Abstract
Retroviruses preferentially package full-length genomic RNA over spliced viral messages. For most retroviruses, this preference is likely due to the absence of all or part of the packaging signal on subgenomic RNAs. In avian leukosis-sarcoma virus, however, we have shown that the minimal packaging signal, MΨ, is located upstream of the 5′ splice site and therefore is present on both genomic and spliced RNAs. We now show that an MΨ-containing heterologous RNA is packaged only 2.6-fold less efficiently than genomic Rous sarcoma virus RNA. Thus, few additional packaging sequences and/or structures exist outside of MΨ. In contrast, we found that env mRNA is not efficiently packaged. These results indicate that either MΨ is not functional on this RNA or the RNA is somehow segregated from the packaging machinery. Finally, deletion of sequences from the 3′ end of MΨ was found to reduce the packaging efficiency of heterologous RNAs.
Packaging of the full-length genomic RNA is a necessary step in the assembly of replication-competent retroviral particles. This process is thought to involve a specific binding event between viral proteins and a signal on the viral genome, alternatively referred to as Ψ, for packaging signal, or E, for encapsidation signal. Core packaging signals have been identified in the 5′ end of most retroviral genomes, and stem-loop structures formed within these sequences have been shown to play a crucial role in protein recognition (reviewed in references 6 and 10). The regions of the viral protein responsible for packaging in retroviruses are found exclusively on the Gag polyprotein (48). In particular, the Cys-His box RNA binding motifs and surrounding basic regions in the C-terminal domain of Gag have been shown to play a role in both specific and nonspecific binding of the viral RNA (10).
Of the simple retroviruses, the best-studied packaging signals are those of avian leukosis-sarcoma virus (ALSV), murine leukemia virus (MLV), and spleen necrosis virus (SNV). The core packaging signal for these viruses is located between the R/U5 region and the gag initiation codon. In ALSV, we have previously identified a 160-nucleotide (nt) signal, MΨ, located between the primer binding site and the gag initiation codon, that is sufficient to confer packaging of a heterologous RNA (7). Additionally, a deletion of 153 nt in the leader of ALSV which encompasses the first 99 nt of MΨ causes a reduction in packaging of genomic RNA (55). In MLV a 350-nt region, Ψ, located between the 5′ splice site (5′ss) and the gag initiation codon, is necessary for packaging of a retroviral vector (41). The packaging efficiency is increased, however, in the presence of sequences from the 5′ end of the gag gene (1, 3, 8). Similarly, in SNV a core 185-nt packaging signal, E, is located between the 5′ss and the gag initiation codon (59). In contrast to the relatively discrete packaging signals of these simple retroviruses, the signal of complex retroviruses appears to be dispersed throughout the 5′ end of the virus (reviewed in reference 10). Efficient human immunodeficiency virus type 1 (HIV-1) vector packaging, for example, requires all of the regions from the 5′ end of the RNA including some gag sequences (11, 14, 42, 49, 52).
Secondary structures formed within retroviral encapsidation sequences have been shown to play a critical role in packaging. In both MLV and SNV, two stem-loop structures, each containing a GACG tetraloop, are important for efficient packaging (27, 44, 45, 61). Computer models of the ALSV packaging signal also predict two stem-loops structures, named O3 and L3. Unlike the stem-loops of MLV and SNV, however, neither O3 nor L3 contains a GACG tetraloop. Mutating either half of the O3 stem greatly reduces the packaging efficiency, while compensatory mutations that restore the base pairing restore packaging to almost wild-type (wt) levels (7, 23, 35). In contrast, mutants retaining neither the primary sequence nor secondary structure of L3 stem were shown to be efficiently packaged (23). These mutants also contained a spontaneous point mutation in the O3 region which appeared to compensate for the mutations in L3.
In retroviruses, spliced RNAs are not packaged at high levels (12, 40, 52). In SNV, MLV, and HIV-1, this can be explained by the absence of all or some of the packaging signal on subgenomic messages. In contrast to these viruses, the 5′ss in ALSV is located downstream of the gag start codon (60). Thus, MΨ is found on both genomic and spliced RNAs. We previously hypothesized that additional ALSV packaging signals might preside in intronic regions that augment packaging of the unspliced genomic RNA (7). In the present study, we addressed this possibility by comparing the packaging efficiency of a heterologous RNA containing MΨ with that of wt Rous sarcoma virus (RSV) RNA. We found that the heterologous RNA is packaged only 2.6-fold less efficiently than genomic RSV RNA, suggesting that few additional packaging sequences and/or structures are likely to exist outside of MΨ. In contrast, we found that env mRNA is not packaged as efficiently as the MΨ-containing heterologous RNA. In this report, we also show that heterologous RNAs with deletions in the 3′ end of MΨ, including sequences from the L3 stem, are packaged less efficiently than RNAs containing the entire signal.
MATERIALS AND METHODS
Plasmids.
