Abstract
Sex differences are widespread during neurodevelopment and play a role in neuropsychiatric conditions such as autism, which is more prevalent in males than females. In humans, males have been shown to have larger brain volumes than females with development of the hippocampus and amygdala showing prominent sex differences. Mechanistically, sex steroids and sex chromosomes drive these differences in brain development, which seem to peak during prenatal and pubertal stages. Animal models have played a crucial role in understanding sex differences, but the study of human sex differences requires an experimental model that can recapitulate complex genetic traits. To fill this gap, human induced pluripotent stem cell–derived brain organoids are now being used to study how complex genetic traits influence prenatal brain development. For example, brain organoids from individuals with autism and individuals with X chromosome–linked Rett syndrome and fragile X syndrome have revealed prenatal differences in cell proliferation, a measure of brain volume differences, and excitatory-inhibitory imbalances. Brain organoids have also revealed increased neurogenesis of excitatory neurons due to androgens. However, despite growing interest in using brain organoids, several key challenges remain that affect its validity as a model system. In this review, we discuss how sex steroids and the sex chromosomes each contribute to sex differences in brain development. Then, we examine the role of X chromosome inactivation as a factor that drives sex differences. Finally, we discuss the combined challenges of modeling X chromosome inactivation and limitations of brain organoids that need to be taken into consideration when studying sex differences.
Keywords: Autism, Brain organoids, Sex chromosomes, Sex differences, Steroids, X chromosome inactivation
Plain Language Summary
Sex differences are a contributing factor in neuropsychiatric conditions such as autism, which is more prevalent in males. Sex differences occur through interactions between sex steroid hormones such as estrogen and testosterone and sex chromosomes (chrX and chrY). Human stem cell–derived brain organoids are laboratory models that mimic brain development. For example, in individuals with neurodevelopmental conditions, brain organoids have revealed an imbalance of neuron populations compared with neurotypical individuals. In this review, we discuss sex steroid and sex chromosome influences on brain development and challenges of this model that need to be taken into account when studying sex differences.
Plain Language Summary
Sex differences are a contributing factor in neuropsychiatric conditions such as autism, which is more prevalent in males. Sex differences occur through interactions between sex steroid hormones such as estrogen and testosterone and sex chromosomes (chrX and chrY). Human stem cell–derived brain organoids are laboratory models that mimic brain development. For example, in individuals with neurodevelopmental conditions, brain organoids have revealed an imbalance of neuron populations compared with neurotypical individuals. In this review, we discuss sex steroid and sex chromosome influences on brain development and challenges of this model that need to be taken into account when studying sex differences.
Steroid hormones such as estrogens, androgens, and progestins that regulate sex-based development of tissues and organs (henceforth called sex steroid hormones) and the sex chromosomes (chromosomes X [chrX] and Y [chrY]) are known to influence sex differences in brain development, both independently and through their interaction (1, 2, 3). Together, they are responsible for global effects on behavior, neuroanatomy, and cellular and molecular mechanisms (4, 5, 6, 7). Independent human behavioral studies have identified sex differences in language, literacy, social skills, and empathy (8). Human sex differences are also well established in neuropsychiatric conditions such as autism (9), attention-deficit/hyperactivity disorder (9), and early-onset obsessive-compulsive disorder (10, 11, 12).
There is considerable evidence in humans of neuroanatomical differences between males and females. Brain volume, which is a global measure of structural differences, is larger in males than females on average (13, 14, 15). However, gray matter density is higher in females across the cerebral cortex (16,17). Sex differences have also been found in specific brain regions. Large studies have reported sex differences in hippocampal and amygdala volumes (18, 19, 20), and the relative size differences between males and females in both structures have been found to depend on age and pubertal development. Females showed larger hippocampal volumes postpuberty (19), while males showed overall larger amygdala volumes during both youth and adulthood (18). Although considerable variation has been reported in size differences between male and female cortical substructures, all studies have consistently reported the existence of sex differences in age-related developmental trajectories (18, 19, 20, 21). Sex differences have also been found in noncortical regions; in the cerebellum, females have larger cerebellar lobules such as Crus II connected with nonmotor regions of the cortex while males have larger motor region–connected lobules (H V and H VIIIA/B) (22).
However, understanding of molecular mechanisms that influence sex differences is predominantly informed by animal models (23, 24, 25). Because most sex differences in human behavioral phenotypes are more complex and often diverge from those of other mammals (26), there is a need for human-specific model systems to study mechanisms. Human-derived stem cells including embryonic stem cells and induced pluripotent stem cells (iPSCs) are able to capture unique human phenotypes driven by complex genetic polymorphisms. Because iPSCs are derived by the reprogramming of cells from individuals who carry specific polymorphisms associated with health and disease (27), in theory, this model enables direct correlation of individual behavioral traits with the molecular mechanisms conferred by their genetic background (28). By growing stem cell–derived, 3-dimensional (3D), self-organizing, and self-patterning neural organoids (hereafter referred to as brain organoids) (29), it is possible to study aspects of molecular mechanisms associated with human brain development and the factors that may disrupt them.
In this review, we focus on the biological basis of sex differences in the brain, specifically highlighting the cellular and molecular mechanisms involved and how human brain organoids are being used to dissect these mechanisms. First, we explore the nature of sex differences and how sex steroid hormones and sex chromosomes act as biological drivers. Then, we take an in-depth look into sex chromosomal regulatory mechanisms that are less well understood in the context of sex differences. Then, we discuss the advantages of using a human stem cell–derived brain organoid model to study sex differences in cellular and molecular traits. Finally, we discuss limitations of using stem cells and brain organoids in the context of developmental phenotypes associated with sex differences.
Framework and Biological Drivers of Sex Differences in Human Behavior
Sex differences cannot be defined strictly based on their biological mechanisms. It is generally believed that sex steroid hormones and sex chromosomes are evolutionarily selected biological factors that influence sex differences (henceforth, we will call these factors biological drivers) (30). However, for reasons of experimental design, scientists have categorized sex differences into 4 broad types: qualitative, quantitative, latent, and population-level, with evidence of these 4 types found in both humans and animals (30,31) (Table 1). These sex differences may be a result of gonadal sex steroids that cause early-life programming during a sensitive prenatal period of brain development and during an additional sensitive period during puberty (32). These sex differences may also be the result of unequal activity of chrX and chrY driving sex differences independent of the gonads and long before their formation, and there is evidence of sexually differentiated gene expression by the 8-cell embryo stage (33).
Table 1.
Framework for Sex Differences in Human Behavior
| Sex Differences Classification | Description | Example |
|---|---|---|
| Qualitative | Sexual dimorphisms related to reproduction (143) | During spatial navigation performance tasks, men tended to rely more on geometric knowledge while women relied on rote learning (144). |
| Quantitative | Average sex differences, occurring on a continuum with a different mean for males and females or where male and female traits differ in the size of their response (145) | In acute and chronic pain response, men show greater sensitivity than women on average (145). |
| Latent | Hidden sex differences that are less well understood, but where the end point traits in males and females are the same, but there are underlying mechanistic differences that may only appear during environmental challenges such as stress, injury, or disease (146) | During eye-blink conditioning, male and female rats do not demonstrate sexual dimorphism in performance. However, performance in male rats is significantly improved upon response to stress, whereas the same stress response impairs performance in females (147). |
| Population-Level | Reflects differences in the distribution of individual traits (46) | In autism, representative behavioral traits are the same in males and females, but there is a skewed male-to-female prevalence of autistic traits (46). |
However, our understanding of how these biological drivers influence molecular mechanisms is informed primarily by animal studies (34), and animal models do not always resemble humans because there is significant species-dependent divergence in brain development (35, 36, 37). For example, overall brain size and connectivity is greater in humans than in rodents and other primates. The human cerebral cortex and cerebellum are significantly larger than those of other primates (38). Proportions of neuronal populations are different in humans; 40% of neurons in humans are upper-layer pyramidal neurons compared with 25% in mice (39). Interneuron numbers are also higher in humans (25%–30%) than in rodents (∼20%) (40). Some human cortical circuits are driven by unique genetic modifiers called human accelerated regions that are not found in rodents (37). Human accelerated regions are regulatory elements on the genome that are enriched for genes that influence neural and cortical development (41). Even steroid hormone response varies between humans and rodents; for example, in humans, androgens are responsible for masculinization of the brain, while in rodents, this is influenced by estrogen (42). Brain size, connectivity, and population of neuronal types are all biological markers influenced by sex differences, as we will discuss in later sections.
