Abstract
Background
Primary Carnitine Deficiency (PCD) is a potentially life-threatening autosomal recessive monogenic disorder arising from mutations in the organic cation transporter 2 (OCTN2) gene. Dilated cardiomyopathy (DCM) is a prevalent symptom associated with this condition, and episodes of metabolic disturbance may lead to sudden death. However, the pathogenic mechanism remains unclear. Here, we sought to investigate the response of cardiomyocytes and alterations in the intercellular communication in individuals with PCD DCM.
Methods
The GSE211650 dataset was downloaded. Subsequently, modular analysis was performed using hdWGCNA. SCENIC was employed for transcription factor analysis. Monocle2 and SCP were applied to conduct trajectory inference and characterize dynamic features. CellChat was used to investigate intercellular interactions.
Results
OCTN2-deficient cardiomyocytes displayed transcriptomic alterations indicative of reduced contractility, developmental abnormalities, and fibrosis. The reduced expression of genes encoding troponin, myosin, and calcium ion transporters may underlie the observed decrease in contractility. Suppressed Wnt signaling and downregulated transcription factors associated with myocardial development suggest potential developmental disturbances in cardiomyocytes. Growth arrest-specific 6 (GAS6) secreted by TNNC1 high cardiomyocytes is implicated in myocardial inflammation and fibrosis. Macrophages-derived secreted phosphoprotein 1 (SPP1) promotes the activation of fibroblasts. Furthermore, there was a reduction in neuronal genes in the OCTN2-deficient group.
Conclusions
Our research has unveiled, for the first time, the responses of cardiomyocytes and alterations in the intercellular communication in PCD DCM, offering valuable insights for the precision treatment of this condition.
Keywords: Single-nucleus RNA seq, Primary carnitine deficiency, Dilated cardiomyopathy, Organic cation transporter 2
Highlights
-
•
First report focus on cardiomyocytes in primary carnitine deficiency (PCD).
-
•
Cardiomyocytes display reduced contractility, abnormal development, and fibrosis in PCD.
-
•
TNNC1 high cardiomyocytes play a pivotal role in cardiac fibrosis and hypertrophy.
-
•
Growth arrest-specific 6 is implicated in myocardial inflammation and fibrosis.
-
•
Macrophages-derived secreted phosphoprotein 1 promotes the activation of fibroblasts.
1. Introduction
Primary Carnitine Deficiency (PCD, OMIM 212140) is an autosomal recessive hereditary condition among fatty acid oxidation disorders (FAODs). The prevalence of PCD shows variation across countries, with estimated rates of 1:142,000 in the United States, 1:40,000 in Japan, and even 1:300 in the Faroe Islands [[1], [2], [3]]. PCD arises from pathogenic variations in SLC22A5 (NC_ 000005.10), resulting in the impairment of the encoded organic cation transporter novel family member 2 (OCTN2) and subsequent loss of function [4,5]. OCTN2 is highly expressed in the myocardium, skeletal muscle, fibroblasts, renal tubules, placenta, and intestine [6]. It functions as both an organic cation transporter and a sodium-dependent high-affinity carnitine transporter, facilitating the uptake of carnitine into cells by co-transporting one molecule of carnitine with one molecule of sodium [7]. Mutations in the SLC22A5 gene impair OCTN2's ability to anchor to the cell membrane, causing it to remain in the cytoplasm or suffer structural and functional domain damage. These mutations lead to defects in OCTN2's transport function, reducing carnitine transfer from the intestines to the blood and from the blood to the cells, and impairing carnitine reabsorption in the renal proximal tubule system [8]. Consequently, carnitine levels in the blood and cells decrease, hindering the β-oxidation of fatty acids. Myocardial cells, which rely heavily on fatty acids, are particularly vulnerable to the pathological mechanisms of PCD.
PCD manifests with symptoms such as dilated cardiomyopathy (DCM), reduced cardiac function, muscle weakness, decreased muscle tone, and abnormal liver function [9,10]. Patients with PCD-related DCM exhibit diminished myocardial contractility and endocardial fibrosis, along with significant lipid accumulation in both ventricles. This was accompanied by cardiac hypertrophy and dilation, particularly noticeable in the left ventricle [11,12]. While an early diagnosis and treatment involving high-dose L-carnitine supplementation are life-saving and can reverse the pathology of PCD, it's worth noting that not all cases are shielded from the development of cardiomyopathy. Cardiac events have been documented in a small number of infants and asymptomatic adult patients with PCD. Lethal arrhythmias and progressive exacerbation of PCD DCM, leading to heart failure, are common causes of sudden cardiac death [9,[13], [14], [15]]. To better follow-up and treat PCD patients, thereby reducing potential lethal risks, it is essential to understand the detailed pathogenic mechanisms underlying PCD DCM.
Recently, single-cell and single-nucleus RNA sequencing (scRNA-seq and snRNA-seq, respectively) have been extensively applied in DCM research, playing a crucial role in revealing specific cellular responses and intercellular communication alteration [16,17]. Unfortunately, there is a notable absence of reports related to PCD DCM. Malte Loos et al. developed the first human induced pluripotent stem cell (hiPSC) model of PCD in vitro and uploaded the single-nucleus RNA sequencing datasets (GSE211650). Their research successfully established the hiPSC PCD DCM model, revealing ferroptosis correlated with fibroblast activation as a novel disease mechanism in PCD [18]. The single-nucleus RNA sequencing results provided partial insights, focusing primarily on fibroblasts. However, there remains a significant gap in the in-depth study of cardiomyocytes, which are critical for understanding the pathophysiology of PCD DCM.
Our study conducted a thorough examination of the response of cardiomyocytes and alterations in the intercellular communication in PCD DCM. We downloaded and utilized the GSE211650 dataset for a comprehensive analysis. To the best of our knowledge, this study represents the first detailed exploration, at the single-cell level, of the pathogenic mechanisms underlying PCD DCM, with a particular focus on the response of cardiomyocytes and intercellular communicational alterations This also represents the first report at the single-cell level of the pathological alterations in cardiomyocytes and intercellular communicational changes in FAODs cardiomyopathy. Our research forms the foundation for precise diagnosis and treatment of PCD DCM, offering valuable insights for research on cardiomyopathy in other FAODs.
2. Materials and methods
2.1. Data collection
Single-nucleus RNA sequencing(snRNA-seq) data was downloaded from GSE211650 in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo). This single-nucleus datasets comprises three groups: healthy controls, SLC22A5 (OCTN2) homozygous point mutation (c.95A > G, p.N32S), and SLC22A5 (OCNT2) knockdown. The SLC22A5 c.95A > G (N32S) mutation is the characteristic PCD mutation on the Faroe Islands, known to be particularly susceptible to potentially lethal cardiac arrhythmias [2]. In our study, the healthy control group is denoted as WT, the SLC22A5 (OCTN2) homozygous point mutation (c.95A > G, p.N32S) group is abbreviated as N32S, and the SLC22A5 (OCNT2) knockdown group is referred to as KO. Collectively, N32S and KO are designated as the OCTN2-deficient group. All the mentioned data are freely available in public databases. Following the initial integration of these three sets of data, a gene expression and phenotype matrix were developed for 15,448 snRNA-seq nuclei.
2.2. Identification and visualization of cell types
Using the Seurat package (V4.3.0.1) within R software (R-4.3.1), Seurat objects were established for both the total and individual cell types from the snRNA-seq gene expression matrix based on their acquisition. Subsequently, the top 3000 genes, identified as the most variable features, served as the foundation for normalizing the snRNA-seq data for each cell using the SCTransform package (V0.3.5). Additionally, we applied the RunPCA function to determine the number of principal components (PC) based on the Seurat objects. FindNeighbors and FindClusters were employed for unsupervised cell clustering, utilizing resolutions ranging from 0.1 to 1 to enhance the delineation of sub-clusters. Potential marker genes were identified using the FindAllMarkers function, and dimensionality reduction was carried out using Uniform Manifold Approximation and Projection (UMAP). The CellCycle Scoring function in the Seurat package was utilized to score each nucleus [19], aiming to mitigate clustering influenced by cell cycle gene expression profiles. S.Score and G2M.Score from SCTransform were applied for re-dimensionality reduction clustering following the aforementioned method. Finally, typical marker genes were employed to categorize cell clusters into known cell lineages.
2.3. Quantify the enrichment of cell cluster
To assess the enrichment of cell clusters among different groups, we conducted a comparison of the observed and expected cell numbers within each cluster. The Ro/e value was computed using the epitools (v.0.5–10.1) R package, applying the formula Ro/e = Observed/Expected [20]. The expected cell numbers for each combination of cell types were derived from the χ2 test. We considered a cluster to be enriched in a specific group if the Ro/e value exceeded 1.
