Abstract
The African swine fever virus (ASFV) gene E165R, which is homologous to dUTPases, has been characterized. A multiple alignment of dUTPases showed the conservation in ASFV dUTPase of the motifs that define this protein family. A biochemical analysis of the purified recombinant enzyme showed that the virus dUTPase is a trimeric, highly specific enzyme that requires a divalent cation for activity. The enzyme is most probably complexed with Mg2+, the preferred cation, and has an apparent Km for dUTP of 1 μM. Northern and Western blotting, as well as immunofluorescence analyses, indicated that the enzyme is expressed at early and late times of infection and is localized in the cytoplasm of the infected cells. On the other hand, an ASFV dUTPase-deletion mutant (vΔE165R) has been obtained. Growth kinetics showed that vΔE165R replicates as efficiently as parental virus in Vero cells but only to 10% or less of parental virus in swine macrophages. Our results suggest that the dUTPase activity is dispensable for virus replication in dividing cells but is required for productive infection in nondividing swine macrophages, the natural host cell for the virus. The viral dUTPase may play a role in lowering the dUTP concentration in natural infections to minimize misincorporation of deoxyuridine into the viral DNA and ensure the fidelity of genome replication.
The processes that preclude the presence of uracil in DNA represent basic maintenance operations in living cells. The uracil misincorporation into DNA seems to be the origin of the so-called “thymineless death”: the abasic site created after uracil excision by an uracil-DNA glycosylase cannot be completely repaired with the correct thymine base if the dUTP/TTP ratio is too high (13). The futile cycle of excision and repair with a new uracil provokes multiple DNA strand breaks and, finally, cell death, through a mechanism that, at least in colon carcinoma cells, is mediated via fas signaling (25).
The presence of uracil in DNA alters specific DNA-protein interactions. Thus, it has been described (21) that interactions between the HSV-1-encoded origin binding protein and its target DNA sequence are altered by the presence of uracil residues in the central region of this sequence. The uracil present in regulatory sequences (such as promoters, origins of replication, enhancers, etc.) can also affect the regulation of gene expression and DNA replication (42).
Although dUMP can be introduced into DNA during the process of DNA synthesis, the spontaneous deamination of cytosine residues can also lead to the presence of uracil in the DNA. In the first case, dU-dA pairs are created, while the deamination of cytosine originates dU-dG pairs. Both of these DNA lesions are highly mutagenic and are ultimately lethal to the cell (26, 27). The deoxyuridine-containing DNA can be repaired by a base excision repair (BER) process (6) that begins with the removal of the wrong base by a uracil-DNA glycosylase. The utilization of dUTP instead of TTP to fill the gap created by the BER process is prevented by the action of a dUTPase (EC 3.6.1.23). This ubiquitous enzyme not only eliminates the dUTP from the deoxynucleoside triphosphate (dNTP) pool, which is important to reduce the presence of uracil in DNA, but also generates dUMP, the precursor for the production of TMP by thymidylate synthase (29).
The presence of dUTPase in several virus families, such as retroviruses, herpesviruses, and poxviruses (9, 18, 55), suggests that the strict control of the dUTP levels is critical in the replication of many viruses (44, 50). It has been shown in the caprine arthritis-encephalitis virus that the viral dUTPase prevents G-to-A substitutions in the virus DNA (52), thus acting as an antimutator agent by promoting a low dUTP concentration. On the other hand, infections in vivo with dUTPase mutants showed a strong reduction in virus production in the case of equine lentivirus (33) and reduced neurovirulence, neuroinvasiveness, and reactivation from latency in herpes simplex virus type 1 (43).
African swine fever virus (ASFV) is an icosahedral virus with a double-stranded DNA genome of about 170 kbp that causes a fatal disease in domestic pigs (53). Morphologically, ASFV is very similar to the iridoviruses that infect vertebrates (3, 11), but the structure of its DNA resembles that of the poxvirus genome (53). Due to its special features, ASFV has been assigned to a new family, Asfarviridae, as a member of the genus Asfivirus (16).
ASFV encodes two systems that may be crucial to ensure the virus genome integrity. One is a set of enzymes necessary for DNA precursor synthesis, such as ribonucleotide reductase (8), thymidine kinase (7), and thymidylate kinase (56). Most probably, this set of enzymes constitutes a system both to bypass the regulation of the corresponding cellular enzymes and to target the viral activities to cytoplasmic sites of viral DNA replication in the infected cells. The other is a BER system (38) that could play a role in correcting the base damages introduced in the viral DNA. In addition, ASFV encodes a protein homologous to dUTPases (58) that could also play an essential role in maintaining the integrity of the viral DNA both by reducing the dUTP levels and by providing the substrate for the thymidylate synthase.
In the present work, we have cloned the ASFV dUTPase and characterized the enzyme as an homotrimer that requires Mg2+ and shows a high specificity for dUTP. Using a dUTPase-deletion mutant, it has been found that the enzyme is required for efficient replication of the virus in nondividing swine macrophages. The importance of maintaining a high cytosolic TTP/dUTP ratio for the viral cycle is discussed in relation to the biochemical characteristics of the enzyme and the growth properties of the dUTPase-defective virus in cultured cells.
MATERIALS AND METHODS
Nucleotides.
The unlabeled nucleotides dCTP, dATP, dGTP, and TTP were purchased from Pharmacia P-L Biochemicals. dUMP and dUTP were purchased from Sigma Chemical Co. [5-3H]dUTP (17 Ci/mmol) and [γ-32P]ATP (3,000 Ci/mmol) were obtained from Amersham International Plc.
Oligonucleotides.
Oligonucleotides DUT-1 (5′-GCGCGCGGATCCATGGCAACAAATTTTTTTATTCAAC) and DUT-2 (5′-CGCGCGCTGCAGTTAAGTTCTCATAATCCCGGCCTCG) were used for PCR amplification of ASFV gene E165R. Oligonucleotide DUT-3 (5′-AAGGTGGGTAGTATGCTTCAGCTTCTTGGG), complementary to nucleotides +61 to +32 of the E165R gene noncoding strand, was used as probe in Northern blot and primer extension analyses. Oligonucleotides DUT-4 (5′-GGCACAATACTGGCTATACGC) and DUT-5 (5′-CGACTGCTTCTGAGTTGCGTT) were used for PCR amplification of E165R gene and flanking sequences.
Amino acid sequence comparisons and phylogenetic analysis.
We utilized the BLAST2 (4, 5, 28) search facilities at the European Molecular Biology Laboratory to detect the amino acid sequences homologous to the ASFV dUTPase. The sequences were then retrieved from public databases and multiple alignment was performed online by using the CLUSTALW program (24) at the European Bioinformatics Institute. Final refinements were done manually. Phylogenetic analysis was performed and analyzed for statistical confidence by using distance, parsimony, and bootstrapping programs from the PHYLIP package (version 3.55c) (20).
Expression of ASFV protein pE165R in E. coli.