Biological Drivers of Sex Differences Are Also Autism Likelihood Factors
The biological drivers of sex differences—sex steroid hormones and sex chromosome complement—are associated with neuropsychiatric conditions. Autism is a typical example of a neuropsychiatric condition with behavioral sex differences (43). Current estimates suggest a 3:1 male-to-female ratio even after accounting for diagnostic bias (44, 45, 46). Atypical prenatal sex steroid levels are associated with increased prevalence of autism and autistic traits (47,48), with both testosterone and estrogen found to be elevated during the critical period of prenatal development in boys with autism (47,49). In females, a higher burden of genetic mutations has been shown to be associated with autism and autistic traits (50,51). This suggests that the combination of sex steroids and genes may increase the likelihood of autism in males. Gene dosage from sex chromosomes are believed to contribute to brain sex differences and autism likelihood (52) (Table 2). For example, the number of autistic traits is increased in individuals with sex chromosomal aneuploidies such as Klinefelter syndrome (KS) (XXY, 47), XYY syndrome (XYY, 47), and Turner syndrome (TS) (XO, 45) (53). Rett syndrome, where there is a mutation in the X-linked gene MECP2 (54), and fragile X syndrome, with a mutation in the X-linked FMR1 gene, are two of the most common syndromic forms of autism (55). Mutations in X-linked genes such as KDM5C (56), ARHGEF9 (57), and SYN1 (58) have recently been shown to confer a high likelihood of autism. According to the SFARI gene database, a total of 22 X-linked genes are classified as high-likelihood autism genes (https://gene.sfari.org/database/human-gene/).
Table 2.
Genotype-Phenotype Relationships of X-Linked Conditions in Humans
| X-Linked Condition | Genotype | Sex Bias | Cognitive Symptoms | Evidence From Brain Organoids |
|---|---|---|---|---|
| Rett Syndrome | Heterozygous mutations in MECP2 | Males die before birth or early infancy | Autism, epilepsy (148) | Altered interneuron populations in MECP2 mutant, with more VIP+ and CALB2+ cells in the mutant Increase in density of excitatory puncta in the mutant Recurring epileptiform-appearing spikes and high-frequency oscillations (120) |
| FXS | Heterozygous mutations in FMR1 gene | Females show milder symptoms | Intellectual disability, autism (149) | Accelerated neural progenitor cell cycle exit in FXS Reduced neural progenitor cell proliferation Number of GABAergic neurons decreased in FXS Accelerated synapse formation and hyperexcitability of synapses (121) |
| Klinefelter Syndrome | Males with extra X chromosome | Only affects males | Reading disabilities, autism (113,150) | Cellular phenotypes unknown. No evidence from brain organoids. |
| Turner Syndrome | Females missing part or all of 1 X chromosome | Only affects females | Intellectual disability, visual-spatial and cognitive weaknesses, autism (151,152) | Cellular phenotypes unknown. No evidence from brain organoids. |
FXS, fragile X syndrome.
Molecular Pathways Conferring Steroid- and Sex Chromosome–Mediated Sex Differences in the Brain
McCarthy and Arnold (25) have proposed 2 models of sexual differentiation, the classic model and the parallel model. According to the classic model, sex differences in brain and behavior are due to the developmental action of gonadal sex steroids. According to the parallel model, sex differences are the result of an imbalance of genes on the sex chromosomes, which suggests a more direct influence of X and Y chromosomal genes on sex differences.
Although present in both males and females, steroid hormone levels usually differ by sex (Figure 1) (59,60). During development, sex steroids originate from the placenta and the maternal circulation (61), the fetal gonads (testes in males), and the brain (25). Most steroid hormone receptors are transcription factors that directly regulate gene activity; however, receptors for estrogens and androgens may also act via second messenger pathways. In the brain, estradiol is known to bind with ER⍺ and ERβ (estrogen receptors ⍺ and β), which act as transcription factors while second messenger pathways are mediated through GPER1 (G protein–coupled estrogen receptor 1) (62,63). Much less is known about the mechanism of androgen activity in the brain. Androgens are known to act solely through the X-linked androgen receptor, with well-established transcription factor activity and second messenger activity that is less well understood (64).
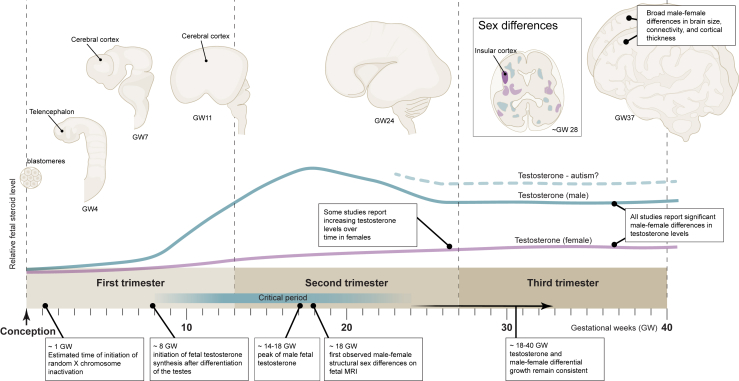
Figure 1.
Timeline of human brain development from conception to birth showing human brain morphology over developmental time (top), predicted testosterone dynamics (middle), and key events in sex-biased development (bottom). The box shows an axial section with examples of regions with growth trajectory (blue, male-biased; purple, female-biased) sex differences found in fetal MRI (141). The insular cortex is an example of a focal region that was larger in males than in females. The sex chromosomes begin exerting sex-specific effects soon after conception. Testosterone is given here as a classic example of a steroid sex hormone with levels that differ by sex. Fetal testosterone increases after the differentiation of the testes in males. Male testosterone is thought to peak between GW14 and GW18 and then remain relatively consistent (141). Studies have generally reported higher male testosterone, and some studies have reported increasing testosterone in females over time (142). GW, gestational week; MRI, magnetic resonance imaging.
There is now growing appreciation that the sex chromosome complement influences sex differences more directly (25,65) (Figure 2). In humans, most tissues show sex-biased gene expression in a subset of sex chromosomal and autosomal genes (66,67). One study found that there was more male-biased gene expression during prenatal brain development (67). The researchers observed changes in sex-differential gene expression based on developmental stage, where a subset of chrY genes (∼12 genes: KDM5D, DDX3Y, ZFY, PCDH11Y, USP9Y, RPS4Y1, CYorf15B, TMSB4Y, NLGN4Y, UTY, EIF1AY, and GYG2P) appeared to be consistently expressed over time while chrX genes seemed to follow a dynamic expression pattern (67). For example, via their histone demethylase activity, KDM5C and KDM6A were identified as major X-linked mediators of sex differences, and the XIST gene mediated X chromosome inactivation (XCI), thereby offsetting an imbalance in gene expression in females versus males (68,69). Associated with prenatal development, the X-linked OGT expressed in the placenta was found to have an indirect but significant effect on the brain (70) (Figure 2). Sex chromosomes may also exert influence through orthologs (genes that have evolved from a common ancestor during speciation) of chrX genes that have been retained on chrY during evolution. For example, KDM5C on chrX and KDM5D on chrY have subtle functional differences (71). The role of XCI, which regulates X-linked gene activity such as that of KDM5C and KDM6A described above (52), will be discussed in more detail in later sections. In the following section, we will summarize the cellular phenotypes affected as a result of sex steroid and sex chromosome activity.
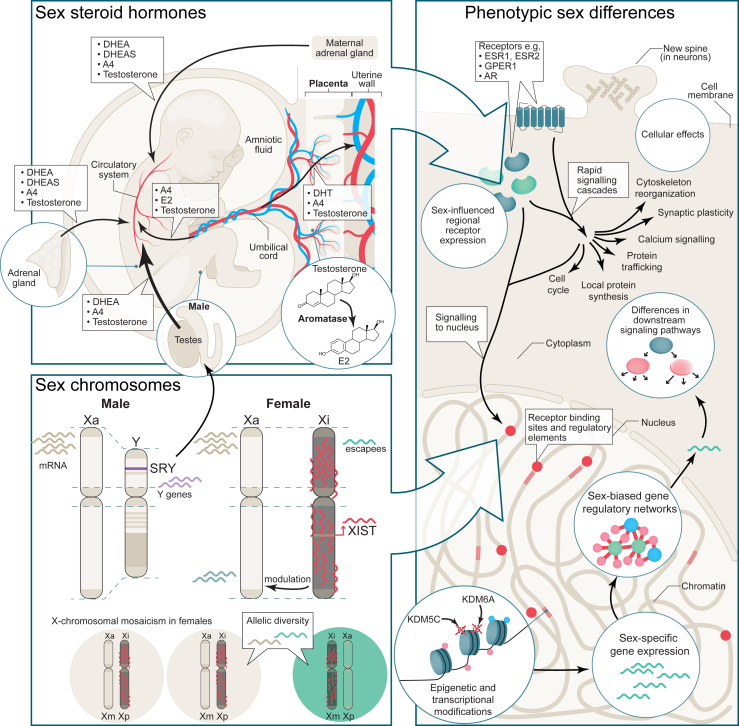
Figure 2.
