Abstract
Background & Aims
Heterozygous SPINK1 mutations are strong risk factors for chronic pancreatitis in humans, yet heterozygous disruption of mouse Spink1 yielded no pancreatic phenotype. To resolve this contradiction, we used CRISPR/Cas9-mediated genome editing to generate heterozygous Spink1-deleted mice (Spink1-KOhet) in the C57BL/6N strain and studied the effect of this allele in trypsin-independent and trypsin-dependent pancreatitis models.
Methods
We investigated severity of acute pancreatitis and progression to chronic pancreatitis in Spink1-KOhet mice after transient (10 injections) and prolonged (2 × 8 injections) cerulein hyperstimulation. We crossed Spink1-KOhet mice with T7D23A and T7D22N,K24R mice that carry strongly autoactivating trypsinogen mutants and exhibit spontaneous chronic pancreatitis.
Results
Prolonged but not transient cerulein stimulation resulted in increased intrapancreatic trypsin activity and more severe acute pancreatitis in Spink1-KOhet mice relative to the C57BL/6N control strain. After the acute episode, Spink1-KOhet mice developed progressive disease with chronic pancreatitis-like features, whereas C57BL/6N mice recovered rapidly. Trypsinogen mutant mice carrying the Spink1-KOhet allele exhibited strikingly more severe chronic pancreatitis than the respective parent strains.
Conclusions
Heterozygous Spink1 deficiency caused more severe acute pancreatitis after prolonged cerulein stimulation and promoted chronic pancreatitis after the cerulein-induced acute episode, and in two strains of trypsinogen mutant mice with spontaneous disease. In contrast, acute pancreatitis induced with limited cerulein hyperstimulation was unaffected by heterozygous Spink1 deletion, in agreement with recent observations that trypsin activity does not mediate pathologic responses in this model. Taken together, the findings strongly support the notion that loss-of-function SPINK1 mutations in humans increase chronic pancreatitis risk in a trypsin-dependent manner.
Keywords: Acute Pancreatitis, Cerulein, Chronic Pancreatitis, Trypsin, Trypsin Inhibitor, Trypsinogen
Summary.
Heterozygous loss-of-function variants in the SPINK1 gene that encodes a trypsin inhibitor are strong risk factors for chronic pancreatitis in humans. This study demonstrates that heterozygous loss of SPINK1 in preclinical mouse models promotes pancreatitis onset and progression in a trypsin-dependent manner.
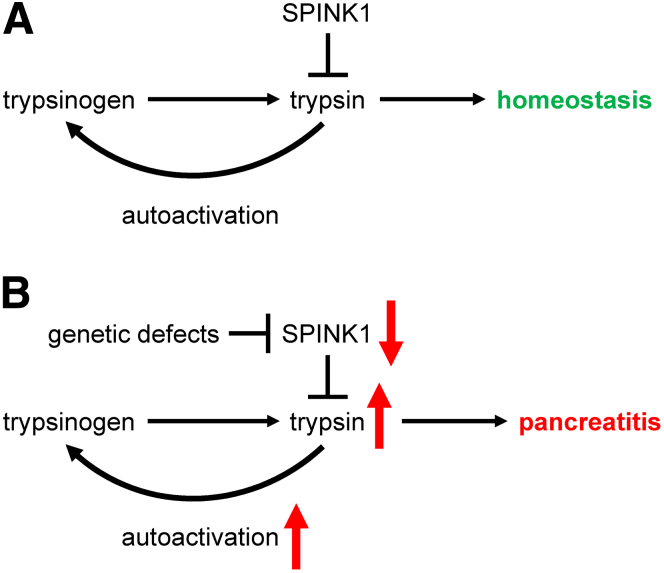
The pancreas secretes a variety of protease precursors, which become activated in the small intestine and catalyze digestion of dietary proteins and peptides.1 The most abundant protease proenzyme is trypsinogen, which has the unique ability to undergo so-called autoactivation; a self-amplifying, bimolecular reaction of trypsin activating trypsinogen.2 Trypsinogen autoactivation inside the pancreas can trigger the inflammatory disorder pancreatitis. To guard against unwanted intrapancreatic trypsin activity, acinar cells produce the serine protease inhibitor Kazal type 1 (SPINK1), a 6.2-kDa secretory trypsin inhibitor. SPINK1 represents approximately 0.1% to 0.8% of the total pancreatic juice protein, which is equivalent to about 2% to 13% of the trypsinogen content in molar terms.2 SPINK1 binds to the major human and mouse trypsin isoforms with high affinity, and forms non-covalent complexes.3,4 The equilibrium dissociation constant (KD) of the binding reaction is in the picomolar range.3,4 Curiously, post-translational tyrosine sulfation of human trypsins weakens inhibitor binding.3 In the presence of excess trypsin, the inhibitory complexes slowly dissociate due to proteolytic inactivation of SPINK1, a phenomenon termed temporary inhibition.4 In the pancreas, the inhibition of trypsin by SPINK1 not only neutralizes any potential harmful effects of the protease, but also prevents trypsinogen autoactivation. Pancreatitis may occur when intrapancreatic trypsin activity overwhelms SPINK1 reserves, and trypsinogen autoactivation can proceed unimpeded (Figure 1).
Figure 1.
Role of SPINK1 in pancreas health. (A) SPINK1 maintains homeostasis through trypsin inhibition and control of trypsinogen autoactivation. (B) Loss-of-function SPINK1 mutations reduce protective SPINK1 levels, leading to increased trypsinogen autoactivation, higher intrapancreatic trypsin activity, and increased risk of pancreatitis.
Evidence that SPINK1 maintains pancreas health came from human genetic studies that demonstrated that loss-of-function variants in SPINK1 strongly increase the risk for chronic pancreatitis (CP).2,5,6 Two relatively frequent variants (p.N34S and c.194+2T>C) and numerous rare variants have been described and replicated in several cohorts.2,5, 6, 7, 8, 9 Although the mechanism of action of the p.N34S variant is still unsettled, the splice-site variant c.194+2T>C offered compelling evidence for a loss-of-function disease mechanism. Multiple studies demonstrated that c.194+2T>C caused exon skipping and loss of SPINK1 mRNA expression.10, 11, 12 Many rare SPINK1 variants also support the loss-of-function pathogenic model. These include promoter variants, translation initiator methionine variants, signal peptide variants, splice-site variants, frame-shift variants, and missense variants that impair SPINK1 secretion.2 It is interesting to note that almost all pathogenic SPINK1 variants described to date diminish expression and/or secretion of the inhibitor. Variants that affect trypsin inhibitory activity are exceptions.3 In the majority of cases, SPINK1 variants are detected in the heterozygous state. Homozygosity is relatively rare and can be considered disease-causing.
Mice express a single orthologue of human SPINK1; the secreted forms of the 2 proteins are 63% identical in their amino-acid sequence. Ohmuraya et al described the first mouse model with deletion of the Spink1 gene (named Spink3 at the time).13 Homozygous Spink1-deficient mice died shortly after birth, and acinar cells showed extensive vacuolization apparently due to autophagy. There was spontaneous trypsin activity detectable in isolated acini, which could be inhibited by nafamostat.14,15 Heterozygous mice showed no spontaneous phenotype. Cerulein-induced trypsin activity and acute pancreatitis were unchanged relative to wild-type littermates. The unexpected finding that heterozygous Spink1-deleted mice exhibited no change in severity of cerulein-induced acute pancreatitis was later confirmed by Rodger Liddle’s group.16 Thus, heterozygous loss of Spink1 in mice does not replicate the human condition where heterozygous SPINK1 variants have a strong effect on CP risk.
Transgenic overexpression of a rat SPINK1 paralog was found to protect against cerulein-induced acute pancreatitis, cerulein-induced CP, and CP elicited by pancreatic overexpression of interleukin-1β.17, 18, 19 These observations were surprising because the experimental models employed were not trypsin-dependent. Furthermore, the findings were difficult to reconcile with the results of the Spink1 deletion studies. The Ohmuraya laboratory generated a mouse model where the human SPINK1 gene was integrated into the X chromosome in homozygous Spink1-deleted mice.20 Human SPINK1 rescued the lethal phenotype of Spink1-deficient mice, and due to random inactivation of one of the X chromosomes, showed a mosaic pattern of expression in females. Acinar cells not expressing SPINK1 suffered extensive vacuolization and died, whereas SPINK1-expressing acinar cells were normal. Even though the mice developed CP as a result of the mosaic cell injury, the human relevance of this elegant model is uncertain. Finally, preliminary data published in 2020 suggested that mice with the c.194+2T>C Spink1 mutation might develop CP, but full characterization of this potentially exciting model is still lacking.21
Recent years saw a surge in the development of genetically engineered mouse models where increased activation of mutant trypsinogens drive pancreatitis onset and progression.22, 23, 24, 25, 26, 27, 28 The availability of these models offers a fresh approach to address the role of heterozygous Spink1 deficiency in trypsin-mediated CP. In the present study, we resolved the long-existing contradiction between human genetic studies and animal experiments by demonstrating that heterozygous loss of Spink1 worsens CP in a trypsin-dependent manner.
