lamina propria
For details, see the supplementary material.
Flow cytometry measurement
For details, see the supplementary material.
Histology
For details, see the supplementary material.
PICRUST 2.0
The sequences of the zOTUs that exhibited rhythmicity after restricted feeding in the IL-10−/−Sv129 group were used to construct the metagenome using PICRUST2.0 [54] for the prediction of metagenomic functionality. The corrected zOTU 16 S rRNA gene copy number was multiplied by the predicted functionality to predict the metagenome. The resulting enzymatic genes classified according to the Enzyme Commission (EC) numbers were mapped to Metacyc pathways. Superclasses were removed, and the metabolic pathway abundance was used for statistical analysis using STAMP (2.1.3). Statistical differences were calculated based on White’s nonparametric t test and the Benjamini–Hochberg–Dales discovery rate to adjust for multiple testing.
Statistical analyses
Statistical analyses were performed using GraphPad Prism, version 9.0.0 (GraphPad Software), JTK_cycle v3.1. R [51] or R. Between-sample microbiota diversity was calculated by generalized UniFrac using GUniFrac v1.1. distances within the Rhea [55] pipeline. Microbiota composition comparison was calculated by the PERMANOVA test on a generalized UniFrac dissimilarity matrix and illustrated by MDS [55]. Circadian profile graphs and phase calculations were analyzed by fitting a cosine-wave equation: y = baseline + (amplitude∙cos(2 ∙ π ∙ ((x-[phase shift)/24]))). The nonparametric algorithm JTK_cycle was used for overall rhythmicity of all zOTUs and transcripts. Connected curves in the circadian profile graphs within the figures indicate significant rhythmicity based on cosine analyses, whereas connected straight lines indicate nonsignificant cosine fits. Comparison of rhythms between datasets was performed by the adjusted version of compareRhythms as previously described [11]. The amplitude calculations depicted in the Manhattan plots are based on the output of JTK_cycle, and the phase was calculated by cosine-wave regression. Comparisons between two groups were performed using the nonparametric Mann‒Whitney test (two-sided). Two-way ANOVA followed by Benjamini‒Hochberg correction was used to compare datasets with two categorical variables. Three-way ANOVA following Benjamini–Krieger–Yekutieli correction was used to compare datasets containing three categorical factors. A p value ≤ 0.05 was assumed to indicate statistical significance.
Supplementary information
Author contributions
SK conceived and coordinated the project. YN performed the mouse experiments and fecal sample collection. YN sampled tissue and performed gene expression analysis and FACS analysis. YN analyzed the 16 S rRNA gene sequencing data and conducted the bioinformatics analysis. BA performed the transfer experiments and the predicted microbiota functionality analysis. MH, CM and KK performed the untargeted and targeted metabolomics and data analysis. MB generated the preliminary flow cytometry data. SK analyzed activity behavior and supervised the work and data analysis. SK and DH secured the funding. YN and SK wrote the manuscript. All authors reviewed and revised the manuscript.
Funding
SK was supported by the German Research Foundation (DFG, project KI 19581) and the European Crohn´s and Colitis Organisation (ECCO, grant 5280024). SK and DH received funding from the Deutsche Forschungsgemeinschaft Fund ((DFG, German Research Foundation) – Projektnummer 395357507 – SFB 1371).
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-024-01189-z.
References
- 1.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120:2600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiessling S, Beaulieu-Laroche L, Blum ID, Landgraf D, Welsh DK, Storch KF, et al. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017;15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiessling S, Dubeau-Laramee G, Ohm H, Labrecque N, Olivier M, Cermakian N. The circadian clock in immune cells controls the magnitude of Leishmania parasite infection. Sci Rep. 2017;7:10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gombert M, Carrasco-Luna J, Pin-Arboledas G, Codoner-Franch P. The connection of circadian rhythm to inflammatory bowel disease. Transl Res. 2019;206:107–18. [DOI] [PubMed] [Google Scholar]
- 6.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–27. [DOI] [PubMed] [Google Scholar]
- 8.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–29. [DOI] [PubMed] [Google Scholar]
- 9.Polidarova L, Sladek M, Sotak M, Pacha J, Sumova A. Hepatic, duodenal, and colonic circadian clocks differ in their persistence under conditions of constant light and in their entrainment by restricted feeding. Chronobiol Int. 2011;28:204–15. [DOI] [PubMed] [Google Scholar]
- 10.Caruso R, Lo BC, Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20:411–26. [DOI] [PubMed] [Google Scholar]