Sex steroid hormones from various sources (top left) and sex chromosome effects (bottom left) give rise to phenotypic sex differences at the cellular level. Note that although we show the placenta in the context of hormones, the placenta can influence sex differences both through sex steroid hormones or sex chromosome complement. Sex steroid hormones in the fetus can originate from the maternal adrenal gland, the placenta, and from the fetal adrenal gland. After entering the fetal circulatory system, hormones can cross the blood-brain barrier to affect brain development. In males, the testes develop and are a source of androgens. DHEA, DHEAS, A4, E2 are examples of key steroid sex hormones. Aromatase (bottom right) can aromatize testosterone to E2. The sex chromosomes (bottom left) can be a source of gene expression differences. In males, there is 1 active X chromosome and the Y chromosome. The SRY gene on the Y chromosome is responsible for testes development. Expression of other Y genes (purple) that have diverged from their X chromosome homologs in evolution can be a source of sex differences. Females have 1 Xa and 1 Xi. Sex-specific gene expression can originate from genes that are expressed from the Xi (escapees) or from modulation of gene expression on the Xa by the Xi. Finally, X-chromosomal mosaicism in females results in allelic diversity (bottom). The placenta is one example of a tissue where sex chromosome complement has an important role in sex differences. Examples of how various cellular and molecular brain sex differences may arise are shown (right). Exposure to steroid hormones can have rapid cellular effects mediated by rapid signaling cascades or canonical gene expression effects through nuclear signaling. Sex chromosome gene expression effects combined with gene expression in response to steroid hormone signaling can modulate regional receptor expression and steroid signaling pathways, resulting in further sex-specific cellular responses. The combined action of the sex chromosomes and steroid hormones can establish sex-biased gene expression, gene regulatory networks, and epigenetic modifications. One example of epigenetic modifications is demethylation, such as by the KDM5C and KDM6A demethylases, which are discussed. A4, androstenedione; AR, androgen receptor; DHEA, dehydroepiandrosterone; DHEAS, dehydroepiandrosterone sulfate; E2, estradiol; ESR, estrogen receptor; GPER1, G protein–coupled estrogen receptor 1; Xa, active X chromosome; Xi, inactive X chromosome; Xm, X maternal; Xp, X paternal.
Effect of Sex Steroids and Chromosomal Sex on Cellular Phenotypes in the Brain
A number of studies that have investigated sex steroid effects have shown marked hormone-mediated sex differences in the brain, specifically in the hippocampus (72, 73, 74). Androgens (both testosterone and dihydrotestosterone) have been found to increase dendritic spine density (73,75,76) while also influencing synaptic long-term depression via a postsynaptic mechanism (77,78). Estrogens have also been shown to increase spine density but have been observed to influence synaptic long-term potentiation (72,79, 80, 81), and this was more prominent in females than males (79,80). In the hypothalamus, estrogen increased survivability of GABAergic (gamma-aminobutyric acidergic) neurons (82,83) and GABAergic projection size in males (84). A wide range of studies that have investigated sex steroid-mediated cellular and molecular mechanisms are summarized in Table S1.
There is accumulating evidence that chromosomal sex may also influence cellular traits independent of effects mediated through sex steroids (25). Mouse models that allow for the separation of gonadal sex and sex chromosomes by the deletion of the Sry gene from chrY and insertion of a Sry transgene into an autosome (known as the four core genotypes model) (2) have found that the presence of only 1 chrY or the absence of 2 chrXs irrespective of gonadal sex increased male bias in a maternal antibody–induced model of autism (85). More sex steroid–independent mechanisms were found using four core genotype mice; for example, XY hypothalamic neurons had greater axonal length than XX hypothalamic neurons (86), while higher numbers of dopaminergic neurons were observed only in male neuronal cultures (87). Beyond the four core genotypes model, hippocampal neurons have been found to develop larger pools of synaptic vesicles in male versus female rats via a sex steroid–independent mechanism (88). Nonneuronal cells such as astrocytes located in the arcuate nucleus of the hypothalamus showed more complex stellate features only in female rats (89). Microglia also showed sex differences in rodents such as in response to environmental factors such as microbiota (90).
XCI Plays a Unique Role in Mediating Sex Chromosomal Effects
Sex chromosomes may have additional influences on sex differences via the X-linked gene regulatory mechanism known as XCI. XCI is a biological process that typically silences one of the 2 chrXs in females (XX individuals) (91). Males (XY individuals) only have 1 chrX, which is always active. Most genes on the inactivated chrX (Xi) are silent, except for ∼20% of Xi genes, which escape XCI on the pseudoautosomal regions of chrX, resulting in higher expression of these genes in XX individuals than in XY individuals (92). Xi modulates gene expression both in cis (i.e., genes found on the same Xi chromosome) and in trans genes (i.e., genes on the other chrX); this means that in XX individuals, Xi is likely to have an influence on gene expression from the active chrX (Xa) (52).
XCI is first seen following implantation of the embryo in humans, and inactivation of chrX occurs randomly so that each chrX is active in approximately half of all cells (93,94) (Figure 3). This phenomenon is known as X-chromosomal mosaicism, and XCI in these cells are stably maintained down cell lineages (95). The chrX-linked long noncoding RNA XIST is responsible for XCI initiating a dampening of transcription via the reduction of gene-activating histone marks and a concurrent increase in gene-repressive histone marks (96, 97, 98). This process is triggered by XIST RNA spreading over cis-regulatory regions across the X chromosome, where it recruits a wide range of chromatin-modifying factors (98,99). The Xa is also transiently coated by a long noncoding RNA, XACT, which interacts with XIST antagonistically to maintain chrX activity (100). It is essential for the survival of the embryo that XIST expression be monoallelic, and a region on the X chromosome called the X inactivation center maintains this monoallelic expression of XIST (90,101).
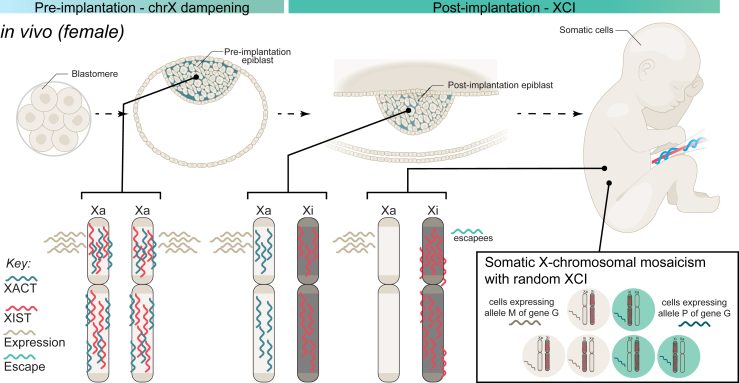
Figure 3.
Typical chrX states in female human embryo development. Human genome activation and expression from the X chromosomes occurs by the 8-cell stage (far left). In the preimplantation embryo (left), gene expression occurs from both of the X chromosomes, which are coated with the long noncoding RNAs XIST and XACT. Gene expression is modulated through chrX dampening. Sometime after implantation (middle), random XCI occurs. Gene expression becomes restricted to only Xa, and Xa is coated with XACT. Xi is coated with XIST, and gene expression is silenced. In somatic cells, Xi remains silenced except for escapee genes. Random XCI is preserved in somatic cells (box), resulting in X-chromosomal mosaicism in females, where a subset of cells will express the maternal allele of a gene, and the remaining cells will express the paternal alleles of a gene. chrX, X chromosome; G, gene; M, maternal; P, paternal; Xa, active chrX; XCI, X chromosome inactivation; Xi, inactive chrX.
Non-pseudoautosomal genes of chrX provide a causal link between XCI and sex differences in the brain. For example, for non-pseudoautosomal chrX genes with chrY orthologs, the brain developmental genes NLGN4X and ZFX showed similar expression levels in males and females, but expression of the orthologous NLGN4Y and ZFY that only occurred in males offset an overall higher dosage of the NLGN4 and ZF gene species in males (102). Although studies have shown that alteration of a single residue in NLGN4Y hindered its ability to mediate synaptogenesis (103), the likelihood of autism, which was maintained through several generations, only appeared to increase significantly upon a mutation in NLGN4X. This resulted in greater susceptibility in males than females who carried a nonmutant NLGN4X allele (104).
The Role of XCI in X-Chromosomal Aneuploidies
X-chromosomal aneuploidies in humans have revealed the significance of XCI in brain development. In the case of TS (45, XO), the most common female sex chromosome aneuploidy, 99% of pregnancies spontaneously terminate within the first trimester (105). In children with TS, it was found that the single chrX did not undergo inactivation, and there was downregulation of pseudoautosomal and XCI escape genes (106). Another study observed that TS-associated embryonic lethality was a result of haploinsufficiency of pseudoautosomal genes (107). The 1% who survived to term demonstrated significant mental and physical disabilities (105). These include deficits in visual-spatial functions and social skills and comorbidities such as attention-deficit/hyperactivity disorder and autism (105). In KS (47, XXY), the most common male sex chromosome aneuploidy, the presence of an additional X chromosome results in variable symptomatology, with only an estimated 25% of XXY genotypes being diagnosed with KS (108,109). This was believed to be due to the additional X chromosome being inactive and variable gene expression of pseudoautosomal escapee genes from this chromosome (108,110). Cognitive symptoms associated with KS were found to be driven by hypogonadism and included social impairments and verbal deficits and co-occurring conditions such as learning disability, autism, and epilepsy (108,111, 112, 113, 114, 115). A recent study identified a set of ∼12 genes dysregulated as a result of the additional Xi in individuals with KS; this included XCI escape genes (KDM6A, KDM5C, SMC1A, ZFX, RBBP7, DDX3X, CDK16, DLG3, USP9X, and BCOR) and pseudoautosomal genes (SLC25A6 and SHOX) (52). However, the XCI and chrX gene dosage–mediated mechanisms associated with chrX aneuploidies remain mostly unknown.
