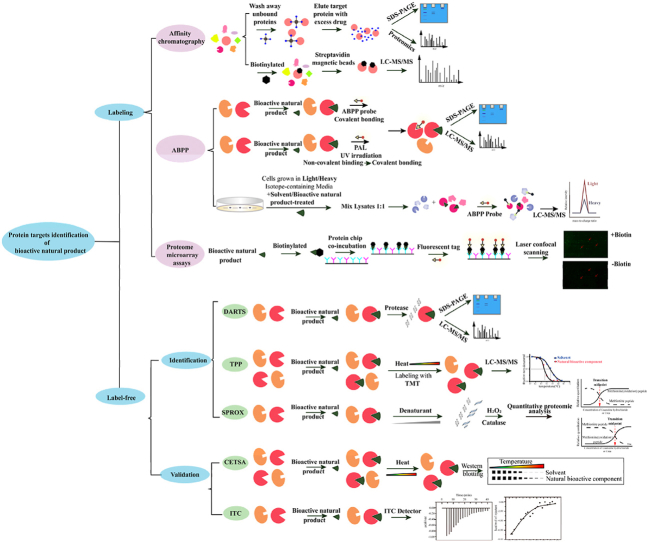
Abstract
Background
Natural products exhibit structural complexity, diversity, and historical therapeutic significance, boasting attractive functions and biological activities that have significantly influenced drug discovery endeavors. The identification of target proteins of active natural compounds is crucial for advancing novel drug innovation. Currently, methods for identifying targets of natural products can be categorized into labeling and label-free approaches based on whether the natural bioactive constituents are modified into active probes. In addition, there is a new avenue for rapidly exploring the targets of natural products based on their innate functions.
Aim
This review aimed to summarize recent advancements in both labeling and label-free approaches to the identification of targets for natural products, as well as the novel target identification method based on the natural functions of natural products.
Methods
We systematically collected relevant articles published in recent years from PubMed, Web of Science, and ScienceDirect, focusing on methods employed for identifying protein targets of bioactive natural products. Furthermore, we systematically summarized the principles, procedures, and successful cases, as well as the advantages and limitations of each method.
Results
Labeling methods allow for the direct labeling of target proteins and the exclusion of indirectly targeted proteins. However, these methods are not suitable for studying post-modified compounds with abolished activity, chemically challenging synthesis, or trace amounts of natural active compounds. Label-free methods can be employed to identify targets of any natural active compounds, including trace amounts and multicomponent mixtures, but their reliability is not as high as labeling methods. The structural complementarity between natural products and their innate receptors significantly increase the opportunities for finding more promising structural analogues of the natural products, and natural products may interact with several structural analogues of receptors in humans.
Conclusion
Each approach presents benefits and drawbacks. In practice, a combination of methods is employed to identify targets of natural products. And natural products' innate functions-based approach is a rapid and selective strategy for target identification. This review provides valuable references for future research in this field, offering insights into techniques and methodologies.
Keywords: Protein targets, Natural products, Labeling methods, Label-free approaches, Natural products' innate functions-based approach
Graphical abstract
This review systematically summarizes recent advancements in target identification methods and technologies for natural products, providing a comprehensive overview of principles, procedures, successful cases, as well as the advantages and limitations of each approach.
Highlights
-
•
Summarizing the recent progress in identifying targets of natural products.
-
•
These methods can be categorized into labeling, label-free approaches, and natural products' innate functions-based avenue.
-
•
Elucidation of the principle, workflow, successful cases of each method.
-
•
Comparing the pros and cons of different approaches.
1. Introduction
Compared to small-molecule drugs synthesized chemically, natural products offer significant advantages in terms of structural novelty, diversity, biocompatibility, and functional versatility. Moreover, they have undergone natural selection and optimization through long-term evolutionary processes [1]. Approximately 70 % of all approved therapeutic agents originate from natural products or their derivatives [2]. Thus, natural products are crucial sources of new drugs [3]. Notably, the 2015 Nobel Prize in Physiology or Medicine awarded to Youyou Tu for discovering the plant natural product artemisinin, which revolutionized malaria treatment, brought immense pride and optimism to the natural product community worldwide. However, unclear modes of action and challenges in identifying target proteins hinder the development of natural products in drug discovery. Thus, exploring and identifying targets of natural products is essential for understanding their mechanisms of action, identifying potential toxicities, and guiding structure-activity relationship studies. Furthermore, it contributes to the discovery of novel disease-relevant targets, playing a critical role in innovative drug research based on natural products.
Currently, with the advancement in chemical biology and biophysics, an increasing array of methods has emerged for identifying targets of natural bioactive molecules. These methods can be broadly classified into labeling and label-free approaches based on whether natural bioactive constituents are modified into active probes. In probe-based methods, natural bioactive molecules are transformed into active probes to explore their interactions with targets, whereas label-free methods rely on changes in the biophysical properties of the target protein upon their binding with the natural bioactive molecule for the identification of the target protein. In addition, a novel avenue for rapidly exploring the targets of natural products based on their innate functions has also been developed. This review primarily summarizes these two methodological categories as well as the new approach based on the innate functions of natural products, providing a systematic elucidation of their principles, workflows, and successful cases, as well as the advantages and disadvantages associated with each approach. This review aims to inspire and provide guidance to researchers in this field.
2. Target identification of natural bioactive components using labeling techniques
Labeling techniques entail chemically modifying natural bioactive components to generate probes with specific markers, such as fluorescent dyes or biotin conjugates. These labeled probes enable the identification of target proteins through corresponding fluorescence imaging or enrichment techniques.
2.1. Affinity chromatography
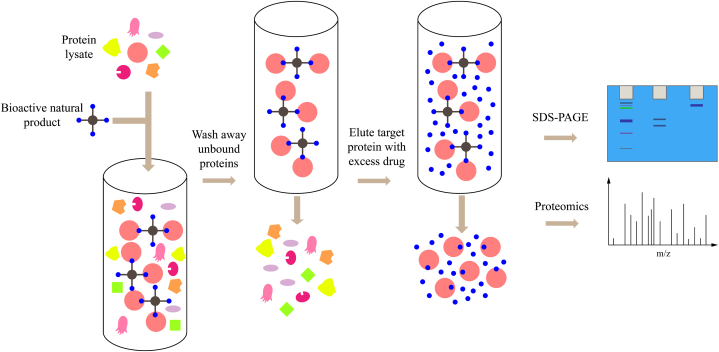
Affinity chromatography stands out as the most traditional method of labeling. As shown in Fig. 1, the typical procedure involves initially identifying the active functional groups of natural bioactive components and then immobilizing them onto a solid support matrix, such as agarose beads or resin. Once immobilized, these probes act as baits. Subsequently, the cell or tissue lysate is either incubated with the solid support matrix loaded with active compounds (when agarose beads are used as the solid support matrix) or passed through the solid support matrix (when resin materials are used as the solid support matrix). Following this, multiple washes with inert solvents are performed to remove non-specifically bound proteins, leaving the proteins that exhibit affinity binding with the immobilized natural bioactive components attached to the solid support matrix. Finally, the bound proteins are eluted through various means, such as heating, using high ionic strength solvents, or adding an excess of free drug to compete with the target protein binding. The eluted proteins are then subjected to quantitative proteomic analysis for identification. This method has been utilized for the discovery of target proteins for numerous well-known compounds. For instance, affinity chromatography assays have enabled the identification of the targets of artesunate [4], arzanol [5], baicalein [6], 20(S)-protopanaxadiol [7], and the herb-derived sesquiterpene lactone compound IJ-5 [8] (Table 1). A notable example is the study by Ito et al. [9], where immobilized probes of thalidomide were employed to identify its target protein, cereblon (CRBN). CRBN forms an E3 ubiquitin ligase complex with damaged DNA binding protein 1 and Cul4A, thereby inhibiting ubiquitin ligase activity crucial for limb outgrowth in zebrafish and chicks, thus contributing to thalidomide teratogenicity. This research was published in the journal “Science”.
Fig. 1.
The typical process of affinity chromatography. Initially, bioactive natural products are immobilized onto a solid support matrix (such as agarose beads or resin), serving as baits. Subsequently, cell lysates or tissue homogenates are either incubated with the active compound-loaded solid support matrix (when agarose beads serve as the solid support matrix) or passed through the solid support matrix (when resin materials serve as the solid support matrix). Afterward, multiple washes with inert solvents are performed to remove unbound proteins. Finally, the retained proteins are eluted either by heating, utilizing high-ionic-strength solvents, or adding an excess of a free drug to compete with the target protein binding. Subsequently, these proteins are identified through quantitative proteomic analysis.
Table 1.
Recent studies that conducted affinity chromatography-based target identification of natural products.
| No. | Natural product | Structure | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|---|
| 1 | Artesunate | 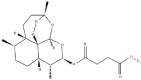 |
Quaternary ammonium salts | Regeneration of pancreatic β cell mass from α cells | Gephyrin | [4] |
| 2 | Arzanol | 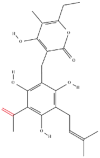 |
Terpenoids | Anti-inflammatory activity | Brain glycogen phosphorylase | [5] |
| 3 | Baicalein | 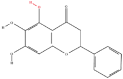 |
Flavonoids | Elimination of liver tumor-initiating stem cell-like cells resistant to mTORC1 inhibition | SAR1B guanosine triphosphatase | [6] |
| 4 | 20(S)-Protopanaxadiol |  |
Saponins | Reduction of fatigue | CK-MM | [7] |
| 5 | IJ-5 | 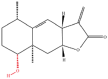 |
Terpenoids | Anti-inflammatory activity | Ubiquitin-conjugating enzyme UbcH5 | [8] |
| 6 | Sappanone A |  |
Ketones | Anti-neuroinflammatory activity | IMPDH2 | [10] |
| 7 | Celastrol | 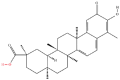 |
Terpenoids | Intervention of the progression of hepatocellular carcinoma | ROCK2 | [11] |
| 8 | Ainsliadimer A | 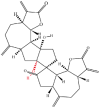 |
Terpenoids | Anticancer and anti-inflammatory activity | IKKβ | [12] |
| 9 | Pateamine A |  |
Alkaloids | Inhibition of eukaryotic translation initiation | Eukaryotic initiation factor 4A | [13] |
| 10 | Adenanthin |  |
Terpenoids | Induction of leukemic cell differentiation | Peroxiredoxins I and II | [14] |
Note: The red-marked portion in the structure refers to the labeling binding site.
