The new NCI60 cell line screen HTS384 shows robust patterns of response to oncology agents and substantial overlap with the classic screen, providing an updated tool for studying therapeutic agents.
Abstract
The NCI60 human tumor cell line screen has been in operation as a service to the cancer research community for more than 30 years. The screen operated with 96-well plates, a 2-day exposure period to test agents, and following cell fixation, a visible absorbance endpoint by the protein-staining dye sulforhodamine B. In this study, we describe the next phase of this important cancer research tool, the HTS384 NCI60 screen. Although the cell lines remain the same, the updated screen is performed with 384-well plates, a 3-day exposure period to test agents, and a luminescent endpoint to measure cell viability based upon cellular ATP content. In this study, a library of 1,003 FDA-approved and investigational small-molecule anticancer agents was screened by the two NCI60 assays. The datasets were compared with a focus on targeted agents with at least six representatives in the library. For many agents, including inhibitors of EGFR, BRAF, MEK, ERK, and PI3K, the patterns of GI50 values were very similar between the screens with strong correlations between those patterns within the dataset from each screen. However, for some groups of targeted agents, including mTOR, BET bromodomain, and NAMPRTase inhibitors, there were limited or no correlations between the two datasets, although the patterns of GI50 values and correlations between those patterns within each dataset were apparent. Beginning in January 2024, the HTS384 NCI60 screen became the free screening service of the NCI to facilitate drug discovery by the cancer research community.
Significance: The new NCI60 cell line screen HTS384 shows robust patterns of response to oncology agents and substantial overlap with the classic screen, providing an updated tool for studying therapeutic agents.
Introduction
The NCI’s 60 human tumor cell line screen (classic NCI60) has been in operation for more than 30 years and has proven to be a very useful drug discovery tool for the cancer research community (1–3). Initially, establishing the feasibility of the NCI60 screen focused on the development of the cell line panel, evaluation of various assays for in vitro drug sensitivity, as well as data analysis and presentation (4–6). The 60 human tumor cell lines were derived from nine cancer types including lung, colon, breast, prostate, melanoma, renal, ovarian, brain, and leukemia. In addition to the criteria that the cells span broad classes of human cancer, the specific cell lines were selected to grow in a single culture medium and demonstrate excellent reproducibility in their growth and response to test agents. Cryopreserved batches of each line were prepared and stored in the NCI Developmental Therapeutics Program Tumor Repository. At the time the NCI60 screen was developed, practical assays for cell growth in 96-well plates were limited. The protein-staining dye sulforhodamine B (SRB) was selected for use in the screen because the chemical fixation step enabled batch processing of plates in a time-independent manner. Additionally, SRB gave the best combination of stain intensity, signal-to-noise ratio, and linearity with cell number (7, 8). The standard range of concentrations tested in the screen was set from 100 μmol/L to 10 nmol/L based on empirical determinations that, for a majority of compounds used during the development of the screen the growth response data across that range captured both the concentration causing 50% inhibition of growth (GI50) and often captured the concentration causing total growth inhibition (TGI).
In April 1990, the fully operational NCI60 screen was rolled out. Initially, approximately 20,000 samples/year including pure small molecules, pure natural products, and partial isolate fractions of natural products were tested in concentration response (1, 9). The mean graph display visualizing differential cell line response was developed as was the COMPARE pattern recognition methodology that could identify common response patterns across the cell lines, independent of the absolute potencies of the tested compounds (4, 5, 10). The mean graph patterns of cell line responses and COMPARE correlations were highly reproducible and enhanced understanding of potential cellular processes targeted by new agents. These advances facilitated studies of structure–activity relationships to direct chemical analog synthesis. For each of the last 5 years, approximately 6,000 compounds were tested in the NCI60 screen in a single concentration prescreen, of which, ∼20% met the criteria for further testing in the five-concentration screen (internal statistics from the Screening and Drug Prep Labs). In that same timeframe, more than 500 peer-reviewed publications have used NCI60 screen data and NCI60 cell line characteristics as a critical component of the study.
The NCI60 screen is moving into a new era. The cell lines remain the same; however, the format has been updated to a fully automated 384-well assay with a 3-day test agent exposure period and a luminescent endpoint for cell viability. This report enumerates some of the similarities and differences between the classic NCI60 screen and the new HTS384 NCI60 screen using a library of 1,003 FDA-approved anticancer drugs and investigational agents tested in both assays. In addition to generating a core set or baseline of data for the HTS384 screen, the patterns of cell line response from the HTS384 screen are compared with the cell line response data from the NCI60 classic screen. The library of 1,003 FDA-approved anticancer drugs and investigational agents were tested at the standard five concentrations used in the classic NCI60 screen to allow comparison of concentration response between the screens and so that targeted compound groups with well-established response patterns in both screens could be analyzed by COMPARE.
Materials and Methods
Submission of compounds for NCI60 HTS384 screening
The standard screening assay is performed across the concentration range of 100 μmol/L to 10 nmol/L in one-log increments. If in vitro data are available when investigators submit new compounds to the HTS384 NCI60 screen, they can request a concentration range starting lower than the standard 100 μmol/L, though the dilutions will still be a series of four serial log dilutions. In addition, if the results from an initial screen of a new compound suggest that the GI50 occurs at a concentration lower than the standard range, then the compound can be retested across an adjusted concentration range if sufficient compound is available (https://dtp.cancer./dscb/compoundSubmission/submissionProcedures.htm).
Compounds
Compounds from the investigational oncology agents (IOA) and approved oncology drugs library were acquired by internal/external synthesis or acquisition from external vendors (3). FDA-approved oncology drugs are available from the NCI at https://dtp.cancer.gov/organization/dscb/obtaining/available_plates.htm. All agents were demonstrated to be >95% pure by proton nuclear magnetic resonance (1H NMR) and LC/MS. In some cases, additional analytical techniques were employed to ensure the integrity of the library (e.g., chiral HPLC, optical rotation, X-ray crystallography, and 13C NMR). Compound stock solutions were prepared in DMSO (Sigma-Aldrich, cat. D2650), except for the platinum compounds, which were prepared in saline (Quality Biological Inc., cat. 114-055-101), at 400-fold the tested concentration and stored at −70°C prior to their use. All agents were prepared for testing as a 5-point concentration series from a high concentration of 100 μmol/L (final) and decreasing in one-log increments. The DMSO stock solutions were prepared manually. For the NCI60 classic screen, the assay plates were prepared manually with inspection and annotation of solubility problems when diluting from the stock. For the HTS384 screen, compound-only plates are included to optically identify solubility problems. Additionally, in both screens, solubility problems at the highest concentration can manifest as a reversal in the progression of growth response values between the two highest concentrations, which are flagged during data processing. Endpoint values will be interpolated as long as they are reached before the reversal.