Brain Organoids Can Be Used to Study Biological Drivers of Sex Differences
Brain organoids have been reliable models for studying cell proliferation and excitation-inhibition (E-I) imbalance and cellular phenotypes affected in neuropsychiatric conditions (35) (Figure 4). Mariani et al. showed increased proliferation of GABAergic neurons in iPSC-derived brain organoids from individuals with autism giving rise to atypical neuronal hyperpolarization associated with Nav1.1, a sodium channel preferentially expressed in GABAergic neurons (116). In another study that examined mutations in 3 high-likelihood autism genes (SUV420H1, ARID1B, and CHD8), the authors found that mutations in any one of these genes resulted in atypical development of GABAergic interneurons and excitatory projection neurons but through distinct molecular pathways (117), and this resulted in a reduction in the frequency and duration of spontaneous activity of neurons resembling network activity associated with the developing brain. More advanced brain organoid systems such as air-liquid interface cortical organoids (118) and cortical spheroids (119) capable of achieving greater neuronal maturity can effectively model E-I phenotypes associated with later prenatal and early postnatal periods.
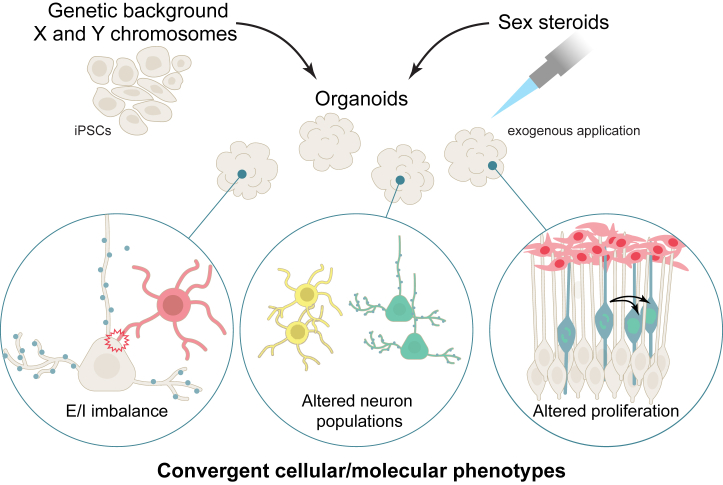
Figure 4.
Examples of convergent cellular and molecular neuropsychiatric condition phenotypes revealed by studies using human iPSC-derived brain organoids: E/I imbalance, altered cell proliferation, and phenotypes that affect specific neuron classes. Future organoid studies can combine the study of genetic background, sex chromosome complement, and exogeneous steroid hormone application. E/I, excitation/inhibition; iPSC, induced pluripotent stem cell.
Mutations in X-linked genes such as MECP2 (which causes Rett syndrome) and FMRP, which causes fragile X syndrome, present with more severe neuropsychiatric symptoms in males than in females, and Rett syndrome is usually lethal in males, with only a few males known to have survived until infancy (54,55). In Rett syndrome brain organoids, the electrophysiological activity of inhibitory interneurons was significantly reduced compared with controls, even though no differences were found in the ratio of glutamatergic to GABAergic neuronal fates (120). In this case, E-I imbalances were attributable to altered neuronal fate. In fragile X syndrome, loss of the FMRP gene triggered a reduction in global proliferation rate in brain organoids coupled with premature neural differentiation (121). This produced fewer inhibitory neurons, which resulted in increased excitability of excitatory neurons. In typical brain organoids from males and females without mutations in X-linked genes, administration of elevated levels of sex steroids was found to increase glutamatergic (excitatory) neuronal output via increased cell proliferation, suggesting another mechanism for E-I imbalances (122). The ability of brain organoids to model neuronal activity associated with neuropsychiatric conditions and demonstrate atypical cellular phenotypes in X-linked conditions and in elevated steroid level conditions adds to their validity as a model for studying brain sex differences in humans.
Limitations of Stem Cell Models Due to Atypical XCI States
Sex chromosomes and XCI play a key role in typical brain development (108). XCI is first observed during implantation of the embryo (93). In theory, stem cell models such as human embryonic stem cells or iPSCs may be most suited to studying the effect of XCI during human brain development. However, few studies have explored XCI traits using stem cell models. This is because in vitro embryonic stem cells and iPSCs demonstrate atypical XCI states that do not resemble in vivo XCI states (123). Four in vitro XCI states are known to exist: 1) XiXIST+Xa, where 1 X chromosome is coated by XIST and is inactive; 2) XiXa, where the inactive X is not coated with XIST; 3) XaXa, where both X chromosomes are active; and 4) XeXa, where the Xi has undergone erosion (Xe) or a partial loss of inactivation (123).
While in humans XiXIST+Xa is the typical XCI state, the XiXa state is functionally similar to XiXIST+Xa, although it is not observed in vivo (124). XaXa and XeXa represent atypical XCI states of full or partial reactivation, respectively, of the inactive chrX due to Xi erosion, and they cannot be reversed during differentiation (125,126), which results in a permanent burden of atypical chrX gene expression during the lifetime of differentiated cells. XCI mosaicism, which balances allelic expression in case of heterozygous disease-causing X-linked nonsynonymous mutations, also cannot be achieved in these stem cell models because they maintain clonal X activation status (125). The Xi erosion–induced atypical XCI states affect chrX gene dosage, influencing the validity of female iPSCs as model systems in studying X-linked conditions (124,125,127). For example, in Lesch-Nyhan syndrome, caused by a heterozygous mutation in the X-linked HPRT gene, iPSC-derived neurons showed variable traits due to reactivation of the functional HPRT gene from Xi. Similar anomalies have also been observed in iPSCs from patients with Rett syndrome with a mutation in the X-linked MECP2 gene (125,127). Thus, to study sex differences, care must be taken to select stem cell lines that do not demonstrate atypical XCI states and to devise strategies to mimic XCI mosaicism in vitro.
Limitations of Brain Organoids as an In Vitro Model of the Human Brain
Brain organoids have limitations that must be considered when using this model system. The most significant limitation is the absence of typical cortical tissue architecture. Although we observe deep-layer and upper-layer neurons, they do not have in vivo–like activity patterns, and their spatial organization is not finely tuned (35). Neuropsychiatric conditions often involve structural differences in specific cortical regions (128, 129, 130), but brain organoid models are only able to model gross dorsal and ventral forebrain traits (131). Although this works fine for studying E-I balance because excitatory and inhibitory neurons are derived from dorsal and ventral forebrain regions, respectively, during development, mechanisms that underlie human behavior such as social cognition, which is driven by subcortical regional activity, is currently beyond the scope of this method. Currently, the secondmost important limitation of brain organoids is the lack of nonneuronal cell types such as microglia. Microglia are the most significantly altered nonneuronal cell type in the autistic brain (132,133), but the development and maintenance of microglial homeostasis in the brain cannot be modeled using brain organoids because microglia are derived from the mesoderm (as opposed to the neuroectoderm for brain organoid) and do not appear to survive protracted developmental periods in vitro when cocultured (134,135). Other limitations of brain organoids include progenitor pool variability (136) and formation of necrotic tissue in larger organoids (137), but both of these limitations now have workarounds. Single-cell transcriptomic methods can help to resolve the variability in organoid progenitor pools through machine learning methods (136,138,139), while slice culture brain organoids at the air-liquid interface have been developed to overcome the influence of necrotic tissue formation (118,140). Other 3D models of brain development may also present unique advantages over brain organoids in studying sex differences (Table S2). As a result, careful consideration must be given to observable traits such as cell proliferation and E-I imbalances when using brain organoids, and if necessary, validations in other 3D model systems should be undertaken.
Conclusions
In this review, we discussed the roles of sex steroid hormones and sex chromosomes on sex differences in brain development. Sex differences are prevalent across behavioral and neuroanatomical modalities in humans. Cellular/molecular markers of sex differences, which are more challenging to study in humans, have been revealed through animal studies. These studies have revealed multiple mechanisms of sex differences influenced by sex steroids and sex chromosomes. However, cellular phenotypes such as cell proliferation, a mechanism for brain volume differences between males and females, and E-I balance, a mechanism commonly affected in neuropsychiatric conditions with a sex bias, show species-specific variations. This has resulted in the need to develop a human model system such as stem cell–derived brain organoids. Brain organoids have already been proven effective in modeling these cellular phenotypes in X-linked neuropsychiatric conditions as well as conditions of elevated sex steroids. However, the influence of sex chromosomes on sex differences, especially chrX, is complex and remains less well understood, partially due to the regulatory activity of XCI. Recent studies have shown that XCI is only partial, and stem cell systems may be prone to atypical XCI states that affect X-linked gene dosage. The usefulness of brain organoids is also limited by their inability to recapitulate complexities of in vivo brain development; thus, other 3D in vitro models may be needed. Nonetheless, it is a flexible system that is helping us understand the biological basis of typical sex differences and sex bias in neuropsychiatric conditions.