Deletions were made in the 3′ end of the Prague C strain MΨ sequence (Genbank accession no. J02342, nt 389 to 548) by PCR using pASY191 as a template. pASY191 is a clone in pBluescript II SK+ (Stratagene) that contains MΨ (7). The sequence corresponding to the first 140 nt of MΨ was amplified by using oligonucleotides 5′Mlu1 (5′-CGACGCGTGATCCTGCCCTCATCCGTCTCGCTTATTC-3′) and 3′140Mlu1 (5′-CGACGCGTGAGTTCTCGGTAGGGTATCTGGGCTC-3′). The underlined sequence is the restriction site for MluI. The sequence corresponding to the first 120 nt of MΨ was amplified by using oligonucleotides 5′Mlu1 and 3′120Mlu1 (5′-CGACGCGTGGGCTCCCTGCAGTAGAGCTCCCTCC-3′). The MluI-digested PCR products were inserted into the unique MluI site of pASY161, a pCMVneo derivative, in which an MluI linker was inserted in the unique SmaI site, downstream of the neomycin phosphotransferase gene (neo) (5, 7, 39). pASY194, which contains the entire 160 nt of MΨ inserted in the MluI site of pASY161, was used as the positive control (7). pASY161 with no retroviral sequences was used as a negative control.
For the experiments comparing the packaging efficiencies of CMVneo-MΨ and genomic RSV RNAs, pASY194 was used to express the heterologous RNA. The viral genomic RNA was expressed from plasmid pRCASBPpuro (a generous gift from Stephen Hughes, Frederick Cancer Research Center). This plasmid contains an infectious RSV proviral sequence (strain Schmidt-Ruppin A, with the exception of pol sequences, which are from the Bryan high-titer strain [25, 31, 50]) in which src has been replaced by the selectable marker, the puromycin acetyltransferase gene (puro). pRCASBPneo served as the template for producing virus for the measurement of env mRNA packaging. This plasmid is similar to pRCASBPpuro except that src has been replaced by neo (2).
pASY185 was used as the template for in vitro transcription of antisense neo probe. This plasmid contains a neo fragment cloned into pBluescript II SK+. pGEM1-GAPDH was used as the template for the antisense glyceraldehyde-3-phosphate dehydrogenase (gapdh) probe. This plasmid contains a fragment from pGAD-28, a chicken gapdh clone inserted in pGEM1 (Promega) (24, 57). pBSPuro, which contains a puro fragment inserted in pBluescript II SK+, was used as the template for the antisense puro probe. pENV3′SS was used as the template for the antisense 3′ss probe. This plasmid contains a fragment overlapping the 3′ss for env from pATV8R (34), an RSV clone, inserted into pBluescript SK+.
Cells and virus.
The quail packaging cell line Q2bn-4D (55) was grown in GM+D+CK (Ham’s F10 medium containing 10% tryptose phosphate broth, 5% calf serum [Gemini Bio-Products], 1% heat-inactivated chick serum [GibcoBRL], and 1% dimethyl sulfoxide). Cells were maintained in 6% CO2 at 37°C. Plasmids were transfected by the modified calcium phosphate method (17) on cells seeded in Dulbecco’s modified Eagle medium supplemented with 10% calf serum. Cells transfected with pASY161 derivatives were selected for drug resistance with GM+D+CK containing G418 (0.1 mg/ml). Cells transfected with pRCASBPpuro were selected for drug resistance with GM+D+CK containing puromycin (2.5 μg/ml). Cells transfected with both constructs were kept under both G418 and puromycin selection. Mass cultures of drug-resistant cells were obtained after approximately 3 weeks under selection. Infected QT6 cells (43) were used to assay the packaging of env mRNA. The virus used for infection was obtained from QT6 cells transfected with pRCASBPneo as described above. Ten milliliters of supernatant from a 100-mm-diameter plate of transfected cells was filtered through a 0.45-μm-pore-size filter and placed on fresh QT6 cells. These infected cells were then selected with G418 as described above, and mass cultures of drug-resistant cells were obtained.
Protein analysis.
For radiolabeling viruses, cells were plated at a density of 5 × 106 cells per 100-mm-diameter plate in GM+D+CK at least 18 h before labeling. The cells were washed twice with phosphate-buffered saline and once with serum-free Dulbecco’s modified Eagle medium minus methionine and cysteine (DME−Met−Cys). The cells were then labeled with 250 μCi of [35S]methionine (EXPRESS 35S protein labeling mix; NEN Research Products) in 2 ml of DME−Met−Cys. After five h of incubation at 37°C in 6% CO2, 3 ml of DME−Met−Cys supplemented with 10% dialyzed fetal bovine serum was added. The following day, supernatants were collected and labeled viral particles were concentrated by high-speed centrifugation through a 20% sucrose cushion. Half of the concentrated virion were set aside for RNase protection analysis (RPA); the remaining half was immunoprecipitated. The labeled viral particles were incubated in 1.0 ml of Ab buffer (20 mM Tris [pH 7.4], 50 mM NaCl, 1 mM EDTA [pH 8.0], 0.5% NP-40, 0.5% deoxycholic acid, 0.5% sodium dodecyl sulfate [SDS], 0.5% aprotinin) with 3 μl of polyclonal rabbit RSV PrB-specific antibody and 30 μl of protein A-Sepharose beads for 90 min at room temperature. The antigen-antibody complexes were washed twice in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% deoxycholic acid, 0.1% SDS, 0.5% aprotinin), once in high-salt buffer (10 mM Tris-HCl [pH 7.4], 2 M NaCl, 1% NP-40, 0.5% deoxycholic acid), and once more with RIPA buffer. The bound proteins were eluted in SDS sample buffer and loaded onto an SDS–12.5% polyacrylamide gel. Following electrophoresis, the gel was dried and, after an overnight exposure, scanned directly by a Molecular Dynamics PhosphorImager. Radioactive bands corresponding to the capsid (CA) protein were quantitated, in machine units, with ImageQuant (Molecular Dynamics) software.