NCI60 cell lines
Working stock vials of the NCI60 cell lines were obtained from the NCI Developmental Therapeutics Program Tumor Repository. Growth characteristics and seeding numbers for both the NCI60 classic screen and the NCI HTS384 screen are shown in Table 1. The disease and major genetic alterations in the cell lines are shown in Supplementary Table S1. The six leukemia cell lines included CCRF-CEM (ATCC, catalog no. CCL-119, RRID:CVCL_0207), HL-60(TB; ATCC, catalog no. CCL-240, RRID:CVCL_A794), K-562 (ATCC, catalog no. CCL-243, RRID:CVCL_0004), MOLT-4 (ATCC, catalog no. CRL-1582, RRID:CVCL_0013), RPMI-8226 (ATCC, catalog no. CCL-155, RRID:CVCL_0014), and SR (ATCC, catalog no. CRL-2262, RRID:CVCL_1711). The nine non–small cell lung cancer cell lines included A-549/ATCC (ATCC, catalog no. CCL-185, RRID:CVCL_0023), EKVX (RRID:CVCL_1195), HOP-62 (RRID:CVCL_1285), HOP-92 (RRID:CVCL_1286), NCI-H226 (ATCC, catalog no. CRL-5826, RRID:CVCL_1544), NCI-H23 (ATCC, catalog no. CRL-5800, RRID:CVCL_1547), NCI-H322M (RRID:CVCL_1557), NCI-H460 (ATCC, catalog no. HTB-177, RRID:CVCL_0459), and NCI-H522 (ATCC, catalog no. CRL-5810, RRID:CVCL_1567). The seven colon cancer cell lines included COLO 205 (ATCC, catalog no. CCL-222, RRID:CVCL_0218), HCC-2998 (RRID:CVCL_1266), HCT-116 (ATCC, catalog no. CCL-247, RRID:CVCL_0291), HCT-15(ATCC, catalog no. CCL-225, RRID:CVCL_0292), HT-29 (ATCC, catalog no. HTB-38, RRID:CVCL_0320), KM12 (RRID:CVCL_1331), and SW620 (ATCC, catalog no. CCL-227, RRID:CVCL_0547). The six central nervous system (CNS) cell lines included SF-268 (RRID:CVCL_1689), SF-295 (RRID:CVCL_1690), SF539SF-539 (RRID:CVCL_1691), SNB-19 (RRID:CVCL_0535), SNB-75 (RRID:CVCL_1706), and U251 (RRID:CVCL_0021). The nine melanoma cell lines included LOX-IMVI (RRID:CVCL_1381), Malme-3M (ATCC, catalog no. HTB-64, RRID:CVCL_1438), M14 (RRID:CVCL_1395), MDA-MB-435 (RRID:CVCL_0417), SK-MEL-2 (ATCC, catalog no. HTB-68, RRID:CVCL_0069), SK-MEL-28 (ATCC, catalog no. HTB-72, RRID:CVCL_0526), SK-MEL-5 (ATCC, catalog no. HTB-70, RRID:CVCL_0527), UACC-257 (RRID:CVCL_1779), and UACC-62 (RRID:CVCL_1780). The seven ovarian cancer cell lines included IGROV-1 (RRID:CVCL_1304), OVCAR-3 (ATCC, catalog no. HTB-161, RRID:CVCL_0465), OVCAR-4 (RRID:CVCL_1627), OVCAR-5 (RRID:CVCL_1628), OVCAR-8 (RRID:CVCL_1629), NCI/ADR-RES(RRID:CVCL_1452), and SK-OV-3 (ATCC, catalog no. HTB-77, RRID:CVCL_0532). The eight renal cancer cell lines included 786-0 (ATCC, catalog no. CRL-1932, RRID:CVCL_1051), A-498 (ATCC, catalog no. HTB-44, RRID:CVCL_1056), ACHN (ATCC, catalog no. CRL-1611, RRID:CVCL_1067), CAKI-1 (ATCC, catalog no. HTB-46, RRID:CVCL_0234), RXF 393L (RRID:CVCL_1673), SN12C (RRID:CVCL_1705), TK-10 (RRID:CVCL_1773), and UO-31 (RRID:CVCL_1911). The two prostate cancer cell lines were PC-3 (ATCC, catalog no. CRL-1435, RRID:CVCL_0035) and DU-145 (ATCC, catalog no. HTB-81, RRID:CVCL_0105). The six breast cancer cell lines included MCF-7 (ATCC, catalog no. HTB-22, RRID:CVCL_0031), MDA-MB-231 (ATCC, catalog no. HTB-26, RRID:CVCL_0062), MDA-MB-468 (ATCC, catalog no. HTB-132, RRID:CVCL_0419), Hs 578T (ATCC, catalog no. HTB-126, RRID:CVCL_0332), BT-549 (ATCC, catalog no. HTB-122, RRID:CVCL_1092), and T-47D (ATCC, catalog no. HTB-133, RRID:CVCL_0553). All 60 human tumor cell lines were cultured in complete media containing RPMI 1640 medium (Gibco, Thermo Fisher Scientific, cat. 21870100), 5% (v/v) defined FBS (HyClone Laboratories Inc., cat. SH30070.03), and 2 mmol/L L-glutamine (Cytiva, cat. SH30034.01). The cell lines were maintained in an incubator at 37°C and 5% CO2 with 95% humidity. After recovery from cryopreservation, the duration in culture depended upon the cell line and ranged from 20 to 30 passages. However, in culture, samples of all cell lines were collected at regular intervals for short tandem repeat profiling and Mycoplasma testing by Labcorp (Laboratory Corporation of America Holdings, formerly known as Genetica DNA Laboratories) to confirm their authenticity and integrity.
Table 1.
NCI60 cell lines showing the NCI/DTP cell line name, disease panel tumor type of origin, the cell line name from Cellosaurus (https://www.cellosaurus.org/), the cell line designation from Cellosaurus based upon historical data classic NCI60 cell line population doubling time in the 96-well format, classic NCI60 cells plated per well in the 96-well format, NCI60 cell line population doubling time in the 384-well format, and NCI60 cells plated per well in the HTS384 NCI60 assay.
| NCI/DTP cell line name | Disease panel name | Cellosaurus line name | Cellosaurus ID | Classic NCI60 doubling time, hours | Classic NCI60 screen plated/96-well cells/well | NCI60 HTS384 screen doubling time, hours | NCI60 HTS384 cells plated/well |
|---|---|---|---|---|---|---|---|
| CCRF-CEM | Leukemia | CCRF-CEM | CVCL_0207 | 26.7 | 40,000 | 32.9 | 2,500 |
| HL-60(TB) | Leukemia | HL-60(TB) | CVCL_A794 | 28.6 | 30,000 | 31.1 | 1,500 |
| K-562 | Leukemia | K-562 | CVCL_0004 | 19.6 | 5,000 | 20.4 | 313 |
| MOLT-4 | Leukemia | MOLT-4 | CVCL_0013 | 27.9 | 30,000 | 30.8 | 1,500 |
| RPMI-8226 | Leukemia | RPMI-8226 | CVCL_0014 | 33.5 | 20,000 | 44.2 | 1,250 |
| SR | Leukemia | SR | CVCL_1711 | 28.7 | 20,000 | 38.9 | 2,500 |
| A-549/ATCC | Non–small cell lung | A-549 | CVCL_0023 | 22.9 | 7,500 | 23.6 | 500 |
| EKVX | Non–small cell lung | EKVX | CVCL_1195 | 43.6 | 20,000 | 55.0 | 1,250 |
| HOP-62 | Non–small cell lung | HOP-62 | CVCL_1285 | 39 | 10,000 | 44.0 | 600 |
| HOP-92 | Non–small cell lung | HOP-92 | CVCL_1286 | 79.5 | 20,000 | 78.2 | 1,000 |
| NCI-H226 | Non–small cell lung | NCI-H226 | CVCL_1544 | 61 | 20,000 | 77.8 | 1,250 |
| NCI-H23 | Non–small cell lung | NCI-H23 | CVCL_1547 | 33.4 | 20,000 | 52.9 | 1,250 |
| NCI-H322M | Non–small cell lung | NCI-H322M | CVCL_1557 | 35.3 | 20,000 | 47.1 | 1,250 |
| NCI-H460 | Non–small cell lung | NCI-H460 | CVCL_0459 | 17.8 | 7,500 | 18.6 | 250 |
| NCI-H522 | Non–small cell lung | NCI-H522 | CVCL_1567 | 38.2 | 20,000 | 69.0 | 1,250 |
| COLO 205 | Colon | COLO 205 | CVCL_0218 | 23.8 | 15,000 | 35.8 | 1,500 |
| HCC-2998 | Colon | HCC-2998 | CVCL_1266 | 31.5 | 15,000 | 43.6 | 750 |
| HCT-116 | Colon | HCT-116 | CVCL_0291 | 17.4 | 5,000 | 20.8 | 300 |
| HCT-15 | Colon | HCT-15 | CVCL_0292 | 20.6 | 10,000 | 23.3 | 500 |
| HT-29 | Colon | HT-29 | CVCL_0320 | 19.5 | 5,000 | 25.4 | 500 |
| KM12 | Colon | KM12 | CVCL_1331 | 23.7 | 15,000 | 29.3 | 750 |
| SW620 | Colon | SW620 | CVCL_0547 | 20.4 | 10,000 | 28.1 | 750 |
| SF-268 | CNS | SF-268 | CVCL_1689 | 33.1 | 15,000 | 57.2 | 1,000 |
| SF-295 | CNS | SF-295 | CVCL_1690 | 29.5 | 10,000 | 52.3 | 1,000 |
| SF-539 | CNS | SF-539 | CVCL_1691 | 35.4 | 15,000 | 36.5 | 500 |
| SNB-19 | CNS | SNB-19 | CVCL_0535 | 34.6 | 15,000 | 40.7 | 1,000 |