Acknowledgments and Disclosures
This work was supported by grants from The Simons Foundation Autism Research Initiative, the Autism Centre for Excellence. AP is supported by the Medical Research Council (MRC)-Sackler ITND (Institute for Translational Neurodevelopment) studentship as part of the MRC Centre for Neurodevelopmental Disorders (Grant No. MR/P502108/1). DA is supported by the Simons Foundation, Autism Centre of Excellence, and Cambridgeshire and Peterborough NHS Foundation Trust. This work was supported by the UK MRC (Grant Nos. MR/L021064/1 and MR/X004112/1 [to DPS]); Royal Society UK (Grant No. RG130856 [to DPS]); UK MRC Centre for Neurodevelopmental Disorders (Grant No. MR/N026063/1 [to ACV and DPS]). DPS is also a recipient of an Independent Researcher Award from the Brain and Behavior Foundation (formally National Alliance for Research on Schizophrenia and Depression) (Grant No. 25957). ACV and DS received financial support from the National Centre for the Replacement, Refinement and Reduction of Animals in Research (Grant No. NC/S001506/1). ACV received funding from UKRI/EC HE guarantee collaborative Grant No. HORIZON-HLTH-2021-STAYHLTH-01-02: 10053515. SB-C received funding from the Wellcome Trust (Grant No. 214322\Z\18\Z). For the purpose of open access, the author has applied a CC BY public copyright license to any author accepted manuscript version that arises from this submission. SB-C also received funding from the Autism Centre of Excellence, SFARI (Simons Foundation Autism Research Initiative), the Templeton World Charitable Fund, and the MRC. All research at the Department of Psychiatry at the University of Cambridge was supported by the National Institute for Health and Care Research (NIHR) Cambridge Biomedical Research Centre (Grant No. NIHR203312) and the NIHR Applied Research Collaboration East of England. Any views expressed are those of the author(s) and not necessarily those of the funders, IHU-JU2, the NIHR, or the Department of Health and Social Care.
ACV has been a speaker and currently receives funding for academic collaborations from bit.bio and GSK. DPS has been a speaker and currently receives funding for academic collaborations from Astra Zeneca, bit.bio, and GSK and has been a consultant for the Wellcome Trust. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
APEX consortium authors and affiliations: Simon Baron-Cohen (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Carrie Allison (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Varun Warrier (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Alex Tsompanidis (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Dwaipayan Adhya (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Rosie Holt (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Paula Smith (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Tracey Parsons (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Joanna Davis (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Matthew Hassall (Autism Research Centre, University of Cambridge, Cambridge, United Kingdom); Daniel H Geschwind (UCLA, Los Angeles, California); Alexander EP Heazell (Tommy’s Maternal and Fetal Research Centre, University of Manchester, Manchester, United Kingdom); Jonathan Mill (University of Exeter Medical School, College of Medicine & Health, University of Exeter, Exeter, United Kingdom); Alice Franklin (University of Exeter Medical School, College of Medicine & Health, University of Exeter, Exeter, United Kingdom); Rosie Bamford (University of Exeter Medical School, College of Medicine & Health, University of Exeter, Exeter, United Kingdom); Jonathan Davies (University of Exeter Medical School, College of Medicine & Health, University of Exeter, Exeter, United Kingdom); Matthew E. Hurles (Wellcome Trust Sanger Institute, Hinxton, United Kingdom); Hilary C. Martin (Wellcome Trust Sanger Institute, Hinxton, United Kingdom); Mahmoud Mousa (Wellcome Trust Sanger Institute, Hinxton, United Kingdom); David H. Rowitch (Department of Pediatrics, University of Cambridge, Cambridge, United Kingdom); Kathy K. Niakan (Centre for Trophoblast Research, University of Cambridge, Cambridge, United Kingdom); Graham J Burton (Centre for Trophoblast Research, University of Cambridge, Cambridge, United Kingdom); Fateneh Ghafari (Centre for Trophoblast Research, University of Cambridge, Cambridge, United Kingdom); Deepak P. Srivastava (MRC Centre for Neurodevelopmental Disorders, King’s College London, London, United Kingdom); Lucia Dutan-Polit (MRC Centre for Neurodevelopmental Disorders, King’s College London, London, United Kingdom); Adam Pavlinek (MRC Centre for Neurodevelopmental Disorders, King’s College London, London, United Kingdom); Madeline A. Lancaster (MRC Laboratory of Molecular Biology, University of Cambridge, Cambridge, United Kingdom); Ilaria Chiaradia (MRC Laboratory of Molecular Biology, University of Cambridge, Cambridge, United Kingdom); Tal Biron-Shental (Department of Obstetrics and Gynecology, Meir Medical Center, Israel); Lidia V Gabis (Child Development Division, Maccabi Health Services, Israel).
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2024.100343.
Contributor Information
Dwaipayan Adhya, Email: da361@medschl.cam.ac.uk.
APEX Consortium:
Simon Baron-Cohen, Carrie Allison, Varun Warrier, Alex Tsompanidis, Dwaipayan Adhya, Rosie Holt, Paula Smith, Tracey Parsons, Joanna Davis, Matthew Hassall, Daniel H. Geschwind, Alexander EP. Heazell, Jonathan Mill, Alice Franklin, Rosie Bamford, Jonathan Davies, Matthew E. Hurles, Hilary C. Martin, Mahmoud Mousa, David H. Rowitch, Kathy K. Niakan, Graham J. Burton, Fateneh Ghafari, Deepak P. Srivastava, Lucia Dutan-Polit, Adam Pavlinek, Madeline A. Lancaster, Ilaria Chiaradia, Tal Biron-Shental, and Lidia V. Gabis
Supplementary Material
References
- 1.Dewing P., Shi T., Horvath S., Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- 2.De Vries G.J., Rissman E.F., Simerly R.B., Yang L.Y., Scordalakes E.M., Auger C.J., et al. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wierman M.E. Sex steroid effects at target tissues: Mechanisms of action. Adv Physiol Educ. 2007;31:26–33. doi: 10.1152/advan.00086.2006. [DOI] [PubMed] [Google Scholar]
- 4.Alexander G.M., Wilcox T., Woods R. Sex differences in infants’ visual interest in toys. Arch Sex Behav. 2009;38:427–433. doi: 10.1007/s10508-008-9430-1. [DOI] [PubMed] [Google Scholar]
- 5.Arnold A.P., Chen X. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;30:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archer J. The reality and evolutionary significance of human psychological sex differences. Biol Rev Camb Philos Soc. 2019;94:1381–1415. doi: 10.1111/brv.12507. [DOI] [PubMed] [Google Scholar]
- 7.Kassam I., Wu Y., Yang J., Visscher P.M., McRae A.F. Tissue-specific sex differences in human gene expression. Hum Mol Genet. 2019;28:2976–2986. doi: 10.1093/hmg/ddz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig I.W., Harper E., Loat C.S. The genetic basis for sex differences in human behaviour: Role of the sex chromosomes. Ann Hum Genet. 2004;68:269–284. doi: 10.1046/j.1529-8817.2004.00098.x. [DOI] [PubMed] [Google Scholar]
- 9.Ramtekkar U.P., Reiersen A.M., Todorov A.A., Todd R.D. Sex and age differences in attention-deficit/hyperactivity disorder symptoms and diagnoses: Implications for DSM-V and ICD-11. J Am Acad Child Adolesc Psychiatry. 2010;49:217–228.e1. [PMC free article] [PubMed] [Google Scholar]
- 10.Mathes B.M., Morabito D.M., Schmidt N.B. Epidemiological and clinical gender differences in OCD. Curr Psychiatry Rep. 2019;21:36. doi: 10.1007/s11920-019-1015-2. [DOI] [PubMed] [Google Scholar]
- 11.Benatti B., Girone N., Celebre L., Vismara M., Hollander E., Fineberg N.A., et al. The role of gender in a large international OCD sample: A Report from the International College of Obsessive-Compulsive Spectrum Disorders (ICOCS) Network. Compr Psychiatry. 2022;116 doi: 10.1016/j.comppsych.2022.152315. [DOI] [PubMed] [Google Scholar]
- 12.Baron-Cohen S., Lombardo M.V., Auyeung B., Ashwin E., Chakrabarti B., Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bellis M.D., Keshavan M.S., Beers S.R., Hall J., Frustaci K., Masalehdan A., et al. Sex differences in brain maturation during childhood and adolescence. Cereb Cortex. 2001;11:552–557. doi: 10.1093/cercor/11.6.552. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie S.J., Cox S.R., Shen X., Lombardo M.V., Reus L.M., Alloza C., et al. Sex differences in the adult human brain: Evidence from 5216 UK Biobank participants. Cereb Cortex. 2018;28:2959–2975. doi: 10.1093/cercor/bhy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sowell E.R., Peterson B.S., Kan E., Woods R.P., Yoshii J., Bansal R., et al. Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cereb Cortex. 2007;17:1550–1560. doi: 10.1093/cercor/bhl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good C.D., Johnsrude I., Ashburner J., Henson R.N., Friston K.J., Frackowiak R.S. Cerebral asymmetry and the effects of sex and handedness on brain structure: A voxel-based morphometric analysis of 465 normal adult human brains. Neuroimage. 2001;14:685–700. doi: 10.1006/nimg.2001.0857. [DOI] [PubMed] [Google Scholar]