RNA analysis.
In all packaging assays except where noted, RPAs were performed on viral and whole-cell lysates by using a Direct Protect kit (Ambion). For making the antisense neo probe for the heterologous packaging assay, pASY185 was linearized with RsrII and in vitro transcribed with T7 RNA polymerase to produce a probe which protects 166 nt of neo. For making the antisense neo probe for comparing heterologous and genomic RNA packaging, pASY185 was linearized with NcoI and in vitro transcribed with T7 RNA polymerase to produce a probe which protects 249 nt of neo. For making the antisense gapdh probe, pGEM1-GAPDH was linearized with HindIII and in vitro transcribed with T7 RNA polymerase to produce a probe which protects 169 nt of gapdh. For making the antisense puro probe to detect the genomic RSV RNA, pBSpuro was digested with BstEII and in vitro transcribed with T7 RNA polymerase, producing a probe which protects 195 nt of puro.
For RPA of cytoplasmic RNAs, lysates were obtained by treating pelleted cells in lysis buffer (50 mM Tris-Cl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40) and spinning out nuclei and cell debris. The cytoplasmic fraction was used in RPA using the Direct Protect kit. For making the antisense probe to the env 3′ss, pENV3′SS was digested with HindIII and in vitro transcribed with T3 RNA polymerase, producing a probe which protects 302 nt of the unspliced message and 221 nt of the spliced env mRNA.
Probes were gel purified on a 6% polyacrylamide gel. After RNase treatment, protected RNAs were separated on a 6% polyacrylamide gel. The dried gel was scanned directly by a Molecular Dynamics PhosphorImager after an overnight exposure. RNA bands were quantitated, in machine units, with ImageQuant software.
Calculation of packaging efficiency.
The packaging efficiency of each RNA was determined by calculating the amount of that RNA in virions (as measured by RPA) normalized to the level of that RNA in cells, relative to a cellular message, gapdh (as measured by RPA of whole-cell lysates). This calculation of the RNA was then normalized to the number of virions used in the viral RPA (as measured by RIPA). Note that while the antisense neo and puro probes are of different lengths and different specific activities, the normalization of the viral RNA to cellular RNA makes any adjustment for this difference unnecessary. To compare packaging efficiencies between different experiments, each time the assay was performed, the calculated packaging efficiencies were normalized to that of CMVneo-MΨ. In the cells expressing both RCASBPpuro and CMVneo-MΨ RNA, the packaging efficiency of the viral genomic RNA was normalized to that of CMVneo-MΨ in the same cells. The packaging efficiency of viral genomic RNA in control cells not expressing CMVneo-MΨ was normalized to the packaging efficiency of CMVneo-MΨ in control cells not expressing the viral genomic RNA.
To calculate the packaging efficiency of the full-length RNA relative to env mRNA, the level of each RNA in virions (as measured by RPA) was normalized to the level of that RNA in cytoplasm (as measured by RPA on cytoplasmic lysates). In each experiment, the calculated packaging efficiencies were normalized to that of env. Although the protected bands for the two species are of different sizes and specific activities, the normalization of the viral RNA to the cellular RNA makes adjustment for this difference unnecessary.
RESULTS
Deletions of the 3′ end of MΨ.
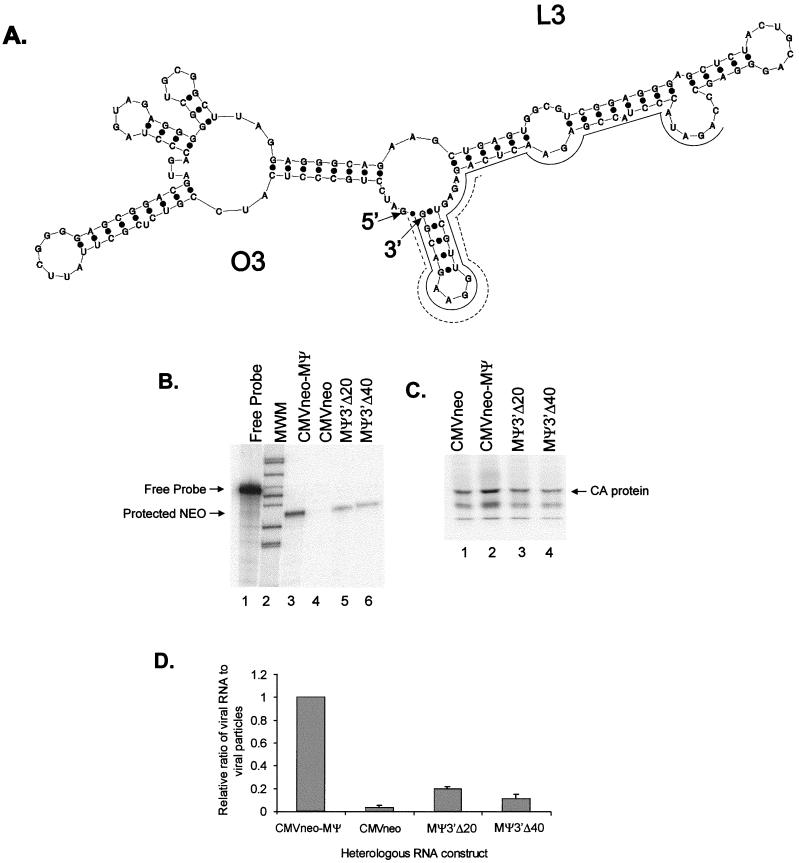
Sequences corresponding to the 3′ end of MΨ are predicted in computer models to be part of a large stem loop, L3, in two ALSV strains, Prague C (Fig. 1A) and Schmidt-Ruppin A (7, 23). Additionally, a computer model of the consensus sequence for 13 ALSV strains shows a similar structure (30). We previously reported that the deletion of 55 nt from the 3′ end of MΨ (MΨΔ3′) reduced packaging only sixfold in our heterologous system, indicating that no essential packaging sequences are found in this region (7).