- 11.Heddes M, Altaha B, Niu Y, Reitmeier S, Kleigrewe K, Haller D, et al. The intestinal clock drives the microbiome to maintain gastrointestinal homeostasis. Nat Commun. 2022;13:6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giebfried J, Lorentz A. Relationship between the biological clock and inflammatory bowel disease. Clocks Sleep. 2023;5:260–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manoogian ENC, Chow LS, Taub PR, Laferrere B, Panda S. Time-restricted eating for the prevention and management of metabolic diseases. Endocr Rev. 2022;43:405–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ella K, Sudy AR, Bur Z, Koos B, Kisiczki AS, Mocsai A, et al. Time restricted feeding modifies leukocyte responsiveness and improves inflammation outcome. Front Immunol. 2022;13:924541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. [DOI] [PubMed] [Google Scholar]
- 17.Halfvarson J, Brislawn CJ, Lamendella R, Vazquez-Baeza Y, Walters WA, Bramer LM, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metwaly A, Dunkel A, Waldschmitt N, Raj ACD, Lagkouvardos I, Corraliza AM, et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat Commun. 2020;11:4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–70.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–37. [DOI] [PubMed] [Google Scholar]
- 22.Morampudi V, Dalwadi U, Bhinder G, Sham HP, Gill SK, Chan J, et al. The goblet cell-derived mediator RELM-beta drives spontaneous colitis in Muc2-deficient mice by promoting commensal microbial dysbiosis. Mucosal Immunol. 2016;9:1218–33. [DOI] [PubMed] [Google Scholar]
- 23.Aamann L, Vestergaard EM, Gronbaek H. Trefoil factors in inflammatory bowel disease. World J Gastroenterol. 2014;20:3223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun E, Lavoie S, Fonseca-Pereira D, Bae S, Michaud M, Hoveyda HR, et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity. 2019;51:871–84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–67. [DOI] [PubMed] [Google Scholar]
- 26.Pazos M, Siccardi D, Mumy KL, Bien JD, Louie S, Shi HN, et al. Multidrug resistance-associated transporter 2 regulates mucosal inflammation by facilitating the synthesis of hepoxilin A3. J Immunol. 2008;181:8044–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allaire JM, Morampudi V, Crowley SM, Stahl M, Yu H, Bhullar K, et al. Frontline defenders: goblet cell mediators dictate host-microbe interactions in the intestinal tract during health and disease. Am J Physiol Gastrointest Liver Physiol. 2018;314:G360–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimba A, Cui G, Tani-Ichi S, Ogawa M, Abe S, Okazaki F, et al. Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity. 2018;48:286–98.e6. [DOI] [PubMed] [Google Scholar]
- 29.He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, et al. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity. 2018;49:1175–90.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jochum SB, Engen PA, Shaikh M, Naqib A, Wilber S, Raeisi S, et al. Colonic epithelial circadian disruption worsens dextran sulfate sodium-induced colitis. Inflamm Bowel Dis. 2022;29:444–57. [DOI] [PMC free article] [PubMed]
- 31.Kim JJ, Shajib MS, Manocha MM, Khan WI Investigating intestinal inflammation in DSS-induced model of IBD. J Vis Exp. 2012;3678. [DOI] [PMC free article] [PubMed]
- 32.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pick R, He W, Chen CS, Scheiermann C. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol. 2019;40:524–37. [DOI] [PubMed] [Google Scholar]
- 34.Leithauser F, Trobonjaca Z, Moller P, Reimann J. Clustering of colonic lamina propria CD4(+) T cells to subepithelial dendritic cell aggregates precedes the development of colitis in a murine adoptive transfer model. Lab Invest. 2001;81:1339–49. [DOI] [PubMed] [Google Scholar]
- 35.de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, et al. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe. 2020;28:258–72.e6. [DOI] [PubMed] [Google Scholar]
- 37.Rutgeerts P, Ghoos Y, Vantrappen G. Bile acid studies in patients with Crohn’s colitis. Gut. 1979;20:1072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrieta MC, Madsen K, Doyle J, Meddings J. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut. 2009;58:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deota S, Lin T, Chaix A, Williams A, Le H, Calligaro H, et al. Diurnal transcriptome landscape of a multi-tissue response to time-restricted feeding in mammals. Cell Metab. 2023;35:150–65.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Chen L, Bai M, Wang S, Ye X, Lin Y, et al. Time-restricted feeding ameliorates dextran sulfate sodium-induced colitis via reducing intestinal inflammation. Front Nutr. 2022;9:1043783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakradeo PS, Keshavarzian A, Singh S, Dera AE, Esteban JPG, Lee AA, et al. Chronotype, social jet lag, sleep debt and food timing in inflammatory bowel disease. Sleep Med. 2018;52:188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jud C, Schmutz I, Hampp G, Oster H, Albrecht U. A guideline for analyzing circadian wheel-running behavior in rodents under different lighting conditions. Biol Proced Online. 2005;7:101–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pelikan A, Herzel H, Kramer A, Ananthasubramaniam B. Venn diagram analysis overestimates the extent of circadian rhythm reprogramming. FEBS J. 2022;289:6605–21. [DOI] [PubMed] [Google Scholar]
- 54.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lagkouvardos I, Fischer S, Kumar N, Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ. 2017;5:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.