2.4. Enrichment analysis
The gene sets from various sources, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Hallmarks, WikiPathways, and Reactome, were obtained from the Bader Lab gene set database (https://download.baderlab.org/EM_Genesets/current_release/Human/symbol/). These gene sets were then consolidated into a list named Genesets for subsequent enrichment analysis. Gene Set Variation Analysis (GSVA) was conducted for cell subsets using the AddModuleScore function of Seurat. Differential analysis was carried out using the wilcox.test. Pathway over-representation analysis (ORA) for specific gene sets was performed using the clusterProfiler package (V4.8.3). Results with a P-value <0.05 and an adjusted P-value (Q value) < 0.05 were considered statistically significant.
2.5. High-dimensional weighted correlation network Analysis(hdWGCNA)
The analysis of snRNA-seq data was carried out using the hdWGCNA package (V0.2.18), even though it is primarily designed for scRNA-seq data [21]. Genes expressed in at least 5 % of the samples were considered for the analysis. Metacells were constructed using cardiomyocytes from the WT group. The pickSoftThreshold function was employed to empirically determine a soft threshold of 8 that best suited the network structure. Subsequent standard downstream analyses were performed following the official pipeline, available at https://smorabit.github.io/hdWGCNA/articles/basic_tutorial.html.
2.6. Estimate the cell cycle
The tricycle R package (V1.8.0) was employed to assess the cell cycle by assigning scores to individual cells based on the expression of G2/M [22] and S-phase marker genes. Utilizing the CellCycle Scoring function in the Seurat package, each cell was scored according to the cell cycle marker genes. Subsequently, each cell was categorized into G1, G2M, and S phases based on its cell cycle score.
2.7. Pathway visualization
The pathview package (V1.40.0) was utilized to visualize KEGG pathways [23]. Healthy controls were used as a baseline. In the gene tag of the pathway, the changes in associated genes for N32S are shown on the left side of the gene tag, while on the other side are those for KO.
2.8. Trajectory analysis
The monocle2 package (V2.28.0) was employed for pseudotime analyses, aiming to identify the differentiation trajectory in cell development. Highly variable genes were determined based on specific filtering criteria: mean expression ≥0.1 and dispersion_empirical ≥1 * dispersion_ft. Dimensionality reduction was achieved using the DDRTree method. Subsequently, the plot_pseudotime_heatmap function was used to generate heatmaps illustrating the transformation of cardiomyocytes. Additionally, the SCP (V0.4.8) package was utilized for trajectory inference and delineation of dynamic features. Standard downstream analyses were carried out following the official pipeline, available at https://github.com/zhanghao-njmu/SCP#trajectory-inference and https://github.com/zhanghao-njmu/SCP#dynamic-features.
2.9. SCENIC analysis
We utilized SCENIC with the pySCENIC package (0.12.1) to explore the prominent transcription factors in distinct clusters of cardiomyocytes using Python. SCENIC reconstructs regulons (comprising transcription factors and their target genes), assesses the activity of these regulons in individual cells, and defines meaningful clusters of cells [24]. pySCENIC involves two key processes: establishing a co-expression network through GENIE3 and identifying targeted genes through motif analysis using the RcisTarget database. The matrix of cardiomyocytes generated using Seurat served as input data. After running GENIE3, motif datasets (hg38_500bp_up_100bp_down_full_tx_v10_clust.genes_vs_motifs.rankings.feather, hg38_10kbp_up_10kbp_down_full_tx_v10_clust.genes_vs_motifs.scores.feather, hg38_500bp_up_100bp_down_full_tx_v10_clust.genes_vs_motifs.scores.feather, hg38_10kbp_up_10kbp_down_full_tx_v10_clust.genes_vs_motifs.rankings.feather) were used to construct regulons for each transcription factor. The AddModuleScore function of Seurat was employed to score transcription factors, and differential analysis was performed using the wilcox.test.
2.10. Cell-cell communication analysis using CellChat
Intercellular communication was determined using the CellChat package (V1.6.1), employing ligand-receptor interaction databases available at http://www.CellChat.org/ [25]. The Seurat data, normalized and transformed into a CellChat object, utilized CellChatDB.human as the receptor-ligand interaction database. Communication probabilities were computed using the computeCommunProb function, showcasing cell interactions in terms of both the number and weight of interactions. The extractEnrichedLR function was employed to extract all pertinent interacting ligand-receptor (L-R) pairs and associated signaling genes for a given signaling pathway, illustrating cell-cell communication mediated by a single L-R pair.
2.11. Statistical analysis
The statistical analyses of snRNA-seq data were conducted using Seurat, CellChat, Monocle, SCENIC, Slingshot, and GSVA. Differential analysis was executed using the wilcox.test. All statistical analyses were carried out using R software (R-4.3.1), except for the SCENIC analysis. In this research, results with a P-value <0.05 and an adjusted P-value (Q value) < 0.05 were deemed statistically significant.
3. Results
3.1. The changes in cell types and transcriptomic profiles of OCTN2-deficient heart cells
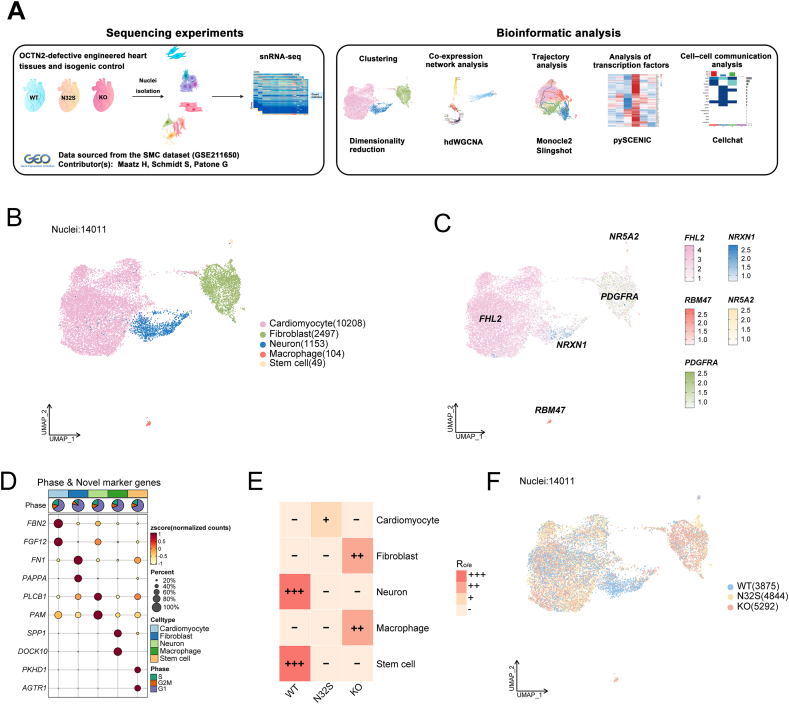
10 × snRNA-seq data were obtained from GSE211650, comprising three distinct groups: healthy controls (WT), SLC22A5 (OCTN2) homozygous point mutation (c.95A > G, p.N32S), and SLC22A5 (OCNT2) KO. The experimental design is illustrated in a schematic chart presented in Fig. 1A. The integration of the three datasets was carried out, followed by the removal of batch effects and the influence of the cell cycle. Subsequently, low-quality nuclei were filtered out (Figs. S1A and B), resulting in a total of 14,011 nuclei for further analysis. UMAP clustering was performed, categorizing these nuclei into 5 subgroups, namely cardiomyocyte, fibroblast, neuron, macrophage, and stem cell (Fig. 1B), as identified by marker genes (Fig. 1C). Heatmaps illustrating the expression levels of discriminative genes for each cluster, along with results from gene ontology biological process (GOBP) and KEGG enrichment analyses, are presented in Fig. S1C. The identification of novel marker genes for each cluster is visualized through a bubble plot, and cell cycles are summarized in a pie chart (Fig. 1D). The distribution of each cluster in WT and the OCTN2-deficient group can be observed in Fig. 1E, F and Fig. S1D. Taken together, PCD caused alterations in cell types and transcriptomic profiles hearts cells. Specifically, significant aggregation of macrophage and fibroblast occurred in the OCT2-deficient group, while neurons exhibited a significant reduction.
Fig. 1.
The changes in cell types and transcriptomic profiles of OCTN2-deficient heart cells. (A) Schematic diagram explaining the workflow of the experimental design. (B) Cluster annotations by using the uniform manifold approximation and projection (UMP) plots. (C) UMP plots showing the expression levels of representative marker genes for cell types, color-coded by gene expression levels. (D) Cell cycle pie chart and novel marker genes of each cell types. (E) The distribution of each cluster in WT and the OCTN2-deficient group is revealed by Ro/e (ratio of observed cell number to expected cell number). (F) UMP plots showing the distribution of each cluster in WT and the OCTN2-deficient group.