Furthermore, natural bioactive components can be biotinylated. By exploiting the strong affinity interaction between streptavidin and biotin, the biotinylated natural bioactive components can be captured, enriched, and purified using streptavidin-coated agarose solid support carriers. This facilitates the capture of target proteins associated with natural bioactive components, which can subsequently be identified via mass spectrometry (MS). Tu and colleagues transformed sappanone A, an anti-neuroinflammatory active component from the traditional Chinese medicine Su-Mu, into a chemical probe conjugated with biotin. This probe was employed to “fish out” the direct target proteins from glial cells, leading to the identification of inosine-5′-monophosphate dehydrogenase 2 (IMPDH2) as a key target of sappanone A [10]. This breakthrough was recognized as one of the “Top 10 Medical Advances in China” in 2017. In addition, biotin affinity purification systems have been employed to identify the targets of celastrol, an active component from the medicinal plant Tripterygium wilfordii Hook F [11], the natural anti-inflammatory active molecule ainsliadimer A [12], the marine natural product pateamine A [13], and adenanthin, a diterpenoid derived from the leaves of Isodon adenanthus [14] (Table 1).
Affinity chromatography facilitates rapid and large-scale enrichment of target proteins. Nevertheless, it faces three primary limitations [15]. Firstly, discerning between high-affinity target proteins and abundant proteins with low affinity poses a challenge. Secondly, the washing process relies on empirical conditions, wherein overly stringent conditions may significantly decrease the number of identified targets, while weak washing conditions could yield false-positive results. Lastly, it fails to capture the interaction between drugs and proteins within live cells under physiological conditions. Additionally, the biotin affinity tag introduces steric hindrance, potentially diminishing or even abolishing the activity of natural bioactive components and hindering the entry of probe molecules into cells.
2.2. Activity-based protein profiling (ABPP)
Given the limitations of affinity chromatography, chemists and biologists have focused on exploring target identification methods suitable for targets exhibiting low abundance and live cells. Currently, the most widely employed and rapidly advancing technique is ABPP, which was introduced by Dr. Cravatt's research team at the Scripps Research Institute in the United States in 1999 [16].
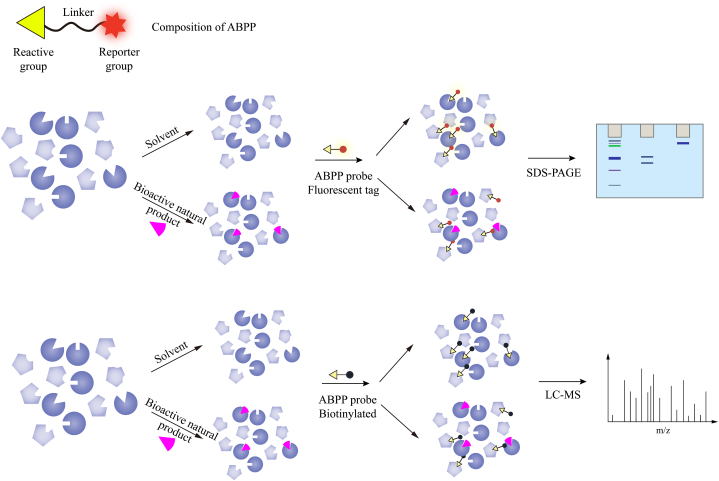
Activity-based probes (ABPs) form the cornerstone of ABPP technology. Unlike immobilized probes, ABPs do not necessitate pre-fixation to a matrix material and can be directly applied to live cells, elucidating drug–target interactions under physiological conditions [17]. ABPs comprise three components: a reactive group, a reporter group, and a linker connecting the reactive and reporter groups (Fig. 2). The reactive group serves as the core of ABPs. Depending on the reactive group, ABPs can be classified as either covalent or non-covalent probes for natural bioactive components. For example, natural products containing electrophilic moieties, such as Michael acceptors, β-lactones, β-lactams, epoxides, haloalkyl carbonyl compounds, and isothiocyanates, can form covalent bonds with the nucleophilic groups of target proteins, facilitating the covalent binding of the probe molecule to the target protein [18]. The design of such probes merely entails incorporating a reporter group via a linker in a region that does not affect the activity of the natural products.
Fig. 2.
The structure of ABPs and the workflow of competitive ABPP. The upper panel depicts the structure of ABPs, which include a reactive group, a reporter group, and a linker facilitating their connection. The middle and lower panels denote the workflow of a competitive ABPP. Initially, cell lysates or live cells are incubated with either the native bioactive natural product or solvent and subsequently labeled with an ABPP probe. Following this, SDS-PAGE or LC–MS/MS are employed to either visualize or identify the targets labeled by the ABPP probe. Targets of the bioactive natural product exhibit diminished signals in the active compound-treated samples compared to those treated with solvent controls.
Fluorescent dyes or biotin are commonly employed as reporter groups in ABPs. However, these labeling molecules suffer from issues such as large size and poor solubility, significantly hampering the ability of ABPs to penetrate the cell membrane [15]. To overcome this limitation, Speers et al. [19] integrated click chemistry with ABPs. In this method, inert reactive groups, such as alkynes and azides, replace the original reporter groups. These inert reactive groups exhibit minimal interference with the chemical properties of natural bioactive components, allowing them to bind to target proteins in live cells or cell lysates. Subsequently, through click chemistry reactions, fluorescent dyes or biotin can be appended to the probe–target complex, facilitating the enrichment and identification of the target proteins [15].
The typical workflow of ABPP entails incubating probes with live cells or cell lysates, facilitating the binding of the small-molecule probe to the active site of its target enzyme. The probe forms a complex with the enzymes through enzymatic reactions or covalent binding with the target protein, induced by ultraviolet (UV) irradiation using photoaffinity groups. Subsequently, the labeled protein is enriched and purified using the probe's reporter group, such as fluorescence or biotin. Finally, target identification is achieved using techniques such as MS.
In ABPP experiments, competitive ABPP groups may be configured by pre-incubating the natural bioactive component with either cell lysates or live cells prior to introducing the natural bioactive component probe (Fig. 2). Should the natural bioactive component effectively compete for binding to the enzymatic active site of the target protein, it would competitively impede the labeling process of the target protein by the probe. Compared to the control group treated solely with the probe, the competitive group exhibits diminished fluorescence signal intensity or reduced peak intensity in MS, facilitating the identification of potential target proteins [20].
Spradlin et al. [21] utilized ABPP platforms to reveal that nimbolide, a terpenoid natural product derived from the Neem tree, binds to a novel functional cysteine crucial for substrate recognition in the E3 ubiquitin ligase RNF114. This interaction inhibits the ubiquitination and degradation of tumor suppressors, thereby impeding cancer pathogenicity. Similarly, Zhao et al. [22] identified the covalently modified target of parthenolide, another terpenoid natural product with potent antitumor activity, using ABPP platforms. Moreover, ABPP has been employed to explore the mechanisms of action of dankastatin B, which exhibits anticancer activity [23], spongiolactones, marine natural products with potential anticancer properties [24], the antimalarial natural product salinipostin A [25], illudin, an anticancer natural product family [26], withangulatin A, a natural small molecule that inhibits the serine synthesis pathway and proliferation of colon cancer cells [27], and piperlongumine, a cancer-selective killing natural product [28], all of which possess electrophilic functional groups (Table 2).
Table 2.
Recent studies that conducted ABPP-based target identification of natural products.
| No. | Natural products | Structure | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|---|
| 1 | Nimbolide | 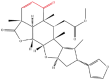 |
Terpenoids | Anticancer activity | E3 ubiquitin ligase RNF114 | [21] |
| 2 | Parthenolide | 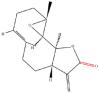 |
Terpenoids | Anticancer activity | Ubiquitin carboxyl-terminal hydrolase 10 (USP10) | [22] |
| 3 | Dankastatin B |  |
Terpenoids | Anticancer activity | Mitochondrial VDAC3 | [23] |
| 4 | Salinipostin A |  |
Cyclopeptide | Antimalarial activity | Essential α/β serine hydrolases | [25] |
| 5 | Illudin |  |
Terpenoids | Anticancer activity | DNA modification and unselective protein binding | [26] |
| 6 | Withangulatin A |  |
Alkaloids | Antitumor activity | phosphoglycerate dehydrogenase (PHGDH) | [27] |
| 7 | Piperlongumine |  |
Alkaloids | Anticancer activity | GSTO1 | [28] |
Note: The red-marked portion in the structure refers to the labeling binding site.
This method offers the advantage of robust specificity. ABPs eliminate the need for pre-fixation to a substrate material and can be directly applied to live cells, elucidating drug–target interactions under physiological conditions. However, its effectiveness is constrained to capturing target proteins that covalently bind with small molecules, thus limiting its applicability.
2.3. Photoaffinity labeling probes (PAL probes)
When designing probes for natural active compounds that bind to targets through non-covalent interactions, it is necessary to introduce photoreactive groups in addition to linker groups and reporter groups. These photoreactive groups, upon UV irradiation, generate highly reactive intermediates known as carbenes, which can form stable covalent bonds between the probe and the target protein. Such probes are categorized as PAL probes. Commonly employed photoaffinity groups encompass diazirine, benzophenone, and aryl azide [29]. Matrine is a plant alkaloid with potent anticancer activities. However, its molecular target(s) and mechanism remain unknown. Wang et al. [30], employing the PAL approach, for the first time, identified annexin A2 as a direct-binding target of matrine in cancer cells. Moreover, photoaffinity probes have been utilized to elucidate the targets of various natural compounds, such as coibamide A, a marine natural product with potent antiproliferative activity against human cancer cells [31], betulinic acid [32], harringtonolide, a bioactive diterpenoid tropone isolated from Cephalotaxus harringtonia with antiproliferation activity [33], pseudolaric acid B possessing anticancer activity [34], the epothilone binding site on β-tubulin [35], quercetin, a flavonoid natural product found in various foods with a wide range of medicinal effects [36], and pladienolide, a naturally occurring antitumor macrolide [37] (Table 3).
Table 3.
Recent studies that performed PAL probe-based target identification of natural products.
| No. | Natural products | Structure | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|---|
| 1 | Matrine | 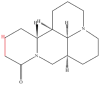 |
Alkaloids | Anticancer activity | Annexin A2 | [30] |
| 2 | Coibamide A | 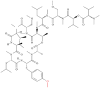 |
Cyclopeptide | Anticancer activity | Sec61 | [31] |
| 3 | Betulinic acid | 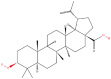 |
Terpenoids | Anticancer activity | Tropomyosin | [32] |
| 4 | Harringtonolide | 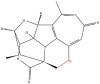 |
Cyclopeptide | Suppression of cancer cell migration | Receptor for activated C kinase 1 | [33] |
| 5 | Pseudolaric acid B | 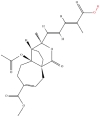 |
Terpenoids | Anticancer activity | CD147 | [34] |
| 6 | Epothilone | 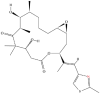 |
Macrolides | Alleviation of CNS injuries | The binding site on β-tubulin | [35] |
| 7 | Quercetin | 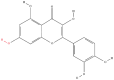 |
Flavonoids | Antitumor activity | HSP70, HSP90, RuvB-like 2 ATPases, and eukaryotic translation initiation factor 3 | [36] |
| 8 | Pladienolide | 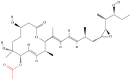 |
Macrolides | Antitumor activity | Splicing factor SF3b | [37] |
Note: The red-marked portion in the structure refers to the labeling binding site.