| SNB-75 | CNS | SNB-75 | CVCL_1706 | 62.8 | 20,000 | 79.9 | 1,250 |
| U251 | CNS | U-251MG | CVCL_0021 | 23.8 | 7,500 | 31.1 | 500 |
| LOX-IMVI | Melanoma | LOX-IMVI | CVCL_1381 | 20.5 | 7,500 | 32.3 | 500 |
| MALME-3M | Melanoma | Malme-3M | CVCL_1438 | 46.2 | 20,000 | 77.7 | 1,250 |
| M14 | Melanoma | M14 | CVCL_1395 | 26.3 | 15,000 | 45.8 | 1,000 |
| MDA-MB-435 | Melanoma | MDA-MB-435 | CVCL_0417 | 25.8 | 15,000 | 46.6 | 1,500 |
| SK-MEL-2 | Melanoma | SK-MEL-2 | CVCL_0069 | 45.5 | 20,000 | 600 | |
| SK-MEL-28 | Melanoma | SK-MEL-28 | CVCL_0526 | 35.1 | 10,000 | 66.7 | 1,000 |
| SK-MEL-5 | Melanoma | SK-MEL-5 | CVCL_0527 | 25.2 | 10,000 | 42.2 | 300 |
| UACC-257 | Melanoma | UACC-257 | CVCL_1779 | 38.5 | 20,000 | 66.3 | 600 |
| UACC-62 | Melanoma | UACC-62 | CVCL_1780 | 31.3 | 10,000 | 40.7 | 300 |
| IGROV-1 | Ovarian | IGROV-1 | CVCL_1304 | 31 | 10,000 | 39.6 | 1,000 |
| OVCAR-3 | Ovarian | OVCAR-3 | CVCL_0465 | 34.7 | 10,000 | 37.6 | 750 |
| OVCAR-4 | Ovarian | OVCAR-4 | CVCL_1627 | 41.4 | 15,000 | 81.5 | 1,000 |
| OVCAR-5 | Ovarian | OVCAR-5 | CVCL_1628 | 48.8 | 20,000 | 57.1 | 2,000 |
| OVCAR-8 | Ovarian | OVCAR-8 | CVCL_1629 | 26.1 | 10,000 | 32.6 | 500 |
| NCI/ADR-RES | Ovarian | NCI-ADR-RES | CVCL_1452 | 34 | 15,000 | 50.9 | 1,250 |
| SK-OV-3 | Ovarian | SK-OV-3 | CVCL_0532 | 48.7 | 20,000 | 43.4 | 500 |
| 786-0 | Renal | 786-O | CVCL_1051 | 22.4 | 10,000 | 35.2 | 500 |
| A-498 | Renal | A-498 | CVCL_1056 | 66.8 | 25,000 | 38.7 | 500 |
| ACHN | Renal | ACHN | CVCL_1067 | 27.5 | 10,000 | 34.9 | 1,000 |
| CAKI-1 | Renal | Caki-1 | CVCL_0234 | 39 | 10,000 | 37.2 | 600 |
| RXF 393 | Renal | RXF 393L | CVCL_1673 | 62.9 | 15,000 | 105.1 | 1,000 |
| SN12C | Renal | SN12C | CVCL_1705 | 29.5 | 15,000 | 48.2 | 1,000 |
| TK-10 | Renal | TK-10 | CVCL_1773 | 51.3 | 15,000 | 52.1 | 1,000 |
| UO-31 | Renal | UO-31 | CVCL_1911 | 41.7 | 15,000 | 38.0 | 1,000 |
| PC-3 | Prostate | PC-3 | CVCL_0035 | 27.1 | 7,500 | 40.5 | 500 |
| DU-145 | Prostate | DU-145 | CVCL_0105 | 32.3 | 10,000 | 32.6 | 1,000 |
| MCF-7 | Breast | MCF-7 | CVCL_0031 | 25.4 | 10,000 | 27.9 | 500 |
| MDA-MB-231/ATCC | Breast | MDA-MB-231 | CVCL_0062 | 41.9 | 20,000 | 48.9 | 1,250 |
| HS 578T | Breast | Hs 578T | CVCL_0332 | 53.8 | 20,000 | 67.2 | 1,000 |
| BT-549 | Breast | BT-549 | CVCL_1092 | 53.9 | 20,000 | 64.0 | 1,000 |
| T-47D | Breast | T-47D | CVCL_0553 | 45.5 | 20,000 | 75.7 | 1,000 |
| MDA-MB-468 | Breast | MDA-MB-468 | CVCL_0419 | 62 | 30,000 | 106.0 | 1,800 |
Classic NCI60 screen
Prior to inoculation into 96-well microplates, suspension cell lines were collected from T flasks and adherent cell lines were removed using TrypLE express (Gibco, Thermo Fisher Scientific, cat. 12605010). The cell lines were harvested by centrifugation for 5 minutes at 212 × g. Following removal of the supernatant, the cells were resuspended in fresh medium and quantified using a Cellometer K2 fluorescent viability cell counter (Nexcelom). Live cells were quantified from dual fluorescence measurements of acridine orange to quantify all cells and propidium iodide to quantify dead cells. Monolayer cell suspensions of 100 μL were dispensed into the wells of 96-well clear flat bottom polystyrene TC-treated microplates (Corning Inc., cat. 3598) using a BioTek MutiFlo FX peristaltic pump with a cassette head (Agilent Technologies, Inc.). The inoculation density for each cell line is shown in Table 1. Following inoculation, the microplates were transferred to an incubator and maintained at 37°C and 5% CO2 with 95% humidity. Twenty-four hours after inoculation, test agents or controls were delivered to the wells of microplates. Test agents and controls prepared as 400× stock solutions were dispensed into 12-channel deep well v-bottom reservoirs (Thermo Fisher Scientific, cat. 1149Q22), and complete media containing 0.1% (50 μg/mL) gentamicin (Gibco, Thermo Fisher Scientific, cat. 15750078) were added to achieve a 1:200 dilution. Final concentrations of the test agents and controls were achieved by transferring 100 μL of solution from the 12-channel reservoirs into the wells of 96-well test microplates (1:2 dilution) using a 12-channel pipette. All test agents were evaluated in technical duplicate, and doxorubicin (NSC123127) was included as a standard in each experiment for quality control. Time zero microplates were also prepared 24 hours after inoculation. For this, 100 μL of complete media containing 0.1% gentamicin was transferred to all wells of the time zero microplates, which were subsequently fixed with cold trichloroacetic acid (TCA; Thermo Fisher Scientific, cat. C987X91). Adherent cell lines were fixed with 50 μL of 50% (w/v) TCA, whereas suspension cell lines were fixed with 50 μL of 80% (w/v) TCA, which was added very slowly to push the suspension cells to the bottom of the plates. After the time zero microplates were refrigerated at 4°C for 1 to 3 hours, the TCA solutions were decanted, the wells were washed multiple times with tap water and the microplates were pounded against paper towels to remove residual water. The microplates were then placed on racks to air dry prior to staining (see below for the staining procedure). After 48 hours of cell line exposure to controls or test agents, the test microplates were removed from incubators and the cells were fixed with the appropriate TCA concentrations, rinsed with water, and dried as described above for the time zero microplates. Next, remaining cellular proteins were stained with sulforhodamine B (SRB, Pylam Products Co. Inc., cat. 74072). Microplates were removed from the drying racks and 100 μL of 0.4% (w/v) SRB in 1% acetic acid were added to the wells of microplates using a Zoom HT LB 920 plate washer (Berthold Technologies). After 10 minutes to 1 hour, the stain solution was washed from the microplate wells three times with 350 μL of a 1% acetic acid (Macron Fine Chemicals, VWR International Holdings, Inc., cat. MK-V193-05) solution (v/v) using a BioTek 405 LS washer (Agilent Technologies, Inc.). The microplates were subsequently pounded against paper towels to remove residual waste and placed on racks to air dry. To solubilize the SRB stain bound to cellular proteins, 100 μL of 10 mmol/L Trizma base (Sigma-Aldrich, cat. T1503) was added to the wells of microplates using a Zoom HT LB 920 plate washer. Next, the microplates were placed on orbital shakers at room temperature. Microplates containing adherent cell lines were agitated for a minimum of 2 hours, whereas those containing suspension cell lines were agitated for a minimum of 5 hours. Finally, absorbance was measured from each well at 515 nm using a BioTek Synergy Neo2 hybrid multimode reader (Agilent Technologies, Inc.; ref. 8).