3.2. Cardiomyocytes in the OCTN2-deficient group undergo pathological alterations
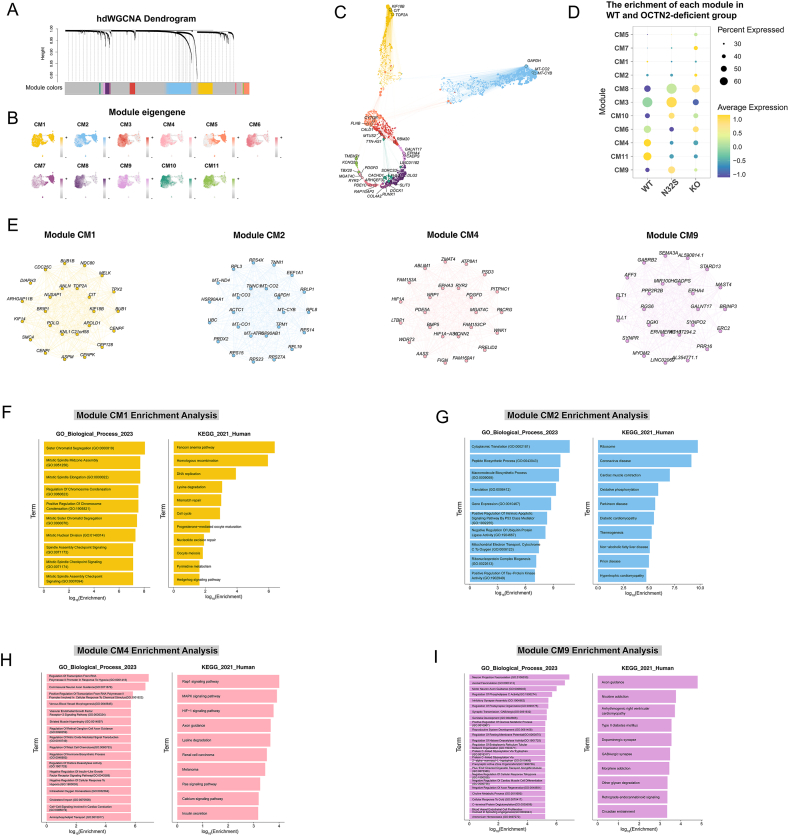
In this study, hdWGCNA was applied to perform co-expression network analysis on single-cell data. Metacells were constructed using cardiomyocytes from the WT sample. Following this, a co-expression network was established, identifying 11 co-expression modules (Fig. 2A) and assigning a unique color to each module for easy identification (Fig. 2B). The UMAP plot of the top3 genes in the co-expression network is shown in Fig. 2C. WT was significantly enriched for cardiomyocytes modules 1, 4 and 11, whereas the OCTN2-deficient group were significantly enriched for module 8, 9 and 10 (Fig. 2D). The ModuleNetworkPlot function was employed to visualize the network underlying the top 25 hub genes for these modules (Fig. 2E, Fig. S2A). Modules 1 and 2 were enriched for genes associated with proliferation and cardiac muscle contraction, respectively (Fig. 2F and G), while modules 4 and 9 were enriched for genes associated with neurons (Fig. 2H and I). The results of enrichment analysis in other modules are presented in Fig. S2B.
Fig. 2.
Single-cell coexpression network analysis by hdWGCNA. (A) Dendrogram showing the hierarchical clustering of genes into co-expression modules based on the topological overlap matrix (TOM). Each leaf on the dendrogram represents a single gene, and the color at the bottom indicates the co-expression module assignment. (B) snRNA-seq UMAP plots as in panel A colored by module eigengenes (MEs) for cardiomyocytes co-expression modules. (C) UMAP plot of cardiomyocytes co-expression network. Each node represents a single gene, and edges represent co-expression links between genes and module hub genes. Point size is scaled by the eigengene-based connectivity (kME). Nodes are colored by co-expression module assignment. The top 3 hub genes per module are labeled. (D) Dot plot for enrichment of modules in WT and OCTN2-deficient. (E) Hub isoform networks for cardiomyocytes co-expression modules 1, 2, 4 and 9. The top 25 hub genes ranked by kMEiso are visualized. Nodes represent isoforms, and edges represent co-expression links. (F–I) Selected genes ontology biological process (GOBP) and kyoto encyclopedia of genes and genomes (KEGG)terms enriched in co-expression modules 1 (F), 2 (G), 4 (H) and 9(I). Bar plots show the log-scaled enrichment of each term.
Cardiac proliferation-related terms were extracted from Gensets for GSVA. Enrichment terms of the OCTN2-deficient group suggested a negative regulation of proliferation in cardiomyocytes (Fig. 3A). Using tricycle, we annotated cell-cycle position for each of the nuclei in UMAP Plot (Fig. 3B). The percentage of G2M phase cells in the OCTN2-deficient group decreased compared to WT (Fig. 3C). Similarly, the G2M scores of the OCTN2-deficient group were significantly reduced compared to WT (Fig. S2C). These results suggest that proliferation may be inhibited in OCTN2-deficient cardiomyocytes. Subsequently, we visualized the differentially expressed genes (DEGs) of cardiac muscle contraction pathway using pathview. In comparison to WT, the OCTN2-deficient group exhibited decreased expression levels of genes encoding troponin, myosin, and calcium ion transporters (Fig. 3D). GSVA results further indicated a reduction in heart contraction in the OCTN2-deficient group (Fig. 3E). Furthermore, we identified that genes associated with cardiac contraction were significantly downregulated in the OCTN2-deficient group, including MYH7 (encoding myosin heavy chain beta isoform), TNNI3 (encoding Troponin I), TNNT2 (encoding cardiac muscle troponin T), ACTC1 (encoding cardiac muscle alpha actin), TPM1 (encoding tropomyosin alpha-1 chain), and RYR2(encoding ryanodine receptor 2) (Fig. 3F). In the process of cardiac calcium-induced calcium release, ryanodine receptor 2 serves as the major mediator for the sarcoplasmic release of stored calcium ions [26]. The reduced expression of genes encoding troponin, myosin, and calcium ion transporters may contribute to the decreased contractility observed in PCD patients.
Fig. 3.
Pathological changes in cardiomyocytes with OCTN2-deficient. (A) Heatmap of GSVA analysis showed the enrichment results for cardiac muscle proliferation-related terms in healthy and OCTN2-deficient. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (B) UMAP embedding of cardiomyocytes data colored by cell-cycle position. (C) Percentage of cells in the G2M phase. (D) Pathview analysis of genes related to cardiac muscle contraction in WT and the OCTN2-deficienct group. N32S and KO were compared separately with WT. (E) Heatmap showed GSVA of myocardial contraction-related terms in WT and the OCTN2-deficient group. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (F) Quantitative analysis of the genes of heart contraction. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, ****p < 0.0001, and ns p > 0.05. (G, H) Pathview analysis of genes related to ECM-receptor interaction pathway (G) and EGFR tyrosine kinase inhibitor resistance pathway (H) in WT and the OCTN2-deficienct group. N32S and KO were compared separately with WT. (I) Heatmap showed GSVA of fibrosis or epithelial-mesenchymal transition (EMT)-related terms in WT and the OCTN2-deficient group. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (J) Quantitative analysis of the genes of fibrosis | EMT. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, ****p < 0.0001, and ns p > 0.05. (L) Heatmap showing the signifcantly diferent activities of TFs in WT and the OCTN2-deficienct group. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (M) Heatmap showing the enriched signaling pathways from KEGG gene sets by GSVA. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (M)Pathview analysis of genes related to hippo signaling pathway in WT and OCTN2-deficiency groups. N32S and KO were compared separately with WT. (N, O) Quantitative analysis of the genes of Hippo(N) and Wnt(O) signaling pathway. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. **p < 0.01, ****p < 0.0001, and ns p > 0.05.
Excessive extracellular matrix (ECM) protein and epithelial-mesenchymal transition (EMT) are key characteristic of cardiac fibrosis [[27], [28], [29]]. In the OCTN2-deficient group, ECM shows the deposition of collagen in the OCTN2-deficient group (Fig. 3G). Additionally, growth arrest-specific 6/AXL receptor tyrosine kinase (GAS6/AXL)-induced EMT occurred in the OCTN2-deficient group (Fig. 3H). The results from GSVA also suggest the possibility of cardiac fibrosis in the OCTN2-deficient group (Fig. 3I). We further identified genes associated with fibrosis, including FN1 (encoding fibronectin), GAS6 (encoding growth arrest-specific 6), COL1A2 (encoding collagen, type I, alpha 2), ACTA2 (encoding actin alpha 2), and POSTN (encoding periostin) [[30], [31], [32], [33]], which were significantly increased in the OCTN2-deficient group (Fig. 3J). Cardiomyocyte hypertrophy-related terms were not enriched in the OCTN2-deficient group (Fig. S2D). The expression of cardiac hypertrophic response genes (NPPA, NPPB, and MYH7) [34,35] was significantly reduced in the OCTN2-deficient group (Fig. S2E).