Results
Heterozygous Deletion of Spink1 in C57BL/6N Mice
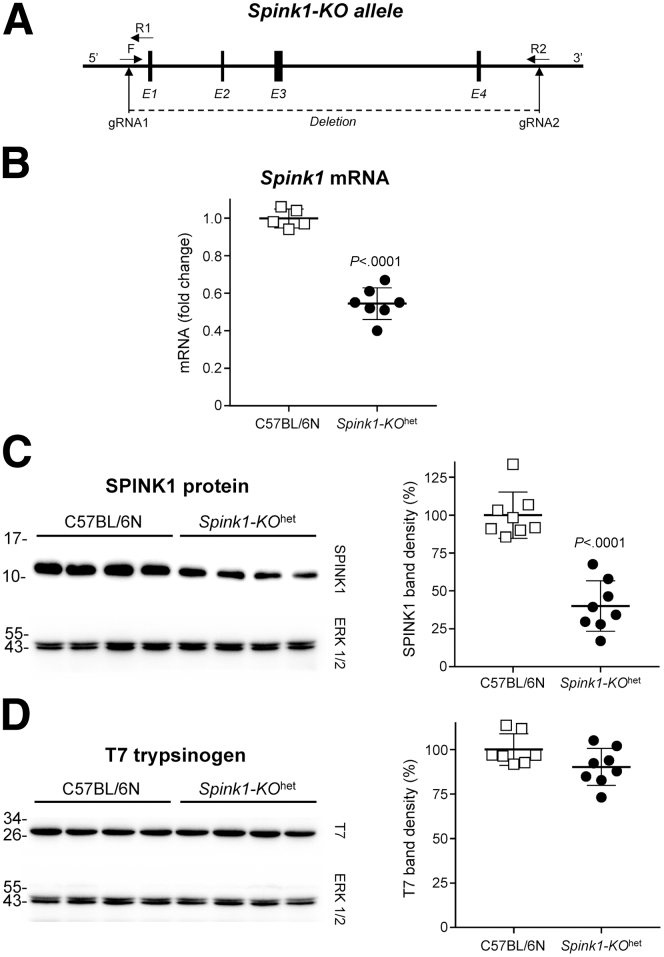
We used CRISPR/Cas9 technology to delete the entire Spink1 gene in the C57BL/6N mouse genome (Figure 2A). Mice heterozygous for the deletion allele (Spink1-KOhet) showed no phenotypic or behavioral changes, bred normally, and pancreas histology was indistinguishable from that of C57BL/6N control mice, up to 6 months of age. Using reverse-transcription quantitative polymerase chain reaction (RT-qPCR) and Western blotting, we confirmed that Spink1 mRNA (Figure 2B) and protein (Figure 2C) levels in the pancreas of Spink1-KOhet mice were reduced by about 50% relative to C57BL/6N control mice, while expression of cationic trypsinogen (isoform T7) was unchanged (Figure 2D). We also replicated the published observation that homozygous deletion of Spink1 resulted in complete pancreas destruction, severe runting, and early death (not shown).13,16
Figure 2.
Effect of heterozygous Spink1 deletion (Spink1-KOhet) on mRNA and protein expression in the mouse pancreas. (A) CRISPR/Cas9-mediated genome engineering was used to delete the entire Spink1 gene. E, Exons; F and R, forward and reverse genotyping primers; gRNA, guide RNAs. (B) Expression of Spink1 mRNA in the pancreas of C57BL/6N and Spink1-KOhet mice was quantified by reverse-transcription quantitative PCR and expressed relative to the average value of the C57BL/6N strain. (C) Protein levels of SPINK1 in pancreas homogenates were determined by Western blotting and densitometry. (D) Protein levels of mouse cationic trypsinogen (T7) were assessed by Western blotting and densitometry. On Western blots, ERK1/2 was measured as loading control. Representative blots from 2 experiments are shown. Densitometry results are expressed as percent of the average band intensity of the C57BL/6N strain. Individual data points with mean and standard deviation are shown. The difference of means between the groups was analyzed by 2-tailed unpaired t-test.
Cerulein-induced Acute Pancreatitis in Spink1-KOhet Mice
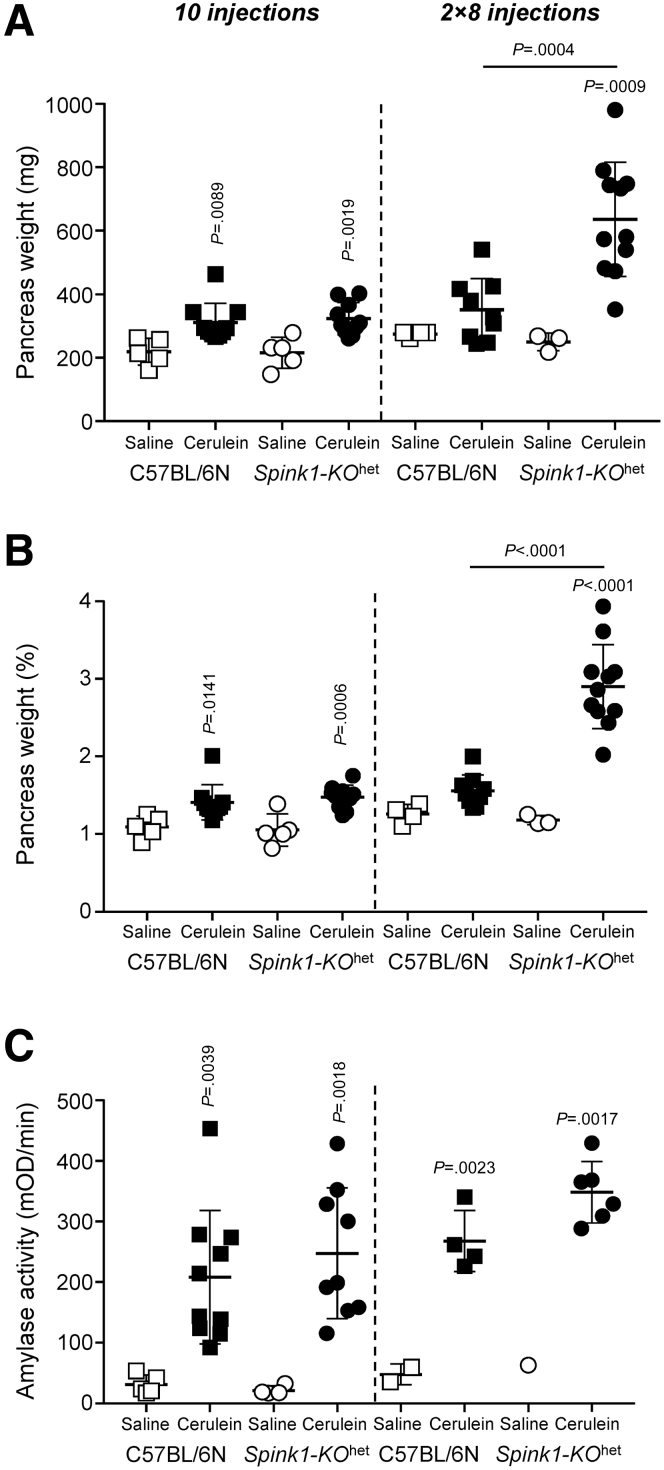
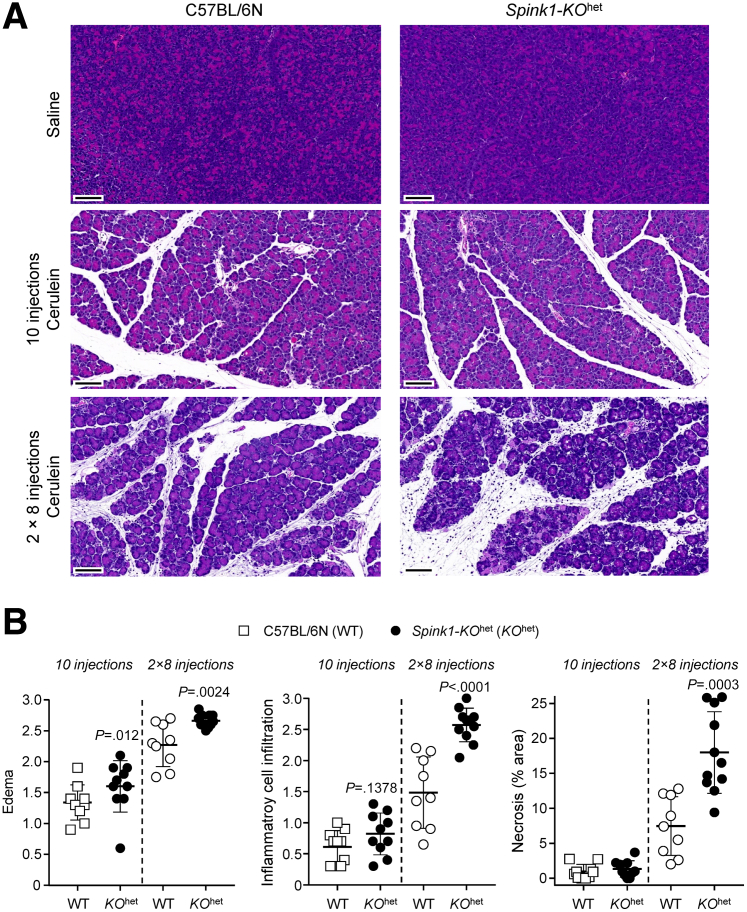
Previous reports found no significant effect of heterozygous Spink1 deletion on the severity of cerulein-induced acute pancreatitis.13,16 Here, we used hourly injections of cerulein (50 μg/kg) to elicit acute pancreatitis in C57BL/6N and Spink1-KOhet mice. We tested the effects of transient hyperstimulation (10 hourly injections on the same day) and prolonged hyperstimulation (2 × 8 injections on consecutive days). When compared with saline-injected controls, cerulein treatment induced significant elevations in pancreas weight (Figure 3A, B), plasma amylase activity (Figure 3C), and histological parameters such as edema, inflammatory cell infiltration, and acinar cell necrosis (Figure 4). With the transient hyperstimulation model, there was no significant difference in disease severity between the 2 strains studied. In contrast, prolonged hyperstimulation with cerulein resulted in significantly higher pancreas weight, histological edema, inflammatory cell infiltration, and necrosis in Spink1-KOhet mice relative to C57BL/6N controls. Plasma amylase activity was also higher in Spink1-deficient animals versus C57BL/6N mice, but the difference did not reach statistical significance.