HTS384 NCI60 screen
The NCI60 cell lines were harvested, and live cells were quantified as described above. Mixed cell suspensions of 40 μL were dispensed into the wells of 384-well white flat bottom polystyrene TC-treated microplates (Greiner Bio-One, cat. 781080) using a Microlab NIMBUS 96 workstation (Hamilton Company). The inoculation density for each cell line is shown in Table 1. Following inoculation, the microplates were transferred to an incubator (LiCONiC, STX500-ICSA) and maintained at 37°C and 5% CO2 with 95% humidity. Twenty-four hours after inoculation, test agents or controls were delivered to the wells of microplates. The test agents and controls were prepared as 400× stock solutions in Echo qualified polypropylene 384-well microtiter plates (Beckman Coulter Life Sciences, cat. 001-14615) and 100 nL was transferred by acoustic dispensing with an Echo 655 Liquid Handler (Beckman Coulter Life Sciences) into the time zero and assay microplates to achieve a 1:400 final dilution. All test agents were evaluated in technical triplicates. Controls in each assay microplate included the DMSO vehicle [0.25% (v/v), final (n = 14)], 100% cytotoxicity [1 μmol/L staurosporin (NSC755774) and 3 μmol/L gemcitabine (NSC613327; n = 7)], and five concentrations of doxorubicin (NSC123127), 25 μmol/L (n = 2), 2.5 μmol/L (n = 1), 250 nmol/L (n = 2), 25 nmol/L (n = 1), and 2.5 nmol/L (n = 1). After delivery of 100 nL DMSO into the wells of the time zero microplates, 40 μL of CellTiter-Glo 2.0 (Promega Corporation, cat. G9243) were dispensed into the wells using a LGR Precise Drop II dispenser (Let’s Go Robotics, Inc.), and luminescence was measured using a PHERAstar FSX (BMG LABTECH), according to Promega’s protocol, to assess cell viability. Following the delivery of controls and test agents to the assay microplates, they were transferred back to the incubator for 72 hours at 37°C with 5% CO2 and 95% relative humidity. After 72 hours of exposure to test agents and controls, 40 μL of CellTiter-Glo 2.0 were dispensed into the wells of the assay microplates and luminescence was measured, according to the manufacturer’s protocol, to assess cell viability. The SK-MEL-2 cell line exhibited inconsistent growth during many of the HTS384 NCI60 assays and was not included in the current data analysis.
The percent treated over control (PTC) was calculated at each of the test agent concentrations using the mean of vehicle control signal values (μVehicle), and the mean of test agent treated signal values (μTreated).
IC50 was calculated as
The percentage growth (%G) was calculated at each of the test agent concentrations using various measurements from the NCI60 screen [mean of time zero signal values (μTzero), mean of vehicle control signal values (μVehicle), and mean of test agent treated signal values (μTreated)].
If μTzero < μTreated, the following equation was used:
If μTzero > μTreated, the following equation was used:
In addition to %G, three response parameters were calculated for each test agent, namely, the GI50, TGI, LC50 (50% lethal concentration) using the following equations:
GI50 was calculated as
TGI was calculated as
LC50 was calculated as
Values were calculated for each of these three parameters if the level of activity was reached in the concentration response and was bracketed by two data points; however, if the effect was not reached or was exceeded, the value for that parameter was expressed as greater or less than the maximum or minimum concentration tested. In both the classic NCI60 screen and the new HTS384 screening system the success of the evaluation of a new compound is based on reaching the GI50 endpoint in at least 40 cell lines, if the compound exhibited concentration dependent growth. A web site (https://ioa.cancer.gov) was established to make available up-to-date data related to the collection of NCI investigational oncology agents and FDA-approved oncology drugs (3). These data are a subset of all public data from the classic NCI60 screen, which are also available (https://wiki.nci.nih.gov/display/NCIDTPdata/NCI-60+Growth+Inhibition+Data).
Study data
The GI50 values from the classic NCI60 and from the HTS384 NCI60 screens were analyzed in several ways. The GI50 values from the classic NCI60 screen were the average by cell line of all GI50 values from replicate experiments in the screen that used the same concentration range as was used in the HTS384 NCI60 screen. In cases in which there was no direct equivalent, the closest concentration range was used. Concentrations are represented as the log10 of the molar concentration whether referencing a concentration response graph or the response endpoint for a GI50 value. Some charts also show concentrations as micromolar. The concentration response data and associated endpoint values from both screens are available for download. Screen endpoint values are determined based on linear interpolation between two concentrations that generate growth above and below the growth response of interest. The processing of the growth response data is illustrated in Supplementary Fig. S1A–S1C (also, see “Data availability”).
COMPARE correlations
COMPARE correlations between the GI50 values for two agents are a modification of a Pearson correlation. This unitless value is a measure of the strength of the linear relationship between two sets of variables (cell line GI50 values), is independent of the order in which the cell lines are considered in the calculation and factors out differences in overall potency in the sets of variables focusing instead on the relative patterns of sensitivities across the cell lines. Cell lines with missing data for one or both agents are ignored in the calculation (5). After the calculation is run, there is a filter that requires at least 40 cell lines to have reported values for both agents and a filter that requires a minimum coefficient of variation of the endpoint values across the cell lines of 0.01. The latter is a default for the COMPARE application, which eliminates from consideration compounds in which the response across all cell lines is the same because there will be no SD. In the IOA compound set, there are 64 compounds with no cell line reaching GI50 at the highest concentration tested (10−4 mol/L, 100 μmol/L). In the classic NCI60 screen, the discrimination among the cell line response resides between 100 and 10 μmol/L for a subset of compounds and is important for identifying compounds that affect target classes, which have been much studied. (1).
Data visualization
To summarize the range of cell line responses for a compound, bar charts for the mean of the GI50s across all the cell lines were used and the SD is a surrogate for the range of values across the cell lines (see Supplementary Fig. S1C). To facilitate interpretation of a collection of Pearson correlations, a correlation map tool was developed that visualizes pairwise correlations among all members of a set of compounds (3). The endpoint data for individual compounds are represented by nodes (circles) on the map and links between the nodes represent the correlations between the endpoint data sets. Links between the nodes are only rendered if the correlation meets user-configurable minimum correlation criteria, and the length of the rendered links is proportional to the correlation with more similar compounds being closer together (3). If the compounds represented by two linked nodes share a common target, then the link carries the color assigned to that target, whereas links between nodes representing different targets render as black.
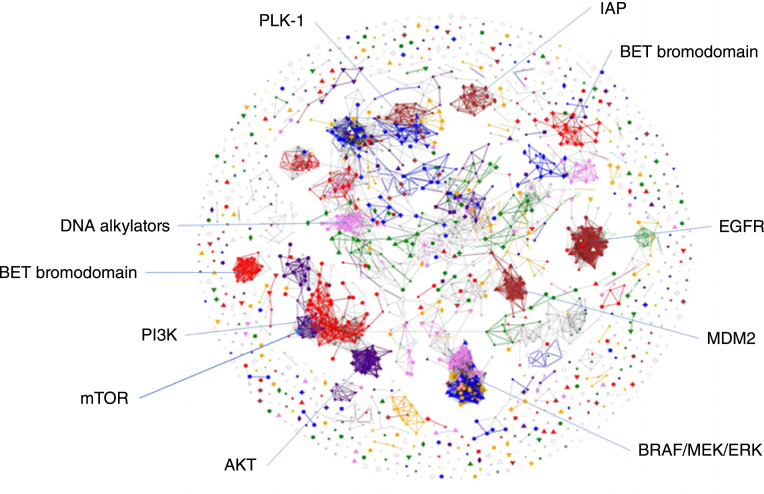
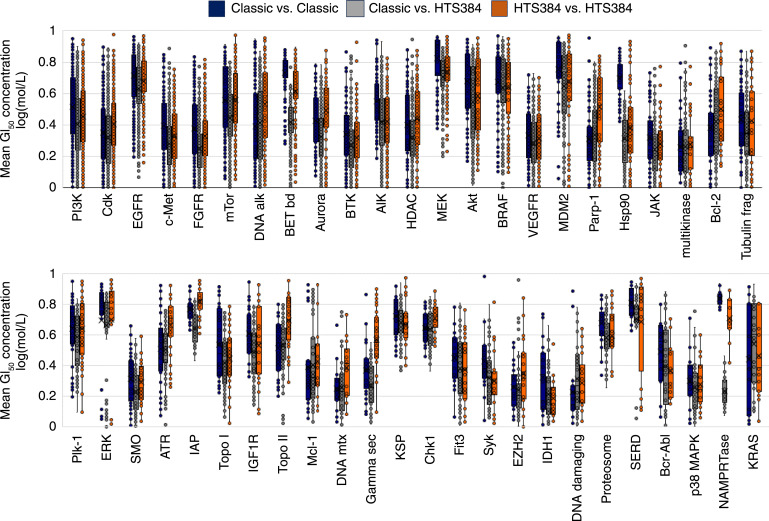
Empirical evaluation of COMPARE correlations indicate that correlations <0.6 are probably not significant indicators of agents acting via similar mechanisms, as reflected by their cell line response patterns, the range of 0.65 to 0.75 are worth consideration, and correlations >0.75 are indicators of likely similarity (5). Correlation maps in the supplementary figures were configured to render links only for correlations at or above 0.75. The correlation map was used to focus on the most significant correlations within a set of correlations; however, it is not suitable for displaying all nonnegative correlations within a set of compounds with a common target (Fig. 1). A box and whisker plot was used to summarize the entire population of positive correlations for all possible pairwise correlations among compounds within a target set. The box represents the range of the second and third quartiles within the distribution of all pairwise correlations, whereas the “whiskers” encompass the first to fourth quartiles (Fig. 2).