To investigate the enrichment of transcription factors in both the WT and the OCTN2-deficient group, we conducted further analysis using SCENIC. In OCTN2-deficient cardiomyocytes, transcription factors crucial for cardiomyocyte development and congenital heart defects, including HAND1, HEY2, FOXM1, MEF2A, NR2F2 and GATA6, are significantly downregulated (Fig. 3K) [[36], [37], [38], [39], [40], [41]]. Additionally, we employed GSVA to study alterations in signaling pathways between WT and the OCTN2-deficient group (Fig. 3L). Hippo and Wnt signaling pathways, closely linked to myocardial proliferation, hypertrophy, and heart size, were found to be enriched in the OCTN2-deficient group [[42], [43], [44]]. DEGs in the Hippo and Wnt pathways are visualized through pathview (Fig. 3M, Fig. S2F). The Hippo signaling pathway was activated, and the expression of YAP1 (encoding yes-associated protein) decreased significantly in the OCTN2-deficient group (Fig. 3N). In Wnt signaling pathway, CTNND2, the gene encoding δ-Catenin, increased significantly in the OCTN2-deficient group. δ-Catenin acts as an inhibitor of the canonical Wnt pathway, while CTNNB1 (encoding β-catenin) significantly decreased. The expression levels of the encoding genes for T cell factor/lymphoid enhancer factor (TCF/LEF), including TCF7, TCF7L1, and LEF1, also showed a significant decrease (Fig. 3O). These results suggest the inhibition of the canonical Wnt signaling pathways. The cGMP-PKG signaling pathway, known for its crucial role in heart disease protection by initiating cardiac protection and inhibiting cardiac hypertrophy [45,46], showed a significant increase in the expression of PRKG1 (encoding cGMP-dependent protein kinase 1) in N32S. Although there was an upward trend in KO, it did not reach statistical significance. However, cGMP-PKG signaling pathway-related cardioprotection is activated in both N32S and KO (Figs. S2G and H).
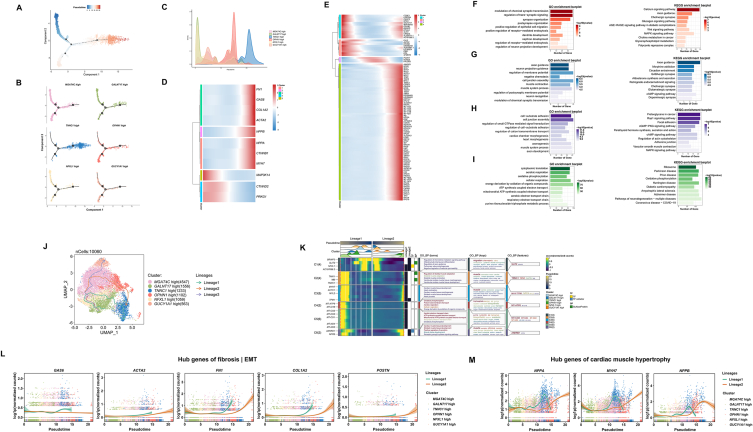
3.3. TNNC1 high clusters play a key role in the pathological alterations of cardiomyocytes in PCD DCM
Unbiased clustering grouped cardiomyocytes into six clusters: MGAT4C high, GALNT17 high, TNNC1 high, OPHN1 high, NFXL1 high, and GUCY1A1 high (Fig. 4A). The Phase and selected marker genes of each cluster are shown in Fig. 4B. There is no apparent specificity in the distribution of cardiomyocytes clusters between WT and the OCTN2-deficient groups (Fig. 4C). The DEGs expression levels of each cluster are presented by heatmaps, along with the results of GOBP and KEGG enrichment analyses (Fig. 4D). Given the pathological changes observed in cardiomyocytes in the preceding analysis, we conducted further cluster analysis. The TNNC1 high cluster exhibited notable enrichment in myocardial hypertrophy-related terms (Fig. 4E). Quantitative analysis confirmed that the TNNC1 high cluster showed higher expression levels of genes associated with myocardial hypertrophy compared to other clusters (Fig. 4F). No significant differences was observed in the expression of YAP1 gene among the various clusters (Fig. 4G). In the TNNC1 high cluster, CTNND2, the inhibitory gene in the canonical pathway, exhibited a significant reduction in expression, while CTNNB1 expression markedly increased. This suggests the potential activation of the Wnt pathway in the TNNC1 high cluster (Fig. 4H). GSVA results indicate enrichment in terms related to the positive regulation of heart contraction in the TNNC1 high cluster (Fig. 4I). Further quantitative analysis also indicates that this cluster overexpresses genes encoding contractile proteins (Fig. 4J). Moreover, the TNNC1 high cluster also plays a crucial role in cardiac fibrosis (Fig. 4K and L). These results suggest the paramount importance of the TNNC1 high cluster in the pathological changes observed in cardiomyocytes.
Fig. 4.
Pathological changes in cardiomyocytes cluster. (A) Cluster annotations by UMP plot. (B) Cell cycle pie chart and selected marker genes of each clusters. (C) Percentage of cell subsets in WT and the OCTN2-deficient group. (D) Heatmap of discriminative genes expression levels and the results of GOBP and KEGG enrichment analysis. (E) Heatmap showed GSVA of myocardial hypertrophy-related terms in cardiomyocytes cluster. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0. (F–H) Quantitative analysis of the genes of myocardial hypertrophy (F), Hippo signaling pathway (G) and Wnt signaling pathway (H). The TNNC1 high cluster was compared with each of the other clusters separately. P value was calculated by wilcox.test. *p < 0.05, **p < 0.01, ****p < 0.0001, and ns p > 0.05. (I) Heatmap showed GSVA of myocardial contraction-related terms in cardiomyocytes cluster. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0. (J) Quantitative analysis of the genes of heart contraction. The TNNC1 high cluster was compared with each of the other clusters separately. P value was calculated by wilcox.test. ****p < 0.0001, and ns p > 0.05. (K) Heatmap showed GSVA of fibrosis or EMT-related terms. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0. (L) Quantitative analysis of the genes of fibrosis |EMT. The TNNC1 high cluster was compared with each of the other clusters separately. P value was calculated by wilcox.test. ***p < 0.001, ****p < 0.0001, and ns p > 0.05.
The pseudotime trajectory analysis of cardiomyocytes clusters was performed with monocle2 to elucidate the process of pathological changes in cardiomyocytes. The TNNC1 high cluster of cardiomyocytes significantly increases at the end of the trajectory (Fig. 5A–C). We used a heatmap to display the dynamic characteristics of genes related to fibrosis and hypertrophy identified in the previous analysis along the pseudotime trajectory (Fig. 5D). Expression levels of these genes increased towards the end of the pseudotime trajectory, while the expression of the protective gene PRKG1 gradually decreased. In addition, there was an increased expression of genes encoding effector proteins of the canonical Wnt pathway and a decreased expression of genes encoding inhibitory proteins, suggesting potential activation of the canonical Wnt pathway. This pathway is closely associated with cardiac hypertrophy and fibrosis [42,47,48]. A set of 3000 differentially expressed genes was obtained from the analysis of pseudotime trajectories and grouped into 4 clusters based on gene classification with similar patterns Top100 genes are presented by heatmaps (Fig. 5E). Results of GO and KEGG enrichment analyses indicate that genes in Cluster 1 and 2 are predominantly involved in signaling pathways related to axon guidance and synapse organization, genes in Cluster 3 are mainly enriched in signaling pathways related to cell-substrate adhesion, and genes in Cluster 4 are primarily associated with signaling pathways implicated in oxidative phosphorylation (Fig. 5F–I). We also reconstructed the pseudo-temporal trajectory through SCP. Lineage 1 and 2 similarly suggest the TNNC1 high cluster was ordered at a later stage along pseudotime trajectory (Fig. 5J). The DEGs inferred along the trajectories of lineage 1 and 2 have been identified and clustered into 6 clusters. Each cluster has been annotated with key genes and subjected to GOBP analysis (Fig. 5K). Cluster C1 shows enrichment for genes involved in chemokine-mediated signaling and axon guidance, indicating a role in cellular migration and neuron development. Cluster C2, C3 and 6 show enrichment in cardiac muscle development and heart contraction. Cluster C4 and C6 are enriched for genes related to oxidative phosphorylation. The dynamic features revealed by SCP also indicate that the expression levels of genes correlated with cardiac fibrosis (Fig. 5L) and hypertrophy (Fig. 5M) gradually increase along the pseudotime trajectory.
Fig. 5.