- 17.Luders E., Narr K.L., Thompson P.M., Woods R.P., Rex D.E., Jancke L., et al. Mapping cortical gray matter in the young adult brain: Effects of gender. Neuroimage. 2005;26:493–501. doi: 10.1016/j.neuroimage.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Koolschijn P.C.M.P., Crone E.A. Sex differences and structural brain maturation from childhood to early adulthood. Dev Cogn Neurosci. 2013;5:106–118. doi: 10.1016/j.dcn.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satterthwaite T.D., Vandekar S., Wolf D.H., Ruparel K., Roalf D.R., Jackson C., et al. Sex differences in the effect of puberty on hippocampal morphology. J Am Acad Child Adolesc Psychiatry. 2014;53:341–350.e1. doi: 10.1016/j.jaac.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wierenga L.M., Sexton J.A., Laake P., Giedd J.N., Tamnes C.K., Pediatric Imaging, Neurocognition, and Genetics Study A key characteristic of sex differences in the developing brain: Greater variability in brain structure of boys than girls. Cereb Cortex. 2018;28:2741–2751. doi: 10.1093/cercor/bhx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goddings A.L., Mills K.L., Clasen L.S., Giedd J.N., Viner R.M., Blakemore S.J. The influence of puberty on subcortical brain development. Neuroimage. 2014;88:242–251. doi: 10.1016/j.neuroimage.2013.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele C.J., Chakravarty M.M. Gray-matter structural variability in the human cerebellum: Lobule-specific differences across sex and hemisphere. Neuroimage. 2018;170:164–173. doi: 10.1016/j.neuroimage.2017.04.066. [DOI] [PubMed] [Google Scholar]
- 23.Shah N.M., Pisapia D.J., Maniatis S., Mendelsohn M.M., Nemes A., Axel R. Visualizing sexual dimorphism in the brain. Neuron. 2004;43:313–319. doi: 10.1016/j.neuron.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Yang C.F., Shah N.M. Representing sex in the brain, one module at a time. Neuron. 2014;82:261–278. doi: 10.1016/j.neuron.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy M.M., Arnold A.P. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naqvi S., Godfrey A.K., Hughes J.F., Goodheart M.L., Mitchell R.N., Page D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science. 2019;365 doi: 10.1126/science.aaw7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Adhya D., Swarup V., Nagy R., Dutan L., Shum C., Valencia-Alarcón E.P., et al. Atypical neurogenesis in induced pluripotent stem cells from autistic individuals. Biol Psychiatry. 2021;89:486–496. doi: 10.1016/j.biopsych.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pașca S.P., Arlotta P., Bateup H.S., Camp J.G., Cappello S., Gage F.H., et al. A nomenclature consensus for nervous system organoids and assembloids. Nature. 2022;609:907–910. doi: 10.1038/s41586-022-05219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy M.M., Arnold A.P., Ball G.F., Blaustein J.D., De Vries G.J. Sex differences in the brain: The not so inconvenient truth. J Neurosci. 2012;32:2241–2247. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker J.B., Koob G.F. Sex differences in animal models: Focus on addiction. Pharmacol Rev. 2016;68:242–263. doi: 10.1124/pr.115.011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sisk C.L., Zehr J.L. Pubertal hormones organize the adolescent brain and behavior. Front Neuroendocrinol. 2005;26:163–174. doi: 10.1016/j.yfrne.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Werner R.J., Schultz B.M., Huhn J.M., Jelinek J., Madzo J., Engel N. Sex chromosomes drive gene expression and regulatory dimorphisms in mouse embryonic stem cells. Biol Sex Differ. 2017;8:28. doi: 10.1186/s13293-017-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S., Seidlitz J., Blumenthal J.D., Clasen L.S., Raznahan A. Integrative structural, functional, and transcriptomic analyses of sex-biased brain organization in humans. Proc Natl Acad Sci USA. 2020;117:18788–18798. doi: 10.1073/pnas.1919091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichmüller O.L., Knoblich J.A. Human cerebral organoids – A new tool for clinical neurology research. Nat Rev Neurol. 2022;18:661–680. doi: 10.1038/s41582-022-00723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genzel L. How to control behavioral studies for rodents-Don’t project human thoughts onto them. eNeuro. 2021;8 doi: 10.1523/ENEURO.0456-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanderhaeghen P., Polleux F. Developmental mechanisms underlying the evolution of human cortical circuits. Nat Rev Neurosci. 2023;24:213–232. doi: 10.1038/s41583-023-00675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herculano-Houzel S. The human brain in numbers: A linearly scaled-up primate brain. Front Hum Neurosci. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutsler J.J., Lee D.G., Porter K.K. Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 2005;1052:71–81. doi: 10.1016/j.brainres.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Bakken T.E., Jorstad N.L., Hu Q., Lake B.B., Tian W., Kalmbach B.E., et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature. 2021;598:111–119. doi: 10.1038/s41586-021-03465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khrameeva E., Kurochkin I., Han D., Guijarro P., Kanton S., Santel M., et al. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 2020;30:776–789. doi: 10.1101/gr.256958.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puts D., Motta-Mena N.V. Is human brain masculinization estrogen receptor-mediated? Reply to Luoto and Rantala. Horm Behav. 2018;97:3–4. doi: 10.1016/j.yhbeh.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Werling D.M., Geschwind D.H. Sex differences in autism spectrum disorders. Curr Opin Neurol. 2013;26:146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnard-Brak L., Richman D., Almekdash M.H. How many girls are we missing in ASD? An examination from a clinic- and community-based sample. Adv Autism. 2019;5:214–224. [Google Scholar]
- 45.Russell G., Steer C., Golding J. Social and demographic factors that influence the diagnosis of autistic spectrum disorders. Soc Psychiatry Psychiatr Epidemiol. 2011;46:1283–1293. doi: 10.1007/s00127-010-0294-z. [DOI] [PubMed] [Google Scholar]
- 46.Loomes R., Hull L., Mandy W.P.L. What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J Am Acad Child Adolesc Psychiatry. 2017;56:466–474. doi: 10.1016/j.jaac.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Baron-Cohen S., Auyeung B., Nørgaard-Pedersen B., Hougaard D.M., Abdallah M.W., Melgaard L., et al. Elevated fetal steroidogenic activity in autism. Mol Psychiatry. 2015;20:369–376. doi: 10.1038/mp.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erdogan M.A., Bozkurt M.F., Erbas O. Effects of prenatal testosterone exposure on the development of autism-like behaviours in offspring of Wistar rats. Int J Dev Neurosci. 2023;83:201–215. doi: 10.1002/jdn.10248. [DOI] [PubMed] [Google Scholar]
- 49.Baron-Cohen S., Tsompanidis A., Auyeung B., Nørgaard-Pedersen B., Hougaard D.M., Abdallah M., et al. Foetal oestrogens and autism. Mol Psychiatry. 2020;25:2970–2978. doi: 10.1038/s41380-019-0454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wigdor E.M., Weiner D.J., Grove J., Fu J.M., Thompson W.K., Carey C.E., et al. The female protective effect against autism spectrum disorder. Cell Genomics. 2022;2 doi: 10.1016/j.xgen.2022.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werling D.M., Geschwind D.H. Recurrence rates provide evidence for sex-differential, familial genetic liability for autism spectrum disorders in multiplex families and twins. Mol Autism. 2015;6:27. doi: 10.1186/s13229-015-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.San Roman A.K., Godfrey A.K., Skaletsky H., Bellott D.W., Groff A.F., Harris H.L., et al. The human inactive X chromosome modulates expression of the active X chromosome. Cell Genomics. 2023;3 doi: 10.1016/j.xgen.2023.100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green T., Flash S., Reiss A.L. Sex differences in psychiatric disorders: What we can learn from sex chromosome aneuploidies. Neuropsychopharmacology. 2019;44:9–21. doi: 10.1038/s41386-018-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carney R.M., Wolpert C.M., Ravan S.A., Shahbazian M., Ashley-Koch A., Cuccaro M.L., et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 55.Hagerman R., Hoem G., Hagerman P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1:12. doi: 10.1186/2040-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adegbola A., Gao H., Sommer S., Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- 57.Machado C.O.F., Griesi-Oliveira K., Rosenberg C., Kok F., Martins S., Passos-Bueno M.R., Sertie A.L. Collybistin binds and inhibits mTORC1 signaling: A potential novel mechanism contributing to intellectual disability and autism. Eur J Hum Genet. 2016;24:59–65. doi: 10.1038/ejhg.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fassio A., Patry L., Congia S., Onofri F., Piton A., Gauthier J., et al. SYN1 loss-of-function mutations in autism and partial epilepsy cause impaired synaptic function. Hum Mol Genet. 2011;20:2297–2307. doi: 10.1093/hmg/ddr122. [DOI] [PubMed] [Google Scholar]
- 59.Bae Y.J., Zeidler R., Baber R., Vogel M., Wirkner K., Loeffler M., et al. Reference intervals of nine steroid hormones over the life-span analyzed by LC-MS/MS: Effect of age, gender, puberty, and oral contraceptives. J Steroid Biochem Mol Biol. 2019;193 doi: 10.1016/j.jsbmb.2019.105409. [DOI] [PubMed] [Google Scholar]