FIG. 1.
(A) Predicted secondary structure of MΨ, obtained from the mfold computer program (32, 33, 63). The O3 and L3 stem-loop structures, as well as the 5′ and 3′ ends of the RNA, are indicated. Solid lines are shown next to sequences deleted in the MΨ3′Δ40 construct; dashed lines are shown next to sequences deleted in the MΨ3′Δ20 construct. (B) RPA of virions released from G418-resistant mass cultures of Q2bn cells transfected with plasmids expressing the heterologous RNA indicated above each lane. RNAs were protected with an antisense neo probe. The expected locations of free probe and of the protected neo bands are indicated. MWM, molecular weight markers. (C) RIPA of viral particles described for panel B. Proteins were precipitated with an α-PrB-specific antibody. The expected size of the CA band is indicated. (D) Average packaging efficiencies of the RNAs from three replications of the experiment relative to CMVneo-MΨ RNA. Error bars represent standard deviations. Packaging efficiencies were calculated as the ratio of neo RNA packaged into particles, as measured by RPA, to the number of viral particles, as measured by RIPA.
In light of this experiment, we hypothesized that mutants with smaller deletions in the 3′ end might confer packaging as efficiently as the full-length MΨ. To test this, we constructed two mutants, MΨ3′Δ20 and MΨ3′Δ40, containing 20- and 40-nt deletions, respectively, from the 3′ end of MΨ (Fig. 1A). We then measured the effect of these deletions on packaging in a heterologous packaging assay (7). The mutated packaging sequences were inserted downstream of the coding sequence for neo in pASY161, a derivative of pCMVneo (5, 7, 39). These plasmids were then transfected into the Q2bn avian packaging cell line (55), and mass cultures of G418-resistant cells were obtained. The heterologous RNAs were expressed at similar levels in the cells, as measured by RPA of cell lysates, using an antisense neo probe (data not shown). Packaging of the heterologous RNAs was detected by RPA of RNA from virions released from the cells, using the antisense neo probe (Fig. 1B). Virion levels were measured by RIPA (Fig. 1C). The packaging efficiency was calculated by normalizing the total heterologous RNA detected in virions to the number of viral particles. The average packaging efficiencies from three repetitions of the packaging assay are shown in Fig. 1D. Each MΨ mutant was packaged more efficiently than CMVneo, the heterologous RNA containing no retroviral sequences (Fig. 1B, lane 4; Fig. 1C lane 1), although neither was packaged as efficiently as the RNA containing the wt MΨ signal, CMVneo-MΨ. MΨ3′Δ20 had a 5-fold decrease in packaging relative to wt MΨ (Fig. 1B, lanes 3 and 5; Fig. 1C, lanes 2 and 3), while MΨ3′Δ40 had a 10-fold decrease in packaging relative to wt MΨ (Fig. 1B, lanes 3 and 6; Fig. 1C, lanes 2 and 4). Thus, while sequences or structures from the 3′ end of MΨ do not appear to be essential for packaging, their presence is necessary to maintain wt levels of packaging.
Packaging of genomic RSV RNA relative to CMVneo-MΨ.
To address the possibility that sequences outside of MΨ might augment packaging in ALSV, we designed an experiment which allows direct measurement of the packaging efficiency of wt replication-competent genomic RSV RNA relative to that of CMVneo-MΨ RNA. We expressed both RNAs in the same cell and determined how frequently each was packaged into virions released from the cells. Any increase in packaging of viral genomic RNA relative to that of CMVneo-MΨ RNA would be indicative that sequences outside of MΨ can enhance the packaging signal.