The open reading frame (ORF) corresponding to the putative dUTPase gene E165R from ASFV (58) was cloned into the pRSET-A bacterial expression vector. This plasmid allows the expression of recombinant proteins as fusions with a multifunctional leader peptide containing a hexahistidyl sequence for purification on Ni2+ affinity resins (30). ORF E165R was PCR amplified from a recombinant plasmid containing the EcoRI E restriction fragment of ASFV strain BA71V (32) by using oligonucleotides DUT-1 (containing a GCGCGC tail and a BamHI restriction site) and DUT-2 (containing a CGCGCG tail and a PstI restriction site). The resulting 0.5-kb PCR product was cloned at the BamHI/PstI sites of vector pRSET-A. E. coli JM109 was used as a host for transformation. The construction of the recombinant expression plasmid, named pRSET-dUTPase, was confirmed by DNA sequencing. Expression of the His-tagged pE165R protein was carried out in E. coli BL21(DE3)/pLysS, which contains the T7 RNA polymerase gene under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter and a plasmid constitutively expressing T7 lysozyme (48). Cells were transformed with plasmid pRSET-dUTPase and grown overnight in Luria-Bertani (LB) medium at 37°C. Flasks containing LB broth were inoculated with a 1/10 volume of an overnight culture of E. coli and incubated in a rotatory shaker at 37°C until the absorbance at 595 nm reached 0.6. Then, IPTG (Sigma) was added to a final concentration of 0.4 mM, and incubation was continued for 4 h at 37°C. Cells were collected by centrifugation for 15 min at 1,900 × g and washed twice with buffer A (50 mM phosphate buffer, pH 7.5; 500 mM NaCl; 20 mM imidazole). After resuspension in the same buffer, the cells were sonicated on ice, and the suspension was then cleared by centrifugation for 10 min at 1,900 × g. The recombinant 21.9 kDa protein was soluble under these conditions, since it remained in the supernatant after centrifugation for 20 min at 14,500 × g. The recombinant protein was analyzed by polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS), in a 7 to 20% polyacrylamide gradient, and visualized by Coomassie blue staining.
Purification of ASFV dUTPase.
Ni-nitriloacetic acid (Ni-NTA) agarose beads (QIAgen), previously equilibrated in buffer A, were added to the soluble fraction containing the recombinant protein, obtained as described above. After being stirred for 1 h at 4°C, the resin was loaded into a column and extensively washed with buffer A. The recombinant ASFV protein pE165R was eluted from the column with buffer B (50 mM phosphate buffer, pH 7.5; 500 mM NaCl; 500 mM imidazole). The eluate was either diluted one-half with 100% glycerol and stored at −20°C or loaded onto a 5-ml glycerol gradient (15 to 30%) containing 50 mM Tris-HCl (pH 7.5), 20 mM ammonium sulfate, 180 mM NaCl, 1 mM EDTA, and 7 mM 2-β-mercaptoethanol; it was then centrifuged at 62,000 rpm (Beckman SW.65 rotor) for 28 h at 4°C. After centrifugation, 25 fractions were collected from the bottom of the tube, examined by SDS-PAGE, and tested for dUTPase activity under the conditions described below.
Antibodies.
Antibodies against the purified His-tagged dUTPase (recombinant dUTPase) were raised in rabbits. The immune serum obtained recognized the recombinant protein on Western blots.
Cells and viruses.
Vero (African green monkey kidney) cells were obtained from the American Type Culture Collection and grown in Dulbecco modified Eagle medium (DMEM) containing 5% newborn calf serum. Swine alveolar macrophages, obtained as described previously (10), were grown in DMEM containing 10% swine serum. The Vero adapted ASFV strain BA71V was propagated and titrated as described previously (19).
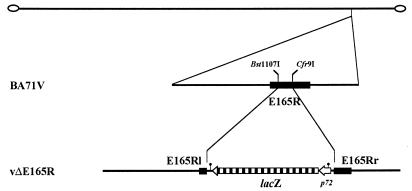
The dUTPase-defective mutant vΔE165R virus was obtained by insertion of the lacZ gene of E. coli into the E165R ORF. A 1,222-bp DNA fragment containing the E165R gene was generated by PCR by using the primers DUT-4 and DUT-5 and VentR DNA polymerase (New England Biolabs). The PCR product was cloned into EcoRV-digested pZErO-2 (Invitrogen), generating the plasmid pE165R. Plasmid p72GAL10T (22) was digested with HindIII, treated with Klenow enzyme and further digested with Cfr9I. The 3.3-kb fragment obtained was cloned into Bst1107I/Cfr9I-digested pE165R to obtain the transfer vector pΔE165R. Recombinant vΔE165R virus was generated as previously described (45). Briefly, Vero cells were infected with BA71V and transfected with pΔE165R. At 48 h postinfection, the cells were harvested, and diluted samples were used to infect Vero cell monolayers. The infected cells were covered with agar and, 4 days later, the β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added to the culture medium. The blue-stained recombinant plaques were selected and used to infect fresh monolayers of Vero cells. In this way, the recombinant virus vΔE165R was purified by three successive rounds of plaque isolation. Since runs of seven or more consecutive thymidylate (7T) residues in the coding strand are signals for 3′-end formation of ASFV mRNAs (2), 7T signals were placed in the transfer vector in order to minimize the risk of causing transcriptional disturbances when chimeric genes were inserted into the virus genome. The position of these signals in the viral genome of the ASFV mutant generated are indicated in Fig. 7.
FIG. 7.
Genomic structure of the ASFV recombinant vΔE165R. The lacZ gene fused to the viral late promoter p72 was inserted into the E165R gene of the ASFV strain BA71V, deleting a 171-bp Bst1107I-Cfr9I fragment from E165R as described in Materials and Methods. Positions of relevant endonuclease restriction sites and signals for 3′-end mRNA formation (•|) are indicated.
Preparation and analysis of RNA.
Vero cells were either mock infected or infected with ASFV (BA71V strain) at a multiplicity of infection of 20 PFU per cell. To obtain early RNA, the cells were infected in the presence of 100 μg of cycloheximide or 40 μg of cytosine arabinoside per ml for 7 h. Late RNA was isolated from cells infected for 18 h in the absence of inhibitors. Whole-cell RNA was prepared by the guanidinium isothiocyanate-cesium chloride extraction procedure (12). Northern blot hybridization was carried out as reported elsewhere (57), with the 32P-end-labeled oligonucleotide DUT-3 as a probe. The same oligonucleotide was used as a probe for primer extension analysis, which was performed essentially as described by Sambrook et al. (47). After 5′ end labeling with 32P, the primer was annealed at 42°C to the different classes of RNA and extended with avian myeloblastosis virus reverse transcriptase for 15 min at 42°C. The primer extension products were then electrophoresed in a 6% polyacrylamide gel and detected, after gel drying, by autoradiography.
Western blotting.
For Western blots, Vero cells were either mock infected or infected with ASFV as before and, at different times postinfection, the cells were lysed in electrophoresis sample buffer. Equivalent amounts of the cell lysates were electrophoresed in SDS-PAGE (7 to 20%) and subsequently transferred to nitrocellulose membranes. The membranes were blocked in T.TBS buffer (20 mM Tris-HCl, pH 7.5; 500 mM NaCl; 0.1% Tween 20) containing 5% dry milk powder for 1 h and then incubated overnight at 4°C with a 1:500 dilution of the antirecombinant dUTPase serum in T.TBS containing 1% dry milk powder. The membranes were washed with T.TBS, incubated with a peroxidase-labeled anti-rabbit serum (Amersham Life Sciences), and the proteins were detected with an electrochemiluminescence system (ECL System; Amersham Life Sciences) according to the manufacturer’s recommendations.
Immunofluorescence.
Vero cells, grown in chamber slides, were either mock infected or infected with ASFV at a multiplicity of infection of 1 PFU per cell and fixed at 14 h postinfection with methanol at −20°C for 5 min. The cells were then incubated with antirecombinant dUTPase serum in 0.1% bovine serum albumin (BSA) at 37°C for 1 h, rinsed three times for 10 min with phosphate-buffered saline (PBS) containing 0.1% BSA, and incubated with fluoresceinated goat anti-rabbit antibodies (Tago, Inc.). Nuclear and viral DNA were visualized by staining with 5 μg of bisbenzimide (Hoechst 33258; Sigma) per ml of PBS for 5 min. Cells were examined under a Zeiss Axiovert microscope.
Enzyme assays.
Unless otherwise indicated, dUTPase activity was determined in a standard 20-μl reaction mixture containing 50 mM Tris-HCl (pH 7.5), 2 mg of BSA per ml, 5 mM MgCl2, 2 mM dithiothreitol, 50 μM [5-3H]dUTP (50 μCi/mmol), and enzyme. Initial velocity studies were performed with dUTP concentrations ranging from 0.2 to 5 μM and 0.5 ng of enzyme. Divalent metal requirements were assessed in the same reaction mixture by substituting MgCl2 with MnCl2, CaCl2, CoCl2, ZnSO4, or NiSO4 at the optimal concentration for each cation. Competition experiments to assess the specificity of the enzyme were done in the standard reaction mixture with the addition of the unlabeled dNTPs (dGTP, dATP, dCTP, TTP, or dUMP) at 20 mM in each case.