- 60.Hines M., Constantinescu M., Spencer D. Early androgen exposure and human gender development. Biol Sex Differ. 2015;6:3. doi: 10.1186/s13293-015-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaludjerovic J., Ward W.E. The interplay between estrogen and fetal adrenal cortex. J Nutr Metab. 2012;2012 doi: 10.1155/2012/837901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava D.P., Penzes P. Rapid estradiol modulation of neuronal connectivity and its implications for disease. Front Endocrinol. 2011;2:77. doi: 10.3389/fendo.2011.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srivastava D.P., Evans P.D. G-protein oestrogen receptor 1: trials and tribulations of a membrane oestrogen receptor. J Neuroendocrinol. 2013;25:1219–1230. doi: 10.1111/jne.12071. [DOI] [PubMed] [Google Scholar]
- 64.Davey R.A., Grossmann M. Androgen receptor structure, function and biology: From bench to bedside. Clin Biochem Rev. 2016;37:3–15. [PMC free article] [PubMed] [Google Scholar]
- 65.Arnold A.P. The organizational–activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 2009;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopes-Ramos C.M., Chen C.Y., Kuijjer M.L., Paulson J.N., Sonawane A.R., Fagny M., et al. Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi L., Zhang Z., Su B. Sex biased gene expression profiling of human brains at major developmental stages. Sci Rep. 2016;6 doi: 10.1038/srep21181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnold A.P. X chromosome agents of sexual differentiation. Nat Rev Endocrinol. 2022;18:574–583. doi: 10.1038/s41574-022-00697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tran N., Broun A., Ge K. Lysine demethylase KDM6A in differentiation, development, and cancer. Mol Cell Biol. 2020;40 doi: 10.1128/MCB.00341-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nugent B.M., O’Donnell C.M., Epperson C.N., Bale T.L. Placental H3K27me3 establishes female resilience to prenatal insults. Nat Commun. 2018;9:2555. doi: 10.1038/s41467-018-04992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellott D.W., Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Cho T.J., et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasegawa Y., Hojo Y., Kojima H., Ikeda M., Hotta K., Sato R., et al. Estradiol rapidly modulates synaptic plasticity of hippocampal neurons: Involvement of kinase networks. Brain Res. 2015;1621:147–161. doi: 10.1016/j.brainres.2014.12.056. [DOI] [PubMed] [Google Scholar]
- 73.Hatanaka Y., Hojo Y., Mukai H., Murakami G., Komatsuzaki Y., Kim J., et al. Rapid increase of spines by dihydrotestosterone and testosterone in hippocampal neurons: Dependence on synaptic androgen receptor and kinase networks. Brain Res. 2015;1621:121–132. doi: 10.1016/j.brainres.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 74.Tozzi A., Bellingacci L., Pettorossi V.E. Rapid estrogenic and androgenic neurosteroids effects in the induction of long-term synaptic changes: Implication for early memory formation. Front Neurosci. 2020;14 doi: 10.3389/fnins.2020.572511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moffat S.D., Zonderman A.B., Metter E.J., Blackman M.R., Harman S.M., Resnick S.M. Longitudinal assessment of serum free testosterone concentration predicts memory performance and cognitive status in elderly men. J Clin Endocrinol Metab. 2002;87:5001–5007. doi: 10.1210/jc.2002-020419. [DOI] [PubMed] [Google Scholar]
- 76.Ooishi Y., Kawato S., Hojo Y., Hatanaka Y., Higo S., Murakami G., et al. Modulation of synaptic plasticity in the hippocampus by hippocampus-derived estrogen and androgen. J Steroid Biochem Mol Biol. 2012;131:37–51. doi: 10.1016/j.jsbmb.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 77.Di Mauro M., Tozzi A., Calabresi P., Pettorossi V.E., Grassi S. Different synaptic stimulation patterns influence the local androgenic and estrogenic neurosteroid availability triggering hippocampal synaptic plasticity in the male rat. Eur J Neurosci. 2017;45:499–509. doi: 10.1111/ejn.13455. [DOI] [PubMed] [Google Scholar]
- 78.Hatanaka Y., Mukai H., Mitsuhashi K., Hojo Y., Murakami G., Komatsuzaki Y., et al. Androgen rapidly increases dendritic thorns of CA3 neurons in male rat hippocampus. Biochem Biophys Res Commun. 2009;381:728–732. doi: 10.1016/j.bbrc.2009.02.130. [DOI] [PubMed] [Google Scholar]
- 79.Fester L., Zhou L., Ossig C., Labitzke J., Bläute C., Bader M., et al. Synaptopodin is regulated by aromatase activity. J Neurochem. 2017;140:126–139. doi: 10.1111/jnc.13889. [DOI] [PubMed] [Google Scholar]
- 80.Vierk R., Glassmeier G., Zhou L., Brandt N., Fester L., Dudzinski D., et al. Aromatase inhibition abolishes LTP generation in female but not in male mice. J Neurosci. 2012;32:8116–8126. doi: 10.1523/JNEUROSCI.5319-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finney C.A., Shvetcov A., Westbrook R.F., Jones N.M., Morris M.J. The role of hippocampal estradiol in synaptic plasticity and memory: A systematic review. Front Neuroendocrinol. 2020;56 doi: 10.1016/j.yfrne.2019.100818. [DOI] [PubMed] [Google Scholar]
- 82.McCarthy M.M. In: Brain Mapping. Toga A.W., editor. Academic Press; Waltham: 2015. Brain sex differences; pp. 27–35. [Google Scholar]
- 83.Davis E.C., Popper P., Gorski R.A. The role of apoptosis in sexual differentiation of the rat sexually dimorphic nucleus of the preoptic area. Brain Res. 1996;734:10–18. [PubMed] [Google Scholar]
- 84.Ibanez M.A., Gu G., Simerly R.B. Target-dependent sexual differentiation of a limbic-hypothalamic neural pathway. J Neurosci. 2001;21:5652–5659. doi: 10.1523/JNEUROSCI.21-15-05652.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gata-Garcia A., Porat A., Brimberg L., Volpe B.T., Huerta P.T., Diamond B. Contributions of sex chromosomes and gonadal hormones to the male bias in a maternal antibody-induced model of autism spectrum disorder. Front Neurol. 2021;12 doi: 10.3389/fneur.2021.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cisternas C.D., Cabrera Zapata L.E., Mir F.R., Scerbo M.J., Arevalo M.A., Garcia-Segura L.M., Cambiasso M.J. Estradiol-dependent axogenesis and Ngn3 expression are determined by XY sex chromosome complement in hypothalamic neurons. Sci Rep. 2020;10:8223. doi: 10.1038/s41598-020-65183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carruth L.L., Reisert I., Arnold A.P. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–934. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- 88.Sertel S.M., Blumenstein W., Mandad S., Shomroni O., Salinas G., Rizzoli S.O. Differences in synaptic vesicle pool behavior between male and female hippocampal cultured neurons. Sci Rep. 2021;11 doi: 10.1038/s41598-021-96846-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mong J.A., Glaser E., McCarthy M.M. Gonadal steroids promote glial differentiation and alter neuronal morphology in the developing hypothalamus in a regionally specific manner. J Neurosci. 1999;19:1464–1472. doi: 10.1523/JNEUROSCI.19-04-01464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carrel L., Brown C.J. When the Lyon(ized chromosome) roars: Ongoing expression from an inactive X chromosome. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tukiainen T., Villani A.-C., Yen A., Rivas M.A., Marshall J.L., Satija R., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boroviak T., Nichols J. Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development. 2017;144:175–186. doi: 10.1242/dev.145177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S.P., Codeluppi S., et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minkovsky A., Patel S., Plath K. Concise review: Pluripotency and the transcriptional inactivation of the female Mammalian X chromosome. Stem Cells. 2012;30:48–54. doi: 10.1002/stem.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janiszewski A., Talon I., Chappell J., Collombet S., Song J., De Geest N., et al. Dynamic reversal of random X-Chromosome inactivation during iPSC reprogramming. Genome Res. 2019;29:1659–1672. doi: 10.1101/gr.249706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marks H., Chow J.C., Denissov S., Françoijs K.J., Brockdorff N., Heard E., Stunnenberg H.G. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loda A., Heard E. Xist RNA in action: Past, present, and future. PLOS Genet. 2019;15 doi: 10.1371/journal.pgen.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sahakyan A., Yang Y., Plath K. The role of Xist in X-chromosome dosage compensation. Trends Cell Biol. 2018;28:999–1013. doi: 10.1016/j.tcb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vallot C., Patrat C., Collier A.J., Huret C., Casanova M., Liyakat Ali T.M., et al. XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell. 2017;20:102–111. doi: 10.1016/j.stem.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 102.Trabzuni D., Ramasamy A., Imran S., Walker R., Smith C., Weale M.E., et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nguyen T.A., Wu K., Pandey S., Lehr A.W., Li Y., Bemben M.A., et al. A cluster of autism-associated variants on X-linked NLGN4X functionally resemble NLGN4Y. Neuron. 2020;106:759–768.e7. doi: 10.1016/j.neuron.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laumonnier F., Bonnet-Brilhault F., Gomot M., Blanc R., David A., Moizard M.P., et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Davenport M.L., Cornea E., Xia K., Crowley J.J., Halvorsen M.W., Goldman B.D., et al. Altered brain structure in infants with Turner syndrome. Cereb Cortex. 2020;30:587–596. doi: 10.1093/cercor/bhz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang X., Hong D., Ma S., Ward T., Ho M., Pattni R., et al. Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage. Proc Natl Acad Sci USA. 2020;117:4864–4873. doi: 10.1073/pnas.1910003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Urbach A., Benvenisty N. Studying early lethality of 45,XO (Turner’s syndrome) embryos using human embryonic stem cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kirk I.K., Weinhold N., Belling K., Skakkebæk N.E., Jensen T.S., Leffers H., et al. Chromosome-wise protein interaction patterns and their impact on functional implications of large-scale genomic aberrations. Cell Syst. 2017;4:357–364.e3. doi: 10.1016/j.cels.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 109.Bojesen A., Juul S., Gravholt C.H. Prenatal and postnatal prevalence of Klinefelter syndrome: A national registry study. J Clin Endocrinol Metab. 2003;88:622–626. doi: 10.1210/jc.2002-021491. [DOI] [PubMed] [Google Scholar]