Figure 1.
A COMPARE correlation map, which comingles 1,003 FDA-approved and investigational oncology agents, run in the classic NCI60 screen and the HTS384 NCI60 screen (2003 GI50 patterns). Nodes represent GI50 determinations across the cell lines of the classic NCI60 screen (small symbols) and the NCI60 HTS384 screen (large symbols). The line length between nodes indicates the magnitude of the Pearson correlation between the GI50 patterns, with lines only rendered for correlations >0.75. Compact clusters are apparent from compounds sharing a molecular target (some are labeled).
Figure 2.
Box and whisker plot distributions (see Supplementary Fig. S2C) of positive correlations from pairwise combinations of mean GI50 values for compounds grouped by target for targets with at least six assigned agents. Blue, correlations within the classic NCI60 GI50 dataset; orange, correlations within the HTS384 NCI60 GI50 dataset; gray, correlations for the comingled mean GI50 values from the classic and HTS384 NCI60 datasets.
Data availability
The data generated in this study are publicly available at the “Download HTS384 data” link at https://dtp.cancer.gov/databases_tools/bulk_data.htm. All other raw data are available upon request from the corresponding author. The data have also been integrated into the existing IOA COMPARE website at https://ioa.cancer.gov. Guidance for using the website is in the “Approved and Investigational Oncology Agents COMPARE” section of the page at https://dtp.cancer.gov/databases_tools/compare.htm.
Results
A higher capacity screen designated HTS384 NCI60 was developed to continue the evaluation of investigator-submitted compounds and biologics as an NCI service to the cancer research community. Development of the NCI60 HTS384 screen required several years from optimizing the inoculation cell number and growth of the cell lines in the 384-well format to assembling and enabling an automated screening system and building a quality control and data processing structure. A 384-well screen using the NCI60 cell lines had already been developed at the University of Pittsburgh (11) as part of the NCI ALMANAC project to identify combinations of approved drugs with therapeutic potential (https://dtp.cancer.gov/ncialmanac), and studies exploring the time dependence of compound effects on the NCI60 cell lines with a luminescence readout were performed (12). The new data handling system for the HTS384 NCI60 leverages automation, modern software, and processing power to allow for complete flexibility in plate layouts, the number of concentrations in a dilution series as well as the number of replicate wells. Before raw data are processed, substantial quality control is applied to look for problematic replicate wells, dilution members, replicates, or trends in the behavior of individual cell lines within each screen run and across multiple screen runs. All parameters are accessible through new graphical interfaces. The raw luminescence data are converted to percent viability by normalizing to the DMSO (vehicle treated) control. Once the screening data pass lab-level QC, the processing pipeline generates the outcome: series of cell growth measurements across a range of concentrations and the GI50, TGI, and LC50 interpolated values from those growth measurements.
The classic NCI60 screen and the new HTS384 NCI60 screen both provide concentration responses for test agents at 100, 10, 1, 0.1, and 0.01 μmol/L as well as the endpoints GI50, TGI, and LC50, which are interpolated from the concentration response data. In this study, a library of 1,003 FDA-approved and investigational small-molecule anticancer agents was screened by the two NCI60 assays. Data for all 1,003 agents were available from the classic NCI60 screen, and for 1,003 agents from the HTS384 NCI60 screen. As a basis for assessing the comparability of the screens, we evaluated COMPARE analyses of the mean GI50 values for the entire set of individual agents as well as several subgroups of agents with common targets.
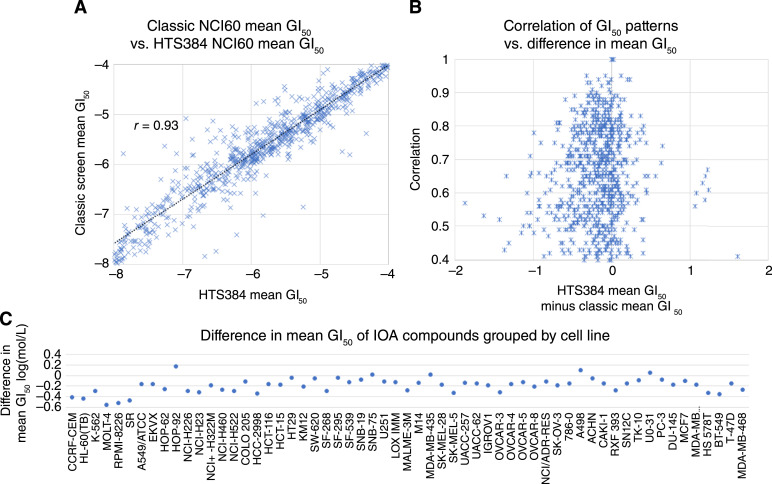
On a compound-by-compound basis, in Fig. 3A the mean GI50 values across all cell lines in the classic screen are plotted against the mean GI50 values in the HTS384 screen for the same compound. There is an overall shift of the HTS384 NCI60 data toward apparent greater potency (74% of the compounds report a lower mean GI50 value) as would be expected due to the longer compound exposure time before the determination is made. On a compound-by-compound basis, in Fig. 3B, the correlation between the HTS384 and classic GI50 values are plotted against the difference in the mean GI50 values for each compound (HTS384 mean GI50 minus the classic mean GI50, negative values indicate that the mean GI50 from the HTS384 screen results was less than the mean GI50 from the classic screen results). For many compounds, even when there are substantial differences in the overall mean GI50 values, there are high correlations between the patterns of GI50 values for the cell lines. Figure 3C shows the difference in the mean GI50 values when data were grouped by cell line across all compounds and suggests that the aggregate differences between the mean GI50 values for the compounds are not being driven by substantial changes in individual cell line behaviors between the two screens but rather reflect overall changes in the measured responses to the compounds.
Figure 3.
A, Scatter plot of the mean GI50 values from the HTS384 NCI60 screen plotted vs. the mean GI50 values from the classic NCI60 screen for individual compounds run in both assays (r = 0.93). B, A scatter plot of the Pearson correlations between the HTS384 NCI60 dataset and classic NCI60 dataset for GI50 data for individual compounds run in both assays plotted against the difference in the mean GI50 values. A total of 228 compounds correlated at 0.75 or greater and 470 compounds correlated at 0.6 or better. C, Mean GI50 for 1003 compounds by cell line across the compounds.
Figure 1 is a graphical summary of what was observed when considering subsets of the collection of compounds grouped by target shown as a correlation map of the GI50 values for the two screens (see Supplementary Fig. S2A–S2D for an illustration of the derivation of the correlation map from sets of COMPARE correlations). Compounds with highly correlated patterns of GI50 values cluster together when links are rendered for Pearson correlations greater than 0.75. Higher correlations between the response patterns bring the nodes closer together (3). With this stringent requirement, many agents form clustered groups based on their assignments to cellular targets or mechanisms of action, and results from the classic NCI60 screen and from the HTS384 screen comingle within many, but not all, of these groups. For some agents, there were cases in which the HTS384 NCI60 demonstrated a lower mean GI50 for a targeted group than the classic NCI60. Examples included some inhibitors of the BET bromodomain, HSP90, IAP, and NAMPRTase. Figure 2 shows the distribution of positive correlations for all possible pairwise combinations of mean GI50 values were assessed for targeted groups with at least six agents. Supplementary Table S2 lists the aggregate GI50 Pearson correlations for 91 targets with more than a single representative within the compound set.
Doxorubicin was used as the experimental quality control compound for both screens. Eighteen runs of the HTS384 NCI60 screen were undertaken to gather the data presented here. The concentration–response growth curves for doxorubicin in each of the runs are shown in Supplementary Fig. S3A and the associated cell line legends are shown in Supplementary Fig. S3B. The correlations of the doxorubicin GI50 patterns among these independent replicates of the HTS384 NCI60 screen indicated a high level of quality and reproducibility, comparable to that observed across nearly 3,000 classic NCI60 screens (Supplementary Fig. S3C).
The grouping of targeted agents indicates that overall, most correlations from the classic NCI60 and HTS384 NCI60 screens are similar. All positive correlations for pairwise combinations of mean GI50 values were assessed for targeted groups with at least six agents and no cutoff correlation (Fig. 2). It is important to note that the whisker plots of all positive correlations show that within the sets of compounds assigned to common targets not all pairwise combinations meet even the reduced stringency of a correlation of 0.6 within the classic NCI60 assay and within the HTS384 assay. A contributing factor to this is that a single target assignment was made for each agent based on literature references without capturing any off-target effects, at least some of which may be known. For some agents, there were cases in which the HTS384 NCI60 demonstrated a lower mean GI50 for a targeted group than the classic NCI60. Examples included some inhibitors of the BET bromodomain, HSP90, IAP, and NAMPRTase. The mean GI50 Pearson correlations for 91 targets with more than single representative are shown in Supplementary Table S2.