All assay mixtures were incubated at 37°C. Reactions were terminated by adding EDTA to a final concentration of 25 mM. Then, 2-μl portions of the reaction mixtures were spotted on PEI-cellulose plates together with 2 μl of a solution containing 10 mM dUTP, 100 mM dUDP, and 100 mM dUMP as nucleotide markers. The plates were developed by using 0.5 M LiCl–2 M acetic acid as a solvent. The sheets were washed with 95% ethanol, air dried at room temperature, and examined under UV irradiation. The spots corresponding to the nucleotide markers were excised for liquid scintillation counting.
To visualize the reaction products, the thin-layer plates were exposed in some cases on a Fujifilm BAS-TR2040S imaging plate, which was analyzed with a Fuji BAS 1500 analyzer. Densitometric analysis of the bands was performed by using TINA 2.0 software.
RESULTS
ASFV protein pE165R belongs to the family of dUTPases.
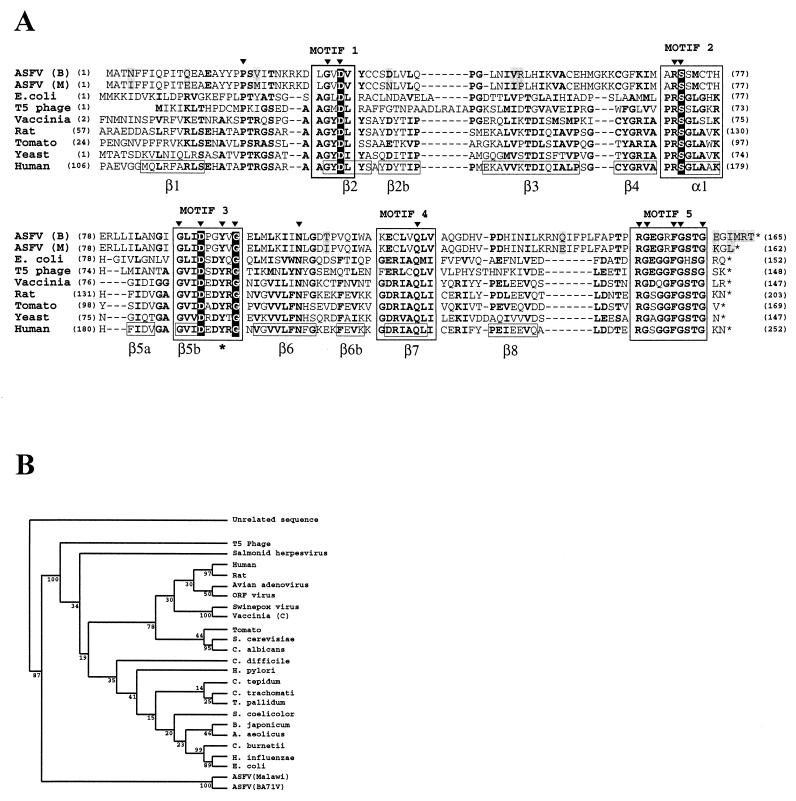
The nucleotide sequence of a 55-kbp region from the right end of the genome of a pathogenic ASFV isolate (Malawi LIL20/1) (15) shows the presence of a putative new member of the dUTPase family. Also, by complete DNA sequencing of the avirulent ASFV BA71V strain and subsequent data base searches for amino acid sequence similarities, it has been reported that the ASFV ORF designated E165R corresponds, with minor variations, to the ORF described in the Malawi strain (58). The prediction of a virus encoded dUTPase was tested by the multiple alignment of these sequences with some of the members of the dUTPase family (Fig. 1A), including two of the dUTPases, the E. coli (14, 31) and human (35) proteins, whose crystal structures have been described. Previously published comparisons of the primary structures of dUTPases from different sources (34) indicate the presence of five conserved motifs along the alignment, which are numbered and boxed in Fig. 1A. According to the human dUTPase structural data (35), the overall structure of the trimeric enzyme consists of three eight-stranded-β-barrels (the β-strands and the α-helix are numbered in Fig. 1A) with the C-terminal β-strands interchanged among the subunits. The enzyme has three active sites which occur at the subunit interfaces. Both ASFV ORFs corresponding to the dUTPase conserve the five motifs deduced from the sequence comparisons. In all sequences, 14 of the 16 invariant residues fall into the defined motifs. The specific recognition pocket for the uracil deoxyribonucleoside is formed by hydrogen bonding within the β5-β6 hairpin, around motif 3, which contains the invariant tyrosine that discriminates between the deoxyribose and the ribose of the incoming nucleotide (indicated with an asterisk at the bottom of the aligned sequences). Motif 5 acts as a barrier that protects the active site and is very well conserved in all of the proteins shown in the alignment. On the other hand, it has been shown that the dUTPase-encoding genes and the dCTP-deaminase encoding genes constitute a paralogous gene family (35). Four amino acids universally conserved throughout all these genes (indicated in white letters in Fig. 1A) are also found in the ASFV sequences [Asp33, Ser72, Asp91, and Gly96 in the ASFV (B) and (M) sequences]. The most variable region in the multiple alignment corresponds to that between motifs 4 and 5, where the ASFV protein presents an intervening sequence. Altogether, these observations suggest that the ASFV dUTPase could function as a trimer since it shows the characteristic sequence patterns of trimeric dUTPases, which were structurally determined in crystallographic studies.
FIG. 1.
(A) Multiple amino acid sequence alignment of protein pE165R and selected members of the dUTPase family. Numbers between parentheses indicate the amino acid position relative to the N terminus of each dUTPase. Five very conserved motifs can be recognized in the family (34), and they are named motifs 1 to 5 at the top of the alignment and shown in boxes. The invariant Tyr residue that discriminates, in the human dUTPase, between deoxyribose and ribose (35) is in boldface type and indicated with an asterisk at the bottom of the aligned sequences. Four amino acids universally conserved throughout the dUTPase-encoding genes and the dCTP-deaminase encoding genes (40) are indicated in white letters. The 16 invariant residues in all sequences are in boldface and are indicated by full triangles at the top of the sequences. Other invariant or conserved residues in at least 5 of the 9 sequences compared are in boldface. According to human dUTPase structural data (35), the α-helix and β-strands in the human dUTPase are indicated, boxed, and numbered at the bottom of the aligned sequences. The residues that differ in the ORFs of the two ASFV isolates are indicated over a gray background. Sequences: ASFV (B), ASFV BA71V strain (SwissProt Q65199); ASFV (M), ASFV Malawi strain (TREMBL Q65243); E. coli, E. coli (SwissProt P06968); T5 phage, T5 bacteriophage (SwissProt O48500); Vaccinia, vaccinia virus strain Copenhagen (SwissProt P21035); Rat, Rattus norvegicus (SwissProt P70583); Tomato, Lycopersicon esculentum (SwissProt P32518); Yeast, S. cerevisiae (SwissProt: P33317); Human (SwissProt P33316). (B) Phylogenetic analysis of ASFV dUTPase. The phylogenetic analysis was performed as described in Materials and Methods with the PHYLIP package. The numbers indicate the statistical confidence of the corresponding branches determined by bootstrapping (200 rounds).
The two ASFV ORFs differ in 13 residues (boxed in gray in the alignment) that are not located in the conserved motifs that define the family. The protein encoded by ORF E165R of ASFV BA71V is three amino acids longer than the equivalent protein of the Malawi virulent strain, as a result of an extension at the carboxyl end.