Developmental trajectories of cardiomyocytes in PCD DCM. (A) Monocle2 analyses showing cardiomyocytes in pseudotime. Each color indicates a time point. (B, C) Cell density variation of cardiomyocytes clusters during the pseudotime. (D) Heatmap depicting the changes of genes along the pseudotime trajectory. (E) Pseudo-heatmap of the top 100 representative DEGs in differentiation branches, and their distribution in the four subgroups after the reclustering of DEGs. (F–I) The results of GO and KEGG enrichment analysis for clusters 1 (F), 2 (G), 3 (H), and 4 (I). (J) Cellular trajectory reconstructed for the process of pathological changes in cardiomyocytes by SCP. The arrow shows the direction of cellular state changes. (K) The heatmap displays the variations of DEGs along the trajectories of lineage 1 and 2, DEGs were clustered into 6 clusters. For each cluster, the enriched terms, key words, and key genes from the GOBP analysis. (L, M) Dynamic analysis of genes related to fibrosis (L) and hypertrophy (M) along pseudotime trajectories.
3.4. Cardiomyocytes in the OCTN2-deficient group demonstrated enhanced modulation of macrophages
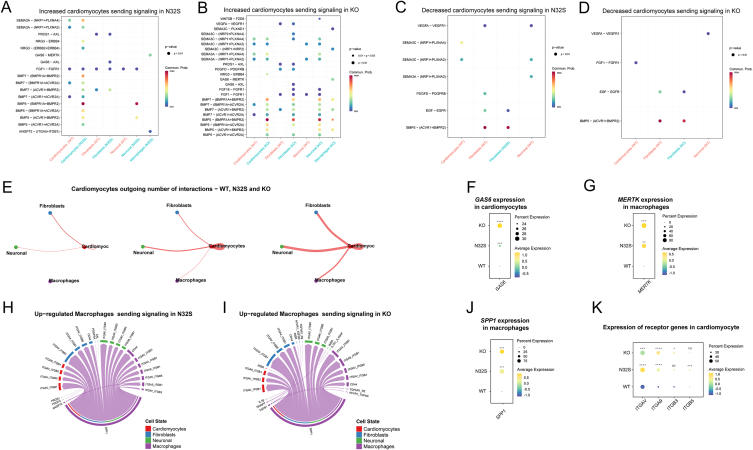
CellChat was utilized to comprehensively evaluate the interactions in terms of number and strength of cell communication between cardiomyocytes, macrophages, fibroblasts, and neurons. Compared to WT, increased cardiomyocyte signaling in N32S and KO is illustrated in Fig. 6A and B, while decreased signaling is shown in Fig. 6C and D. Subsequently, we visualized the number of outgoing interactions of cardiomyocytes (Fig. 6E). In WT, there is no communication between cardiomyocytes and macrophages, whereas in the OCTN2-deficient group, mutual interaction between cardiomyocytes and macrophages is observed. We conducted further analysis of the upregulated L-R interactions involved in the interaction between cardiomyocytes and macrophages in N32S and KO. Within the enhanced signaling interaction of cardiomyocytes to macrophages, GAS6 has been reported to be involved in macrophage polarization [49,50]. The levels of GAS6 expression significantly increased in the OCTN2-deficient group, and there was also a significant upregulation of MERTK (encoding proto-oncogene tyrosine-protein kinase MER) in macrophages (Fig. 6F and G). Macrophages exhibited elevated expression of SPP1 (encoding secreted phosphoprotein 1), and their communication with OCTN2-deficient cardiomyocytes was enhanced (Fig. 6H–K). Considering the role of SPP1 in ECM [51], this process may lead to the deposition of proteins in the ECM, further exacerbating cardiomyocytes fibrosis.
Fig. 6.
CellChat analysis of the crosstalk between cardiomyocytes and macrophages. (A, B) Increased ligand-receptor interactions between cardiomyocytes and non-cardiomyocytes in N32S (A) and KO(B), with the dot color reflecting the communication probability, blank indicating the communication probability zero, and dot size representing the p value (C, D) Decreased ligand-receptor interactions between cardiomyocytes and non-cardiomyocytes in N32S (C) and KO (D) with the dot color reflecting the communication probability, blank indicating the communication probability zero, and dot size representing the p value. (E) Cardiomyocytes outgoing number of interactions in WT, N32S and KO. (F) Quantitative analysis of GAS6 expression in cardiomyocytes. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. ***p < 0.001, ****p < 0.0001. (G) Quantitative analysis of MERTK expression in macrophages. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. ****p < 0.0001, and **p < 0.01. GAS6: The gene encoding growth arrest-specific 6. MERTK: The gene encoding Proto-oncogene tyrosine-protein kinase MER. (H, I) Up-regulated macrophages sending signaling in N32S (H) and KO (I) visualized by chord diagram. (J) Quantitative analysis of SPP1 expression in macrophages. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. ***p < 0.001. SPP1: The gene encoding Secreted phosphoprotein 1. (K) Quantitative analysis of SPP1 receptors gene expression in cardiomyocytes. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, ***p < 0.001, ****p < 0.0001, and ns p > 0.05.
3.5. SPP1+ macrophages play a crucial role in the activation of fibroblasts
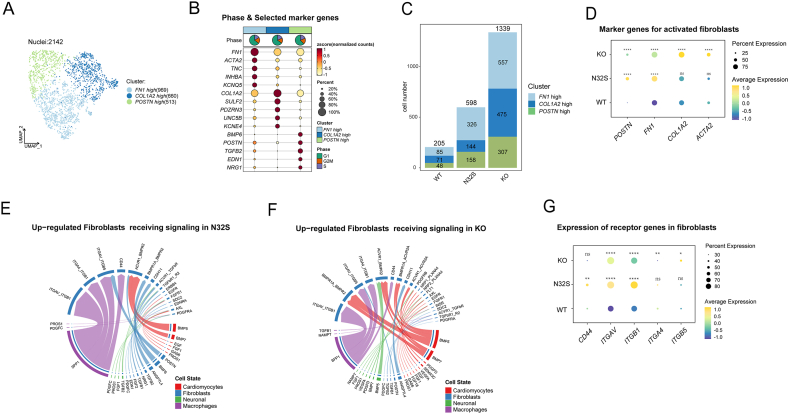
Unbiased clustering revealed three distinct fibroblast clusters characterized by high expression levels of FN1, COL1A2, and POSTN, as illustrated in Fig. 7A. Phase and selected marker genes of each cluster are depicted in Fig. 7B. The number of fibroblasts significantly increased in the OCTN2-deficient group, as shown in Fig. 7C. Additionally, marker genes associated with fibroblast activation exhibited a significant increase in the OCTN2-deficient group, as illustrated in Fig. 7D. Notably, among the increased signals received by fibroblasts, the L-R interactions corresponding to SPP1 had the largest proportion in the OCTN2-deficient group, as indicated in Fig. 7E and F. Moreover, prior in vitro investigations involving cultured human atrial fibroblasts (hAF) have shown that SPP1 promotes fibroblast proliferation and enhances the production of collagen I and fibronectin [52]. Genes associated with the SPP1 receptor also exhibited increased expression in fibroblasts, particularly ITGAV (encoding Integrin alpha-V) and ITGB1 (encoding Integrin beta 1), as shown in Fig. 7G. Overall, SPP1+ macrophages play a significant role in the activation of fibroblasts.
Fig. 7.
Fibroblasts regulatory changes in PCD DCM. (A) Cluster annotations by using the Seurat the UMP plot. (B) Cell cycle pie chart and selected marker genes of each clusters. (C) Cell number of fibroblast clusters in WT and the OCTN2-deficient group. (D) Quantitative analysis of the marker genes for activated fibroblasts. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. ****p < 0.0001, and ns p > 0.05. (E, F) Up-regulated fibroblasts receiving signaling in N32S (E) and KO (F) visualized by chord diagram. (G) Quantitative analysis of SPP1 receptors gene expression in fibroblasts. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, **p < 0.01, ****p < 0.0001, and ns p > 0.05.