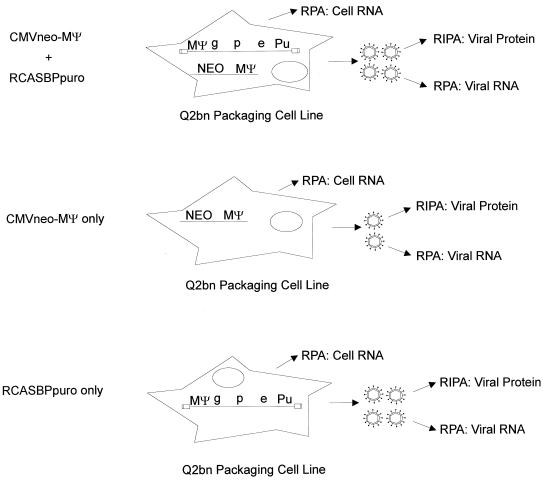
A schematic of the experiment is shown in Fig. 2. We transfected Q2bn packaging cells with both pASY194, the construct expressing CMVneo-MΨ, and pRCASBPpuro, which expresses a full-length infectious clone of RSV in which the src gene has been replaced by the selectable marker puro. Control cells were transfected with pASY194 only or pRCASBPpuro only. Cells were selected with the appropriate drug(s), and mass cultures of drug-resistant cells were obtained. RPAs were performed on cell lysates to determine the expression level of each RNA relative to a cellular RNA, gapdh (Fig. 3A). Cells were metabolically labeled with [35S]methionine, and labeled viral particles were concentrated. One-half of the collected particles was set aside for RPA (Fig. 3B); the remaining half was immunoprecipitated with antiviral antibody (Fig. 3C).
FIG. 2.
Schematic of the assay to determine the packaging efficiency of genomic RSV RNA relative to CMVneo-MΨ RNA. Q2bn packaging cells were transfected with both pASY194, which expressed CMVneo-MΨ, and pRCASBPpuro, which expresses a full-length infectious clone of RSV. Control cells were transfected with pASY194 only or pRCASBPpuro only. RPAs were performed on viral and whole-cell or cytoplasmic lysates. RIPA was performed on viral particles. g, gag gene encoding viral structural proteins; p, pol gene encoding reverse transcriptase and integrase; e, env gene encoding glycoproteins; Pu, puro gene encoding puromycin acetyltransferase; NEO, neo gene encoding neomycin phosphotransferase.
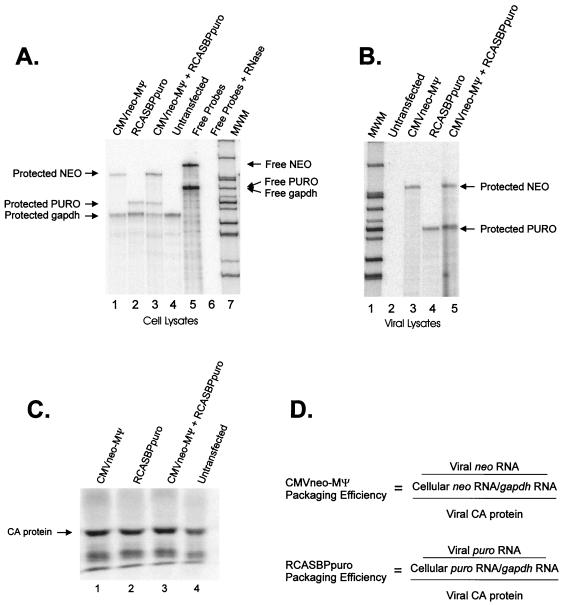
FIG. 3.
(A) RPA of whole-cell lysates from cells described in the legend to Fig. 2. The RNA expressed in the cells is indicated above each lane. Lane 4 contains lysates from untransfected Q2bn cells. The sample in lane 5 had no RNase added; the sample in lane 6 had no cell lysate added. CMVneo-MΨ was detected with an antisense neo probe. RCASBPpuro RNA was detected with an antisense puro probe. A cellular message, gapdh, was detected with antisense gapdh probe. The bands corresponding to free neo, puro, and gapdh probes, as well as the protected bands, are indicated. Free puro and gapdh probes are of similar size and appear as one band in this gel. MWM, molecular weight markers. (B) RPA of viral lysates. The RNA expressed in the cells from which the virions were released is indicated above each lane. The puro and neo probes described for panel A were also used in this assay. (C) RIPA of viral particles released from the cell lines, using PrB-specific antibody. The band corresponding to CA protein is indicated. (D) Formula used for calculating packaging efficiency of each RNA. The level of each RNA in virions was measured by RPA of viral lysates, shown in panel B. The level of the RNA relative to gapdh in cells was measured by RPA of cell lysates, shown in panel A. The number of viral particles was determined by RIPA, shown in panel C.
The formula used to calculate the packaging efficiency of the RNAs is shown in Fig. 3D. The amount of heterologous RNA or viral RNA detected in particles was normalized to the cellular level of that same RNA relative to gapdh RNA. This experiment was initially performed on total cell RNA. However, we later used cytoplasmic lysates and obtained similar results (data not shown). This RNA measurement was then normalized to the number of viral particles detected by RIPA (see Materials and Methods). The average packaging efficiencies from three repetitions of the experiment are shown in Table 1. When both RNAs were expressed in the same cell, the heterologous RNA was packaged only 2.7-fold less efficiently than the viral genomic RNA (Fig. 3A, lane 3; Fig. 3B, lane 5; Fig. 3C, lane 3). This result indicates that many of the sequences and/or structures involved in packaging are contained within MΨ, although sequences outside of the minimal packaging signal may enhance the signal in the full-length message. Similarly, the packaging efficiency of CMVneo-MΨ when expressed by itself in cells was only 3.1-fold less than the packaging efficiency of viral genomic RNA when expressed by itself in cells (Fig. 3A, lanes 1 and 2; Fig. 3B, lanes 3 and 4; Fig. 3C, lanes 1 and 2). Thus, it appears that the packaging machinery is not saturated in the cells expressing both RNAs and that the packaging rate of CMVneo-MΨ is unaffected by the presence or absence of competing RNAs.