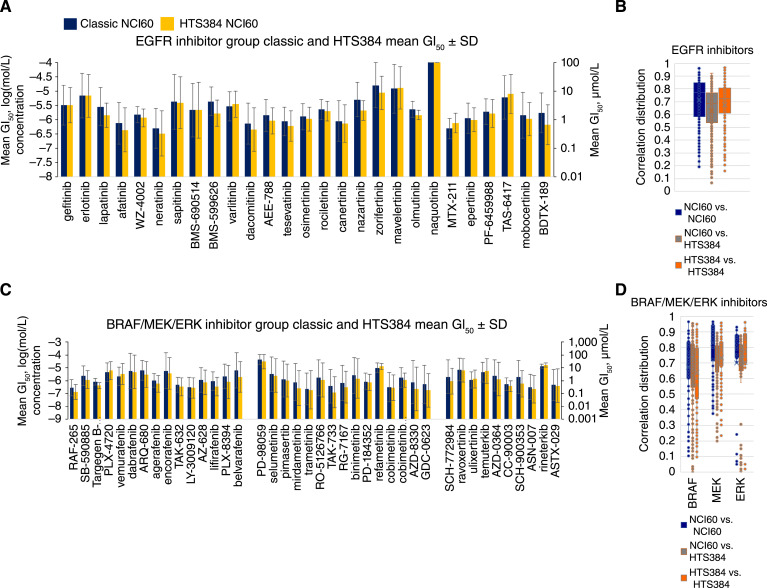
The mean GI50 values for EGFR-targeted agents were similar between the classic and HTS384 NCI60 screens (Fig. 4A). Mean GI50 values for the agents ranged from about −6.3 mol/L (0.5 μmol/L) to −4.88 mol/L (13 μmol/L). The distributions of all positive correlations for EGR-targeted agents are shown in Fig. 4B. When these data were rendered as a COMPARE correlation map at a correlation of >0.75, the cluster shown in Supplementary Fig. S4A resulted. When the data were restricted to cluster as the classic versus the HTS384 NCI60 data, two compact clusters shown in Supplementary Fig. S4B were generated.
Figure 4.
A, Mean GI50 values for compounds targeting the EGFR. Dark blue, data from the classic NCI60 screen; orange, data from the HTS384 NCI60 screen. Error bars indicate SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). B, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of EGFR-targeted agents. The box encompasses the second and third quartiles. Associated correlation maps for correlations >0.75 are shown in Supplementary Fig. S4A and S4B. C, Mean GI50 values for compounds targeting BRAF, MEK, and ERK. Dark blue, data from the classic NCI60 screen; orange, data from the HTS384 NCI60 screen. Error bars indicate SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). D, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of the agents that target BRAF, MEK, and ERK. The box encompasses the second and third quartiles. Associated correlation maps for correlations >0.75 are shown in Supplementary Fig. S5A and S5B.
The mean GI50 values for a large group of inhibitors targeting BRAF, MEK, and ERK indicated more heterogeneity between the classic and HTS384 NCI60 screens (Fig. 4C). The most potent compounds in this group had mean GI50 values of −6.5 mol/L (0.32 μmol/L) and −6.86 mol/L (0.14 μmol/L) in the classic NCI60 screen and the HTS384 NCI60 screen, respectively. The least potent compounds in this group had mean GI50 values of −4.35 mol/L (45 μmol/L) and −4.5 mol/L (31.7 μmol/L) in the classic NCI60 screen and the HTS384 NCI60 screen, respectively. The distributions of all positive correlations for these agents are shown in Fig. 4D. COMPARE correlation map presentations of these data are shown in Supplementary Fig. S5A and S5B. With comingling of data from the classic and HTS384 NCI60 screens (Supplementary Fig. S5A), there was extensive clustering across the targets for several agents as evident by the links shown as black lines. When data from the two screens were prevented from comingling, two similar complex clusters resulted (Supplementary Fig. S5B). Extensive cross target linking is evident (black lines) in both the classic NCI60 cluster and the HTS384 NCI60 cluster. This indicates that the overall cellular responses (GI50 values) reflect the perturbation of targets involved in common pathways.
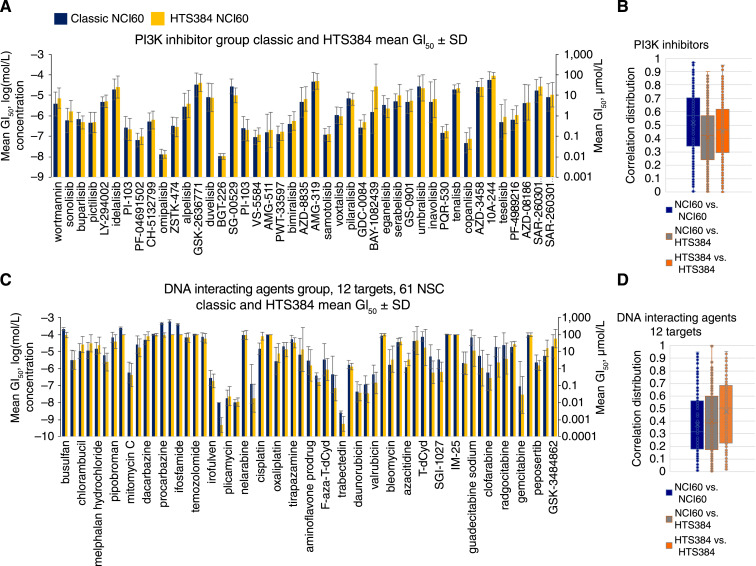
The PI3K-targeted group includes 32 compounds, the largest group in the IOA library. The mean GI50 values for agents in the PI3K-targeted group were generally similar between the classic and HTS384 NCI60 datasets (Fig. 5A). The most potent compound in this group was BTG-226 with mean GI50 values of −7.96 mol/L (0.0109 μmol/L) and −7.97 mol/L (0.0107 μmol/L) in the classic and HTS384 NCI60 datasets, respectively. The least potent compound in this group was IOA-244 with mean GI50 values of −4.25 M (56.23 μmol/L) and −4.03 mol/L (93.32 μmol/L) in the classic and HTS384 NCI60 datasets, respectively. The distributions of all positive correlations for PI3K-targeted agents are shown in Fig. 5B. The PI3K-targeted group included several nonclustered and tightly clustered agents when analyzed as a COMPARE correlation map at a correlation of >0.75 and the classic and HTS384 NCI60 datasets were allowed to comingle. (Supplementary Fig. S6A). When the data from the two screens were prevented from comingling, two complex clusters resulted with many unlinked compounds from both assays (Supplementary Fig. S6B). This result indicates that some of the compounds may be nonselective.
Figure 5.
A, Mean GI50 values for the PI3K-targeted agents. Dark blue, mean GI50 values from the classic NCI60 screen; orange, those from the HTS384 NCI60 screen. Error bars indicate SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). B, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of PI3K-targeted agents. The box encompasses the second and third quartiles. Associated correlation maps for correlations >0.75 are shown in Supplementary Fig. S6A and S6B. C, Mean GI50 values for the DNA interacting group of drugs and compounds. Dark blue, mean GI50 values from the classic NCI60 screen; orange, mean GI50 values from the HTS384 NCI60 screen. Error bars indicate SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). D, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of PI3K-targeted agents. The box encompasses the second and third quartiles.
DNA interacting agents form another large heterogeneous group of drugs and compounds (Fig. 5C) with 12 different specific targets represented. These compounds span a large range of potency from cyclophosphamide, ifosfamide, and temozolomide, which are prodrugs with little or no activity in cell culture, to exquisitely potent compounds such as trabectedin and lurbinectedin. The distributions of all positive correlations for the DNA interacting agents are shown in Fig. 5D.