Figure 3.
Cerulein-induced acute pancreatitis in Spink1-KOhetmice. C57BL/6N and Spink1-KOhet mice were treated with either transient (10 hourly injections, left panels) or prolonged (2 × 8 hourly injections, right panels) hyperstimulation with cerulein. Control mice were given saline injections. Animals were euthanized 1 hour (10 injections) or 30 minutes (2 × 8 hourly injections) after the last injection. (A) Pancreas weight in mg units. (B) Pancreas weight expressed as percent bodyweight. (C) Plasma amylase activity. Individual data points with mean and standard deviation are shown. The difference of means between the groups was analyzed by 1-way analysis of variance followed by Tukey’s post-hoc test.
Figure 4.
Histology of cerulein-induced acute pancreatitis in Spink1-KOhetmice. C57BL/6N and Spink1-KOhet mice were treated with either transient (10 hourly injections) or prolonged (2 × 8 hourly injections) hyperstimulation with cerulein. Control mice were given saline injections. Animals were euthanized 1 hour (10 injections) or 30 minutes (2 × 8 hourly injections) after the last injection. (A) Representative hematoxylin-eosin-stained pancreas sections. Scale bars are 100 μm. (B) Histology scoring of pancreas sections from cerulein-treated mice for edema, inflammatory cell infiltration, and acinar cell necrosis. The pancreas of saline-treated mice showed no visible edema, inflammatory cells, or necrosis. Individual data points were graphed with the mean and standard deviation indicated. The difference of means between the groups was analyzed by 2-tailed unpaired t-test.
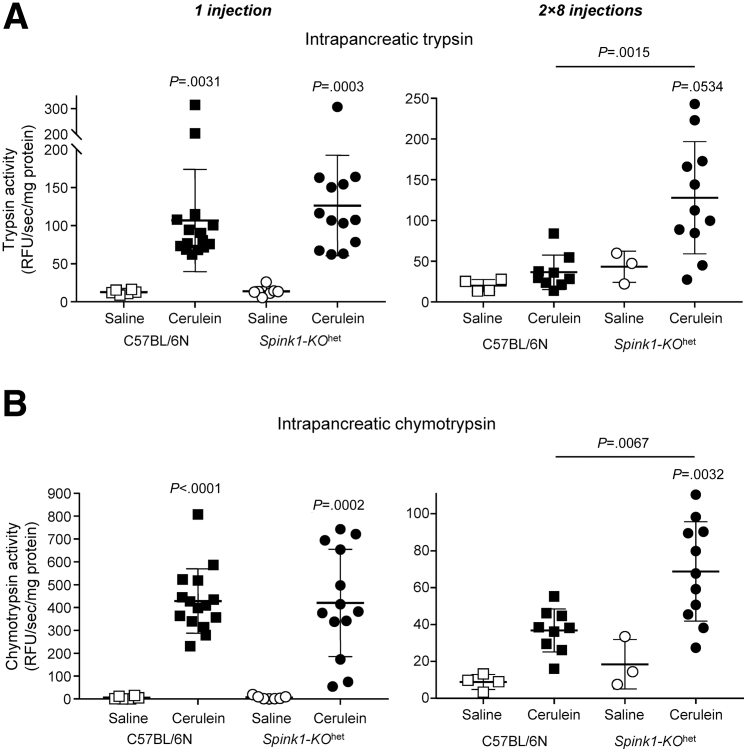
We measured cerulein-induced intrapancreatic protease activation 30 minutes after a single cerulein injection and after prolonged (2 × 8 injections) cerulein hyperstimulation (Figure 5). Relative to saline controls, cerulein caused significant increases in trypsin (Figure 5A) and chymotrypsin (Figure 5B) activity of pancreas homogenates. No appreciable difference was observed in the extent of protease activation between the C57BL/6N and Spink1-KOhet mice after a single injection of cerulein. In sharp contrast, prolonged hyperstimulation resulted in significantly higher trypsin and chymotrypsin activity in the pancreas of Spink1-KOhet mice versus C57BL/6N controls.
Figure 5.
Cerulein-induced intrapancreatic protease activation in Spink1-KOhetmice. C57BL/6N and Spink1-KOhet mice were administered a single injection (left panels) or 2 × 8 hourly injections (right panels) of cerulein. Control animals were given saline injections. Mice were euthanized 30 minutes after the single/last injection, the pancreas was homogenized, and protease activities were measured as described in Methods. (A) Intrapancreatic trypsin activity. (B) Intrapancreatic chymotrypsin activity. Individual values with the mean and standard deviation were graphed. The difference of means was analyzed by 1-way analysis of variance with Tukey’s post-hoc test.