This method enables the analysis of proteins that cannot be targeted by ABPs. Nevertheless, photoaffinity groups may still display non-specific binding to abundant cellular proteins.
2.4. ABPP coupled with the stable isotope labeling by amino acids in cell culture (SILAC) technique
To enhance quantitative accuracy, Cravatt et al. integrated the SILAC technique and ABPP, yielding ABPP–SILAC. SILAC operates by substituting specific amino acids in cell culture media with either naturally occurring isotope (light) or stable isotope (heavy)-labeled amino acids. Following five to six cell cycles, stable isotope-labeled amino acids are fully incorporated into newly synthesized proteins, replacing endogenous amino acids. Lysates from light- and heavy-isotope-labeled cells are equitably combined, separated via gel electrophoresis, and subsequently identified via MS. Each peptide manifests as a pair in MS data, with the light-labeled peptide containing the light amino acid and the heavy-labeled peptide containing the heavy amino acid. A 1:1 ratio in SILAC peptide pairs signifies unaltered protein abundance in the proteome. If the SILAC peptide pair ratio (light vs. heavy) is large, it indicates a potential target of the natural product [38].
By integrating competitive ABPP with SILAC, the heavy amino acid-labeled group is subjected to treatment with the natural active component within the cell lysate (or within live cells), serving as the competitive group, while the light amino acid-labeled group remains untreated. Subsequently, the two groups are combined in accordance with cell number or protein quantity, followed by the addition of the natural active component probe to label the target proteins. MS is then employed for target identification purposes (Fig. 3). A diminished intensity of the heavy-labeled peptide peak implies potential binding of the natural product to the target protein. Wang and colleagues employed the ABPP–SILAC technique to explore the targets involved in the lipid-lowering effects of baicalin, the principal active component of Scutellaria baicalensis. They synthesized a baicalin-based photoaffinity probe and employed competitive ABPP–SILAC to capture its targets. LC–MS/MS analysis, complemented by bioinformatics analysis and reverse validation, revealed carnitine palmitoyltransferase 1A as a target of baicalin in the context of obesity amelioration [39]. In addition, Chen et al. [40] identified peroxiredoxin 6 as a direct target of withangulatin A in the alleviation of non-small cell lung cancer (NSCLC) via ABPP–SILAC (Table 4).
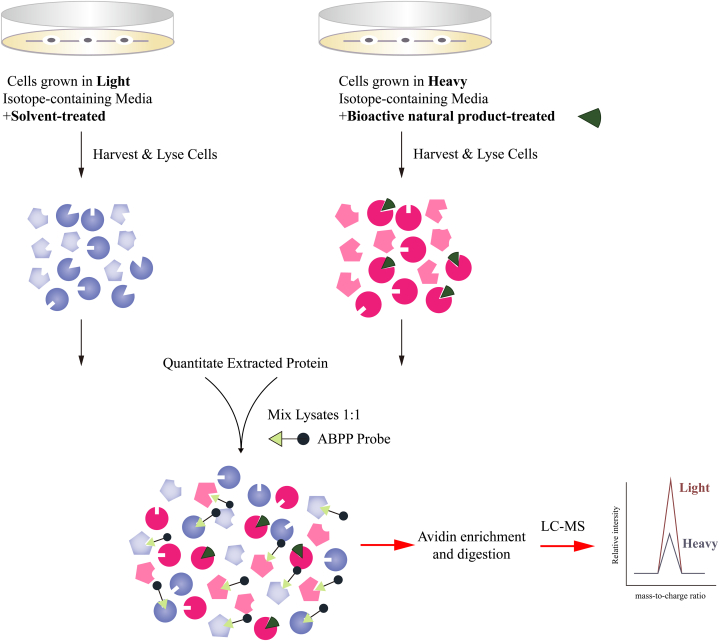
Fig. 3.
The typical process of competitive ABPP–SILAC. The heavy amino acid-labeled group is treated with the natural active compound, serving as the competitive group, while the light amino acid-labeled group remains untreated. Subsequently, the two groups are combined in accordance with either cell number or protein quantity, followed by the addition of the bioactive natural product probe for labeling the target proteins. LC–MS/MS is subsequently employed to identify the proteins captured by the probe. The SILAC ratio (light vs. heavy) quantified for each protein serves as an indicator of the potential target of the active natural compound, with a higher ratio suggesting a stronger likelihood of interaction.
Table 4.
Recent studies that performed ABPP–SILAC-based target identification of natural products.
| No. | Natural products | Structure | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|---|
| 1 | Baicalin | 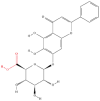 |
Flavonoids | Alleviation of obesity | Carnitine palmitoyltransferase 1A | [39] |
| 2 | Withangulatin A | 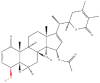 |
Alkaloids | Anti-NSCLC effects | Peroxiredoxin 6 | [40] |
Note: The red-marked portion in the structure refers to the labeling binding site.
2.5. Proteome microarray assays
Firstly, biotin or fluorescent-labeling natural active components were co-incubated with a protein chip. After washing away unbound components, the fluorescence intensity of each spot on the chip is determined via fluorescence scanning or laser confocal scanning. When biotin-labeling natural active components are employed, they are co-incubated with fluorescent-labeling streptavidin and subjected to fluorescence scanning to screen potential targets of the natural active components. Chen and colleagues utilized this technique to explore the targets of arsenic [41]. Arsenic demonstrates excellent efficacy in acute promyelocytic leukemia treatment and possesses significant therapeutic potential for various tumors. However, its broad anticancer mechanisms remain unclear. Researchers co-incubated biotinylated arsenic with a protein chip containing 16,368 proteins, utilizing Cy3-labeling streptavidin to identify arsenic-binding proteins. Fluorescence scanning revealed enhanced fluorescence signals in the probe group compared to the negative control group. They identified 360 proteins directly interacting with arsenic, including the well-known arsenic target PML. Previously, fewer than 20 proteins directly interacting with arsenic had been identified worldwide, underscoring the protein chip platform's efficacy in rapidly and comprehensively identifying drug targets. The most significantly enriched proteins are implicated in the glycolysis pathway. Detailed biochemical and metabolomics analyses have identified hexokinase-2 (HK2) as a key target of arsenic, functioning as a rate-limiting enzyme in glycolysis. This research was published in “Proceedings of the National Academy of Sciences of the United States of America”. Liu et al. [42] identified bufalin, a natural product-derived molecular glue, which targets E2F2 degradation, leading to the transcriptional suppression of multiple oncogenes and the growth of hepatocellular carcinoma in vitro and in vivo. In addition, proteome microarray assays have been employed to identify the targets of coelonin, an active anti-inflammatory component of Bletilla striata [43] (Table 5).
Table 5.
Recent studies that conducted proteome microarray-based target identification of natural products.
| No. | Natural products | Structure | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|---|
| 1 | Arsenic | 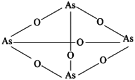 |
Elemental | Broad anticancer activity | 360 proteins, among which HK2 is a key target | [41] |
| 2 | Bufalin | 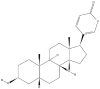 |
Steroids | Suppression of hepatocellular carcinoma growth | E2F2 | [42] |
| 3 | Coelonin | 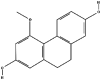 |
Alkaloids | Anti-inflammatory activity | PTEN | [43] |
This method offers the advantages of necessitating minimal sample volumes and boasting high throughput, enabling the simultaneous analysis of thousands to millions of proteins. Additionally, protein chips are relatively user-friendly. However, they entail high costs due to the need for a series of expensive, high-end instruments. Furthermore, standardization issues are observed with protein chips.
3. Target identification and target validation of natural bioactive components using label-free techniques
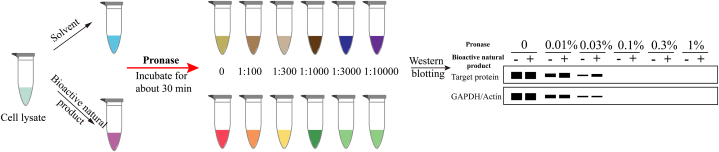
Despite significant advancements in affinity-based methods for screening natural product targets, limitations persist [44]. Some natural products lack suitable modification sites or synthetic methods to introduce such sites. Additionally, exogenous modification groups may affect the activity of natural active compounds or hinder their binding to true targets, thereby compromising target identification accuracy. To overcome these challenges, researchers have devised various target screening methods that obviate the need for structural modifications of natural active molecules. Instead, they employ biophysical approaches to assess the impact of the binding of natural active compounds to target proteins on protein stability, thus facilitating the discovery of targets for natural active molecules. Moreover, label-free techniques can be categorized into target identification and target validation methods. For example, drug affinity responsive target stability (DARTS), thermal proteome profiling (TPP), and stability of proteins from rates of oxidation (SPROX) methods are primarily used for target validation, while cellular thermal shift assays (CETSA) and isothermal titration calorimetry (ITC) are often used for target identification.
3.1. Target identification methods
3.1.1. DARTS
In 2009, Lomenick et al. introduced DARTS [45]. The method's principle posits that upon the binding of a small molecule to a target protein, the small molecule occupies the recognition site of proteolytic enzymes, impeding their interaction with the cleavage site on the target protein. Consequently, the target protein exhibits enhanced resistance to proteolytic cleavage and increased stability. Changes in the resistance to proteolysis pre- and post-small molecule binding can be discerned via electrophoresis. Subsequent identification of distinct bands on the protein gel can be achieved through MS to ascertain the target proteins of the small molecule. As a proof-of-principle, researchers selected the FKBP12 protein, a known target of rapamycin, and FK506. Proteolysis was conducted using subtilisin protease, and the resistance of FKBP12 to proteolysis subsequent to binding with the compound was observed via SDS-PAGE, thus affirming the scientific validity and feasibility of this method [45].
The typical procedure of this method involves cell lysis, followed by dividing the lysate into two groups (Fig. 4): the active natural compound group and the solvent group. The lysates of the active natural compound group are incubated with the bioactive compound, while the lysates of the solvent group are treated with the solvent alone. This allows the bioactive compound to bind to the target proteins. Subsequently, both the solvent and the bioactive compound groups are further divided into five to six subgroups, and each subgroup is treated with a gradient concentration of proteolytic enzyme for protein digestion. Commonly used proteolytic enzymes include subtilisin protease, thermolysin, and protease from Streptomyces griseus. Finally, various methods, such as SDS-PAGE, 2D-PAGE, gel staining techniques, and gel or non-gel MS, are employed to detect or identify the bound proteins.