A phylogenetic analysis was done based on the multiple alignment by using the PHYLYP package (see Materials and Methods). The branching pattern was determined with a maximum parsimony method statistically tested by bootstrapping analysis. The global branching pattern is coincident with other previously published phylogenetic analysis (40), where the ASFV protein was not included. The presence of the new sequences does not change the global pattern (Fig. 1B), since the ASFV dUTPase sequences group together in a separate branch.
Expression and purification of the recombinant protein pE165R.
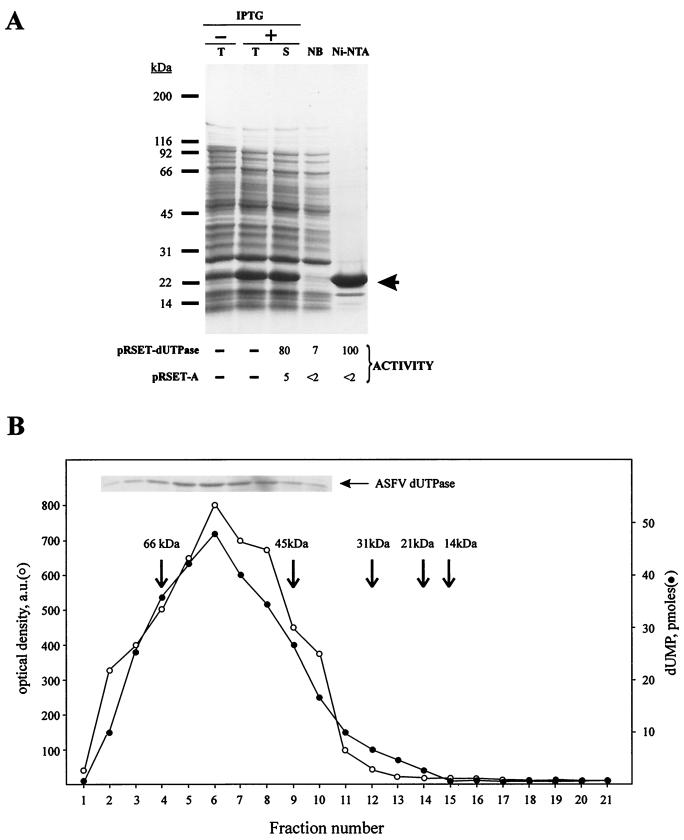
E. coli cells were transformed with plasmid pRSET-dUTPase that contains the ASFV BA71V gene E165R, potentially coding for a dUTPase protein. The plasmid expresses a recombinant dUTPase protein consisting of an N-terminal hexa-His tag (3.6 kDa) fused to protein pE165R (18.3 kDa). A polypeptide with a size corresponding approximately to that expected for the recombinant protein (21.9 kDa) was found in larger amounts in the crude extracts from cells induced with IPTG (Fig. 2A, lane +T) than in those from non-induced cells (lane −T). This protein, making up to 9% of the total E. coli protein (calculated after substracting the densitometric value of the protein band with the same mobility which is present in the noninduced cells), remained soluble under the extraction conditions used (Fig. 2A, lane S). Based on the His tag present at its N-terminal end, the recombinant dUTPase protein could be purified in a single step by Ni-NTA affinity chromatography (see Materials and Methods), although some polypeptide contaminants could be observed when loading high amounts (∼10 μg) of the purified fraction (Fig. 2A, lane Ni-NTA). A similar purification process was followed with extracts from cells transformed with plasmid pRSET-A (not shown) as a control for the dUTPase activity assays. The different fractions were assayed for dUTPase activity (Fig. 2A, bottom) by using equal amounts of protein (0.2 μg). The endogenous dUTPase activity of the E. coli cells transformed with the control vector (pRSET-A) is very low compared to the dUTPase activity of pRSET-dUTPase transformed cells. After purification, both the unbound fraction (NB) and the final fraction (Ni-NTA) corresponding to cells carrying the empty vector exhibit the same low activity than the initial fraction (S). By contrast, the purified fraction (but not the NB fraction) from pRSET-dUTPase transformed cells showed a strong dUTPase activity (Fig. 2A, bottom). We conclude that the pRSET-dUTPase transformed cells produce an active dUTPase that copurifies with the induced protein.
FIG. 2.
(A) Expression and purification of ASFV recombinant dUTPase. (Top) Coomassie blue staining after SDS-PAGE separation of total extracts from control non-induced (−T) and IPTG-induced (+T) E. coli BL21(DE3)/pLysS cells transformed with the recombinant plasmid pRSET-dUTPase. The soluble (S) fraction of the latter extract, prepared as described in Materials and Methods, was also analyzed. The electrophoretic analysis of the unbound fraction (NB) and the highly purified fraction (Ni-NTA) obtained after Ni-NTA chromatography of the soluble fraction is also shown. The electrophoretic migration of molecular weight markers is indicated on the left. The arrow shows the expected position for the recombinant protein. (Bottom) dUTPase activity present in the different fractions. dUTPase activity was determined in the soluble extracts from IPTG-induced bacterial cells containing either the control plasmid pRSET-A or the plasmid pRSET-dUTPase, as well as in the unbound and Ni-NTA fractions of the Ni-NTA column chromatography. Equal amounts of protein (200 ng) were used in all cases. The activities are indicated as the percentage of dUTP transformed into dUMP under the corresponding Coomassie blue-stained lanes in the case of pRSET-dUTPase and the equivalent positions in the case of the control pRSET-A. (B) Cosedimentation of dUTPase activity with recombinant ASFV dUTPase. The recombinant ASFV dUTPase protein eluted from the Ni-NTA column was sedimented through a glycerol gradient (15 to 30%) as described in Materials and Methods. The inset shows an SDS-PAGE analysis followed by Coomassie blue staining of gradient fractions 2 to 10. The stained dUTPase band in each fraction was quantified by densitometric analysis, and the values obtained are expressed as optical density arbitrary units (a.u.). The dUTPase activity in the different fractions is expressed as picomoles of dUMP generated. Arrows indicate the sedimentation position of several molecular mass markers centrifuged in the same gradient. The markers used were BSA (66 kDa), ovoalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21 kDa), and lysozyme (14 kDa).
Subunit structure of ASFV dUTPase.
In order to determine the molecular mass of the native enzyme and as an additional purification step, the Ni-NTA fraction was sedimented through a glycerol gradient, and the collected fractions were individually assayed for dUTPase activity. As shown in Fig. 2B, the observed single activity peak overlapped with the protein peak, sedimenting with an apparent molecular mass of ∼55 kDa, which corresponds, most probably, to a trimeric form of ASFV dUTPase. This result is in agreement with the trimeric structure described for dUTPases (14, 31, 35). Fractions 5 to 7 of the glycerol gradient were pooled and used for further in vitro analysis of ASFV dUTPase activity.
Enzymatic properties of ASFV dUTPase.
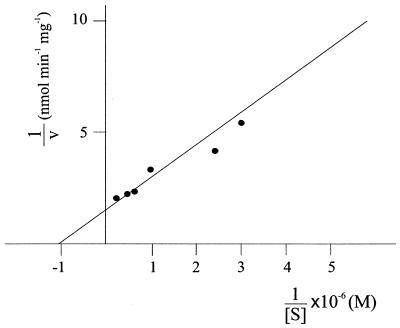
Initial velocity studies of the reaction catalyzed by the ASFV dUTPase were performed with 0.5 ng of enzyme and by varying the dUTP concentration between 0.2 and 5 μM, as described in Materials and Methods. The obtained Michaelis-Menten-type substrate kinetics indicated an apparent Km of 1 μM (Fig. 3).
FIG. 3.
Double-reciprocal plot of initial velocity of dUTPase reaction. Enzyme assays were done as described in Materials and Methods, with 0.5 ng of dUTPase and by varying the dUTP concentration between 0.2 and 5 μM.