TABLE 1.
Packaging of viral genomic RNA relative to CMVneo-MΨ RNAa
| RNA expressed in Q2bn cells | Packaging efficiency (mean ± SD)b
|
||
|---|---|---|---|
| CMVneo-MΨ | RCASBPpuro | RCASBPpuro adjusted for envc | |
| CMVneo-MΨ + RCASBPpuro | 1.00 | 2.76 ± 0.81 | 2.6 |
| CMVneo-MΨ | 1.00 | NDd | ND |
| RCASBPpuro | ND | 3.11 ± 0.13 | 2.9 |
Packaging efficiency was calculated as the ratio of neo RNA packaged in virions to the amount of neo RNA in the cell, as measured by RPA. This was normalized to the number of virions, as measured by RIPA. All packaging efficiencies were normalized to CMVneo-MΨ.
Results are from at least three independent experiments.
The packaging efficiency of the env mRNA is 1/16 that of the genomic RNA (Fig. 4B). To adjust for the packaging of env in the competition cells, we multiplied 2.76 by 15/16. The same calculation was performed on the control cells.
ND, not determined.
Packaging of env mRNA.
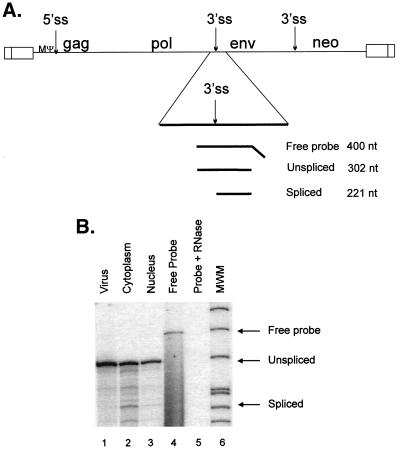
Since puro sequences are on both the genomic and subgenomic RNAs expressed from RCASBPpuro, it was important to determine whether subgenomic RNAs are efficiently packaged. This does not appear to be the case for retroviruses (12, 40, 62). However, since packaging of subgenomic RNA could alter our calculations in the previous assay, we have tested this directly in our system. Our lab had previously measured a less than sevenfold increase in packaging of ALSV genomic RNA relative to env mRNA in total cell RNA (4). Since viral assembly occurs in the cytoplasm, we hypothesized that measuring the ratio of the two RNAs in the cytoplasm would give us a more precise calculation of the packaging efficiencies. For this experiment, we infected QT6 cells with replication-competent RSV virus, RCASBPneo, in which the src gene has been replaced by neo. Mass cultures of G418-resistant cells were obtained. RPAs were performed on cytoplasmic RNAs and on virions released from the infected cells. To detect both unspliced and env RNAs, we used an antisense probe corresponding to the 3′ss for env that differentially protects the two RNAs (Fig. 4A). Using this probe, we found that the ratio of unspliced to env mRNA in the cytoplasm was 12.6 to 1 (Fig. 4B, lane 2) and that the ratio of unspliced to spliced env RNA in virions was 195.3 to 1 (Fig. 4B, lane 1). Several unexpected bands appear in both lanes and were detected in three independent repetitions of the assay. We believe these bands may represent aberrantly spliced or initiated RNAs. The packaging efficiency of the two RNAs was calculated by normalizing the amount of each RNA detected in virus to the amount of the same RNA in the cytoplasm. Thus, although the env RNA contains MΨ, it is packaged 15.5-fold less efficiently than the unspliced RNA. The results of this experiment were used to adjust the calculated packaging efficiencies of genomic RNA in the previous assay, to remove the effects of packaging env RNA (Table 1).
FIG. 4.
(A) The RCASBPneo genome showing the location of the minimal packaging signal, MΨ, relative to the viral splice sites. Unspliced and spliced viral mRNAs were detected in RCASBPneo-infected cells and viral particles by RPA, using a probe antisense to the env 3′ss. The regions of the two RNAs protected by the probe, as well as the predicted size of the protected region, are shown. gag, gene encoding viral structural proteins; pol, gene encoding reverse transcriptase and integrase; env, gene encoding glycoproteins; neo, gene encoding neomycin phosphotransferase. (B) RPA of viral, cytoplasmic, and nuclear lysates, using the antisense 3′ss probe described for panel A. The sample in lane 4 was not treated with RNase; the sample in lane 5 had no lysate added. The expected locations of free probe and of protected unspliced and spliced RNA bands are indicated. MWM, molecular weight markers.
DISCUSSION
We previously defined a 160-nt packaging sequence, MΨ, which confers efficient packaging to a heterologous RNA. We now show that the deletion of sequences from the 3′ end of MΨ, including part of the L3 stem, results in a reduction in packaging of the heterologous RNA. While the effects on packaging are significant, these mutants are still packaged more efficiently than CMVneo, which contains no packaging sequences. The simplest explanation of these findings is that these sequences or structures are directly involved in packaging. However, Doria-Rose and Vogt recently showed that viral mutants retaining neither the primary sequence nor secondary structure of L3 can be efficiently packaged (23). Taken together, our current hypothesis is that L3 does not directly interact with trans-acting factors in packaging. L3 may be entirely dispensable for packaging or may instead serve to stabilize the O3 element which directly interacts with these proteins. In our deletion mutants, in the absence of sequences required for L3 formation, alternative structures may form such that O3 sequences and/or structures are no longer in the proper context for packaging. We are currently performing experiments to address these hypotheses.