Fig. 4.
The typical process of DARTS. Initially, cell lysates are incubated either with a solvent or with a natural active compound. Following this, both groups are subdivided into five to six subgroups. Subsequently, gradient proteolysis is performed for each subgroup. Finally, the samples are subjected to SDS-PAGE for the detection of the target molecule.
The DARTS technique, which does not necessitate chemical modification of small molecules, offers a convenient means to identify direct targets of natural products [46]. It has seen widespread application in recent years. Huang et al. [47] employed the DARTS method and discerned that the antitumor natural active compound batzelladine A targets HSP90. The target of the primary active compound, andrographolide, in the traditional Chinese medicinal Andrographis paniculata, known for its heat-clearing and detoxifying properties, was also identified using the DARTS technique. As the concentration of proteolytic enzymes increased, proteins in the solvent group gradually degraded, while andrographolide inhibited the hydrolysis of the target protein, dynamin-related protein 1 (DRP1), compared to the control group [48]. Moreover, DARTS confirmed the targets of polyphyllin D, a natural product effective against NSCLC [49], ergolide, a sesquiterpene lactone natural product with anti-inflammatory and anticancer activities used to treat inflammatory diseases [50], aconitine, an active compound exhibiting cardiotoxic effects found in Aconitum species [51], crellastatin A, a cytotoxic sulfated bis-steroid isolated from the Vanuatu Island marine sponge Crella sp. [52], cryptotanshinone, isolated from the roots of Salvia miltiorrhiza inhibiting the terminal differentiation of human keratinocytes [53], and grape seed extract in anti-colorectal cancer applications [54] (Table 6).
Table 6.
Recent studies that conducted DARTS-based target identification of natural products.
| No. | Natural products | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|
| 1 | Batzelladine A | Alkaloids | Suppression of lung cancer tumorigenesis | HSP90 | [47] |
| 2 | Andrographolide | Terpenoids | Alleviation of parkinsonism | DRP1 | [48] |
| 3 | Polyphyllin D | Saponins | NSCLC therapy | Distruption of HSC70–LAMP2A interaction | [49] |
| 4 | Ergolide | Terpenoids | Alleviation of inflammatory diseases | NLRP3 | [50] |
| 5 | Aconitine | Alkaloids | Cardiotoxicity | Cytosolic phospholipase A2 | [51] |
| 6 | Crellastatin A | Macrocyclic depsipeptides | Anticancer activity | PARP-1 | [52] |
| 7 | Cryptotanshinone | Diterpenoid quinone | Alleviation of keratinopathic ichthyosis | FKBP1A | [53] |
| 8 | Grape seed extract | Polyphenolic compounds | Anticancer activity | Endoplasmic reticulum stress response proteins | [54] |
The limitations of DARTS encompass the following: certain small molecules may bind to the target protein without inducing significant conformational changes of the target; certain proteins could exhibit reduced sensitivity to proteolytic enzymes, thereby resulting in imperceptible alterations in DARTS outcomes; and the capacity to identify targets characterized by low protein abundance remains limited.
3.1.2. TPP
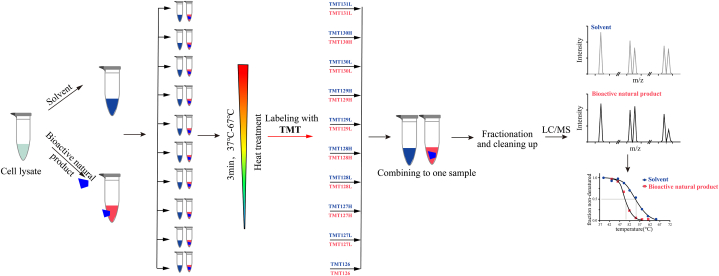
TPP, a combination of CETSA and multiplexed quantitative MS, enables the monitoring of global protein thermal stability changes during interactions with small molecules, thereby facilitating high-throughput screening of target proteins [55]. TPP operates on two principles: the principles of CETSA and multiplexed quantitative MS. Unlike CETSA, TPP characterizes protein thermal stability based on the melting temperature (Tm), denoting the temperature at which 50 % of the protein unfolds during thermal denaturation. Typically, proteins bound to small molecules exhibit increased stability and higher Tm values compared to unbound proteins. By comparing the Tm values and melting curves of proteins with and without small-molecule binding, target proteins can be identified. Multiplexed quantitative MS primarily refers to tandem mass tag (TMT), a peptide labeling technology developed by Thermo Fisher in the United States. TMT facilitates the labeling of peptides with up to 10 isotopic tags, enabling the simultaneous comparison of relative protein levels across 10 different samples. Therefore, TMT serves as a high-throughput analysis method that is particularly suitable for differential protein analysis of samples subjected to various treatments or treatment durations. Furthermore, through stable isotope labeling of different samples, all labeled samples can be pooled for subsequent processing, significantly reducing experimental errors and considerably improving quantification accuracy.
The typical protocol of this method involves treating cells or cell lysates with or without an active natural compound, followed by dividing the samples into 10 equal portions and heating them at increasing temperatures to induce protein denaturation and aggregation. The supernatant is then collected and digested using trypsin to generate peptides, which are subsequently labeled using TMT10. These labeled samples are mixed and analyzed through LC–MS/MS to calculate the Tm values of each protein, thereby screening for target proteins (Fig. 5). Building upon the potent efficacy of the natural product vioprolide A against human acute lymphoblastic leukemia cells, Kirsch et al. [56] conducted TPP, confirming nucleolar protein 14, essential in ribosome biogenesis, as a specific target of vioprolide A in Jurkat cells. Furthermore, in a study published in the journal “Science Translational Medicine”, TPP was utilized to identify the targets of antimalarial drugs, revealing that the antimalarial drug quinine directly binds to the parasite's intracellular purine nucleotide phosphorylases [57]. Chen et al. [58] demonstrated that rhodojaponin VI, a grayanotoxin from Rhododendron molle, alleviates neuropathic pain by targeting N-ethylmaleimide-sensitive fusion protein through the TPP approach. Moreover, TPP has been employed to elucidate the targets of tutin-induced epilepsy [59], the natural sesquiterpene lactone alantolactone, which inhibits NSCLC [60], kurarinone, which attenuates MPTP-mediated neuroinflammation [61], and arone, which protects against neuroinflammation [62] (Table 7).
Fig. 5.
Schematic workflow of TPP. Initially, cell lysates are incubated either with or without the active natural compound. Subsequently, aliquots are incubated at 10 distinct temperatures and then centrifuged. Following this, the proteins are digested with trypsin to generate peptides. After TMT10 labeling, the samples are mixed, fractionated, and subjected to LC–MS/MS analysis to ascertain the Tm values of individual proteins, thereby facilitating the screening of target proteins.
Table 7.
Recent studies that conducted TPP-based target identification of natural products.
| No. | Natural products | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|
| 1 | Vioprolide A | Cyclopeptide | Anti-human acute lymphoblastic leukemia | Nucleolar protein 14 | [56] |
| 2 | Quinine | Alkaloids | Antimalarial activity | Malarial parasite's intracellular purine nucleotide phosphorylase | [57] |
| 3 | Rhodojaponin VI | Saponins | Alleviation of neuropathic pain | N-ethylmaleimide-sensitive fusion protein | [58] |
| 4 | Tutin | Ketones | Induction of epilepsy | Calcineurin | [59] |
| 5 | Alantolactone | Terpenoids | Inhibition of NSCLC | AKR1C1 | [60] |
| 6 | Kurarinone | Flavonoids | Attenuation of MPTP-mediated neuroinflammation | Soluble epoxide hydrolase enzyme | [61] |
| 7 | Arone | Terpenoids | Protection against neuroinflammation | Histone-remodeling chaperone ASF1a | [62] |
This method offers the advantage of monitoring global thermal stability changes in proteins during drug action, enabling high-throughput detection of target proteins. However, it is unsuitable for heat-insensitive proteins, some of which require extreme temperature conditions to exhibit changes in thermal stability. Additionally, certain proteins may not display significant alterations in thermal stability upon ligand binding, rendering their identification unfeasible through TPP. Furthermore, extracting additional thermodynamic parameters from TPP data, aside from the Tm value, poses challenges. Moreover, the magnitude of the Tm shift following protein–ligand binding does not consistently correlate with ligand affinity.
3.1.3. SPROX
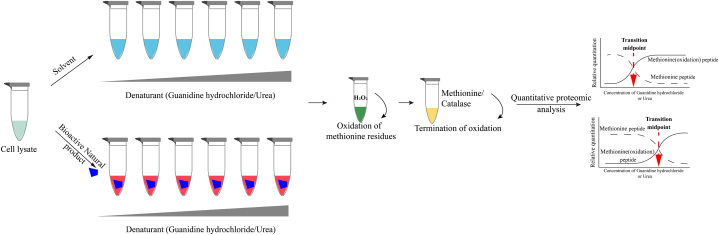
SPROX, introduced by Fitzgerald and colleagues from Duke University's Department of Chemistry in 2008 [63], represents another method for detecting natural product-induced target stability. This method capitalizes on the enhanced antioxidant capacity observed in target proteins upon binding with natural products. Protein oxidation capacity is determined by assessing the oxidation rate of methionine residues (most proteins typically contain at least one methionine residue, which can serve as a crucial marker for assessing the global dynamics of protein folding and unfolding equilibrium). The underlying principle is that as the concentration of chemical denaturants (such as guanidine hydrochloride or urea) increases, the protein unfolds, and increased exposure of its methionine residues to solvent renders them susceptible to oxidation in the unfolded state. Target proteins bound to natural products exhibit heightened structural stability. They exhibit slower unfolding in the presence of chemical denaturants. The addition of hydrogen peroxide, aimed at oxidizing methionine residues, results in a decelerated rate of methionine residue oxidation. The oxidation level of methionine residues is subsequently assessed via MS, and a concentration-dependent curve is generated, depicting the variation in oxidation and non-oxidized product. The intersection of the two curves denotes the transition midpoint. Binding of the natural product to the target protein enhances stability, necessitating a relatively higher denaturant concentration to achieve the equivalent oxidation product levels. Consequently, the transition midpoint shifts towards higher denaturant concentrations. Therefore, the potential target protein can be inferred by comparing the transition midpoints between the drug and control groups.