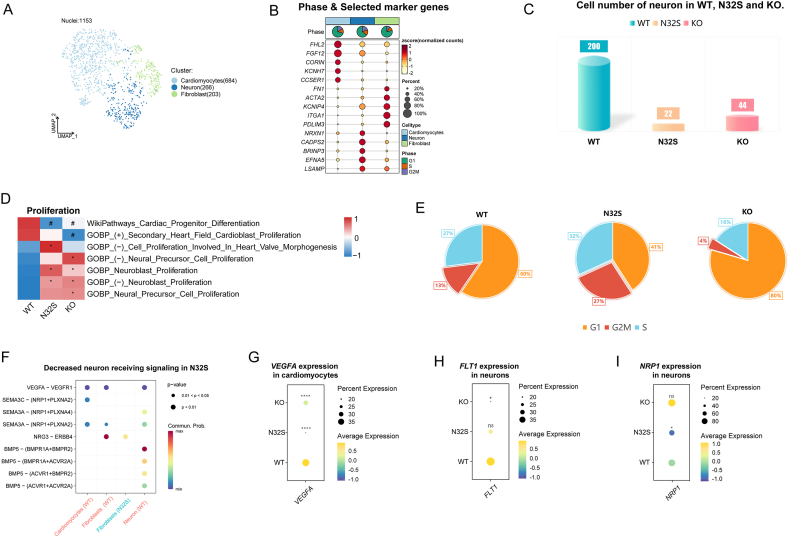
3.6. Cell number of neurons significantly decreased in the OCTN2-deficient group
Further dimensionality reduction and clustering of neurons were illustrated through UMAP plots (Fig. 8A). The phase and selected marker genes of each cluster are presented in Fig. 8B. Non-neuronal cells were excluded from subsequent analysis. A notable decrease in the number of neurons was observed in the OCTN2-deficient group compared to WT, with the number of neurons in N32 even lower than that in KO (Fig. 8C). GSVA results suggest that neuronal proliferation in the OCTN2-deficient group may be negatively regulated (Fig. 8D). Further analysis revealed a significant decrease in the percentage of G2M-phase cells in KO, while there was an increase in the percentage of G2M-phase cells in N32S (Fig. 8E). This suggests that the reduction in neurons in KO is likely due to negative regulation of proliferation. To explore the reasons behind the diminished neurons in the OCTN2-deficient group, CellChat was employed to study interactions between cardiomyocytes and neurons. In the L-R interactions between cardiomyocytes with OCTN2 deficiency and neurons, both the number and strength of interactions involving vascular endothelial growth factor A (VEGFA) and vascular endothelial growth factor receptor 1 (VEGFR1) (L-R) were diminished (Fig. 8F, Fig. S3A). The expression of VEGFA gene (encoding VEGFA) in cardiomyocytes of the OCTN2-deficient group showed a significant decrease (Fig. 8G). In KO, the expression of FLT1 (encoding VEGFR1) in neurons was significantly reduced (Fig. 8H), while in N32S, it was the expression of NRP1 (encoding Neuropilin 1) that showed a significant decrease (Fig. 8I). NRP1 is a receptor for specific subtypes of semaphorin-3A (SEMA3A) and VEGFA c [53,54]. Therefore, despite the significant increase in the secretion of SEMA3A by cardiomyocytes in the OCTN2-deficient group (Fig. S3B), the decrease in NRP1 on neurons resulted in a reduction in both the number and strength of SEMA3A/NRP1 (L-R) interactions. The expression levels of PLXNA2 (encoding plexin-A2) and PLXNA4 (encoding plexin-A4), which encode receptors for SEMA3A, did not show significant differences in neurons (Fig. S3C). These analyses suggest that the reduction in VEGFA secretion by cardiomyocytes, combined with the decrease in neuronal VEGFR1 and NRP1, may be the cause of the reduction in neurons in the OCTN2-deficient group.
Fig. 8.
Cell number of neurons significantly decreased in the OCTN2-deficient group. (A) Cluster annotations by using the UMP clustering. (B) Cell cycle pie chart and selected marker genes of each clusters. (C) Cell number of neurons in WT and the OCTN2-deficient group. (D) Heatmap showed GSVA of proliferation-related terms in healthy and OCTN2-deficient. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05 & avg_log2FC > 0; #p < 0.05 & avg_log2FC < 0. (E) Percentage of cells in the G2M phase. (F) Decreased neurons receiving signaling in N32S, with the dot color reflecting the communication probability, blank indicating the communication probability zero, and dot size representing the p value. (G) Quantitative analysis of VEGFA expression in neurons. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. ****p < 0.0001. VEGFA: The gene encoding vascular endothelial growth factor A. (H) Quantitative analysis of FLT1 expression in neurons. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, and ns p > 0.05. FLT1: The gene encoding vascular endothelial growth factor receptor 1. (I) Quantitative analysis of NRP1 expression in neurons. N32S and KO were compared separately with WT. P value was calculated by wilcox.test. *p < 0.05, and ns p > 0.05. NRP1: The gene encoding neuropilin 1.
4. Discussion
Understanding the specific cellular responses and changes in the intercellular communication under disease conditions is critical for diagnosing, treating, and assessing the prognosis of the disease. Although PCD DCM involves the collaborative interaction of various cell types, cardiacmyocytes remain the most critical determinant of cardiac status. This study aimed to investigate the response of cardiomyocytes and alterations in the intercellular communication in PCD DCM, proposing potential mechanisms for weakened contractility, developmental disorders, and fibrosis in cardiomyocytes. The study revealed the crucial role of TNNC1-high cardiomyocytes in myocardial fibrosis and hypertrophy. Additionally, key genes involved in communication changes between cardiomyocytes, macrophages, fibroblasts, and neurons were identified. Our study provides the first detailed report at the single-cell level of the cardiomyocyte response and intercellular communication alterations within PCD DCM.
The transcriptome of cardiomyocytes deficient in OCTN2 exhibits pathological alterations, suggesting reduced contractility, developmental disturbances, and fibrosis. The decreased expression of genes encoding troponin, myosin, and ryanodine receptor 2 in OCTN2-deficient cardiomyocytes may contribute to the observed reduction in contractility. Signal pathway analysis results suggest the potential inhibition of the Wnt pathway in cardiomyocytes of the OCTN2-deficient group, which may be a contributing factor to the decreased number of cardiomyocytes in the G2/M phase. Moreover, transcription factors associated with myocardial development and congenital heart defects, including HAND1, HEY2, FOXM1, MEF2A, NR2F2, and GATA6, are significantly downregulated in OCTN2-deficient cardiomyocytes. These findings collectively indicate potential developmental disturbances in cardiomyocytes with OCTN2 deficiency. In the context of cardiac fibrosis, crucial pathological processes involve the deposition of proteins in the extracellular matrix and epithelial-to-mesenchymal transition. In the cardiomyocytes of the OCTN2-deficient group, the upregulation of the GAS6 gene significantly contributes to the initiation of myocardial fibrosis. GAS6 serves as a trigger for myocardial fibrosis by inducing EMT through the GAS6/AXL pathway and enhancing macrophage regulation via the GAS6/MERTK pathway [49,50,55]. SPP1+ macrophage cells play a pivotal role in this process, participating not only in the deposition of proteins within ECM but also contributing to fibroblast activation [51,56,57]. Activated fibroblasts exhibit increased expression of fibrosis-promoting genes such as FN1, POSTN, COL1A2, ACTA2, further exacerbating protein deposition in the ECM and promoting the progression of myocardial fibrosis. In conclusion, GAS6 initiation of EMT and excessive protein deposition in the ECM constitutes crucial steps in myocardial fibrosis. Blocking the secretion of GAS6 from cardiomyocytes may represent an effective strategy to reduce the occurrence of cardiac fibrosis in PCD DCM.
Pseudotime ordering analysis was employed to discern the transcriptomic dynamics as cardiomyocytes transitioned into the failing state. Multiple independent analyses provided a list of potential key genes, with the majority being linked to myocardial hypertrophy, including MYH7, NPPA, and NPPB [34,35]. These genes all exhibit increased expression in the later stages of the pseudo-temporal trajectory. Examining the distribution of cardiomyocyte clusters along the pseudo-temporal trajectory reveals that the TNNC1 high cluster predominantly resides in the later stages. Furthermore, within the TNNC1 high cluster, the Wnt/β-catenin signaling pathway is activated, representing a common pathogenic mechanism underlying myocardial hypertrophy and fibrosis [42,47,48]. In summary, the TNNC1 high cluster overexpresses genes related to myocardial fibrosis and hypertrophy and is predominantly distributed at the end of the pseudotime trajectory. These findings underscore the critical role of the TNNC1 high cluster in cardiomyocytes in PCD DCM.
Sudden cardiac death resulting from events is the most severe symptom of PCD. In our study, a notable decrease in neurons was observed within the OCTN2-deficient group. Findings from hdWGCNA suggest that cardiomyocytes may play a role in the negative regulation of neuronal proliferation. Further analysis with CellChat suggests this regulation may be associated with the attenuation of the VEGFA/VEGFR1 pathway. While VEGFA is well-known for its essential roles in blood vessel growth, emerging evidence suggests its involvement in various neuronal functions both in vitro and in vivo. These functions include neurogenesis, neuronal migration, neuronal survival, and axon guidance. Notably, the removal of VEGFA expression from neural progenitor cells and their descendants has been shown to impair brain vascularization, leading to widespread neuronal death in mouse embryos [47,48,58]. These reports primarily focus on the central nervous system, especially the brain, with limited research investigating the impact of VEGFA on myocardial neurons. This gap underscores the need for further exploration. In comparison to KO, N32S not only exhibited a decrease in the VEGFA/VEGFR1 pathway but also a decline in the NRP1 receptor. The absence of the NRP1 receptor leads to abnormal development of the cardiac sympathetic nervous system, leading to sinus node bradycardia, akin to mice lacking SEMA3A, a secreted glycoprotein regulating axon guidance [59]. In summary, our research proposes a potential mechanism for arrhythmias caused by PCD. The supplementation of VEGFA emerges as a crucial intervention for treating PCD-induced arrhythmias and reducing the risk of sudden death.