Chronic Progression After Cerulein-induced Acute Pancreatitis in Spink1-KOhet Mice
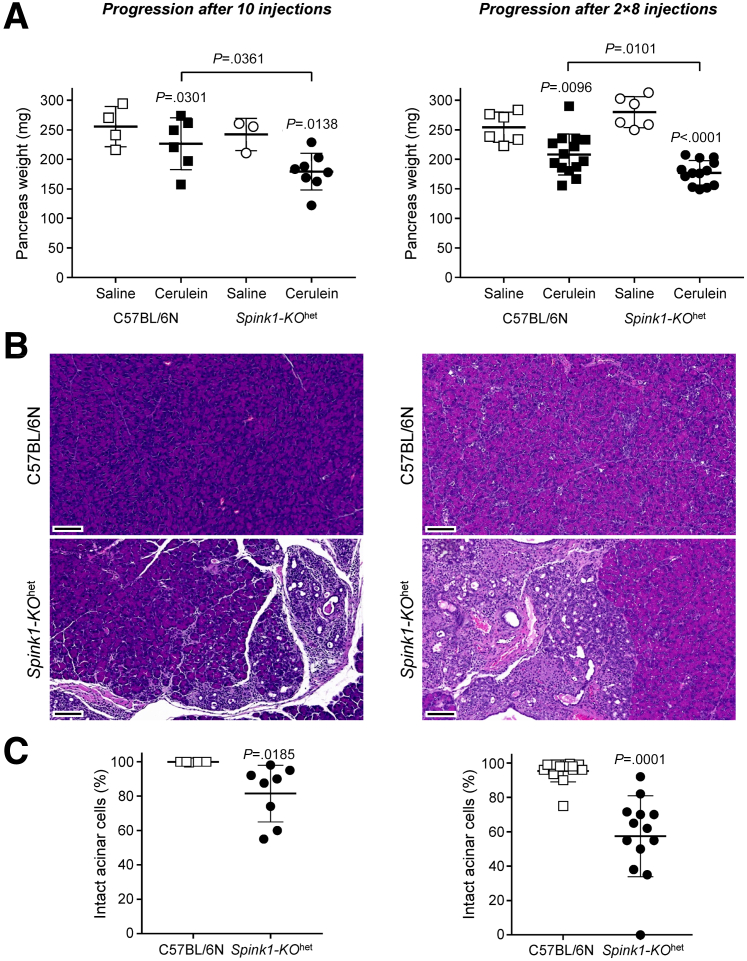
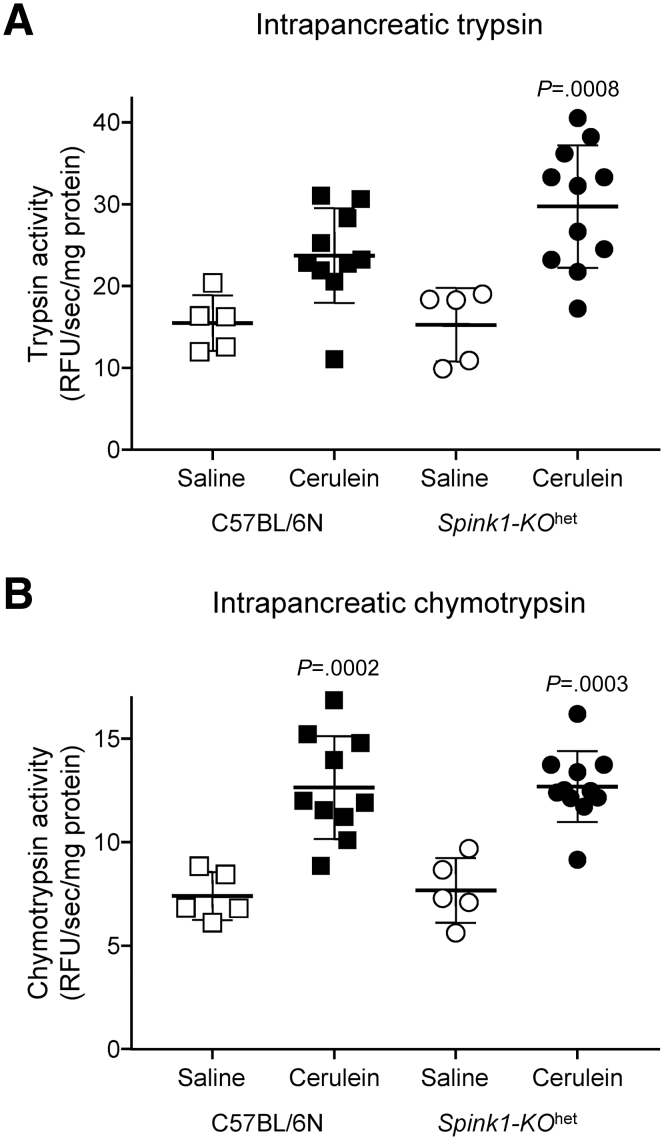
C57BL/6N mice rapidly recover after a cerulein-induced pancreatitis attack with essentially complete normalization of pancreas histology within a few days.29 In contrast, mice carrying trypsinogen mutants exhibit CP-like, progressive, atrophic changes after an acute episode of cerulein-induced pancreatitis, which can be prevented by administration of a small-molecule trypsin inhibitor.25,28, 29, 30 Thus, progression to CP after an acute attack is driven by trypsin. When acute pancreatitis in Spink1-KOhet mice was elicited by transient hyperstimulation with cerulein (10 injections), and animals were euthanized a week after the last injection, the pancreas weight was significantly lower than those of C57BL/6N controls (Figure 6A) and histological analysis showed areas of acinar atrophy, dilated ducts, and inflammatory cells (Figure 6B). Semi-quantitative scoring for intact acini indicated about 20% histological atrophy on average (Figure 6C). A similar but more severe outcome was observed when prolonged hyperstimulation was performed according to the protocol by Jensen et al,31 where mice were given 8 hourly injections on 2 consecutive days and were euthanized 3 days after the last injection. Again, we observed reduced pancreas weight (Figure 6A), somewhat more extensive chronic changes on histology (Figure 6B), and more than 40% acinar cell atrophy in Spink1-KOhet mice (Figure 6C). Importantly, chronic progression in Spink1-KOhet mice was accompanied by a noticeable trend for increased intrapancreatic trypsin activity relative to C57BL/6N mice, whereas chymotrypsin activity was similar in the 2 strains (Figure 7). The findings are consistent with the notion that development of CP after a cerulein-induced acute episode is trypsin-dependent.
Figure 6.
Disease progression after cerulein-induced acute pancreatitis in Spink1-KOhetmice. To assess progression after transient hyperstimulation with cerulein, C57BL/6N and Spink1-KOhet mice were given 10 hourly injections of saline or cerulein and euthanized 1 week later (left panels). To evaluate progression after prolonged hyperstimulation with cerulein, mice were given 8 hourly saline or cerulein injections on two consecutive days, and euthanized 3 days later (right panels). (A) Pancreas weight. (B) Representative hematoxylin-eosin stained sections from cerulein-treated mice. Scale bars are 100 μm. (C) Histology scoring for intact acinar cells. Individual values with the mean and standard deviation were plotted. The difference of means between 2 groups and multiple groups was analyzed by 2-tailed unpaired t-test and 1-way analysis of variance with Tukey’s post-hoc test, respectively.
Figure 7.
Intrapancreatic protease activity during disease progression after cerulein-induced acute pancreatitis. C57BL/6N and Spink1-KOhet mice were given 2 × 8 hourly injections of saline or cerulein on consecutive days and euthanized 3 days later. The pancreas was homogenized, and protease activities were measured as described in Methods. (A) Intrapancreatic trypsin activity. (B) Intrapancreatic chymotrypsin activity. Individual values with the mean and standard deviation were graphed. The difference of means was analyzed by 1-way analysis of variance with Tukey’s post-hoc test. The P values indicate pairwise comparisons between the saline and cerulein groups. The difference between the cerulein groups was not significant.
Heterozygous Spink1 Deficiency Increases Severity of Spontaneous Chronic Pancreatitis in T7D23A and T7D22N,K24R Mice
Mouse models carrying rapidly autoactivating cationic trypsinogen mutants develop progressive CP characterized by significant atrophy, diffuse fibrosis, dilated ducts, inflammatory cell infiltrates, and adipose tissue replacement.22,28 In T7D23Ahet mice, the disease typically starts at the age of 3 to 4 weeks, often with a brief episode of acute pancreatitis followed by rapid progression to chronic changes.22 In T7D22N,K24Rhom mice, the age of onset is around 6 weeks and progression to end-stage CP is slower.28 To test whether trypsin-dependent CP in these mouse models is affected by heterozygous Spink1 deficiency, we generated T7D23Ahet × Spink1-KOhet and T7D22N,K24Rhom × Spink1-KOhet crosses.