An alternative role for the L3 stem-loop in packaging may be through its involvement in dimer formation. Dimerization domains colocalize with the packaging signal in many retroviruses, including MLV (20, 51, 58) and HIV (9, 36, 53). Thus, it has long been postulated that dimerization of genomic RNAs is necessary for retroviral encapsidation (reviewed in reference 10). A dependence of genomic RNA packaging on dimer formation has never been definitively shown in RSV. There is some evidence that nascent RSV virions contain monomeric RNA which is only later dimerized (15, 18, 37, 48). In contrast, Stoltzfus and Snyder were able to detect unstable dimeric RNAs in nascent RSV virions (56). While studies of ALSV dimerization localize the dimer linkage structure (DLS) to the 5′ end of the genome, there is little consensus as to its precise location. Most of these putative DLSs are located downstream of MΨ (13, 37, 47). However, Fosse et al. have shown that palindromic sequences in the L3 loop play a critical role in ALSV dimer formation in vitro (28). Interestingly, it has been recently demonstrated that RSV mutants that do not contain a palindromic L3 loop can be efficiently packaged (23). Additionally, our MΨΔ3′ mutant does not contain L3 loop sequences, but is packaged only sixfold less efficiently than CMVneo-MΨ (7). Thus, if the L3 loop is responsible for RSV dimer formation, it would appear that this process is not absolutely essential for packaging. While these results are suggestive, we cannot rule out a role for dimerization in packaging without further investigation. It is possible that additional sequences or structures in MΨ function can function as a DLS.
As described here, a heterologous RNA containing the 160-nt MΨ is packaged only 2.6-fold less efficiently than replication-competent, viral genomic RNA. This indicates that MΨ itself contains most of the sequences and/or structures necessary for directing ALSV genomic RNA into viral particles. These results are in agreement with recent three-hybrid experiments in our lab which show that ALSV Gag binds specifically to MΨ with high affinity (38). Our results also suggest that additional sequences outside of MΨ may make minor contributions to packaging. MΨ confers packaging as efficiently as Ψ+, the original 683-nt ALSV packaging signal identified by our lab, which spans the entire 5′ end of the genome through the first 250 nt of gag (4, 5, 7). Thus, any additional packaging sequences must be located downstream of this region.
It is possible that the greater packaging efficiency of the full-length RNA relative to CMVneo-MΨ RNA is due not to the presence of additional packaging sequences but to the different context of MΨ in the two constructs. ALSVs have three short upstream open reading frames (uORFs) 5′ of the gag ORF. The third uORF is located within MΨ (7). Many, but not all, mutations that affect uORF3 translation also affect packaging (21, 22, 46, 54). There is a debate in the literature as to whether this correlation indicates a functional coupling of packaging and uORF3 translation (21, 22), or whether there are instead important RNA packaging structures that overlap uORF3 that are disrupted in these mutants (54). In our heterologous construct, uORF3 is downstream of the neo ORF and is therefore not likely to be efficiently translated. Thus, if the processes of packaging and uORF3 translation are indeed coupled, this could explain why CMVneo-MΨ is packaged less efficiently than the viral RNA.
We have also shown that in contrast to CMVneo-MΨ, env mRNA, which also contains MΨ, is not efficiently packaged. Other retroviruses also preferentially package genomic RNA over spliced RNAs; however, in most of these viruses, all or part of the packaging signal is located in the intron and is thus absent in the spliced RNAs. Since this is not the case in ALSV, other mechanisms must exist that prevent packaging of the spliced RNA. It is possible that MΨ is not folded properly on the env mRNA. Sequences downstream of the splice site on the subgenomic RNA may interact with sequences within MΨ, leading to the folding of an alternative, packaging-incompetent structure. While these putative inhibitory sequences would necessarily be found on the unspliced RNA, they would likely be too distal to disrupt folding of the packaging signal. Alternatively, env mRNA might be segregated from the packaging machinery, such that it is not readily accessible to assembling virions. Importantly, this putative segregation does not appear to be the result of interaction of the env mRNA with the splicing machinery. Our lab has previously shown that when the ALSV Ψ+ signal (which spans the 5′ss) was placed on a heterologous RNA, the spliced RNAs were packaged as efficiently as the unspliced RNAs (4). The compartmentalization of the RNA might instead be at the level of mRNA translation. env mRNAs are translated on the rough endoplasmic reticulum, while genomic RNAs are translated on polyribosomes in the cytoplasm (19). This mechanism, though, cannot explain the exclusion of other spliced retroviral messages in viral particles.