Different divalent metals were added to the reaction mixture to assess whether the purified enzyme (1 ng in all cases) requires a specific divalent cation (Table 1). Although the enzyme shows a considerable activity in the absence of any added divalent cation, the addition of 5 mM MgCl2 to the reaction significantly increased the activity. However, the presence of other divalent cations, in particular Ni2+ and Ca2+, was inhibitory. Addition of 1 mM EDTA, a chelator of Mg2+ and Ca2+, resulted in the complete inhibition of dUTPase activity. This inhibitory effect could be reversed by the addition of MgCl2, or, less efficiently, by the addition of MnCl2 or ZnSO4. Also, the addition of CoCl2 or NiSO4 partially restored the activity. These results indicate that (i) the enzyme requires a divalent cation for its activity, (ii) the purified enzyme probably retains some Mg2+ that is sequestered by the addition of EDTA, and (iii) the preferred cation for activity is Mg2+.
TABLE 1.
Effect of added divalent cations on dUTPase activitya
| Added cationb (mM) | Relative activityc with:
|
|
|---|---|---|
| No EDTA | 1 mM EDTA | |
| None | 60 | 0 |
| Mg2+ (5) | 100 | 100 |
| Mn2+ (5) | 40 | 57 |
| Co2+ (5) | 50 | 26 |
| Zn2+ (5) | 50 | 52 |
| Ni2+ (25) | 35 | 30 |
| Ca2+ (10) | 15 | 0 |
The assay was as described in Materials and Methods by using 1 ng of enzyme in each reaction.
The effect of each cation was evaluated at its respective optimal concentration either directly or after incubation with 1 mM EDTA for 5 min. In all cases, incubation after the addition of the cation proceeded for 15 min at 37°C.
The results are expressed as the percentage of activity relative to that obtained with Mg2+. The average of three independent experiments is shown.
Substrate specificity was tested in competition experiments with other deoxynucleotides (Table 2). None of the deoxynucleotides showed a significant competition with dUTP despite the great excess (400-fold) of the unlabeled nucleotides used.
TABLE 2.
Effect of unlabeled deoxynucleotides on dUTPase activitya
| Added deoxynucleotide | Relative activityb |
|---|---|
| None | 100 |
| TTP | 98 |
| dGTP | 93 |
| dCTP | 92 |
| dATP | 83 |
| dUMP | 97 |
The assays were done in the presence of 50 μM [5-3H]dUTP (50 μCi/mmol) and 20 mM concentrations of each unlabeled deoxynucleotide by using the standard conditions described in Materials and Methods. Each reaction contained 1 ng of enzyme and was incubated for 15 min at 37°C.
The results are expressed as the percentage of activity relative to that obtained in the absence of competitor. The average of three independent experiments is shown.
Transcriptional analysis of ORF E165R.
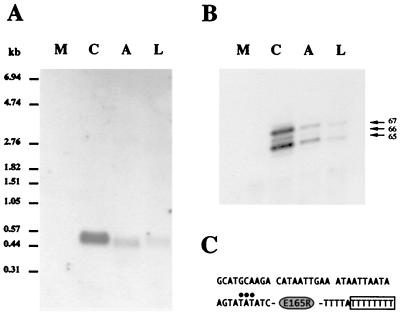
To study the expression of the ASFV BA71V E165R gene during the viral infection and to determine the transcription initiation site, Northern blot and primer extension analyses were carried out with the 32P-labeled oligonucleotide DUT-3 (see Materials and Methods), specific for this ORF, as a hybridization probe or as a primer. Hybridization of this oligonucleotide to Northern blots containing RNA from mock-infected Vero cells and early (cycloheximide and cytosine arabinoside) and late RNA from Vero cells infected with ASFV revealed a single RNA species of ca. 0.5 kb in the infected cells which was more abundant in the case of cycloheximide RNA (Fig. 4A).
FIG. 4.
Transcriptional analysis of E165R gene. (A) Northern blot analysis of E165R transcripts. Total RNA from mock-infected cells (M), early RNA from cells infected with ASFV in the presence of cycloheximide (C) or cytosine arabinoside (A), and late RNA from cells infected in the absence of inhibitors (L). The 32P-labeled oligonucleotide DUT-3 (see Materials and Methods) was used as a probe. The sizes (in kilobases) of RNA molecular mass markers are shown. (B) Primer extension analysis of the 5′ end of E165R transcripts. The same classes of RNA used for Northern blot analysis were hybridized with DUT-3 oligonucleotide and extended with avian myeloblastosis virus reverse transcriptase. The sizes (in nucleotides) of the major bands, calculated by using as markers an irrelevant DNA sequencing reaction, are indicated. (C) Sequence corresponding to the regions surrounding the E165R gene. Positions of transcription initiation sites are indicated by full circles. An empty box encloses the run of eight thymidylate residues potentially used for 3′-end formation of mRNA.
To map the 5′ ends of the transcription products of ORF E165R, the 32P-labeled oligonucleotide DUT-3 was annealed to the classes of RNA described above and extended with avian myeloblastosis virus reverse transcriptase, as described in Materials and Methods. In agreement with the Northern blot results, the oligonucleotide primer hybridized with early and late RNA. After extension with reverse transcriptase, three main bands were detected, corresponding to initiation of transcription at positions −4, −5, and −6 relative to the first nucleotide of the translation start codon (Fig. 4B and C).
A motif composed of seven consecutive thymidylate residues (the 7T motif), identified as a signal for 3′-end formation of ASFV mRNAs (2) was found five nucleotides downstream of the translation stop codon of ORF E165R (Fig. 4C). Transcription termination at this site would produce an RNA of approximately 570 bases, which is roughly the size of the RNA band detected by Northern blot hybridization.
ASFV dUTPase induction during infection.
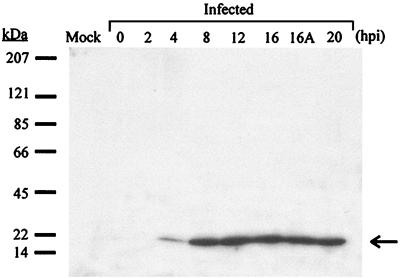
Antibodies raised against the recombinant dUTPase were used in Western blot analysis to determine the induction of ASFV dUTPase in infected Vero cells at different times postinfection, as described in Materials and Methods. A polypeptide band migrating at the expected position for protein pE165R (∼18.3 kDa) was observed at 4 hours postinfection and later, with maximal amounts between 8 and 20 h after infection (Fig. 5). The presence of the protein in extracts from cytosine arabinoside-treated cells (lane 16A) indicates that its expression begins before the onset of DNA replication, a finding in agreement with the transcriptional analysis.
FIG. 5.
ASFV dUTPase expression in infected cells analyzed by Western blot. Mock-infected or ASFV-infected Vero cells were lysed at different times postinfection and subjected to Western blot analysis by using an antirecombinant dUTPase serum. The number of hours postinfection (hpi) corresponding to each infected cell extract is indicated. The lane 16A corresponds to cells infected with ASFV in the presence of cytosine arabinoside and lysed at 16 hours postinfection. The arrow shows the expected position for ASFV dUTPase (18.3 kDa). The electrophoretic migration of molecular mass markers is shown on the left.
Immunolocalization of ASFV dUTPase in infected cells.
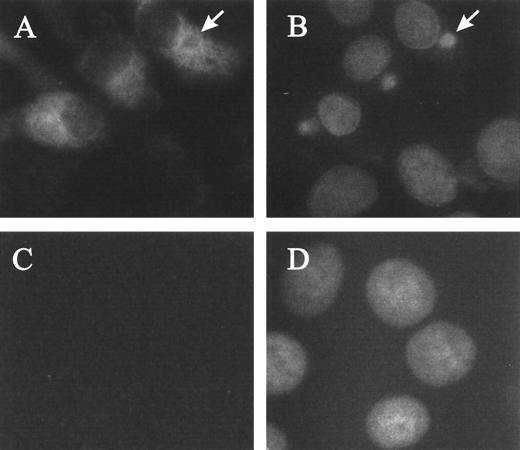
Immunofluorescence experiments were carried out to determine the subcellular localization of the protein encoded by ORF E165R. As illustrated in Fig. 6, infected cells (panel A) show a dispersed cytoplasmic staining pattern, while essentially no signal is detected in mock-infected cells (panel C). In some cases, the exclusion of the protein from the viral factory, as in the factory indicated by an arrow in panels A and B, is evident.