- 110.Deng X., Berletch J.B., Nguyen D.K., Disteche C.M. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Høst C., Skakkebæk A., Groth K.A., Bojesen A. The role of hypogonadism in Klinefelter syndrome. Asian J Androl. 2014;16:185–191. doi: 10.4103/1008-682X.122201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Skakkebaek A., Gravholt C.H., Chang S., Moore P.J., Wallentin M. Psychological functioning, brain morphology, and functional neuroimaging in Klinefelter syndrome. Am J Med Genet C Semin Med Genet. 2020;184:506–517. doi: 10.1002/ajmg.c.31806. [DOI] [PubMed] [Google Scholar]
- 113.Cederlöf M., Ohlsson Gotby A., Larsson H., Serlachius E., Boman M., Långström N., et al. Klinefelter syndrome and risk of psychosis, autism and ADHD. J Psychiatr Res. 2014;48:128–130. doi: 10.1016/j.jpsychires.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 114.Viswanathan V., Eugster E.A. Etiology and treatment of hypogonadism in adolescents. Pediatr Clin North Am. 2011;58:1181–1200. doi: 10.1016/j.pcl.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foland-Ross L.C., Ghasemi E., Lozano Wun V.L., Aye T., Kowal K., Ross J., Reiss A.L. Executive dysfunction in Klinefelter syndrome: Associations with brain activation and testicular failure. J Clin Endocrinol Metab. 2023;109:e88–e95. doi: 10.1210/clinem/dgad487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paulsen B., Velasco S., Kedaigle A.J., Pigoni M., Quadrato G., Deo A.J., et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–273. doi: 10.1038/s41586-021-04358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Giandomenico S.L., Mierau S.B., Gibbons G.M., Wenger L.M.D., Masullo L., Sit T., et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gordon A., Yoon S.J., Tran S.S., Makinson C.D., Park J.Y., Andersen J., et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci. 2021;24:331–342. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samarasinghe R.A., Miranda O.A., Buth J.E., Mitchell S., Ferando I., Watanabe M., et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat Neurosci. 2021;24:1488–1500. doi: 10.1038/s41593-021-00906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kang Y., Zhou Y., Li Y., Han Y., Xu J., Niu W., et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat Neurosci. 2021;24:1377–1391. doi: 10.1038/s41593-021-00913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kelava I., Chiaradia I., Pellegrini L., Kalinka A.T., Lancaster M.A. Androgens increase excitatory neurogenic potential in human brain organoids. Nature. 2022;602:112–116. doi: 10.1038/s41586-021-04330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patel S., Bonora G., Sahakyan A., Kim R., Chronis C., Langerman J., et al. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 2017;18:54–67. doi: 10.1016/j.celrep.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bansal P., Ahern D.T., Kondaveeti Y., Qiu C.W., Pinter S.F. Contiguous erosion of the inactive X in human pluripotency concludes with global DNA hypomethylation. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., et al. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 127.Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beacher F.D.C.C., Radulescu E., Minati L., Baron-Cohen S., Lombardo M.V., Lai M.C., et al. Sex differences and autism: Brain function during verbal fluency and mental rotation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hernandez L.M. Sex-differential neuroanatomy in autism: A shift toward male-characteristic brain structure. Am J Psychiatry. 2023;180:8–10. doi: 10.1176/appi.ajp.20220939. [DOI] [PubMed] [Google Scholar]
- 130.Ecker C., Bookheimer S.Y., Murphy D.G.M. Neuroimaging in autism spectrum disorder: Brain structure and function across the lifespan. Lancet Neurol. 2015;14:1121–1134. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 131.Birey F., Li M.Y., Gordon A., Thete M.V., Valencia A.M., Revah O., et al. Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell. 2022;29:248–264.e7. doi: 10.1016/j.stem.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 132.Xu Z.X., Kim G.H., Tan J.W., Riso A.E., Sun Y., Xu E.Y., et al. Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat Commun. 2020;11:1797. doi: 10.1038/s41467-020-15530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Su L., Zhang M., Ji F., Zhao J., Wang Y., Wang W., et al. Microglia homeostasis mediated by epigenetic ARID1A regulates neural progenitor cells response and leads to autism-like behaviors. Mol Psychiatry. 2022 doi: 10.1038/s41380-022-01703-7. [published online Jul 20] [DOI] [PubMed] [Google Scholar]
- 134.Haenseler W., Sansom S.N., Buchrieser J., Newey S.E., Moore C.S., Nicholls F.J., et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Rep. 2017;8:1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Popova G., Soliman S.S., Kim C.N., Keefe M.G., Hennick K.M., Jain S., et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell. 2021;28:2153–2166.e6. doi: 10.1016/j.stem.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Qian X., Su Y., Adam C.D., Deutschmann A.U., Pather S.R., Goldberg E.M., et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26:766–781.e9. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Studholme C., Kroenke C.D., Dighe M. Motion corrected MRI differentiates male and female human brain growth trajectories from mid-gestation. Nat Commun. 2020;11:3038. doi: 10.1038/s41467-020-16763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gitau R., Adams D., Fisk N.M., Glover V. Fetal plasma testosterone correlates positively with cortisol. Arch Dis Child Fetal Neonatal Ed. 2005;90:F166–F169. doi: 10.1136/adc.2004.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.DeCasien A.R., Guma E., Liu S., Raznahan A. Sex differences in the human brain: A roadmap for more careful analysis and interpretation of a biological reality. Biol Sex Differ. 2022;13:43. doi: 10.1186/s13293-022-00448-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lövdén M., Herlitz A., Schellenbach M., Grossman-Hutter B., Krüger A., Lindenberger U. Quantitative and qualitative sex differences in spatial navigation. Scand J Psychol. 2007;48:353–358. doi: 10.1111/j.1467-9450.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- 145.Mogil J.S. Qualitative sex differences in pain processing: Emerging evidence of a biased literature. Nat Rev Neurosci. 2020;21:353–365. doi: 10.1038/s41583-020-0310-6. [DOI] [PubMed] [Google Scholar]
- 146.De Vries G.J. Minireview: Sex differences in adult and developing brains: Compensation, compensation, compensation. Endocrinology. 2004;145:1063–1068. doi: 10.1210/en.2003-1504. [DOI] [PubMed] [Google Scholar]
- 147.Shors T.J., Chua C., Falduto J. Sex differences and opposite effects of stress on dendritic spine density in the male versus female hippocampus. J Neurosci. 2001;21:6292–6297. doi: 10.1523/JNEUROSCI.21-16-06292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vignoli A., Fabio R.A., La Briola F., Giannatiempo S., Antonietti A., Maggiolini S., Canevini M.P. Correlations between neurophysiological, behavioral, and cognitive function in Rett syndrome. Epilepsy Behav. 2010;17:489–496. doi: 10.1016/j.yebeh.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 149.Huddleston L.B., Visootsak J., Sherman S.L. Cognitive aspects of fragile X syndrome. Wiley Interdiscip Rev Cogn Sci. 2014;5:501–508. doi: 10.1002/wcs.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Boada R., Janusz J., Hutaff-Lee C., Tartaglia N. The cognitive phenotype in Klinefelter syndrome: A review of the literature including genetic and hormonal factors. Dev Disabil Res Rev. 2009;15:284–294. doi: 10.1002/ddrr.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hong D., Scaletta Kent J., Kesler S. Cognitive profile of Turner syndrome. Dev Disabil Res Rev. 2009;15:270–278. doi: 10.1002/ddrr.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wolstencroft J., Skuse D. Social skills and relationships in Turner syndrome. Curr Opin Psychiatry. 2019;32:85–91. doi: 10.1097/YCO.0000000000000472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.