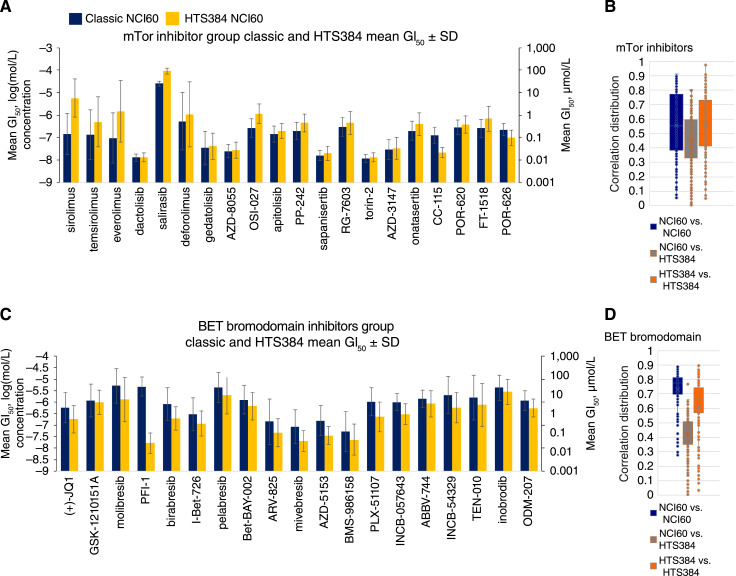
The first generation allosteric rapalog mTOR inhibitors sirolimus (rapamycin), temsirolimus, and everolimus were about two logs less potent in the HTS384 NCI60 screen than in the classic NCI60 screen (Fig. 6A). The pairwise distribution of correlations in the HTS384 and classic screens were similar (Fig. 6B); however, the allosteric rapalogs did not cluster with the other compounds in the HTS384 NCI60 dataset at a correlation of 0.75 or higher. (Supplementary Fig. S7A and S7B). As with other kinase inhibitor groups, the more recent mTOR competitive kinase inhibitors sapanisertib, onatasertib, and apitolisib produced similar mean GI50 values (Fig. 6A) and formed compact clusters both in the comingled classic and HTS384 NCI60 datasets (Supplementary Fig. S7A) as well as when the data from each screen were examined individually (Supplementary Fig. S7B). When the stringency of the Pearson correlation clustering was decreased to 0.65, more connections were made between the two datasets; however, the allosteric rapalogs remained independent of the main cluster in the classic NCI60 dataset (not shown). Growth–response curves in the representative cell lines are shown for the rapalogs (Supplementary Fig. S8A) and for the competitive inhibitors (Supplementary Fig. S8B).
Figure 6.
A, Mean GI50 values for mTOR-targeted agents. Dark blue, mean GI50 values from the classic NCI60 screen ; orange, those from the HTS384 NCI60 screen. Error bars indicate SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). B, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of mTOR-targeted agents. The box encompasses the second and third quartiles. Associated correlation maps for correlations >0.75 are shown in Supplementary Fig. S7A and S7B. C, Mean GI50 values for agents targeting the BET bromodomain. Dark blue, data from the classic NCI60 screen; orange, data from the HTS384 NCI60 screen. Error bars are SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). D, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of agents that target the BET bromodomain. The box encompasses the second and third quartiles. Associated correlation maps for correlations >0.75 are shown in Supplementary Fig. S8A and S8B.
The mean GI50 values for BET bromodomain targeted agents were uniformly lower in the HTS384 NCI60 screen than in the classic NCI60 screen (Fig. 6C). The largest difference observed was nearly three logs from the compound PFI-1. Unlike several of the kinase targeted groups, the BET bromodomain inhibitors did not link when allowed to form a comingled cluster (Supplementary Fig. S9A). The clusters remained the same when comingling was prevented (Supplementary Fig. S9B). This disconnect is clear from inspection of the pairwise combinations of mean GI50 values for the BET bromodomain targeted agents (Fig. 6D). The concentration response of five bromodomain inhibitors in five representative NCI60 cell lines (MDA-MB-468 breast adenocarcinoma, SK-MEL-5 cutaneous melanoma, LOX-IMVI amelanotic melanoma, HOP-92 non–small cell lung carcinoma, and HCC-2998 colon adenocarcinoma) showed increased cytotoxicity in the HTS384 NCI60 screen (Supplementary Fig. S10A).
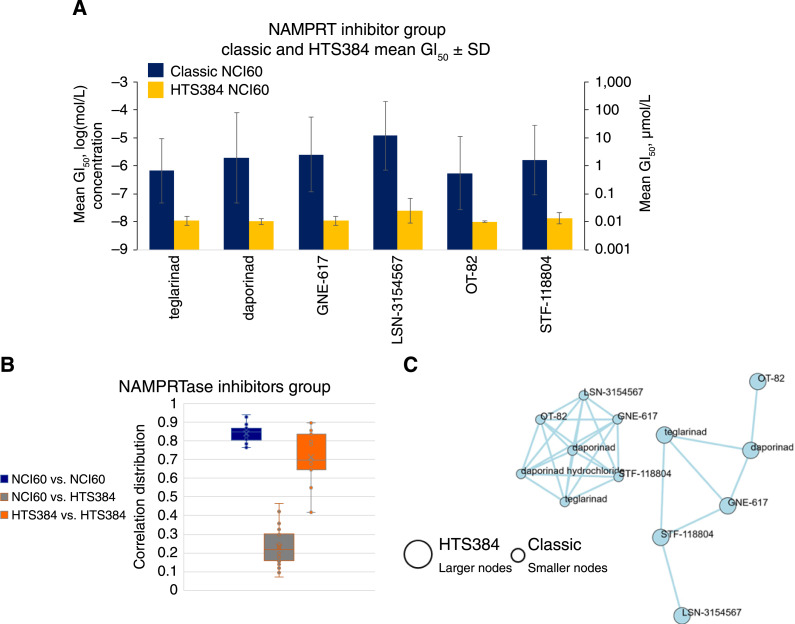
The NAMPRTase-targeted agents uniformly demonstrated lower GI50 values by as much as two logs in the HTS384 NCI60 screen compared with the classic NCI60 screen (Fig. 7A) The NAMPRTase-targeted agents were more tightly correlated within the set of classic NCI60 data (Fig. 7B) and did not form a comingled cluster between the classic and HTS384 NCI60 datasets (Fig. 7C). The two NAMPRTase clusters remained independent even at reduced Pearson stringencies (Fig. 7B). Concentration response for four NAMPRTase inhibitors in five representative NCI60 cell lines (SW620 colon adenocarcinoma, HCC-2998 colon adenocarcinoma, M14 amelanotic melanoma, SN12C renal cell carcinoma, and OVCAR-8 high-grade ovarian serous adenocarcinoma) are shallow in both datasets with 1 to 2 logs more cytotoxicity in the HTS384 NCI60 dataset than in the NCI60 classic dataset (Supplementary Fig. S10B).
Figure 7.
A, Mean GI50 values for the NAMPRTase-targeted agents. Dark blue, data from the classic NCI60 screen; orange, data from the HTS384 NCI60 screen. Error bars are SDs serving as surrogates for the range of individual cell line GI50 values relative to the mean of responses across the 60 cell lines (see Supplementary Fig. S1C). B, Box and whisker representation (see Supplementary Fig. S2C) of all possible pairwise combinations of NAMPRTase-targeted agents. The box encompasses the second and third quartiles. C, At a Pearson correlation threshold of >0.75, NAMPRTase-targeted agents form two independent clusters even when mean GI50 values from the classic and HTS384 NCI60 datasets are allowed to comingle.
Discussion
The NCI60 screen was established based upon the hypothesis that compounds could be found that selectively killed one type of cancer cell compared with the other eight cancers represented in the screening panel. Over time as the genetics of cancer were elucidated, it became evident that patterns of cytotoxic activity across the NCI60 set of human tumor cell lines could serve as fingerprints for the mechanism of action of compounds being screened. The selection of cell lines was based on their availability, growth rates in the selected growth media, formation of a tightly bound monolayer, disease, and responses to clinically active drugs (4, 5, 7, 13). The NCI60 cell line panel includes nine cancer types (leukemia, non–small cell lung, colon, CNS, melanoma, ovarian, renal, prostate, and breast) and has been used to profile potential oncology small-molecule therapeutic agents for more than 30 years. Extensive genomic and proteomic profiling of the NCI60 cell lines makes this among the best characterized collection of human cancer cell lines including studies of gene mutations, amplifications and deletions, proteomics, the methylome, microRNA, exosomes, and more (2, 3). Data from NCI60 cell lines are available from multiple websites including https://dtp.cancer.gov/databases_tools/bulk_data.htm; https://discover.nci.nih.gov/rsconnect/cellminercdb/; https://web.expasy.org/cellosaurus/; https://www.cbioportal.org/; https://cancer.sanger.ac.uk/cosmic; https://discover.nci.nih.gov/rsconnect/cellminercdb/. The NCI60 screen has proved to be a useful tool for drug discovery by the cancer research community and has facilitated the elucidation of molecular targets for potential new oncology agents.