FIG. 6.
Immunofluorescence detection of ASFV dUTPase in infected Vero cells. Mock-infected or ASFV-infected Vero cells were fixed at 14 hours postinfection, incubated with antirecombinant dUTPase serum, and then stained with fluoresceinated goat anti-rabbit antibody and with a fluorescent DNA dye (Hoechst 33258) as described in Materials and Methods. Panels: A, anti-dUTPase staining pattern of ASFV-infected cells; B, DNA staining pattern of the field shown in panel A; C, anti-dUTPase staining pattern of mock-infected cells; D, DNA staining pattern of the field shown in panel C. The arrows in panels A and B indicate a viral factory from which the immunolabeling is excluded.
Construction of a dUTPase deletion mutant of ASFV.
To test the role of the dUTPase gene of ASFV in the infection, we constructed a recombinant E165R gene deletion mutant (vΔE165R), generated by in vivo homologous recombination. The plasmid vector pΔE165R, constructed as described in Materials and Methods, was designed to allow the replacement of an ASFV genomic DNA fragment of 171-bp with the marker gene lacZ fused to the virus promoter p72 (Fig. 7). This replacement would disrupt the E165R ORF and eliminate from the virus genome most of the protein sequence (from Tyr-35 to Asp-91), including motifs 2 and 3. The structure of vΔE165R is shown in Fig. 7, where the lacZ gene is transcribed in the opposite direction from E165R. The deletion of the dUTPase gene was confirmed by DNA hybridization and Western blot analysis of vΔE165R-infected Vero cells (data not shown).
vΔE165R replication kinetics in cultured cells.
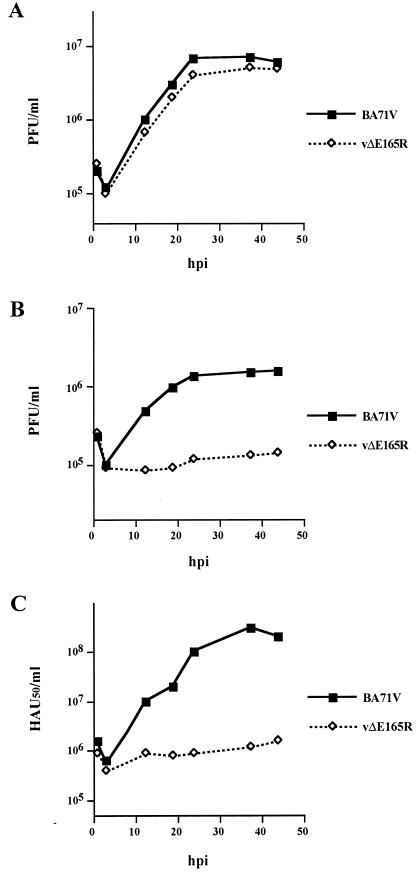
Although the mutant virus was successfully purified from cultured Vero cells, the growth properties of the recombinant vΔE165R in this cell line were examined in more detail and compared to those of the parental BA71V virus. The mutant and parental viruses were found to replicate with the same kinetics and to the same extent in Vero cells (Fig. 8A), confirming that deletion of the dUTPase gene had no effect on ASFV replication in this cell type.
FIG. 8.
Growth curves of parental BA71V and mutant vΔE165R viruses in Vero cells (A) and swine macrophage cultures (B and C). Vero cells or swine macrophages were infected with BA71V or vΔE165R at a multiplicity of infection of 5 PFU per cell. At different times (hours) postinfection, samples were collected and titrated by plaque assay on fresh Vero cells (A and B) or by hemadsorption assay (C) on swine macrophages as previously described (19).
We then investigated the replication kinetics of the parental and mutant viruses in cultured porcine macrophages, which are the main targets in natural ASFV infections. The virus production in these cells was measured both in Vero cells and in macrophages, finding a strong reduction of virus yield for vΔE165R compared with parental virus. Thus, the total yield of the mutant virus was reduced to ca. 10% of parental virus as determined by plaque assay in Vero cells (Fig. 8B) or to ca. 1% by hemadsorption assay in macrophages (Fig. 8C).
DISCUSSION
Sequence similarity comparisons based on the reported DNA sequence of ASFV strain BA71V (58) predicted the existence of viral mechanisms to ensure the integrity of the ASFV genome. ASFV appears to be the only virus described that may encode both a BER system (38), involved in repairing DNA base damages generated by hydrolysis or oxidation, and a dUTPase that would prevent the introduction of deoxyuridine into the viral DNA.
The most prominent feature of the described structure of dUTPases (31, 35, 41) is the fact that the three subunits of these trimeric enzymes collaborate to form each of the three active sites, each subunit supplying residues that are critical to enzyme function and catalysis. The high specificity of the enzyme is dependent on the interaction of all three subunits with the substrate. Thus, in human dUTPase, the uracil and deoxyribose are primarily recognized by one subunit and the phosphate groups by the adjacent subunit (35), whereas the bound substrate is capped by residues of the C-terminal tail of the third subunit. The multiple alignment of Fig. 1 shows that, although the ASFV dUTPase enzyme displays a limited global sequence homology in comparison to other dUTPases, as illustrated by the peripheral position of the enzyme in the phylogenetic branching pattern shown in Fig. 1B, it conserves all of the basic residues involved in the interactions that define the basis for catalysis. Thus, in the first subunit, Tyr193 of human dUTPase, which is invariant in the alignment, and packs against the base, corresponds to Tyr94 in ASFV dUTPase. On the other hand, in the second subunit, the phosphate groups are recognized by Arg173, Ser174, and Gly175 of human dUTPase, corresponding to Arg71, Ser72, and Ser73 in the ASFV enzyme, respectively. Finally, Phe155 in the ASFV protein corresponds to Phe246 located in the C-terminal substrate-capping region in the third subunit of the human dUTPase. This region corresponds to motif 5, the most conserved sequence of the whole alignment, where Phe246 stacks above the bound uracil base providing key hydrophobic interactions.
To characterize biochemically the ASFV dUTPase, we have purified a recombinant enzyme expressed in E. coli. Since an hexa-His tag at the amino terminus of the protein has proved to be useful for obtaining active and highly purified human dUTPase, suitable even for crystallization purposes (35), we utilized the same strategy to facilitate the purification process. Essentially, no dUTPase activity was found in the Ni-NTA fractions from extracts of E. coli cells transformed with plasmid pRSET-A lacking the ASFV E165R gene, suggesting that the purified viral enzyme is free of contaminating endogenous hydrolases. The purified ASFV dUTPase is a trimeric enzyme, as deduced from the observed sedimentation in glycerol gradients, and exhibits many of the characteristics of other dUTPases (46) since it is highly specific for dUTP and shows a high affinity for this substrate (Km = 1 μM). This high specificity probably reflects the necessary selectivity of the systems that control the nucleotide levels in the cells (29). The purified enzyme requires a divalent cation for activity, with a marked preference for Mg2+, and is probably complexed with this cation since it shows activity in the absence of added divalent cations and since this activity can be abolished by EDTA and restored by the addition of Mg2+. These structural and biochemical properties are in agreement with the features expected from the sequence comparisons. On the basis of the global homology (92% identity) and the strict conservation of the regions necessary for the activity, we conjecture that the biochemical properties of the dUTPase from the ASFV virulent strain Malawi are, probably, very similar to those described here.