The typical procedure of SPROX is as follows (Fig. 6): Initially, a protein lysate is treated with a buffer containing varying denaturant concentrations in the presence or absence of the active natural product to establish the unfolding/refolding equilibrium of the protein. Subsequently, hydrogen peroxide is introduced to initiate the oxidation of methionine residues. Upon the completion of oxidation, an excess of either methionine or hydrogen peroxide scavenger is added to quench the oxidation reaction. Subsequently, the sample is digested using trypsin, following which the levels of oxidation and non-oxidation in methionine-containing peptide segments are quantified using MS. Finally, a curve is generated with the quantity of oxidized and non-oxidized methionine-containing peptide segments plotted on the y-axis and the denaturant concentration plotted on the x-axis. The shift in the transition midpoint is utilized to identify potential target proteins. The inventors of this technique, the Fitzgerald team, employed SPROX in 2011 to investigate the target proteins of the active compound resveratrol. They identified six potential target proteins in addition to the known target protein, cytoplasmic aldehyde dehydrogenase [64]. In addition, Fitzgerald employed the SPROX technique to analyze the thermodynamics of the geldanamycin–HSP90 interaction [65], as well as elucidate the targets of manassantin A, which exhibits anticancer activity [66] (Table 8).
Fig. 6.
The workflow of SPROX. Initially, in both the presence and absence of the active natural product, cell lysates are combined with various denaturant-containing buffers. Following equilibration of protein unfolding/refolding, hydrogen peroxide is added to induce the oxidation of methionine residues. Subsequently, the oxidation reaction is quenched using methionine or a hydrogen peroxide scavenger. Finally, proteins in each denaturant-containing buffer are digested into peptides for subsequent quantitative proteomics analysis.
Table 8.
Recent studies that conducted SPROX-based target identification of natural products.
This method offers the advantage of facilitating comprehensive assessment of protein folding states and potentially precise measurement of the binding domains and peptide segments engaged in the interaction between natural products and target proteins. Nonetheless, it is constrained by its ability to solely detect the binding of natural products to proteins containing methionine residues. In addition, generating the standard curve necessitates a large sample volume, thereby limiting the widespread utilization of this method.
3.2. Target validation methods
3.2.1. CETSA
In 2013, Martinez Molina et al. introduced CETSA [67], a ligand-induced target stability detection method similar to DARTS. This technique capitalizes on ligand-induced alterations in target protein, such as alterations in protein conformation, increased hydrophobicity within the protein, and chemical cross-linking within or between subunits of the target protein, resulting in enhanced thermal stability. As the temperature rises, proteins that are not bound to ligands unfold and undergo rapid denaturation, leading to precipitation. In contrast, ligand-bound proteins maintain their folded state and remain relatively stable, thereby bolstering the thermal stability of the target protein. Molina and colleagues applied this method to evaluate the thermal melting profiles of four distinct clinical drug targets in cell lysates, each displaying characteristic melting curves. The introduction of drugs capable of binding to these proteins to the cell lysates resulted in significant alterations in the melting profiles, thereby validating the scientific integrity and practical utility of CETSA.
The standard procedure of CETSA entails incubating equivalent quantities of cellular lysates or tissue homogenates with small molecules, followed by heating at various temperatures to induce protein denaturation and aggregation. Proteins bound to drugs remain relatively stable, whereas unbound proteins undergo rapid denaturation and precipitation with rising temperatures. Relative quantification of target proteins in the supernatant is performed via western blotting, thereby confirming the target proteins of the small molecules. Usenamine A, a novel product, was extracted from the lichen Usnea longissima. Yang et al. [68] identified myosin-9 as the direct target of usenamine A through MS, SPR assays, and CETSA. In addition, CETSA has been employed in combination with other target identification methods to validate the targets of paederosidic acid, which suppresses osteoclast formation and neuropathic pain [69], echinatin, a natural compound isolated from licorice, which effectively inhibits the hemolytic activity of methicillin-resistant Staphylococcus aureus [70], α-asarone, an important component of the Huangxiong formula for treating ischemic stroke [71], nitidine chloride, an extract from Zanthoxylum nitidum, used in multiple myeloma treatment [72], pentoxifylline, which attenuates non-alcoholic fatty liver [73], the primary active constituents of the Xiang-lian pill, which suppresses pancreatic tumor [74], toosendanin, a natural triterpenoid saponin with anti-colorectal cancer activity [75], glytabastan B, a coumestan isolated from Glycine tabacina, which is used for the prevention and treatment of rheumatoid arthritis [76], tubocapsenolide A, a withanolide-type steroid with therapeutic potential for osteosarcoma [77], the primary constituents of the Zuojin capsule, which is used to combat colorectal cancer [78], proanthocyanidin A1, which ameliorates chemotherapy-induced thrombocytopenia [79], cucurbitacin B, which inhibits NSCLC [80], celastrol and gambogic acid, which exhibit anti-breast cancer activity [81], shikonin, a natural bioactive component of Lithospermum erythrorhizon with anti-triple-negative breast cancer activity [82] (Table 9).
Table 9.
Recent studies that conducted CETSA-based target identification of natural products.
| No. | Natural products | Category | Bioactivity | Target protein | Reference |
|---|---|---|---|---|---|
| 1 | Usenamine A | Benzofuran derivatives | Alleviation of parkinsonism | Myosin-9 | [68] |
| 2 | Paederosidic acid | Terpenoids | Suppression of osteoclast formation and neuropathic pain | P2Y14 receptor | [69] |
| 3 | Echinatin | Flavonoids | Inhibition of the hemolytic activity of methicillin-resistant S. aureus | Indirect binding to α-hemolysin | [70] |
| 4 | α-Asarone | Aromatic hydrocarbons | Alleviation of ischemic stroke | PI3K | [71] |
| 5 | Nitidine chloride | Alkaloids | Alleviation of multiple myeloma | ABCB6 | [72] |
| 6 | Pentoxifylline | Methylxanthine derivatives | Alleviation of non-alcoholic fatty liver | Toll-like receptor 4 (TLR4) | [73] |
| 7 | Primary active constituents of the Xiang-lian pill (evodiamine, rutaecarpine, and stigmasterol) | Alkaloids | Anti-pancreatic cancer | PTGS2 and PTGS1 | [74] |
| Steroids | |||||
| 8 | Toosendanin | Terpenoids | Anti-colorectal cancer | Shh | [75] |
| 9 | Glytabastan B | Flavonoids | Prevention and alleviation of rheumatoid arthritis | ERK2, JNK1 and class Ⅰ PI3K catalytic subunit p110 | [76] |
| 10 | Tubocapsenolide A | Terpenoids | Therapeutic potential for osteosarcoma | Src homology 2 phosphatase 2 | [77] |
| 11 | Primary constituents of Zuojin capsule | Alkaloids | Anti-colorectal cancer activity | CDKN1A, Bcl2, E2F1, PRKCB, MYC, CDK2, and MMP9 | [78] |
| Flavonoids | |||||
| Steroids | |||||
| Indole | |||||
| 12 | Proanthocyanidin A1 | Flavonoids | Amelioration of chemotherapy-induced thrombocytopenia | JAK2 | [79] |
| 13 | Cucurbitacin B | Terpenoids | Anti-NSCLC activity | TLR4 | [80] |
| 14 | Celastrol and gambogic acid | Terpenoids | Anti-breast cancer activity | ERα Y537S mutant | [81] |
| 15 | Shikonin | Terpenoids | Anti-triple-negative breast cancer activity | IMPDH2 | [82] |
One limitation of this method is its dependency on a relatively high concentration of the natural active compound for detection. Additionally, quantification primarily hinges on western blotting, which exhibits low throughput. Consequently, this method is typically utilized as a validation tool. Following the capture of small molecule-targeted proteins through alternative techniques, CETSA is employed for validation purposes.
3.2.2. ITC
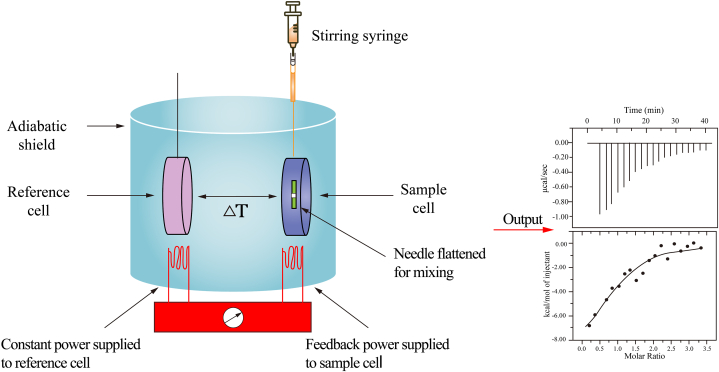
ITC enables the direct quantification of heat released or absorbed during biomolecular binding processes. When a compound binds to a protein, it elicits a heat exchange detectable via ITC. In an ITC experiment, one reactant is placed in a temperature-controlled sample cell, linked to a reference cell through a heat flow circuit, with both cells in the same external environment. A specific titrant (such as a natural active compound) is incrementally injected into the sample cell. Upon reaction between the sample and titrant, heat is either absorbed or released. After each injection of the sample, heat is absorbed or released, causing temperature changes between the sample and reference cells. A heat-sensitive device detects temperature differences between the two cells upon binding, providing feedback to a heater to rectify temperature disparities and equilibrate the sample and reference cells. Subsequent titrant injections gradually increase the molar ratio between the titrant and protein, as the protein reaches saturation. The frequency of titrant binding events decreases, and the heat change decreases until the number of titrations in the sample cell is in excess relative to the protein, indicating saturation. Utilizing software integrated with the isothermal titration calorimeter, parameters such as binding constants, stoichiometry (n), and enthalpy change can be obtained through data fitting, thus yielding comprehensive thermodynamic insights into molecular interactions [83].
Based on the aforementioned principle, the typical procedure entails configuring the reference and sample cells to the desired experimental temperature. The natural active compound is loaded into a highly precise injection device, such as a syringe, which is then inserted into the sample cell containing the target protein. The natural active compound is injected into the sample cell in multiple increments. The slightest temperature variation, on the order of millionths of a degree Celsius, can be detected and quantified if binding occurs between the natural active compound and the protein. During each injection, the microcalorimeter records all released heat until equilibrium is reached in the binding reaction, with the measured heat being proportional to the degree of binding. Utilizing the software provided with the isothermal titration calorimeter, the acquired data can be fitted and processed to determine thermodynamic parameters (Fig. 7). For instance, polyphenols play an essential role in modulating the gut microbiota and enhancing the integrity of the mucus barrier. Feng et al. [84] demonstrated that the pyrogallol-containing polyphenols epigallocatechin gallate and tannic acid strongly bind to the nucleophilic thiol groups of mucins. This binding was shown to serve as cross-linkers within mucin networks, thereby reinforcing the mucus barrier, via ITC. Xue et al. [85] revealed that thymoquinone, a significant phytoconstituent of Nigella sativa used in Alzheimer's disease therapy, binds spontaneously to human transferrin. The binding constant was determined to be 0.22 × 106 M−1, forming a stable complex, using both ITC and spectroscopic methods. In addition, ITC has been employed to elucidate the targets of glycyrrhizin derivatives, which suppress cancer chemoresistance [86], valonea tannin, which exhibits tyrosinase inhibition activity [87], bioactive coffee compounds that protect against serotonin degradation for treating depression and potentially type 2 diabetes [88], neoandrographolide, a novel inhibitor of Rab5 [89], piperine, a bioactive constituent of black pepper, shown to alleviate experimental allergic encephalomyelitis [90], luteolin, which exhibits antibacterial activity [91], red clover isoflavones, which inhibit cancer cell metastasis [92], and the molecular interactions of acetylcholinesterase with potential acetylcholinesterase inhibitors isolated from the root of Rhodiola crenulata [93] (Table 10).