In summary, our study proposes potential mechanisms underlying the observed decline in contractility, developmental disturbances, and fibrosis in cardiomyocytes within PCD DCM. We have identified the crucial role of the TNNC1 high cluster in myocardial fibrosis and hypertrophy. Additionally, our analysis pinpointed potential key genes involved in the intercellular communicational alterations of PCD DCM. These findings offer valuable insights for early diagnosis, precise treatment, and prognosis assessment of PCD DCM.
5. Conclusion
Our study provides the first detailed report at the single-cell level of the cardiomyocyte response and intercellular communication alterations in PCD DCM. Specifically, the transcriptome of OCTN2-deficient cardiomyocytes indicates alterations, implying reduced contractility, developmental disturbances, and fibrosis. The decreased expression of genes encoding troponin, myosin, and calcium ion transporters is likely responsible for the observed decrease in contractility. Suppressed Wnt signaling and downregulated transcription factors related to myocardial development suggest potential developmental disturbances in cardiomyocytes. Furthermore, GAS6 secreted by TNNC1-high cardiomyocytes contributes to myocardial inflammation and fibrosis. SPP1 produced by macrophages promotes fibroblast activation. Furthermore, there was a reduction in neuronal genes in the OCTN2-deficient group. Our research forms the foundation for precise diagnosis and treatment of PCD DCM and offers valuable insights for research on cardiomyopathy in other FAODs.
Limitations
-
1.
The human induced pluripotent stem cell (hiPSC) model of primary carnitine deficiency (PCD) cannot fully replicate all in vivo phenomena.
-
2.
Our research is specific to the c.95A > G (N32S) mutation in the SLC22A5 gene, limiting its generalizability.
-
3.
Due to the rarity of primary carnitine deficiency and the difficulty in obtaining myocardial samples, further validation with clinical samples is currently lacking.
Summary Points.
-
●
First report focuses on cardiomyocytes in primary carnitine deficiency (PCD).
-
●
Cardiomyocytes display reduced contractility, abnormal development, and fibrosis in PCD.
-
●
TNNC1 high cardiomyocytes play a pivotal role in cardiac fibrosis and hypertrophy.
-
●
Growth arrest-specific 6 is implicated in myocardial inflammation and fibrosis.
-
●
Macrophages-derived secreted phosphoprotein 1 promotes the activation of fibroblasts.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo); https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211650.
The data included in article and supplementary material in article. Data associated with my study have not been deposited into a publicly available repository.
Funding
This work was supported by Postdoctoral Program of Chongqing Natural Science Foundation (CSTB2022NSCQ-BHX0706), and Scientific research project of Women and Children's Hospital of Chongqing Medical University (No. 2021YJQN02).
CRediT authorship contribution statement
Yifan Yin: Writing – original draft, Resources, Methodology, Investigation, Formal analysis, Conceptualization. Liang Ye: Writing – review & editing, Investigation, Funding acquisition, Formal analysis, Conceptualization. Min Chen: Investigation, Formal analysis. Hao Liu: Supervision. Jingkun Miao: Writing – review & editing, Resources, Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33581.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Koizumi A., Nozaki J., Ohura T., Kayo T., Wada Y., Nezu J., et al. Genetic epidemiology of the carnitine transporter OCTN2 gene in a Japanese population and phenotypic characterization in Japanese pedigrees with primary systemic carnitine deficiency. Hum. Mol. Genet. 1999;8(12):2247–2254. doi: 10.1093/hmg/8.12.2247. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen J., Nielsen O.W., Janzen N., Duno M., Gislason H., Køber L., et al. Carnitine levels in 26,462 individuals from the nationwide screening program for primary carnitine deficiency in the Faroe Islands. J. Inherit. Metab. Dis. 2014;37(2):215–222. doi: 10.1007/s10545-013-9606-2. [DOI] [PubMed] [Google Scholar]
- 3.Therrell B.L., Jr., Lloyd-Puryear M.A., Camp K.M., Mann M.Y. Inborn errors of metabolism identified via newborn screening: ten-year incidence data and costs of nutritional interventions for research agenda planning. Mol. Genet. Metabol. 2014;113(1–2):14–26. doi: 10.1016/j.ymgme.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang N.L., Ganapathy V., Wu X., Hui J., Seth P., Yuen P.M., et al. Mutations of OCTN2, an organic cation/carnitine transporter, lead to deficient cellular carnitine uptake in primary carnitine deficiency. Hum. Mol. Genet. 1999;8(4):655–660. doi: 10.1093/hmg/8.4.655. [DOI] [PubMed] [Google Scholar]
- 5.Cederbaum S.D., Koo-McCoy S., Tein I., Hsu B.Y., Ganguly A., Vilain E., et al. Carnitine membrane transporter deficiency: a long-term follow up and OCTN2 mutation in the first documented case of primary carnitine deficiency. Mol. Genet. Metabol. 2002;77(3):195–201. doi: 10.1016/s1096-7192(02)00169-5. [DOI] [PubMed] [Google Scholar]
- 6.Wagner C.A., Lükewille U., Kaltenbach S., Moschen I., Bröer A., Risler T., et al. Functional and pharmacological characterization of human Na(+)-carnitine cotransporter hOCTN2. Am. J. Physiol. Ren. Physiol. 2000;279(3):F584–F591. doi: 10.1152/ajprenal.2000.279.3.F584. [DOI] [PubMed] [Google Scholar]
- 7.Longo N., Frigeni M., Pasquali M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta. 2016;1863(10):2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen J., Lund A.M., Risom L., Wibrand F., Gislason H., Nielsen O.W., et al. Residual OCTN2 transporter activity, carnitine levels and symptoms correlate in patients with primary carnitine deficiency. Mol Genet Metab Rep. 2014;1:241–248. doi: 10.1016/j.ymgmr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen J., Dunø M., Lund A.M., Steuerwald U., Hansen S.H., Joensen H.D., et al. Increased risk of sudden death in untreated primary carnitine deficiency. J. Inherit. Metab. Dis. 2020;43(2):290–296. doi: 10.1002/jimd.12158. [DOI] [PubMed] [Google Scholar]
- 10.Wang S.S., Rao J., Li Y.F., Zhang Z.W., Zeng G.H. Primary carnitine deficiency cardiomyopathy. Int. J. Cardiol. 2014;174(1):171–173. doi: 10.1016/j.ijcard.2014.03.190. [DOI] [PubMed] [Google Scholar]
- 11.Bennett M.J., Hale D.E., Pollitt R.J., Stanley C.A., Variend S. Endocardial fibroelastosis and primary carnitine deficiency due to a defect in the plasma membrane carnitine transporter. Clin. Cardiol. 1996;19(3):243–246. doi: 10.1002/clc.4960190320. [DOI] [PubMed] [Google Scholar]
- 12.Tripp M.E., Katcher M.L., Peters H.A., Gilbert E.F., Arya S., Hodach R.J., et al. Systemic carnitine deficiency presenting as familial endocardial fibroelastosis: a treatable cardiomyopathy. N. Engl. J. Med. 1981;305(7):385–390. doi: 10.1056/nejm198108133050707. [DOI] [PubMed] [Google Scholar]
- 13.Roussel J., Labarthe F., Thireau J., Ferro F., Farah C., Roy J., et al. Carnitine deficiency induces a short QT syndrome. Heart Rhythm. 2016;13(1):165–174. doi: 10.1016/j.hrthm.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen J., Nielsen O.W., Lund A.M., Køber L., Djurhuus H. Primary carnitine deficiency and pivalic acid exposure causing encephalopathy and fatal cardiac events. J. Inherit. Metab. Dis. 2013;36(1):35–41. doi: 10.1007/s10545-012-9488-8. [DOI] [PubMed] [Google Scholar]
- 15.Louis L., Margaux G., Claire G., Delphine L., Sandrine R., Emmanuel R., et al. Infantile primary carnitine deficiency: a severe cardiac presentation unresponsive to carnitine supplementation. JIMD Rep. 2023;64(1):35–41. doi: 10.1002/jmd2.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaffin M., Papangeli I., Simonson B., Akkad A.D., Hill M.C., Arduini A., et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature. 2022;608(7921):174–180. doi: 10.1038/s41586-022-04817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichart D., Lindberg E.L., Maatz H., Miranda A.M.A., Viveiros A., Shvetsov N., et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science. 2022;377(6606) doi: 10.1126/science.abo1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loos M., Klampe B., Schulze T., Yin X., Theofilatos K., Ulmer B.M., et al. Human model of primary carnitine deficiency cardiomyopathy reveals ferroptosis as a novel mechanism. Stem Cell Rep. 2023;18(11):2123–2137. doi: 10.1016/j.stemcr.2023.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25(12):1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue R., Zhang Q., Cao Q., Kong R., Xiang X., Liu H., et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022;612(7938):141–147. doi: 10.1038/s41586-022-05400-x. [DOI] [PubMed] [Google Scholar]