Certain viruses have acquired mechanisms, such as the encoding of dUTPases that are free of normal cellular regulatory constraints, to prevent the danger represented by the synthesis of uracil-substituted DNA. The ASFV dUTPase could function in this way. In macrophages the pool of deoxynucleotides in general, and that of TTP and dCTP in particular, is very low (<1 pmol/106 cells), and there is no de novo synthesis of these nucleotides (49). The finding that the viral enzyme is localized in the cytoplasm of ASFV-infected cells and is expressed at both early and late times of infection is consistent with a role in maintaining a high TTP/dUTP ratio to minimize the introduction of uracil into the viral DNA. Although the viral dUTPase has not been found in the nucleus, it is likely that the surveillance role performed by the cytoplasmic enzyme could be sufficient to ensure also the fidelity of the viral DNA synthesis which occurs in the nucleus at earlier postinfection times (23).
In eukaryotic cells, removal of uracil that has been introduced into the DNA is performed by a BER process that also repairs other DNA base damages and DNA strand breaks generated by hydrolysis and oxidation (6). The first step in BER depends on the nature of the DNA damage. The presence of uracil requires the consecutive actions of a specific uracil-N-glycosylase (UNG) and an AP-endonuclease. However, a single-strand DNA break could be directly managed by this latter enzyme. A specific DNA polymerase and a DNA ligase catalyze the next steps in BER. The presence of such a basic set of three enzymes (an AP-endonuclease, a repair-specific DNA polymerase, and a DNA ligase) has been described only in two viruses, both possessing very large genomes: ASFV (38, 58) and a group B entomopoxvirus that infects the migratory grasshopper Melanoplus sanguinipes (MsEPV) (1). Interestingly, MsEPV also possesses a UNG and a photoreactivation system but lacks, in contrast to other poxviruses (36), the genes involved in nucleotide metabolism, including the dUTPase, despite the high susceptibility to dUMP introduction into the genome due to its high A+T content (81.7%). Possibly, in the dUTPase-lacking MsEPV, the UNG could be essential in a BER process to eliminate uracil in DNA.
Since our sequence analysis has not identified any protein with clear similarity to UNG (58), it is tempting to speculate that, in the absence of such an enzyme, an efficient dUTPase would be probably essential to prevent the frequent introduction of deoxyuridine during the replication of the large ASFV DNA genome. The replication kinetics of ASFV recombinant vΔE165R showed that dUTPase activity is necessary for virus growth in macrophages, the natural host cells, but not in Vero cells. The differences in virus replication observed between these two cell types could be due to the levels of cellular dUTPase. In relation to this, it has been shown that dUTPases are both developmental and cell cycle regulated (17, 37, 39), i.e., their levels are high in dividing cells and low in terminally differentiated and/or nondividing cells. The finding that the virus mutant replicated efficiently in actively dividing Vero cells could thus be due to the presence of an endogenous dUTPase activity sufficiently high to compensate for the lack of the virally encoded dUTPase. In contrast, nondividing swine macrophages may have low levels of cellular dUTPase activity and thus are unable to support efficient replication of the mutant. In the light of these results, we hypothesize that ASFV encodes a dUTPase for the establishment of infections in macrophages to compensate for a low cellular activity. A similar hypothesis has been proposed for a number of lentiviruses, where dUTPase mutants replicated deficiently in nondividing monocyte and macrophage cells and normally in dividing cells (50, 51, 54). It should be mentioned, however, that the normal replication kinetics of ASFV dUTPase mutant in Vero cells could also be explained by the existence of a high deoxynucleotide pool in dividing cells (49), as a high concentration of TTP will make the misincorporation of dUTP into the viral DNA unlikely.
Further studies with the vΔE165R mutant should reveal if uracil is incorporated into the viral DNA during virus replication and whether this incorporation results in higher mutation levels. In vivo experiments with pigs infected with dUTPase-defective virulent isolates of ASFV should also be of importance to assess the possible role of dUTPase in viral pathogenesis.
ACKNOWLEDGMENTS
This work was supported by Dirección General de Investigación Científica y Técnica grant PB96-0902-C02-01, European Community grant FAIR5-CT97-3441, Comunidad Autónoma de Madrid grant 07B/0032/1997, Ministerio de Educación y Cultura grant AGF98-1352-CE, and by an institutional grant from Fundación Ramón Areces. Alí Alejo was a fellow of the Ministerio de Educación y Cultura.
REFERENCES
- 1.Afonso C L, Tulman R, Lu Z, Oma E A, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán F, Rodríguez J M, Andrés G, Pérez R, Viñuela E, Rodríguez J F. Transcriptional analysis of multigene family 110 of African swine fever virus. J Virol. 1992;66:6655–6667. doi: 10.1128/jvi.66.11.6655-6667.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida J D, Waterson A P, Plowright W. The morphological characteristics of African swine fever virus and its resemblance to tipula iridescent virus. Arch Gesamte Virusforsch. 1967;20:392–396. doi: 10.1007/BF01241958. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 6.Barnes D E, Lindhal T, Sedgwick B. DNA repair. Curr Opin Cell Biol. 1993;5:424–433. doi: 10.1016/0955-0674(93)90007-d. [DOI] [PubMed] [Google Scholar]
- 7.Blasco R, Lopez-Otín C, Muñoz M, Bockamp E O, Simón-Mateo C, Viñuela E. Sequence and evolutionary relationships of African swine fever virus thymidine kinase. Virology. 1990;178:301–304. doi: 10.1016/0042-6822(90)90409-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boursnell M, Shaw K, Yáñez R J, Viñuela E, Dixon L. The sequences of the ribonucleotide reductase genes from African swine fever virus show considerable homology with those of the orthopoxvirus, vaccinia virus. Virology. 1991;184:411–416. doi: 10.1016/0042-6822(91)90860-e. [DOI] [PubMed] [Google Scholar]
- 9.Broyles S S. Vaccinia virus encodes a functional dUTPase. Virology. 1993;195:863–865. doi: 10.1006/viro.1993.1446. [DOI] [PubMed] [Google Scholar]
- 10.Carrascosa A L, Santarén J F, Viñuela E. Production and titration of African swine fever virus in porcine alveolar macrophages. J Virol Methods. 1982;3:303–310. doi: 10.1016/0166-0934(82)90034-9. [DOI] [PubMed] [Google Scholar]
- 11.Carrascosa J L, Carazo J M, Carrascosa A L, García N, Santisteban A, Viñuela E. General morphology and capsid fine structure of African swine fever virus particles. Virology. 1984;132:160–172. doi: 10.1016/0042-6822(84)90100-4. [DOI] [PubMed] [Google Scholar]
- 12.Chirgwin J M, Przbyla A E, McDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 13.Curtin N J, Harris A L, Aherne G W. Mechanism of cell death following thymidylate synthase inhibition: 2′-deoxyuridine-5′-triphosphate accumulation, DNA damage, and growth inhibition following exposure to CB3717 and dipyridamole. Cancer Res. 1991;51:2346–2352. [PubMed] [Google Scholar]
- 14.Dauter Z, Wilson K S, Larsson G, Nyman P O, Cedergren-Zeppezauer E S. The refined structure of dUTPase from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1998;54:735–749. doi: 10.1107/s0907444997016223. [DOI] [PubMed] [Google Scholar]
- 15.Dixon L K, Twigg S R, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smith G L. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate Malawi LIL20/1. J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 16.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Viñuela, and P. J. Wilkinson. The Asfarviridae. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: Seventh Report of the International Committee for the Taxonomy of Viruses, in press. Academic Press, New York, N.Y.