ALSV MΨ is similar to the core packaging signals of other simple retroviruses in that it is located between the U5/R region and the gag start codon. In several other ways, however, MΨ appears to be unique. First, as we have described here, MΨ is present on subgenomic messages. Second, the predicted secondary structure of MΨ does not bear any obvious resemblance to the signal of the other viruses. In particular, it does not contain a stem-loop with a GACG loop motif, shown to be important for packaging in SNV and MLV, two closely related viruses (27, 44, 45, 61). It would be interesting to determine if the tertiary structure of MΨ is similar to the packaging signals of these other retroviruses. This seems reasonable since the regions of ALSV nucleocapsid protein that have been implicated as important for interaction with viral RNA, including the Cys-His boxes and the surrounding basic regions, are similar to those of other retroviruses (10). While many techniques exist to probe RNA secondary structures, until recently it has been difficult to determine how these structures interact in three dimensions. In the last several years, however, rapid advances have been made in X-ray crystallography of RNAs, including RNAs the size of MΨ (16, 26, 29). It is therefore feasible that in the not so distant future we could have a picture of the three-dimensional structure of MΨ and finally address some of these questions.
ACKNOWLEDGMENTS
This work was supported by grant CA 18282 from the National Cancer Institute to M.L.L. J.D.B. was supported by a National Science Foundation graduate fellowship.
We thank Julie Overbaugh for critique of the manuscript.
REFERENCES
- 1.Adam M, Miller A D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D J, Stone J, Lum R, Linial M L. The packaging phenotype of SE21Q1b provirus is related to high proviral expression and not trans-acting factors. J Virol. 1995;69:7319–7323. doi: 10.1128/jvi.69.11.7319-7323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armentano D, Yu S-F, Kantoff P W, vonRuden R, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronoff R, Linial M L. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banks J D, Beemon K L, Linial M L. RNA regulatory elements in the genomes of simple retroviruses. Semin Virol. 1997;8:194–204. [Google Scholar]
- 7.Banks J D, Yeo A, Green K, Cepeda F, Linial M L. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout B, van Wamel J L. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 11.Berkowitz R D, Hammarskjold M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 12.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieth E, Gabus C, Darlix J-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canaani E, Helm K V, Duesberg P. Evidence for 30-40S RNA as precursor of the 60-70S RNA of Rous sarcoma virus. Proc Natl Acad Sci USA. 1973;70:401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung K S, Smith R E, Stone M P, Joklik W K. Comparison of immature (rapid) and mature Rous Sarcoma virus particles. Virology. 1972;50:851–864. doi: 10.1016/0042-6822(72)90439-4. [DOI] [PubMed] [Google Scholar]
- 19.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1767–1845. [Google Scholar]
- 20.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278–303) and new roles for stem-loops H2 (310–352) and H3 (355–374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 21.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doria-Rose N A, Vogt V M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J Virol. 1998;72:8073–8082. doi: 10.1128/jvi.72.10.8073-8082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dugaiczyk A, Haron J A, Stone E M, Dennison O E, Rothblum K N, Schwartz R J. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- 25.Federspiel M J, Hughes S H. Effects of the gag region on genome stability: avian retroviral vectors that contain sequences from the Bryan strain of Rous sarcoma virus. Virology. 1994;203:211–220. doi: 10.1006/viro.1994.1478. [DOI] [PubMed] [Google Scholar]
- 26.Ferre-D’Amare A R, Zhou K, Doudna J A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 27.Fisher J, Goff S P. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 28.Fosse P, Motte N, Roumier A, Gabus C, Muriaux D, Darlix J L, Paoletti J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996;35:16601–16609. doi: 10.1021/bi9613786. [DOI] [PubMed] [Google Scholar]
- 29.Golden B L, Gooding A R, Podell E R, Cech T R. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 30.Hackett P B, Dalton M W, Johnson D P, Petersen R B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1992;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes S, Greenhouse J J, Petropoulos C J, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 34.Katz R A, Omer C A, Weis J H, Mitsialis S A, Faras A J, Guntaka R V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus on structure of large terminal repeats in circle junctions. J Virol. 1982;42:346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 37.Lear A L, Haddrick M, Heaphy S. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology. 1995;212:47–57. doi: 10.1006/viro.1995.1452. [DOI] [PubMed] [Google Scholar]
- 38.Lee E-G, Yeo A, Kraemer B, Wickens M, Linial M L. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent methods. J Virol. 1999;73:6282–6292. doi: 10.1128/jvi.73.8.6282-6292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987;49:93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- 40.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann R, Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985;54:401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moscovici C, Moscovici M G, Jiminez H, Lai M M C, Hayman M J, Vogt P K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 44.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moustakas A, Sonstegard T S, Hackett P B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oertle S, Spahr P F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petropoulos C J, Hughes S H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prats A C, Roy C, Wang P A, Erard M, Housset V, Gabus C, Paoletti C, Darlix J L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson J H, Child L A, Lever A M L. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6542–6552. [PMC free article] [PubMed] [Google Scholar]
- 55.Stoker A W, Bissell M J. Development of avian sarcoma and leukosis virus-based vector-packaging cell lines. J Virol. 1988;62:1008–1015. doi: 10.1128/jvi.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoltzfus C M, Snyder P N. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975;16:1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tikhonenko A T, Black D J, Linial M L. Viral Myc oncoproteins in infected fibroblasts down-modulate thrombospondin-1, a possible tumor suppressor gene. J Biol Chem. 1996;271:30741–30747. doi: 10.1074/jbc.271.48.30741. [DOI] [PubMed] [Google Scholar]
- 58.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe S, Temin H M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5′ long terminal repeat and the start of the gag gene. Proc Natl Acad Sci USA. 1982;79:5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss R, Teich N, Varmus H, Coffin J. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 61.Yang S, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y Q, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]