Qualitatively, the data from the new screen are the same as for the classic NCI60 screen: cell growth measurements across a set of five log dilutions of the test agent. IC50 values are interpolated from the growth measurements considered a percentage of treated growth relative to control, whereas GI50, TGI, and LC50 values are interpolated from the growth measurements corrected for cell number at the beginning of the assay (GIPRCNT). When the classic NCI60 was being developed, it was thought that the GI50 and TGI endpoints might be more responsive indicators for different categories of agents; so, the standard assay concentration range that was picked was one that most-often captured those two endpoints.
The new HTS384 NCI60 screen was developed to maintain the free screening service for the cancer research community. The updated screen makes extensive use of laboratory automation to test cells in a 384-well format with a 3-day exposure period to test agents and a luminescent readout for cell viability. The data presented provide an initial characterization of the HTS384 NCI60 screen and a comparison with data from the classic NCI60 screen as the NCI transitions to the new service. Of interest, and concern, was whether the large collection of public data from the classic NCI60 screen could be used in conjunction with data from the new HTS384 screen. Accordingly, the concentrations for test agents were maintained with the highest concentration in the HTS384 NCI60 screen is set at a high concentration of 10−4 mol/L effectively asking whether the results of the compounds in an initial uninformed assay in the new HTS384 system would be able to identify the same compound in the historic, classic NCI60 data. This was done even for compounds with an optimum concentration range that started at less than 10−4 mol/L.
The COMPARE (https://ioa.cancer.gov) program allowed the direct comparison of data between the two screens for 1,003 FDA-approved and investigational agents and showed good comparability in patterns of GI50 values for more than 45 molecular target groups with at least six compounds. Although mean GI50 values from the two screens were similar for several kinase targeted agents, non-kinase targeted agents, such as inhibitors of the BET bromodomain, HSP90, and NAMPRTase, generally, had lower mean GI50 values in the HTS384 NCI60 screen. It is possible that the longer compound exposure period in the HTS384 NCI60 screen resulted in some agents showing strong correlations within both the classic and the HTS384 NCI60 screens, but low or no correlations between the two screens. The increased compound exposure period might have allowed cell line differences to more fully manifest or for other off-target effects to play a greater role in affecting the response of the cells.
Summary and conclusions
Figures 1 and 2 provide graphical summaries of our findings: there is substantial overlap between the results from the classic NCI60 screen and from the HTS384 NCI60 screen for many, but not all, of the compounds grouped by target, however, even for targets that do not substantially overlap between the two screens there is clustering within the new screen comparable with that of the classic NCI60 screen.
Similarities and differences were identified in the responses to targeted agents from the classic and HTS384 NCI60 screens. In addition to the focused set of FDA-approved and investigational agents discussed here, there are data for approximately 60,000 public compounds that have been tested in the classic NCI60 screen, most of which have defined chemical structures. That rich dataset can also be used to evaluate results from the new HTS384 NCI60 screen. Given that there is substantial overlap between the two screens for many targeted agents and mechanisms of action, it would not be unreasonable to use results from the new HTS384 NCI60 screen as a reference for COMPARE correlations against classic NCI60 datasets. As a corollary, it would not be unreasonable to run COMPARE against the classic NCI60 data using data from the HTS384 NCI60 screen for new agents with unknown targets or mechanisms of action. As is the case with data from the classic NCI60 screen being probed with the data from the same screen, strong correlations would suggest compounds or mechanisms of action for further consideration, whereas a lack of correlation could indicate a compound that acts via a mechanism not already represented in the NCI/DTP public data or could indicate a mechanism that cannot manifest during the time course of the screen or could indicate that the compound has a new target or mechanism not yet documented in the NCI60 database. For researchers who have built a series of compounds and associated screening data using the classic NCI60 screen for mechanistic or structural categories, focused assays with representative compounds in the HTS384 NCI60 screen could quantify the link between the old and the new screen.
Supplementary Material
Table listing NCI60 cell lines, disease of origin and key genetic alterations
Aggregate correlations of compound pairs grouped by target for 91 targets with more than a single representative compound
Legends for Supplementary Figures S1-S10
Diagram outlining processing of assay data
Diagram outlining COMPARE calculations and data presentation formats.
Doxorubicin control conc/resp
Correlation plots EGFR-targeting agents.
Correlation plots agents targeting BRAF, MEK or ERK.
Correlation plots PI3K-targeting agents
Correlation plots mTor-targeting agents.
Representative conc/resp mTor-targeting agents.
Correlation plots BET-targeting agents.
Representative conc/resp BET-targeting agents and NAMPRTase-targeting agents.
Acknowledgments
In memoriam: This study is dedicated to the memory of Dr. Michael C. Alley, PhD. Mike was a key member of the team that developed the NCI60 screen. He was recruited to DTP in 1986 and authored the article that was the genesis of the NCI60 screen. Mike will be remembered for his attention to detail that served the cancer research community for 30 years and for his kindness to all. This project was funded in whole or in part with federal funds from the NCI, NIH, under contract HHSN261201500003I. This research was supported in part by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the NCI.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
No disclosures were reported.
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authors’ Contributions
M.W. Kunkel: Data curation, software, formal analysis, visualization, writing–original draft, writing–review and editing. N.P. Coussens: Data curation, formal analysis, investigation, methodology, writing–original draft, project administration. J. Morris: Conceptualization, resources. R.C. Taylor: Software. T.S. Dexheimer: Investigation. E.M. Jones: Investigation. J.H. Doroshow: Funding acquisition. B.A. Teicher: Conceptualization, formal analysis, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–23. [DOI] [PubMed] [Google Scholar]
- 2. Holbeck SL, Camalier R, Crowell JA, Govindharajulu JP, Hollingshead M, Anderson LW, et al. The National Cancer Institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res 2017;77:3564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morris J, Kunkel MW, White SL, Wishka DG, Lopez OD, Bowles L, et al. Targeted investigational oncology agents in the NCI-60: a phenotypic systems-based resource. Mol Cancer Ther 2023;22:1270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 1991;83:757–66. [DOI] [PubMed] [Google Scholar]
- 5. Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst 1989;81:1088–92. [DOI] [PubMed] [Google Scholar]
- 6. Shoemaker RH, Monks A, Alley MC, Scudiero DA, Fine DL, McLemore TL, et al. Development of human tumor cell line panels for use in disease-oriented drug screening. Prog Clin Biol Res 1988;276:265–86. [PubMed] [Google Scholar]
- 7. Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res 1988;48:589–601. [PubMed] [Google Scholar]
- 8. Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990;82:1107–12. [DOI] [PubMed] [Google Scholar]
- 9. Evans JR, Akee RK, Chanana S, McConachie GD, Thornburg CC, Grkovic T, et al. National Cancer Institute (NCI) program for natural product discovery: exploring NCI-60 screening data of natural product samples with artificial neural networks. ACS Omega 2023;8:9250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyd MR, Paull KD. Some practical considerations and applications of the national-cancer-institute in-vitro anticancer drug discovery screen. Drug Dev Res 1995;34:91–109. [Google Scholar]
- 11. Close DA, Wang AX, Kochanek SJ, Shun T, Eiseman JL, Johnston PA. Implementation of the NCI-60 human tumor cell line panel to screen 2260 cancer drug combinations to generate >3 million data points used to populate a large matrix of anti-neoplastic agent combinations (ALMANAC) database. SLAS Discov 2019;24:242–63. [DOI] [PubMed] [Google Scholar]
- 12. Evans DM, Fang J, Silvers T, Delosh R, Laudeman J, Ogle C, et al. Exposure time versus cytotoxicity for anticancer agents. Cancer Chemother Pharmacol 2019;84:359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer 2001;84:1424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table listing NCI60 cell lines, disease of origin and key genetic alterations
Aggregate correlations of compound pairs grouped by target for 91 targets with more than a single representative compound
Legends for Supplementary Figures S1-S10
Diagram outlining processing of assay data
Diagram outlining COMPARE calculations and data presentation formats.
Doxorubicin control conc/resp
Correlation plots EGFR-targeting agents.
Correlation plots agents targeting BRAF, MEK or ERK.
Correlation plots PI3K-targeting agents
Correlation plots mTor-targeting agents.
Representative conc/resp mTor-targeting agents.
Correlation plots BET-targeting agents.
Representative conc/resp BET-targeting agents and NAMPRTase-targeting agents.
Data Availability Statement
The data generated in this study are publicly available at the “Download HTS384 data” link at https://dtp.cancer.gov/databases_tools/bulk_data.htm. All other raw data are available upon request from the corresponding author. The data have also been integrated into the existing IOA COMPARE website at https://ioa.cancer.gov. Guidance for using the website is in the “Approved and Investigational Oncology Agents COMPARE” section of the page at https://dtp.cancer.gov/databases_tools/compare.htm.