Fig. 7.
The principle and general workflow of ITC. Initially, the reference and sample cells are equilibrated to the desired temperature. Subsequently, a highly accurate injection device loaded with the active compound is inserted into the sample cell containing the target protein. The natural active compound is titrated into the sample cell through sequential injections. During each injection, the microcalorimeter measures the released heat until the binding reaction reaches equilibrium. Thermodynamic parameters are derived by fitting the titration curve.
Table 10.
Recent studies that performed ITC-based target identification of natural products.
| No. | Natural products | Category | Bioactivity | Target protein | Reference | |
|---|---|---|---|---|---|---|
| 1 | Epigallocatechin gallate and tannic acid | Polyphenols | Reinforcement of the mucus barrier | Nucleophilic thiol groups of mucins | [84] | |
| 2 | Thymoquinone | Isothiocyanates | Alleviation of Alzheimer's disease | Transferrin | [85] | |
| 3 | Glycyrrhizin derivatives | Terpenoids | Suppression of cancer chemoresistance | Progesterone receptor membrane component 1 | [86] | |
| 4 | Valonea tannin | Polyphenols | Tyrosinase inhibition activity | Skin whitening properties | [87] | |
| 5 | Coffee compounds | Alkaloids | Alleviation of depression and potentially type 2 diabetes | Monoamine oxidase A | [88] | |
| Polyphenols | ||||||
| 6 | Neoandrographolide | Terpenoids | Anticancer activity | Rab5 | [89] | |
| 7 | Piperine | Alkaloids | Alleviation of experimental allergic encephalomyelitis | Dihydroorotate dehydrogenase | [90] | |
| 8 | Luteolin | Flavonoids | Antibacterial activity | HK853 | [91] | |
| 9 | Red clover isoflavones | Flavonoids | Inhibition of cancer cell metastasis | Actin | [92] | |
| 10 | Constituents isolated from the root of R. crenulata | Flavonoids | Alleviation of Alzheimer's disease | Acetylcholinesterase | [93] | |
| Terpenoids | ||||||
| Phenols |
This technique boasts several advantages, including its rapidity, accuracy, and minimal sample demand. It does not enforce stringent requirements on the reaction system, such as transparency, turbidity, or viscosity. ITC furnishes detailed and comprehensive thermodynamic insights into molecular interactions, facilitating precise quantification of molecular interaction levels. However, a limitation of this method is its low throughput. Consequently, it is commonly employed as a supplementary screening tool in high-throughput screening protocols.
4. Identification of novel drugs and drug targets based on the innate function of natural products
Notably, there is a novel avenue for rapidly exploring the targets of natural products based on their innate functions [94]. In the course of evolution, organisms produce specialized secondary metabolites to exhibit important functions, such as defence, survival, and reproduction, via interacting with specific target proteins in inter-kingdom entities. The structural complementarity between ligands and their targets provide more opportunities for finding promising ligand analogues. For instance, tabtoxin, a dipeptide monobactam synthesized by Pseudomonas syringae, inhibits glutamine synthetase (GS) in plants [95]. GS, which has a conserved active site in both plants and mammals, is a significant target for cancer therapy [96]. Consequently, structural analogues of tabtoxin hold promise as potent human GS antagonist for cancer treatment [97,98].
Conversely, ligands may actively interact with several structural analogues of the receptors present in different branches of life. Inspired by this concept, we can identify therapeutic targets based on the innate functions of natural metabolites. For example, the steroid hormone ecdysterone and its homologues interact with ecdysone receptors in arthropods. These receptors are composed of EcR proteins and ultraspiracle protein (USP) heterodimers, functioning as nuclear receptors for ecdysone [99]. In humans, there are several structural analogues of ecdysone receptors, including liver X receptor α (LXRα), LXRβ, estrogen receptor α (ERα), ERβ, retinoid X receptor α (RXRα), and bile acid receptors. LXRβ is a promising therapeutic target for inflammation [100], cancer [101], dyslipidemia [102], and diabetes [103], while ER is a target for the treatment of breast cancer [104]. Some ecdysterone analogues have been experimentally validated in cancer, diabetes, and inflammation [105,106]. Therefore, ecdysterone and its analogues may act as selective LXR agonists and ER modulators against human diseases.
Similarly, rhizobitoxin, a plant toxin produced by the fungus Bradyrhizobium elkanii, restrains the activity of β-cystathionase and 1-aminocyclopropane-1-carboxylate (ACC) synthase to exert its natural effects [107,108]. Structural analysis reveals that the plant enzymes β-cystathionase and ACC synthase have structural analogues in human proteins, namely γ-cystathionase and kynurenine aminotransferase, respectively. Over-activity of γ-cystathionase is linked to various diseases [109], while kynurenine aminotransferase activity is associated with schizophrenia and cognitive impairment [110]. Hence, these proteins are potential therapeutic targets. Some known inhibitors share structural similarities with rhizobitoxin [111], suggesting their potential action on these proteins. Although rhizobitoxin is known to bind ACC synthase, there is no evidence of its binding to kynurenine aminotransferase. However, structural analogues of rhizobitoxin have been found to suppress kynurenine aminotransferase.
In addition to the functions of metabolites, enzymes that catalyze their synthesis can also be potential therapeutic targets in humans. For instance, strictosidine synthase, a plant enzyme, has a human structural homolog, regucalcin, which may serve as a therapeutic target [94]. Although agonists or antagonists for regucalcin have not been studied yet, this concept can be applied to develop relevant drugs.
5. Conclusions and prospects
Nature has been continuing to be a major source of medicinal products, and especially providing revolutionized structural leads in the treatment of serious diseases for millennia, such as flavonoids, alkaloids, steroidglycosides, sesquiterpene, essential oil and steroids, all of which play an important role in medicinal chemistry. The identification of natural products' targets is as important as the knowledge of natural products themselves because clarifying the former provides several new avenues for finding better drugs. Target identification is the first step in the early stage of drug discovery, followed by lead optimization.
Based on the foregoing, labeling methods facilitate direct labeling of target proteins while excluding indirectly targeted ones, thereby significantly reducing false-positive results in target identification. Nevertheless, these methods entail a high application threshold and are unsuitable for investigating post-modified compounds with abolished activity, compounds exhibiting chemically challenging synthesis, or analyzing trace amounts of natural active compounds. Conversely, non-labeling methods can be used to identify targets of any natural active compound, even in trace amounts or multicomponent mixtures, albeit with lower reliability compared to labeling methods. Challenges persist in eliminating the influence of other proteins within the protein complex where the natural active compound binds. Each method presents distinct advantages and limitations (Table 11). In practical applications, the selection of appropriate target discovery methods is contingent upon the characteristics of small molecules or target proteins. Typically, a combination of multiple methods is employed to identify the targets of natural products [[112], [113], [114], [115]]. Furthermore, a recent review outlined target identification and validation approaches for natural products categorized according to whether the natural products were labeled [116]. Building upon this, this review clarifies the principles and processes of each method comprehensively. Moreover, this review presents chemical structures and modifying moieties relevant to the labeling techniques.
Table 11.
Benefits and drawbacks of each target identification approach.
| Approach | Benefits | Drawbacks |
|---|---|---|
| Affinity chromatography | 1. The synthesis of immobilized probes is simple. 2. It facilitates the analysis of all adsorbed proteins without discrimination. 3. It can be easily conducted in individual laboratories. |
1. Discrimination between truly high-affinity target proteins and high-abundance proteins with low affinity is necessary. 2. The elution process relies on empirical knowledge; excessively strong washing conditions can significantly reduce the number of identified targets, while weak elution conditions can yield false-positive results. 3. It does not reveal interactions between drugs and proteins in live cells under physiological conditions. 4. Biotin affinity tags introduce steric hindrance, potentially reducing or even abolishing the activity of natural active compounds and impeding probe molecule entry into cells. |
| ABPP | 1. It exhibits strong specificity. 2. ABPs do not require pre-immobilization on matrix material. 3. It can be directly applied to live cells, elucidating drug–target interactions under physiological conditions. |
1. Only proteins capable of forming covalent interactions with bioactive natural products can be captured. 2. Synthesizing ABPs relies on the structure of the bioactive natural product and necessitates expertise in medicinal chemistry. |
| PAL | 1. It can facilitate the analysis of proteins that cannot be targeted by ABPs. | 1. Photoaffinity groups still exhibit non-specific binding to abundant cellular proteins. 2. The synthesis of PAL probes necessitates expertise in medicinal chemistry. |
| Proteome microarray assay | 1. It requires minimal sample volumes. 2. High throughput enables parallel analysis of thousands to millions of proteins. 3. Utilizing protein chips is relatively simple. |
1. It is relatively costly, necessitating a range of expensive, high-end instruments. 2. Standardization issues with the chips. |
| DARTS | 1. No structural modification of natural bioactive compounds is necessary. 2. It can be easily implemented by individual laboratories. |
1. Certain natural products do not significantly affect the conformation of the target protein upon binding. 2. Some proteins are less susceptible to proteases, resulting in undetectable changes in DARTS. 3. The ability to identify targets that exhibit low protein abundance is limited. |
| TPP | 1. It monitors changes in protein thermal stability throughout the proteome during the action of natural bioactive compounds. 2. It enables high-throughput detection of target proteins. |
1. Inapplicable to heat-insensitive proteins. 2. The extraction of thermodynamic parameters besides the Tm value from TPP data is challenging. 3. The magnitude of the Tm shift after protein binding with natural active compounds does not consistently correlate with binding affinity. 4. Time-consuming and costly. |
| SPROX | 1. It enables the large-scale assessment of protein folding states and potentially precise determination of binding domains and peptide segments involved in the interactions between compounds and target proteins. | 1. It can only detect binding between proteins containing methionine residues and small molecules. 2. Generating the standard curve requires a large sample volume. 3. The process is complex, costly, and requires substantial resources. |
| CETSA | 1. No structural modification of natural bioactive compounds is required. 2. It can be easily implemented by individual laboratories. |
1. Detection necessitates relatively high concentrations of the natural active compound. 2. The process exhibits low throughput in discovering new targets and is primarily utilized for target validation. |
| ITC | 1. It provides fast, accurate, and low-sample-volume analysis with minimal requirements for reaction system transparency, turbidity, and viscosity. 2. It enables precise determination of comprehensive thermodynamic information. |
1. It is typically utilized as an auxiliary screening technique in high-throughput screening due to its low-throughput nature. 2. It requires specialized ITC equipment. |
New techniques continuously emerge, leveraging the aforementioned technologies, to aid in the identification of target proteins of active natural products, thus significantly impacting novel drug development.