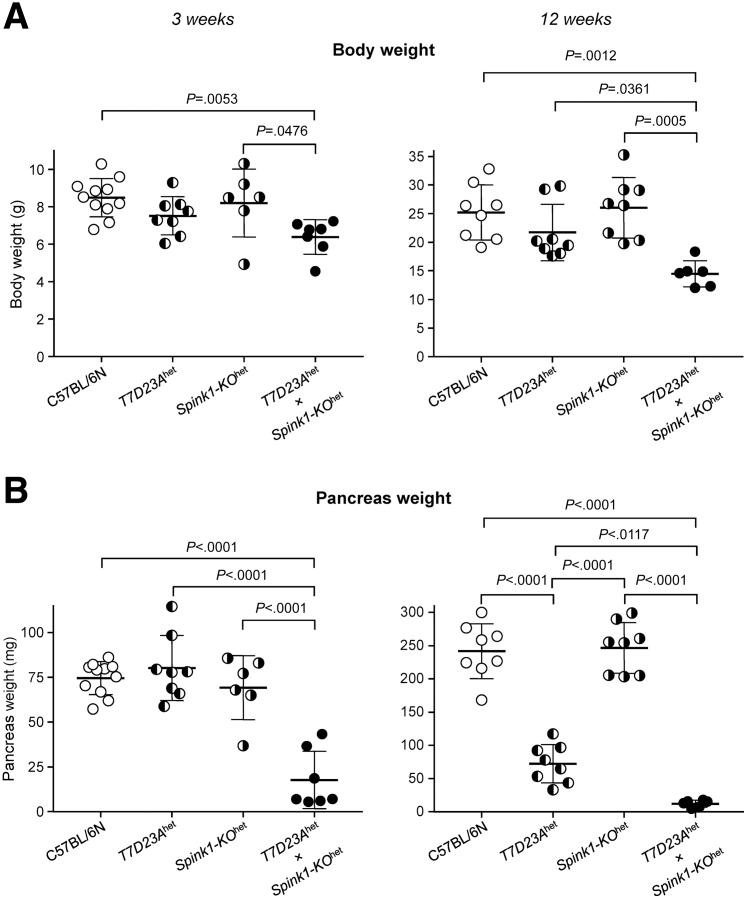
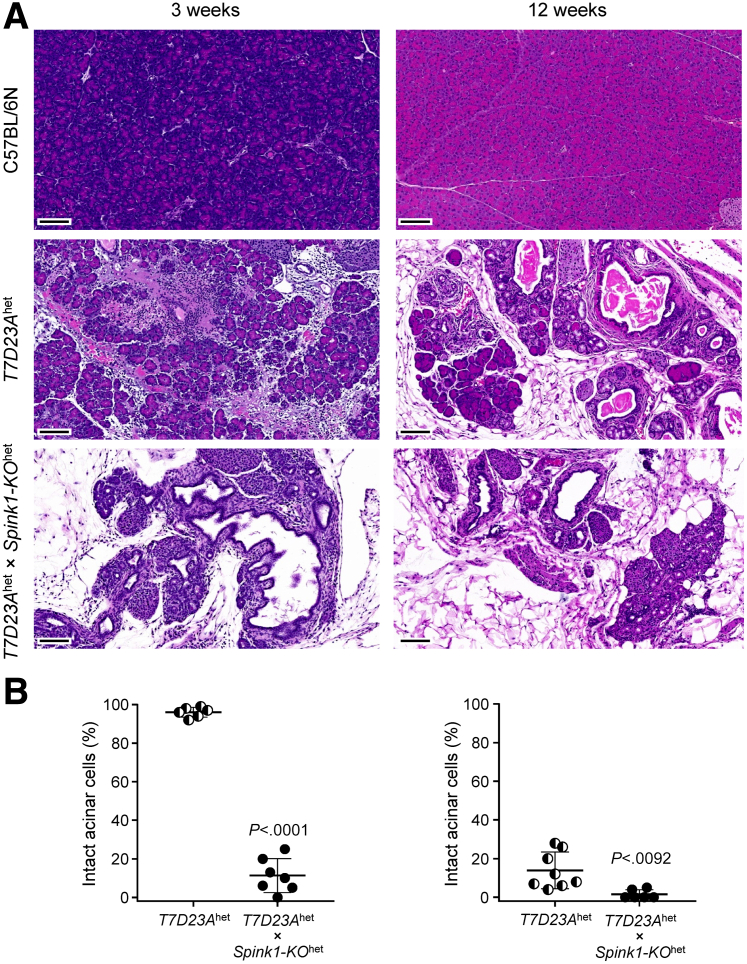
When disease severity was analyzed at 3 weeks and 12 weeks, T7D23Ahet × Spink1-KOhet mice exhibited lower body weight (Figure 8A), more severe pancreas atrophy (Figure 8B), and more pronounced histological changes (Figure 9A) than T7D23Ahet controls, indicating that lower SPINK1 levels caused accelerated progression. At 3 weeks of age, all pancreata from T7D23A mice (n = 6) showed signs of acute pancreatitis (edema, inflammatory cell infiltration, small areas of necrosis) without significant acinar cell loss or chronic changes. In contrast, the pancreas from all T7D23Ahet × Spink1-KOhet mice (n = 7) exhibited acinar cell atrophy (Figure 9B), inflammatory cells, dilated ducts, and fatty replacement. At 12 weeks of age, all T7D23Ahet mice (n = 8) progressed to CP, and all T7D23Ahet × Spink1-KOhet mice had severe CP with increased acinar cell loss relative to T7D23Ahet animals (Figure 9B).
Figure 8.
Effect of heterozygous Spink1 deletion on chronic pancreatitis severity in T7D23A mice. C57BL/6N, T7D23Ahet, Spink1-KOhet, and T7D23Ahet × Spink1-KOhet mice were euthanized at 3 weeks (left panels) and 12 weeks (right panels) of age. (A) Body weight. (B) Pancreas weight. Individual values with the mean and standard deviation were plotted. The difference of means was analyzed by 1-way analysis of variance with Tukey’s post-hoc test.
Figure 9.
Histological evaluation of pancreatitis in T7D23Ahet× Spink1-KOhetmice. C57BL/6N, T7D23Ahet, and T7D23Ahet × Spink1-KOhet mice were euthanized at 3 weeks (left panels) and 12 weeks (right panels) of age. (A) Representative hematoxylin-eosin stained pancreas sections. Scale bars are 100 μm. (B) Histology scoring for intact acinar cells. Individual values with the mean and standard deviation were plotted. The difference of means between the groups was analyzed by 2-tailed unpaired t-test.
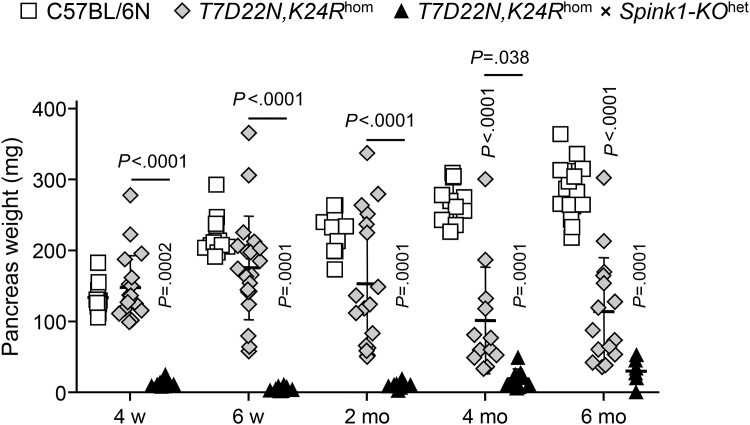
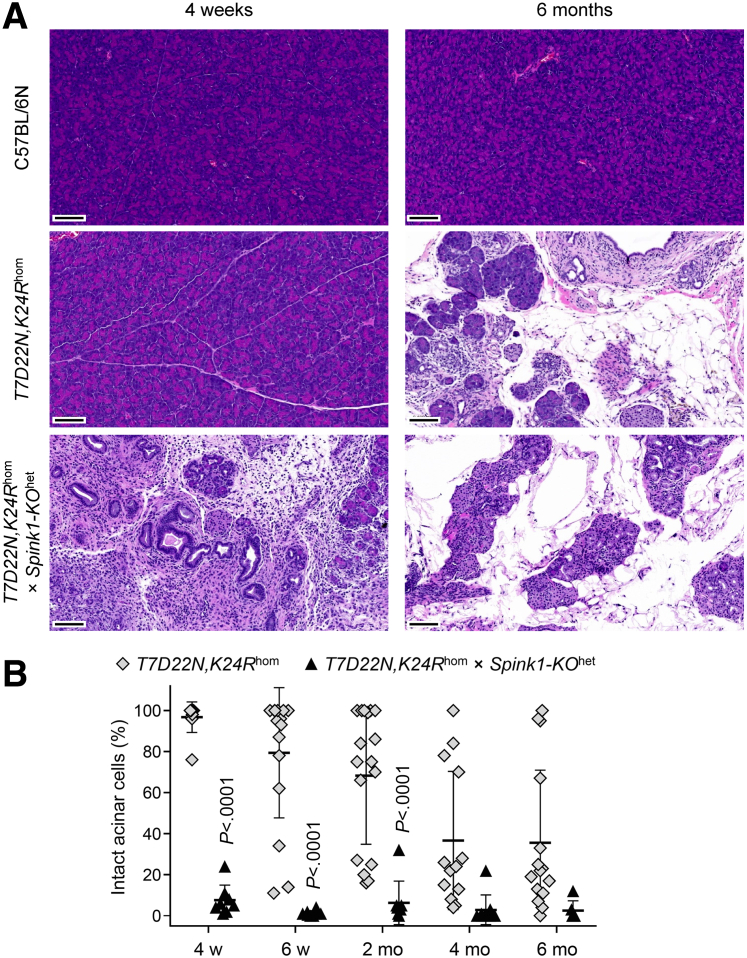
In a similarly dramatic fashion, T7D22N,K24Rhom × Spink1-KOhet mice developed severe CP as early as 4 weeks of age, as judged by the massive pancreas atrophy (Figure 10). The pancreas weight of C57BL/6N control mice increased as a function of time and almost doubled by 6 months of age relative to the 4-week value. T7D22N,K24Rhom mice exhibited decreasing pancreas weight after 2 months of age, as CP with acinar atrophy set in. In contrast, the weight of the pancreas from T7D22N,K24Rhom × Spink1-KOhet mice was severely reduced as early as 4 weeks of age and remained diminished throughout the 6-month time course. Histology analysis confirmed the early-onset CP in T7D22N,K24Rhom × Spink1-KOhet mice relative to T7D22N,K24Rhom animals. As shown in Figure 11A, at 4 weeks of age, the pancreas of T7D22N,K24Rhom mice looked normal, indistinguishable from those of C57BL/6N, whereas T7D22N,K24Rhom × Spink1-KOhet mice showed early CP characterized by massive acinar atrophy, high number of inflammatory cells, and acinar-to-ductal metaplasia. At 6 months of age, end-stage chronic disease was evident in both strains with large areas replaced by adipose tissue (Figure 11A). When multiple pancreas sections from mice euthanized at various ages were scored for intact acinar cells, the results clearly confirmed the drastic, early-onset acinar cell atrophy in T7D22N,K24Rhom × Spink1-KOhet mice relative to T7D22N,K24Rhom mice, which showed slowly progressing disease with more individual variation (Figure 11B). Taken together, the findings indicate that Spink1 deficiency strongly promoted disease onset and progression in mouse strains with trypsinogen mutations that are prone to develop spontaneous CP in a trypsin-dependent manner.