- 21.Morabito S., Reese F., Rahimzadeh N., Miyoshi E., Swarup V. High dimensional co-expression networks enable discovery of transcriptomic drivers in complex biological systems. bioRxiv. 2022;2022 2009. 2022.509094. [Google Scholar]
- 22.Zheng S.C., Stein-O'Brien G., Augustin J.J., Slosberg J., Carosso G.A., Winer B., et al. Universal prediction of cell-cycle position using transfer learning. Genome Biol. 2022;23(1):41. doi: 10.1186/s13059-021-02581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tunwell R.E., Wickenden C., Bertrand B.M., Shevchenko V.I., Walsh M.B., Allen P.D., et al. The human cardiac muscle ryanodine receptor-calcium release channel: identification, primary structure and topological analysis. Biochem. J. 1996;318(Pt 2):477–487. doi: 10.1042/bj3180477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M., López de Juan Abad B., Cheng K. Cardiac fibrosis: myofibroblast-mediated pathological regulation and drug delivery strategies. Adv. Drug Deliv. Rev. 2021;173:504–519. doi: 10.1016/j.addr.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berk B.C., Fujiwara K., Lehoux S. ECM remodeling in hypertensive heart disease. J. Clin. Invest. 2007;117(3):568–575. doi: 10.1172/jci31044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeisberg E.M., Tarnavski O., Zeisberg M., Dorfman A.L., McMullen J.R., Gustafsson E., et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13(8):952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 30.Li L., Zhao Q., Kong W. Extracellular matrix remodeling and cardiac fibrosis. Matrix Biol. 2018;68–69:490–506. doi: 10.1016/j.matbio.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Bellan M., Cittone M.G., Tonello S., Rigamonti C., Castello L.M., Gavelli F., et al. Gas6/TAM system: a key modulator of the interplay between inflammation and fibrosis. Int. J. Mol. Sci. 2019;20(20) doi: 10.3390/ijms20205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikoloudaki G., Creber K., Hamilton D.W. Wound healing and fibrosis: a contrasting role for periostin in skin and the oral mucosa. Am. J. Physiol. Cell Physiol. 2020;318(6):C1065–c1077. doi: 10.1152/ajpcell.00035.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meulendijks E.R., Al-Shama R.F.M., Kawasaki M., Fabrizi B., Neefs J., Wesselink R., et al. Atrial epicardial adipose tissue abundantly secretes myeloperoxidase and activates atrial fibroblasts in patients with atrial fibrillation. J. Transl. Med. 2023;21(1):366. doi: 10.1186/s12967-023-04231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Man J., Barnett P., Christoffels V.M. Structure and function of the Nppa-Nppb cluster locus during heart development and disease. Cell. Mol. Life Sci. 2018;75(8):1435–1444. doi: 10.1007/s00018-017-2737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren Z., Yu P., Li D., Li Z., Liao Y., Wang Y., et al. Single-cell reconstruction of progression trajectory reveals intervention principles in pathological cardiac hypertrophy. Circulation. 2020;141(21):1704–1719. doi: 10.1161/circulationaha.119.043053. [DOI] [PubMed] [Google Scholar]
- 36.Firulli B.A., George R.M., Harkin J., Toolan K.P., Gao H., Liu Y., et al. HAND1 loss-of-function within the embryonic myocardium reveals survivable congenital cardiac defects and adult heart failure. Cardiovasc. Res. 2020;116(3):605–618. doi: 10.1093/cvr/cvz182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Walree E.S., Dombrowsky G., Jansen I.E., Mirkov M.U., Zwart R., Ilgun A., et al. Germline variants in HEY2 functional domains lead to congenital heart defects and thoracic aortic aneurysms. Genet. Med. 2021;23(1):103–110. doi: 10.1038/s41436-020-00939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta A., Kalinichenko V.V., Yutzey K.E. FoxO1 and FoxM1 transcription factors have antagonistic functions in neonatal cardiomyocyte cell-cycle withdrawal and IGF1 gene regulation. Circ. Res. 2013;112(2):267–277. doi: 10.1161/circresaha.112.277442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moustafa A., Hashemi S., Brar G., Grigull J., Ng S.H.S., Williams D., et al. The MEF2A transcription factor interactome in cardiomyocytes. Cell Death Dis. 2023;14(4):240. doi: 10.1038/s41419-023-05665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Turki S., Manickaraj A.K., Mercer C.L., Gerety S.S., Hitz M.P., Lindsay S., et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am. J. Hum. Genet. 2014;94(4):574–585. doi: 10.1016/j.ajhg.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gharibeh L., Yamak A., Whitcomb J., Lu A., Joyal M., Komati H., et al. GATA6 is a regulator of sinus node development and heart rhythm. Proc. Natl. Acad. Sci. U.S.A. 2021;118(1) doi: 10.1073/pnas.2007322118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malekar P., Hagenmueller M., Anyanwu A., Buss S., Streit M.R., Weiss C.S., et al. Wnt signaling is critical for maladaptive cardiac hypertrophy and accelerates myocardial remodeling. Hypertension. 2010;55(4):939–945. doi: 10.1161/hypertensionaha.109.141127. [DOI] [PubMed] [Google Scholar]
- 43.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R.L., et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Liu S., Heallen T., Martin J.F. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 2018;15(11):672–684. doi: 10.1038/s41569-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 45.Inserte J., Garcia-Dorado D. The cGMP/PKG pathway as a common mediator of cardioprotection: translatability and mechanism. Br. J. Pharmacol. 2015;172(8):1996–2009. doi: 10.1111/bph.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koitabashi N., Aiba T., Hesketh G.G., Rowell J., Zhang M., Takimoto E., et al. Cyclic GMP/PKG-dependent inhibition of TRPC6 channel activity and expression negatively regulates cardiomyocyte NFAT activation Novel mechanism of cardiac stress modulation by PDE5 inhibition. J. Mol. Cell. Cardiol. 2010;48(4):713–724. doi: 10.1016/j.yjmcc.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackenzie F., Ruhrberg C. Diverse roles for VEGF-A in the nervous system. Development. 2012;139(8):1371–1380. doi: 10.1242/dev.072348. [DOI] [PubMed] [Google Scholar]
- 48.White A.L., Bix G.J. VEGFA isoforms as pro-angiogenic therapeutics for cerebrovascular diseases. Biomolecules. 2023;13(4):702. doi: 10.3390/biom13040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers K.V., Amend S.R., Pienta K.J. Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the tumor microenvironment. Mol. Cancer. 2019;18(1):94. doi: 10.1186/s12943-019-1022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang F., Wei Y., Han D., Li Y., Shi S., Jiao D., et al. Interaction with CD68 and regulation of GAS6 expression by endosialin in fibroblasts drives recruitment and polarization of macrophages in hepatocellular carcinoma. Cancer Res. 2020;80(18):3892–3905. doi: 10.1158/0008-5472.Can-19-2691. [DOI] [PubMed] [Google Scholar]
- 51.Tan Y., Zhao L., Yang Y.G., Liu W. The role of osteopontin in tumor progression through tumor-associated macrophages. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.953283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin R., Wu S., Zhu D., Qin M., Liu X. Osteopontin induces atrial fibrosis by activating Akt/GSK-3β/β-catenin pathway and suppressing autophagy. Life Sci. 2020;245 doi: 10.1016/j.lfs.2020.117328. [DOI] [PubMed] [Google Scholar]
- 53.He Z., Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell. 1997;90(4):739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 54.Soker S., Takashima S., Miao H.Q., Neufeld G., Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92(6):735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 55.Fernández-Fernández L., Bellido-Martín L., García de Frutos P. Growth arrest-specific gene 6 (GAS6). An outline of its role in haemostasis and inflammation. Thromb. Haemostasis. 2008;100(4):604–610. doi: 10.1160/th08-04-0253. [DOI] [PubMed] [Google Scholar]
- 56.Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54(2) doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoeft K., Schaefer G.J.L., Kim H., Schumacher D., Bleckwehl T., Long Q., et al. Platelet-instructed SPP1(+) macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 2023;42(2) doi: 10.1016/j.celrep.2023.112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon S., Chang M.S., Koh S.H., Choi Y.K. Repair mechanisms of the neurovascular unit after ischemic stroke with a focus on VEGF. Int. J. Mol. Sci. 2021;22(16):8543. doi: 10.3390/ijms22168543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maden C.H., Gomes J., Schwarz Q., Davidson K., Tinker A., Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev. Biol. 2012;369(2):277–285. doi: 10.1016/j.ydbio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo); https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211650.
The data included in article and supplementary material in article. Data associated with my study have not been deposited into a publicly available repository.