In addition, there is a novel avenue for rapidly exploring the targets of natural products based on their innate functions. If a natural metabolite (a) has been biosynthesized to fulfill a specific role through its interaction with protein (A), the natural target of a, it presents an ideal strategy to explore therapeutically important human proteins that are structurally analogous to protein A. The aforementioned target validation approaches can be employed to validate the binding affinity of each target with a. Conversely, the structural analogues of natural functional metabolite a may interact with protein A with varying degrees of affinity, thereby exerting similar effects against human diseases.
Funding statement
This work was supported by the Matching Grant of the National Nature Science Foundation of China from Nanjing University of Chinese Medicine, China (grant number NZY81903857), the National Natural Science Foundation of China (grant number 81903857), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant number 24KJA360005) and the National Natural Science Foundation of China (grant number 81961128020).
Ethics declarations
Not applicable.
Data availability
No data was used for the research described in the article.
CRediT authorship contribution statement
Xuan Jiang: Writing – original draft. Kinyu Shon: Visualization. Xiaofeng Li: Visualization. Guoliang Cui: Writing – review & editing. Yuanyuan Wu: Project administration. Zhonghong Wei: Project administration. Aiyun Wang: Resources, Project administration. Xiaoman Li: Writing – review & editing, Funding acquisition, Conceptualization. Yin Lu: Resources, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Xiaoman Li, Email: lixm@njucm.edu.cn.
Yin Lu, Email: luyingreen@njucm.edu.cn.
References
- 1.Shen B. A new Golden age of natural products drug discovery. Cell. 2015;163(6):1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parthasarathy A., Mantravadi P.K., Kalesh K. Detectives and helpers: natural products as resources for chemical probes and compound libraries. Pharmacol. Therapeut. 2020;216 doi: 10.1016/j.pharmthera.2020.107688. [DOI] [PubMed] [Google Scholar]
- 3.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 4.Shaashua L., Shabat-Simon M., Haldar R., Matzner P., Zmora O., Shabtai M., et al. Perioperative COX-2 and β-adrenergic blockade improves metastatic biomarkers in breast cancer patients in a phase-II randomized trial. Clin. Cancer Res. 2017;23(16):4651–4661. doi: 10.1158/1078-0432.CCR-17-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Gaudio F., Pollastro F., Mozzicafreddo M., Riccio R., Minassi A., Monti M.C. Chemoproteomic fishing identifies arzanol as a positive modulator of brain glycogen phosphorylase. Chem. Commun. 2018;54(91):12863–12866. doi: 10.1039/c8cc07692h. [DOI] [PubMed] [Google Scholar]
- 6.Wu R., Murali R., Kabe Y., French S.W., Chiang Y.M., Liu S., et al. Baicalein targets GTPase-mediated autophagy to eliminate liver tumor-initiating stem cell-like cells resistant to mTORC1 inhibition. Hepatology. 2018;68(5):1726–1740. doi: 10.1002/hep.30071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F., Zhu K., Chen L., Ouyang L., Chen C., Gu L., et al. Protein target identification of ginsenosides in skeletal muscle tissues: discovery of natural small-molecule activators of muscle-type creatine kinase. Journal of ginseng research. 2020;44(3):461–474. doi: 10.1016/j.jgr.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Hua Y., Wang D., Shan L., Zhang Y., Zhu J., et al. A sesquiterpene lactone from a medicinal herb inhibits proinflammatory activity of TNF-α by inhibiting ubiquitin-conjugating enzyme UbcH5. Chem. Biol. 2014;21(10):1341–1350. doi: 10.1016/j.chembiol.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., et al. Identification of a primary target of thalidomide teratogenicity. Science (New York, N.Y.) 2010;327(5971):1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 10.Liao L.X., Song X.M., Wang L.C., Lv H.N., Chen J.F., Liu D., et al. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114(29):E5986–e5994. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du S., Song X., Li Y., Cao Y., Chu F., Durojaye O.A., et al. Celastrol inhibits ezrin-mediated migration of hepatocellular carcinoma cells. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-68238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong T., Li C., Wang X., Dian L., Zhang X., Li L., et al. Ainsliadimer A selectively inhibits IKKα/β by covalently binding a conserved cysteine. Nat. Commun. 2015;6:6522. doi: 10.1038/ncomms7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Rzasa R.M., et al. Isolation and identification of eukaryotic initiation factor 4A as a molecular target for the marine natural product Pateamine A. Methods Enzymol. 2007;431:303–324. doi: 10.1016/S0076-6879(07)31014-8. [DOI] [PubMed] [Google Scholar]
- 14.Liu C.X., Yin Q.Q., Zhou H.C., Wu Y.L., Pu J.X., Xia L., et al. Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells. Nat. Chem. Biol. 2012;8(5):486–493. doi: 10.1038/nchembio.935. [DOI] [PubMed] [Google Scholar]
- 15.Yao T., Xu X., Huang R. Recent advances about the applications of click reaction in chemical proteomics. Molecules. 2021;26(17) doi: 10.3390/molecules26175368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Patricelli M.P., Cravatt B.F. Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 1999;96(26):14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Wang Y., Ma N., Tian J., Shao Y., Zhu B., et al. Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification. Signal Transduct. Targeted Ther. 2020;5(1):72. doi: 10.1038/s41392-020-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gersch M., Kreuzer J., Sieber S.A. Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012;29(6):659–682. doi: 10.1039/c2np20012k. [DOI] [PubMed] [Google Scholar]
- 19.Speers A.E., Cravatt B.F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11(4):535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Galmozzi A., Dominguez E., Cravatt B.F., Saez E. Application of activity-based protein profiling to study enzyme function in adipocytes. Methods Enzymol. 2014;538:151–169. doi: 10.1016/B978-0-12-800280-3.00009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spradlin J.N., Hu X., Ward C.C., Brittain S.M., Jones M.D., Ou L., et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 2019;15(7):747–755. doi: 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W.S., Chen K.F., Liu M., Jia X.L., Huang Y.Q., Hao B.B., et al. Investigation of targets and anticancer mechanisms of covalently acting natural products by functional proteomics. Acta Pharmacol. Sin. 2023;48(8):1701–1711. doi: 10.1038/s41401-023-01072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belcher B.P., Machicao P.A., Tong B., Ho E., Friedli J., So B., et al. Chemoproteomic profiling reveals that anti-cancer natural product dankastatin B covalently targets mitochondrial VDAC3. Chembiochem : a European journal of chemical biology. 2023 doi: 10.1002/cbic.202300111. [DOI] [PubMed] [Google Scholar]
- 24.Wright M.H., Tao Y., Drechsel J., Krysiak J., Chamni S., Weigert-Munoz A., et al. Quantitative chemoproteomic profiling reveals multiple target interactions of spongiolactone derivatives in leukemia cells. Chem. Commun. 2017;53(95):12818–12821. doi: 10.1039/c7cc04990k. [DOI] [PubMed] [Google Scholar]
- 25.Yoo E., Schulze C.J., Stokes B.H., Onguka O., Yeo T., Mok S., et al. The antimalarial natural product salinipostin A identifies essential α/β serine hydrolases involved in lipid metabolism in P. Falciparum parasites. Cell Chem. Biol. 2020;27(2):143–157.e145. doi: 10.1016/j.chembiol.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le P., Nodwell M.B., Eirich J., Sieber S.A. A chemical proteomic analysis of illudin-interacting proteins. Chemistry. 2019;25(54):12644–12651. doi: 10.1002/chem.201902919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Zhu T., Liu X., Zhu D., Zhang Y., Wu S., et al. Identification of a novel PHGDH covalent inhibitor by chemical proteomics and phenotypic profiling. Acta Pharm. Sin. B. 2022;12(1):246–261. doi: 10.1016/j.apsb.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Zhao Y., Cao R., Li L., Cai G., Li J., et al. Activity-based protein profiling reveals GSTO1 as the covalent target of piperlongumine and a promising target for combination therapy for cancer. Chem. Commun. 2019;55(30):4407–4410. doi: 10.1039/c9cc00917e. [DOI] [PubMed] [Google Scholar]
- 29.Sumranjit J., Chung S.J. Recent advances in target characterization and identification by photoaffinity probes. Molecules. 2013;18(9):10425–10451. doi: 10.3390/molecules180910425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D., Cao Y., Zheng L., Lv D., Chen L., Xing X., et al. Identification of Annexin A2 as a target protein for plant alkaloid matrine. Chem. Commun. 2017;53(36):5020–5023. doi: 10.1039/c7cc02227a. [DOI] [PubMed] [Google Scholar]
- 31.Tranter D., Paatero A.O., Kawaguchi S., Kazemi S., Serrill J.D., Kellosalo J., et al. Coibamide A targets Sec61 to prevent biogenesis of secretory and membrane proteins. ACS Chem. Biol. 2020;15(8):2125–2136. doi: 10.1021/acschembio.0c00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martín-Acosta P., Meng Q., Klimek J., Reddy A.P., David L., Petrie S.K., et al. A clickable photoaffinity probe of betulinic acid identifies tropomyosin as a target. Acta Pharm. Sin. B. 2022;12(5):2406–2416. doi: 10.1016/j.apsb.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu T.Y., Wu X.T., Chen C., Liu X.Q., Zhu L., Luo J.G., et al. Photoaffinity probe reveals the potential target of harringtonolide for cancer cell migration inhibition. ACS Med. Chem. Lett. 2022;13(3):449–456. doi: 10.1021/acsmedchemlett.1c00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Di Z., Li X., Shan Y., Li W., Zhang H., et al. Chemical proteomics reveal CD147 as a functional target of pseudolaric acid B in human cancer cells. Chem. Commun. 2017;53(62):8671–8674. doi: 10.1039/c7cc04345g. [DOI] [PubMed] [Google Scholar]