Figure 10.
Effect of heterozygous Spink1 deletion on chronic pancreatitis severity in T7D22N,K24Rhommice. C57BL/6N, T7D22N,K24Rhom, and T7D22N,K24Rhom × Spink1-KOhet mice were euthanized at 4 weeks, 6 weeks, 2 months, 4 months, and 6 months of age, and the pancreas weight was measured to assess bulk acinar atrophy. Note that part of this experiment showing data for C57BL/6N and T7D22N,K24Rhom mice was published previously.28 Individual values with the mean and standard deviation were graphed. The difference of means was analyzed by 1-way analysis of variance followed by Tukey’s post-hoc test.
Figure 11.
Histological evaluation of chronic pancreatitis severity in T7D22N,K24Rhom× Spink1-KOhetmice. C57BL/6N, T7D22N,K24Rhom, and T7D22N,K24Rhom × Spink1-KOhet mice were euthanized at 4 weeks, 6 weeks, 2 months, 4 months, and 6 months of age. (A) Representative hematoxylin-eosin stained pancreas sections from 4-week-old (left panels) and 6-month-old (right panels) mice. Scale bars are 100 μm. (B) Histology scoring for intact acinar cells. Individual values with the mean and standard deviation were plotted. The difference of means was analyzed by 1-way analysis of variance followed by Tukey’s post-hoc test.
Discussion
In the present study, we set out to demonstrate that heterozygous Spink1 deficiency increases severity of trypsin-dependent CP in mice. Heterozygous SPINK1 mutations are strong risk factors for CP in humans; however, animal models to date failed to offer direct evidence for this notion. Here, we utilized not only the classic cerulein-induced pancreatitis model but also recently developed trypsinogen mutant mouse strains.
In agreement with previous reports,13,16 we found that the severity of cerulein-induced acute pancreatitis was unaffected by heterozygous Spink1 deletion when the disease was elicited with transient hyperstimulation (10 hourly injections). Although this finding may have been baffling at the time of the earlier reports, recent developments in the field offer better understanding. Thus, in this model of secretagogue-induced acute pancreatitis, the pathological responses are not mediated by intrapancreatic trypsin activity.32 Consequently, severity parameters should be unaffected by the extent of SPINK1-mediated trypsin inhibition. Furthermore, we also made the somewhat unexpected observation that intrapancreatic trypsin and chymotrypsin activities induced by a single injection of cerulein were unchanged in Spink1-KOhet mice relative to C57BL/6N controls. It appears that in the vesicular compartment where cathepsin B generates trypsin activity, SPINK1 is ineffective, possibly due to degradation. Our results support the notion that transient cerulein hyperstimulation causes trypsin-independent acute pancreatitis where heterozygous loss of Spink1 has no impact on severity.
Interestingly, a different picture emerged when mice were subjected to prolonged hyperstimulation with cerulein (2 × 8 hourly injections). Under these conditions, acute pancreatitis was significantly more severe in Spink1-KOhet mice relative to C57BL/6N animals, and intrapancreatic trypsin and chymotrypsin activities were also significantly higher in the Spink1-deficient mice when measured 30 minutes after the last injection. These observations indicate that under prolonged hyperstimulation with cerulein, acute pancreatitis becomes significantly more trypsin-dependent, and protective SPINK1 levels can have a measurable impact on disease severity.
Using mice carrying rapidly autoactivating trypsinogen mutants, we previously demonstrated that after an acute episode of cerulein-induced pancreatitis, progressive disease develops characterized by CP-like features such as acinar atrophy, macrophage infiltration, acinar-to-ductal metaplasia, duct dilation, fibrosis, and adipose tissue replacement.28,29 Recovery is slow and incomplete; even 3 months after the acute episode, the pancreas is not fully normal. In contrast, C57BL/6N mice recover rapidly and show essentially normal histology within 3 days. This progressive CP phenotype is driven by intrapancreatic trypsin activity and can be prevented by a small-molecule trypsin inhibitor administered orally after the cerulein injections.25,30 Remarkably, Spink1-KOhet mice exhibited a similar phenotype, which was observed after 10 injections of cerulein and even more strongly after 2 × 8 injections. Importantly, the CP-like disease was associated with elevated intrapancreatic trypsin activity in Spink1-KOhet mice relative to wild-type controls. These results indicate that heterozygous deletion of Spink1 facilitates the development of trypsin-dependent CP after cerulein-induced acute pancreatitis.
The strongest evidence that heterozygous Spink1 deficiency promotes CP came from the experiments using the T7D23A and T7D22N,K24R trypsinogen mutant strains. T7D23Ahet and T7D22N,K24Rhom mice develop CP spontaneously, which progresses within months to an atrophic pancreas with fatty replacement of acinar cells.22,28 When T7D23Ahet mice were crossed with Spink1-KOhet mice, the disease course accelerated, and atrophy was so pronounced that the pancreas dysfunction resulted in measurable body weight loss. The effect of the Spink1-KOhet allele was even more dramatic in T7D22N,K24Rhom mice, which normally exhibit CP with a later age of onset and slower progression. However, heterozygous Spink1 deletion resulted in severe pancreas atrophy as early as 4 weeks of age. Although data are not shown, we also crossed the Spink1-KOhet strain with heterozygous T7D22N,K24Rhet mice, which do not develop CP spontaneously.28 Interestingly, 4 of 11 compound heterozygous mice had CP at 6 months of age, indicating that intrapancreatic trypsinogen activation was increased to pathological levels in T7D22N,K24Rhet mice by the heterozygous deletion of Spink1.
It is noteworthy that reduced SPINK1 levels modify disease severity without causing spontaneous pancreatitis. In fact, early genetic literature referred to human SPINK1 variants as disease modifiers in CP.6 The situation is somewhat reminiscent of how lipolysis worsens acute pancreatitis but does not initiate the disease, as documented in a number of recent studies.33, 34, 35
While this paper was under review, a new study was published where adeno-associated virus was used as a vector to overexpress human SPINK1 in the mouse pancreas.36 The authors demonstrated protective effects in acute and chronic pancreatitis induced by cerulein injections or duct ligation, and in the c.194+2T>C Spink1 model. A direct comparison with the present study is difficult due to the different experimental conditions employed, but the overall efficacy of virally transduced SPINK1 against pancreatitis is consistent with our conclusions.
Taken together, the findings presented here provide compelling evidence that heterozygous Spink1 deficiency increases severity and rate of progression of CP in trypsin-dependent mouse models. Thus, heterozygous Spink1 deletion in mice reproduces the loss-of-function effects of human heterozygous SPINK1 mutations and confirms that even partial reduction in protective SPINK1 levels can increase intrapancreatic trypsin activity and promote CP.
Methods
Accession Numbers and Nomenclature
Spink1 (legacy name Spink3) genomic sequence: NC_000084.7, Mus musculus strain C57BL/6J chromosome 18, GRCm39. Spink1 coding DNA: NM_009258.5, Mus musculus serine peptidase inhibitor, Kazal type 1 (Spink1), mRNA.
Animal Studies Approval
Animal experiments were performed at the University of California, Los Angeles (UCLA) with the approval and oversight of the Animal Research Committee, including protocol review and post-approval monitoring. The animal care program is managed in full compliance with the United States (U.S.) Animal Welfare Act, the U.S. Department of Agriculture Animal Welfare Regulations, the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the National Research Council's Guide for the Care and Use of Laboratory Animals. UCLA has an approved Animal Welfare Assurance statement on file with the U.S. Public Health Service, National Institutes of Health, Office of Laboratory Animal Welfare, and it is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Generation of the Spink1-KO Mouse Strain
The gene encoding mouse Spink1 (legacy name Spink3) is located on chromosome 18, it spans ∼9 kb, and comprises 4 exons. En bloc deletion of the Spink1 gene (Spink1-KO) from c.-473 to c.∗1737 (11,200 nucleotides) was accomplished by CRISPR/Cas9-mediated genome engineering (Cyagen US Inc). Spink1-KO mice were maintained on the C57BL/6N genetic background in the heterozygous state (Spink1-KOhet) because homozygous animals died shortly after birth. C57BL/6N mice obtained from Charles River Laboratories or produced in our breeding facility from the same stock were used as experimental controls. Both male and female animals (8–10 weeks of age) were included in the studies.