- 17.Duker N J, Grant C L. Alterations in the levels of deoxyuridine triphosphatase, uracil-DNA glycosylase and AP endonuclease during the cell cycle. Exp Cell Res. 1980;125:493–497. doi: 10.1016/0014-4827(80)90145-7. [DOI] [PubMed] [Google Scholar]
- 18.Elder J H, Lerner D L, Hasselkus-Light C S, Fontenot D J, Hunter E, Luciw P A, Montelaro R C, Phillips T R. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enjuanes L, Carrascosa A L, Moreno M A, Viñuela E. Titration of African swine fever virus. J Gen Virol. 1976;32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 21.Focher F, Verri A, Verzeletti S, Mazzarello P, Spadari S. Uracil in OriS of herpes simplex 1 alters its specific recognition by origin binding protein (OBP): does virus induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma. 1992;102:S67–S71. doi: 10.1007/BF02451788. [DOI] [PubMed] [Google Scholar]
- 22.García R, Almazán F, Rodríguez J M, Alonso M, Viñuela E, Rodriguez J F. Vectors for the genetic manipulation of African swine fever virus. J Biotech. 1995;40:121–131. doi: 10.1016/0168-1656(95)00037-q. [DOI] [PubMed] [Google Scholar]
- 23.García-Beato R, Salas M L, Viñuela E, Salas J. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology. 1992;188:637–649. doi: 10.1016/0042-6822(92)90518-t. [DOI] [PubMed] [Google Scholar]
- 24.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 25.Houghton J A, Harwood F G, Tillman D M. Thymineless death in colon carcinoma cells is mediated via fas signaling. Proc Natl Acad Sci USA. 1997;94:8144–8149. doi: 10.1073/pnas.94.15.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Impellizzeri K J, Anderson B, Burgers P M. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J Bacteriol. 1991;173:6807–6810. doi: 10.1128/jb.173.21.6807-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingraham H A, Dickey L, Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- 28.Karlin S, Altschul S F. Applications and statistics for multiple high-scoring segments in molecular sequences. Proc Natl Acad Sci USA. 1993;90:5873–5877. doi: 10.1073/pnas.90.12.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornberg A, Baker T A. DNA replication. 2nd ed. San Francisco, Calif: Freeman; 1992. [Google Scholar]
- 30.Kroll D J, Abedel-Malek Abdel-Hafiz H, Marcell T, Simpson S, Chen C Y, Gutierrez-Hartmann A, Lustbader J W, Hoeffler J P. A multifunctional prokaryotic protein expression system: overproduction, affinity purification, and selective detection. DNA Cell Biol. 1993;12:441–453. doi: 10.1089/dna.1993.12.441. [DOI] [PubMed] [Google Scholar]
- 31.Larsson G, Svensson L A, Nyman P O. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP) Nat Struct Biol. 1996;3:532–538. doi: 10.1038/nsb0696-532. [DOI] [PubMed] [Google Scholar]
- 32.Ley V, Almendral J M, Carbonero P, Beloso A, Viñuela E, Talavera A. Molecular cloning of African swine fever virus DNA. Virology. 1984;133:249–257. doi: 10.1016/0042-6822(84)90392-1. [DOI] [PubMed] [Google Scholar]
- 33.Lichenstein D L, Rushlow K E, Cook R F, Raabe M L, Swardon C J, Kociba G J, Issel C J, Montelaro R C. Replication in vitro and in vivo of a equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeoch D J. Protein sequence comparisons show that the “pseudoproteases” encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mol C D, Harris J M, McIntosh E M, Tainer J A. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 36.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 37.Nation M D, Guzder S N, Giroir L E, Deutsch W A. Control of Drosophila deoxyuridine triphosphatase: existence of a developmentally expressed protein inhibitor. Biochem J. 1989;259:593–596. doi: 10.1042/bj2590593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveros M, Yáñez R J, Salas M L, Salas J, Viñuela E, Blanco L. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J Biol Chem. 1997;272:30899–30910. doi: 10.1074/jbc.272.49.30899. [DOI] [PubMed] [Google Scholar]
- 39.Pardo E G, Gutierrez C. Cell cycle- and differentiation stage-dependent variation of dUTPase activity in higher plant cells. Exp Cell Res. 1990;186:90–98. doi: 10.1016/0014-4827(90)90214-u. [DOI] [PubMed] [Google Scholar]
- 40.Prangishvili D, Klenk H P, Jakobs G, Schmiechen A, Hanselmann C, Holz I, Zillig W. Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV. J Biol Chem. 1998;273:6024–6029. doi: 10.1074/jbc.273.11.6024. [DOI] [PubMed] [Google Scholar]
- 41.Prasad G S, Stura E A, McRee D E, Laco G S, Hasselkus-Light C, Elder J H, Stout C D. Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus. Protein Sci. 1996;5:2429–2437. doi: 10.1002/pro.5560051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu W T, Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: the GCN4-DNA complex. Nucleic Acids Res. 1992;20:771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyles R B, Sawtell N M, Thompson R L. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol. 1992;66:6706–6713. doi: 10.1128/jvi.66.11.6706-6713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyles R B, Thompson R L. Mutations in accessory DNA replicating functions alter the relative mutation frequency of herpes simplex virus type 1 strains in cultured murine cells. J Virol. 1994;68:4514–4524. doi: 10.1128/jvi.68.7.4514-4524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodríguez J M, Almazán F, Viñuela E, Rodriguez J F. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the β-galactosidase gene. Virology. 1992;188:67–76. doi: 10.1016/0042-6822(92)90735-8. [DOI] [PubMed] [Google Scholar]
- 46.Roseman N A, Evans R K, Mayer E L, Rossi M A, Slabaugh M B. Purification and characterization of the vaccinia virus deoxyuridine triphosphatase expressed in Escherichia coli. J Biol Chem. 1996;271:23506–23511. doi: 10.1074/jbc.271.38.23506. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 49.Terai C, Carson D A. Pyrimidine nucleotide and nucleic acid synthesis in human monocytes and macrophages. Exp Cell Res. 1991;193:375–381. doi: 10.1016/0014-4827(91)90110-g. [DOI] [PubMed] [Google Scholar]
- 50.Turelli P, Pétursson G, Guiguen F, Mornex J-F, Vigne R, Quérat G. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J Virol. 1996;70:1213–1217. doi: 10.1128/jvi.70.2.1213-1217.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Threadgill D S, Steagall W K, Flaherty M T, Fuller F J, Perry S T, Rushlow K E, Le Grice S F, Payne S L. Characterization of equine infectious anemia virus dUTPase: growth properties of a dUTPase-deficient mutant. J Virol. 1993;67:2592–2600. doi: 10.1128/jvi.67.5.2592-2600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turelli P, Guiguen F, Mornex J F, Vigne R, Quérat G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viñuela E. Molecular biology of African swine fever virus. In: Becker Y, editor. African swine fever. Boston, Mass: Martinus Nijhoff Publishing; 1987. pp. 31–49. [Google Scholar]
- 54.Wagaman P C, Hasselkus-Light C S, Henson M, Lerner D L, Phillips T R, Elder J H. Molecular cloning and characterization of deoxyuridine triphosphatase from feline immunodeficiency virus (FIV) Virology. 1993;196:451–457. doi: 10.1006/viro.1993.1501. [DOI] [PubMed] [Google Scholar]
- 55.Wohlrab F, Francke B. Deoxyribopyrimidine triphosphatase activity specific for cells infected with herpes simplex virus type 1. Proc Natl Acad Sci USA. 1980;77:1872–1876. doi: 10.1073/pnas.77.4.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yáñez R J, Rodríguez J M, Rodriguez J F, Salas M L, Viñuela E. African swine fever virus thymidylate kinase gene: sequence and transcriptional mapping. J Gen Virol. 1993;74:1633–1638. doi: 10.1099/0022-1317-74-8-1633. [DOI] [PubMed] [Google Scholar]
- 57.Yáñez R J, Viñuela E. African swine fever virus encodes a DNA ligase. Virology. 1993;193:531–536. doi: 10.1006/viro.1993.1161. [DOI] [PubMed] [Google Scholar]
- 58.Yáñez R J, Rodríguez J M, Nogal M L, Yuste L, Enríquez C, Rodríguez J F, Viñuela E. Analysis of the complete sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]