Genotyping Spink1-KOhet Mice
The following primers were used to genotype mice for the Spink1-KO allele. Common forward primer F1, 5′-CAA ACA CCC TGG GAA AGA TAA TCT GTC-3′, wild-type allele reverse primer R1, 5′-TAC CTG CTA AAC TCA GCA GGG CC-3′, deletion allele reverse primer R2, 5′-GGC CTG TCC AGG GAT CAA TCA TAA-3′. PCR reactions contained all 3 primers. The amplicon size from the deletion allele was 540 bp, whereas the wild-type allele gave an 839 bp product.
Trypsinogen Mutant Mouse Strains
Development and characterization of T7D23A and T7D22N,K24R mice carrying rapidly autoactivating cationic trypsinogen (isoform T7) mutants in the C57BL/6N genetic background were reported recently.22,28 T7D23A mice were maintained in the heterozygous state (T7D23Ahet) because homozygous animals become runted and die early. T7D22N,K24R mice were kept in the homozygous state (T7D22N,K24Rhom).
Western Blot and Densitometry
Mouse pancreas tissue (30–40 mg) was homogenized in 300- to 400-μL ice-cold PBS (pH 7.4) supplemented with Halt protease and phosphatase inhibitor (catalog number 1861284, Thermo Fisher Scientific). The homogenate was cleared by centrifugation (10 minutes, 13,500 rpm, 4 °C) and aliquots of the supernatant (30 μg total protein per lane) were electrophoresed on 15% SDS-PAGE minigels. The proteins were transferred onto an Immobilon-P membrane (catalog number IPVH00010, MilliporeSigma) and the membrane was blocked with 10% solubilized milk powder. Incubations with primary and secondary antibodies were carried out overnight at 4 °C and for 1 hour at 22 °C, respectively. Membranes were first probed for SPINK1 or T7 trypsinogen, then stripped, and re-probed for ERK1/2. The following antibodies were used. Rabbit polyclonal antibody against mouse SPINK1 was used at a dilution of 1:1,000 (catalog number 2744, Cell Signaling Technology); rabbit polyclonal antibody against T7 trypsinogen22 was used at 1:10,000 dilution; rabbit monoclonal antibody against ERK1/2 was used at 1:1,000 dilution (catalog number 4695, Cell Signaling Technology). As secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG (catalog number HAF008, R&D Systems) was used at 1:10,000 dilution. T7 trypsinogen and ERK1/2 were detected with the SuperSignal West Pico chemiluminescent substrate (catalog number 34580, Thermo Fisher Scientific). SPINK1 protein was detected with the SuperSignal West Femto Maximum Sensitivity chemiluminescent substrate (catalog number 34095, Thermo Fisher Scientific). Densitometric evaluation of protein band intensities on Western blots was carried out with the ImageJ software (version 1.52a).
RNA Isolation and Reverse Transcription qPCR
Total RNA was isolated from mouse pancreata (20 mg) using the RNeasy Plus Mini Kit (Qiagen). Two μg RNA was reverse-transcribed with the High Capacity cDNA Reverse Transcription Kit (catalog number 4368814, Thermo Fisher Scientific). Quantitative PCR was performed with the Mm00436765_m1 (Spink1, FAM dye) and Mm01612987_g1 (Rpl13a reference gene, VIC dye) TaqMan probes, using the Taqman Universal PCR Master Mix (catalog number: 4304437, Thermo Fisher Scientific). Relative expression levels were estimated with the ΔΔCT method as described before.23
Cerulein-induced Pancreatitis
To induce acute pancreatitis, mice were administered hourly injections of cerulein (50 μg/kg dose) intraperitoneally causing transient (10 injections) or prolonged (2 × 8 injections on consecutive days) hyperstimulation, as indicated. Control mice were given saline injections. To assess acute pancreatitis, mice were euthanized 1 hour (10 injections) or 30 minutes (2 × 8 injections) after the last injection. To study progression to CP after the acute episode, mice were euthanized 1 week (10 injections) or 3 days (2 × 8 injections) after the last injection.31
Intrapancreatic Protease Activity
Intrapancreatic trypsin and chymotrypsin activity were measured from freshly prepared pancreas extracts according to our published protocol.37 Mice were euthanized 30 minutes after a single injection of cerulein (50 μg/kg dose), 30 minutes after 2 × 8 cerulein injections, or 3 days after 2 × 8 cerulein injections, as indicated. Control mice were given saline injections and were euthanized at the same times. Enzyme activity was expressed in RFU/sec/mg protein units, which represents the rate of substrate cleavage in relative fluorescent units per second, normalized to the total protein content.
Plasma Amylase Assay
Heparinized blood plasma (1 μL) was used to measure amylase activity with a kinetic assay, as described previously.23 The rate of substrate cleavage was expressed in mOD/min unit.
Histology
Pancreas tissue was fixed and stored in 10% neutral buffered formalin, paraffin-embedded, sectioned, and stained with hematoxylin-eosin (UCLA Translational Pathology Core Laboratory and Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center). Pancreas sections of cerulein-treated mice were scored by visual inspection for 3 parameters of acute pancreatitis (tissue edema, inflammatory cell infiltration, and acinar cell necrosis) and for intact acinar cells in CP. Pancreas sections from saline-treated mice were not scored because they exhibited no histological changes. Scoring was performed in a blinded fashion by 2 independent observers, and their results were averaged. Pancreas sections were scanned with the Pannoramic DESK II DW slide scanner (3DHISTECH) and the files were viewed with the SlideViewer (version 2.6, 3DHISTECH) software. Using the 1.5× magnification setting, the entire tissue was visualized and divided into 10 equal squares with the “Annotation” feature. The squares covered about 75% to 80% of the total tissue area. All 10 squares were scored, and the average was used as the final score for a given slide. Edema, inflammatory cells, and intact acini in CP were scored using the 10× magnification setting while acinar cell necrosis was scored at 10 to 20×.
To estimate the degree of tissue edema, an arbitrary scale of 0 to 3 was used, where 0 indicated the absence of visible edema. A score of 1 denoted slight widening of the inter-lobular spaces, corresponding to <10% of the area inspected, with no significant separation of acini due to inter-acinar edema. A score of 2 specified moderate widening of the inter-lobular spaces, corresponding to 10% to 50% of the area analyzed, with slight but visible separation of acini due to inter-acinar edema. A score of 3 indicated marked widening of the inter-lobular spaces, corresponding to >50% of the visual area, together with significant separation of acini due to inter-acinar edema. In cases where the degree of tissue edema was not uniform within a given square, scores of 0.5, 1.5, and 2.5 were used, as appropriate.
To evaluate the extent of inflammatory cell infiltration, an arbitrary scale of 0 to 3 was used, where 0 indicated the absence of visible inflammatory cells. A score of 1 denoted the presence of low-density infiltrates (<5 cells per 0.01 mm2 area) in the inter-lobular spaces, corresponding to <10% of the area analyzed, with no inflammatory cells around the acini. A score of 2 indicated medium-density infiltrates (5–20 cells per 0.01 mm2 area) within the inter-lobular spaces observed in 10% to 50% of the visual field, with some inflammatory cells visible in the inter-acinar spaces. A score of 3 specified high-density infiltrates (>20 cells per 0.01 mm2 area) in the widened inter-lobular spaces visible in >50% of the tissue area inspected. In cases where the inflammatory cell infiltration was not uniform within a given square, scores of 0.5, 1.5, and 2.5 were used, as appropriate.
The degree of acinar cell necrosis in acute pancreatitis and the proportion of intact acinar cells in CP were expressed as percent of the total tissue area examined. Necrosis was defined as partial or complete loss of acinar cell histoarchitecture, including areas with amorphous, pink material, acinar cell ghosts, and swollen, vacuolated acinar cells.32
Statistics
Results were graphed as individual data points with the mean and standard deviation values indicated. The difference of means between 2 groups and within multiple groups was analyzed by 2-tailed unpaired t-test and by 1-way analysis of variance with Tukey’s post-hoc analysis, respectively. P < .05 was considered statistically significant.
Acknowledgments
CRediT Authorship Contribution
Alexandra Demcsák (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Project administration: Supporting; Writing – original draft: Supporting; Writing – review & editing: Equal)
Miklós Sahin-Tóth (Conceptualization: Lead; Data curation: Supporting; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Supporting; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the National Institutes of Health grants R01 DK117809 and R01 DK082412 (to Miklós Sahin-Tóth), the National Pancreas Foundation award 20233127, the Hirshberg Foundation for Pancreatic Cancer Research award 20230501, and the Department of Defense grant HT9425-24-1-0164, PR230469 (to Alexandra Demcsák).
Data Availability All materials are available for research purposes upon request. All data are included in the manuscript.
References
- 1.Rinderknecht H. Activation of pancreatic zymogens. Normal activation, premature intrapancreatic activation, protective mechanisms against inappropriate activation. Dig Dis Sci. 1986;31:314–321. doi: 10.1007/BF01318124. [DOI] [PubMed] [Google Scholar]
- 2.Hegyi E., Sahin-Tóth M. Genetic risk in chronic pancreatitis: the trypsin-dependent pathway. Dig Dis Sci. 2017;62:1692–1701. doi: 10.1007/s10620-017-4601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabó A., Toldi V., Gazda L.D., et al. Defective binding of SPINK1 variants is an uncommon mechanism for impaired trypsin inhibition in chronic pancreatitis. J Biol Chem. 2021;296 doi: 10.1016/j.jbc.2021.100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morales Granda N.C., Toldi V., Miczi M., et al. Inhibition of mouse trypsin isoforms by SPINK1 and effect of human pancreatitis-associated mutations. Pancreatology. 2023;23:358–366. doi: 10.1016/j.pan.2023.04.043. [DOI] [PubMed] [Google Scholar]
- 5.Witt H., Luck W., Hennies H.C., et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 6.Pfützer R.H., Barmada M.M., Brunskill A.P., et al. SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000;119:615–623. doi: 10.1053/gast.2000.18017. [DOI] [PubMed] [Google Scholar]
- 7.Aoun E., Chang C.C., Greer J.B., et al. Pathways to injury in chronic pancreatitis: decoding the role of the high-risk SPINK1 N34S haplotype using meta-analysis. PLoS One. 2008;3:e2003. doi: 10.1371/journal.pone.0002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Leo M., Bianco M., Zuppardo R.A., et al. Meta-analysis of the impact of SPINK1 p.N34S gene variation in Caucasic patients with chronic pancreatitis. An update. Dig Liver Dis. 2017;49:847–853. doi: 10.1016/j.dld.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Tang X.Y., Zou W.B., Yu F.F., et al. Meta-analysis of the impact of the SPINK1 c.194+2T>C variant in chronic pancreatitis. Dig Liver Dis. 2020;52:143–148. doi: 10.1016/j.dld.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Kume K., Masamune A., Kikuta K., Shimosegawa T. [-215G>A; IVS3+2T>C] mutation in the SPINK1 gene causes exon 3 skipping and loss of the trypsin binding site. Gut. 2006;55:1214. doi: 10.1136/gut.2006.095752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kereszturi E., Király O., Sahin-Tóth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut. 2009;58:545–549. doi: 10.1136/gut.2008.164947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang X.Y., Lin J.H., Zou W.B., et al. Toward a clinical diagnostic pipeline for SPINK1 intronic variants. Hum Genomics. 2019;13:8. doi: 10.1186/s40246-019-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohmuraya M., Hirota M., Araki M., et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology. 2005;129:696–705. doi: 10.1016/j.gastro.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 14.Ohmuraya M., Hirota M., Araki K., et al. Enhanced trypsin activity in pancreatic acinar cells deficient for serine protease inhibitor kazal type 3. Pancreas. 2006;33:104–106. doi: 10.1097/01.mpa.0000226889.86322.9b. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto D., Ohmuraya M., Wang J., et al. Effect of low-molecular weight trypsin inhibitor, nafamostat mesilate, on trypsin activity using the pancreatic acinar cells. Pancreas. 2009;38:595–597. doi: 10.1097/MPA.0b013e31819156de. [DOI] [PubMed] [Google Scholar]
- 16.Romac J.M., Ohmuraya M., Bittner C., et al. Transgenic expression of pancreatic secretory trypsin inhibitor-1 rescues SPINK3-deficient mice and restores a normal pancreatic phenotype. Am J Physiol Gastrointest Liver Physiol. 2010;298:G518–G524. doi: 10.1152/ajpgi.00431.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan J.D., Romac J., Peng R.Y., et al. Transgenic expression of pancreatic secretory trypsin inhibitor-I ameliorates secretagogue-induced pancreatitis in mice. Gastroenterology. 2005;128:717–727. doi: 10.1053/j.gastro.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 18.Nathan J.D., Romac J., Peng R.Y., et al. Protection against chronic pancreatitis and pancreatic fibrosis in mice overexpressing pancreatic secretory trypsin inhibitor. Pancreas. 2010;39:e24–e30. doi: 10.1097/MPA.0b013e3181bc45e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romac J.M., Shahid R.A., Choi S.S., et al. Pancreatic secretory trypsin inhibitor I reduces the severity of chronic pancreatitis in mice overexpressing interleukin-1β in the pancreas. Am J Physiol Gastrointest Liver Physiol. 2012;302:G535–G541. doi: 10.1152/ajpgi.00287.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakata K., Araki K., Nakano H., et al. Novel method to rescue a lethal phenotype through integration of target gene onto the X-chromosome. Sci Rep. 2016;6 doi: 10.1038/srep37200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun C., Liu M., An W., et al. Heterozygous Spink1 c.194+2T>C mutant mice spontaneously develop chronic pancreatitis. Gut. 2020;69:967–968. doi: 10.1136/gutjnl-2019-318790. [DOI] [PubMed] [Google Scholar]
- 22.Geisz A., Sahin-Tóth M. A preclinical model of chronic pancreatitis driven by trypsinogen autoactivation. Nat Commun. 2018;9:5033. doi: 10.1038/s41467-018-07347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jancsó Z., Sahin-Tóth M. Mutation that promotes activation of trypsinogen increases severity of secretagogue-induced pancreatitis in mice. Gastroenterology. 2020;158:1083–1094. doi: 10.1053/j.gastro.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H., Swidnicka-Siergiejko A.K., Daniluk J., et al. Transgenic expression of PRSS1R122H sensitizes mice to pancreatitis. Gastroenterology. 2020;158:1072–1082.e7. doi: 10.1053/j.gastro.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui F., Zhang Y., Wan J., et al. Trypsin activity governs increased susceptibility to pancreatitis in mice expressing human PRSS1R122H. J Clin Invest. 2020;130:189–202. doi: 10.1172/JCI130172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan J., Haddock A., Edenfield B., et al. Transgenic expression of human PRSS2 exacerbates pancreatitis in mice. Gut. 2020;69:2051–2052. doi: 10.1136/gutjnl-2019-320399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Wan J., Wang L., et al. Wild-type human PRSS2 and PRSS1R122H cooperatively initiate spontaneous hereditary pancreatitis in transgenic mice. Gastroenterology. 2022;163:313–315.e4. doi: 10.1053/j.gastro.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demcsák A., Sahin-Tóth M. Rate of autoactivation determines pancreatitis phenotype in trypsinogen mutant mice. Gastroenterology. 2022;163:761–763. doi: 10.1053/j.gastro.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jancsó Z., Sahin-Tóth M. Chronic progression of cerulein-induced acute pancreatitis in trypsinogen mutant mice. Pancreatology. 2022;22:248–257. doi: 10.1016/j.pan.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pesei Z.G., Jancsó Z., Demcsák A., et al. Preclinical testing of dabigatran in trypsin-dependent pancreatitis. JCI Insight. 2022;7 doi: 10.1172/jci.insight.161145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen J.N., Cameron E., Garay M.V., et al. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Geisz A., Tran T., Orekhova A., Sahin-Tóth M. Trypsin activity in secretagogue-induced murine pancreatitis is solely elicited by cathepsin B and does not mediate key pathologic responses. Gastroenterology. 2023;164:684–687.e4. doi: 10.1053/j.gastro.2023.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya C., Navina S., Singh V.P. Role of pancreatic fat in the outcomes of pancreatitis. Pancreatology. 2014;14:403–408. doi: 10.1016/j.pan.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel K., Trivedi R.N., Durgampudi C., et al. Lipolysis of visceral adipocyte triglyceride by pancreatic lipases converts mild acute pancreatitis to severe pancreatitis independent of necrosis and inflammation. Am J Pathol. 2015;185:808–819. doi: 10.1016/j.ajpath.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Oliveira C., Khatua B., Noel P., et al. Pancreatic triglyceride lipase mediates lipotoxic systemic inflammation. J Clin Invest. 2020;130:1931–1947. doi: 10.1172/JCI132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y.C., Mao X.T., Sun C., et al. Pancreas-directed AAV8-hSPINK1 gene therapy safely and effectively protects against pancreatitis in mice. Gut. 2024 Mar 29 doi: 10.1136/gutjnl-2023-330788. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 37.Mosztbacher D., Demcsák A., Sahin-Tóth M. Measuring digestive protease activation in the mouse pancreas. Pancreatology. 2020;20:288–292. doi: 10.1016/j.pan.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]