- 35.Ranade A.R., Higgins L., Markowski T.W., Glaser N., Kashin D., Bai R., et al. Characterizing the epothilone binding site on β-tubulin by photoaffinity labeling: identification of β-tubulin peptides TARGSQQY and TSRGSQQY as targets of an epothilone photoprobe for polymerized tubulin. J. Med. Chem. 2016;59(7):3499–3514. doi: 10.1021/acs.jmedchem.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R.E., Hunt C.R., Chen J., Taylor J.S. Biotinylated quercetin as an intrinsic photoaffinity proteomics probe for the identification of quercetin target proteins. Bioorg. Med. Chem. 2011;19(16):4710–4720. doi: 10.1016/j.bmc.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3(9):570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 38.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics : MCP. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Dai J., Liang K., Zhao S., Jia W., Liu Y., Wu H., et al. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc. Natl. Acad. Sci. U. S. A. 2018;115(26):E5896–e5905. doi: 10.1073/pnas.1801745115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., Gong L., Liu X., Zhu T., Zhou W., Kong L., et al. Identification of peroxiredoxin 6 as a direct target of withangulatin A by quantitative chemical proteomics in non-small cell lung cancer. Redox Biol. 2021;46 doi: 10.1016/j.redox.2021.102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H.N., Yang L., Ling J.Y., Czajkowsky D.M., Wang J.F., Zhang X.W., et al. Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. U. S. A. 2015;112(49):15084–15089. doi: 10.1073/pnas.1521316112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu T.T., Yang H., Zhuo F.F., Yang Z., Zhao M.M., Guo Q., et al. Atypical E3 ligase ZFP91 promotes small-molecule-induced E2F2 transcription factor degradation for cancer therapy. EBioMedicine. 2022;86 doi: 10.1016/j.ebiom.2022.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang F., Li M., Wang H., Ding B., Zhang C., Ding Z., et al. Coelonin, an anti-inflammation active component of Bletilla striata and its potential mechanism. Int. J. Mol. Sci. 2019;20(18) doi: 10.3390/ijms20184422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang J., Kim Y., Kwon H.J. Advances in identification and validation of protein targets of natural products without chemical modification. Nat. Prod. Rep. 2016;33(5):719–730. doi: 10.1039/c5np00107b. [DOI] [PubMed] [Google Scholar]
- 45.Lomenick B., Hao R., Jonai N., Chin R.M., Aghajan M., Warburton S., et al. Target identification using drug affinity responsive target stability (DARTS) Proc. Natl. Acad. Sci. U. S. A. 2009;106(51):21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren Y.S., Li H.L., Piao X.H., Yang Z.Y., Wang S.M., Ge Y.W. Drug affinity responsive target stability (DARTS) accelerated small molecules target discovery: principles and application. Biochem. Pharmacol. 2021;194 doi: 10.1016/j.bcp.2021.114798. [DOI] [PubMed] [Google Scholar]
- 47.Huang X.H., Yan X., Zhang Q.H., Hong P., Zhang W.X., Liu Y.P., et al. Direct targeting of HSP90 with daurisoline destabilizes β-catenin to suppress lung cancer tumorigenesis. Cancer Lett. 2020;489:66–78. doi: 10.1016/j.canlet.2020.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Geng J., Liu W., Gao J., Jiang C., Fan T., Sun Y., et al. Andrographolide alleviates Parkinsonism in MPTP-PD mice via targeting mitochondrial fission mediated by dynamin-related protein 1. Br. J. Pharmacol. 2019;176(23):4574–4591. doi: 10.1111/bph.14823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong R.F., Qin C.J., Yin Y., Han L.L., Xiao C.M., Wang K.D., et al. Discovery of a potent inhibitor of chaperone-mediated autophagy that targets the HSC70-LAMP2A interaction in non-small cell lung cancer therapy. Br. J. Pharmacol. 2023 doi: 10.1111/bph.16165. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Ren M., Chen J., Xu H., Li W., Wang T., Chi Z., et al. Ergolide covalently binds NLRP3 and inhibits NLRP3 inflammasome-mediated pyroptosis. Int. Immunopharm. 2023;120 doi: 10.1016/j.intimp.2023.110292. [DOI] [PubMed] [Google Scholar]
- 51.Wei J., Fan S., Yu H., Shu L., Li Y. A new strategy for the rapid identification and validation of the direct targets of aconitine-induced cardiotoxicity. Drug Des. Dev. Ther. 2021;15:4649–4664. doi: 10.2147/DDDT.S335461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morretta E., Tosco A., Festa C., Mozzicafreddo M., Monti M.C., Casapullo A., Crellastatin A. A PARP-1 inhibitor discovered by complementary proteomic approaches. ChemMedChem. 2020;15(3):317–323. doi: 10.1002/cmdc.201900634. [DOI] [PubMed] [Google Scholar]
- 53.Esch S., König S., Bopp B., Jose J., Brandt S., Hensel A. Cryptotanshinone from Salvia miltiorrhiza roots reduces cytokeratin CK1/10 expression in keratinocytes by activation of peptidyl-prolyl-cis-trans-isomerase FKBP1A. Planta Med. 2019;85(7):552–562. doi: 10.1055/a-0660-0441. [DOI] [PubMed] [Google Scholar]
- 54.Derry M.M., Somasagara R.R., Raina K., Kumar S., Gomez J., Patel M., et al. Target identification of grape seed extract in colorectal cancer using drug affinity responsive target stability (DARTS) technique: role of endoplasmic reticulum stress response proteins. Curr. Cancer Drug Targets. 2014;14(4):323–336. doi: 10.2174/1568009614666140411101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tu Y., Tan L., Tao H., Li Y., Liu H. CETSA and thermal proteome profiling strategies for target identification and drug discovery of natural products. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2023;116 doi: 10.1016/j.phymed.2023.154862. [DOI] [PubMed] [Google Scholar]
- 56.Kirsch V.C., Orgler C., Braig S., Jeremias I., Auerbach D., Müller R., et al. The cytotoxic natural product vioprolide A targets nucleolar protein 14, which is essential for ribosome biogenesis. Angew. Chem. 2020;59(4):1595–1600. doi: 10.1002/anie.201911158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dziekan J.M., Yu H., Chen D., Dai L., Wirjanata G., Larsson A., et al. Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay. Sci. Transl. Med. 2019;11(473) doi: 10.1126/scitranslmed.aau3174. [DOI] [PubMed] [Google Scholar]
- 58.Chen K., Wang T., Li Y., Wu J., Zhao C.X., Liu S., et al. Rhodojaponin VI indirectly targets Cav 2.2 channels via N-ethylmaleimide-sensitive fusion protein to alleviate neuropathic pain. Acta Pharm. Sin. B. 2023;13(3):1326–1336. doi: 10.1016/j.apsb.2023.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Q.T., Yang W.Q., Zang C., Zhou L., Zhang C.J., Bao X., et al. The toxic natural product tutin causes epileptic seizures in mice by activating calcineurin. Signal Transduct. Targeted Ther. 2023;8(1):101. doi: 10.1038/s41392-023-01312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Z., Li S., Liu J., Zhang C., Jian C., Wang L., et al. Natural product alantolactone targeting AKR1C1 suppresses cell proliferation and metastasis in non-small-cell lung cancer. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.847906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun C.P., Zhou J.J., Yu Z.L., Huo X.K., Zhang J., Morisseau C., et al. Kurarinone alleviated Parkinson's disease via stabilization of epoxyeicosatrienoic acids in animal model. Proc. Natl. Acad. Sci. U. S. A. 2022;119(9) doi: 10.1073/pnas.2118818119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X.W., Feng N., Wang L.C., Liu D., Hua Y.M., Zhang C., et al. Small-molecule arone protects from neuroinflammation in LPS-activated microglia BV-2 cells by targeting histone-remodeling chaperone ASF1a. Biochem. Pharmacol. 2020;177 doi: 10.1016/j.bcp.2020.113932. [DOI] [PubMed] [Google Scholar]
- 63.West G.M., Tang L., Fitzgerald M.C. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal. Chem. 2008;80(11):4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 64.Dearmond P.D., Xu Y., Strickland E.C., Daniels K.G., Fitzgerald M.C. Thermodynamic analysis of protein-ligand interactions in complex biological mixtures using a shotgun proteomics approach. J. Proteome Res. 2011;10(11):4948–4958. doi: 10.1021/pr200403c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu Y., Wallace M.A., Fitzgerald M.C. Thermodynamic analysis of the geldanamycin-hsp 90 interaction in a whole cell lysate using a mass spectrometry-based proteomics approach. J. Am. Soc. Mass Spectrom. 2016;27(10):1670–1676. doi: 10.1007/s13361-016-1457-2. [DOI] [PubMed] [Google Scholar]
- 66.Geer Wallace M.A., Kwon D.Y., Weitzel D.H., Lee C.T., Stephenson T.N., Chi J.T., et al. Discovery of manassantin A protein targets using large-scale protein folding and stability measurements. J. Proteome Res. 2016;15(8):2688–2696. doi: 10.1021/acs.jproteome.6b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science (New York, N.Y.) 2013;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 68.Yang A., Zeng K., Huang H., Liu D., Song X., Qian Y., et al. Usenamine A induces apoptosis and autophagic cell death of human hepatoma cells via interference with the Myosin-9/actin-dependent cytoskeleton remodeling. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2023;116 doi: 10.1016/j.phymed.2023.154895. [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Li Y., Zhu Y., Ji W., Wang Y., Dong X., et al. Structure-based virtual screening for discovery of paederosidic acid from Paederia scandens as novel P2Y(14)R antagonist. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2023;115 doi: 10.1016/j.phymed.2023.154851. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W., Gong Q., Tang Z., Ma X., Wang Z., Guan J., et al. The natural product, echinatin, protects mice from methicillin-resistant Staphylococcus aureus pneumonia by inhibition of alpha-hemolysin expression. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1128144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao S., Zhang P., Yan Y., Xu W., Li J., Wang L., et al. Network pharmacology-based prediction and validation of the active ingredients and potential mechanisms of the Huangxiong formula for treating ischemic stroke. J. Ethnopharmacol. 2023;312 doi: 10.1016/j.jep.2023.116507. [DOI] [PubMed] [Google Scholar]
- 72.Yin Z., Lv Y., Deng L., Li G., Ou R., Chen L., et al. Targeting ABCB6 with nitidine chloride inhibits PI3K/AKT signaling pathway to promote ferroptosis in multiple myeloma. Free Radic. Biol. Med. 2023;203:86–101. doi: 10.1016/j.freeradbiomed.2023.04.003. [DOI] [PubMed] [Google Scholar]
- 73.Xu D., Zhao W., Feng Y., Wen X., Liu H., Ping J. Pentoxifylline attenuates nonalcoholic fatty liver by inhibiting hepatic macrophage polarization to the M1 phenotype. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2022;106 doi: 10.1016/j.phymed.2022.154368. [DOI] [PubMed] [Google Scholar]
- 74.Jiao J., Cheng C.S., Xu P., Yang P., Zhang K., Jing Y., et al. Mechanisms of pancreatic tumor suppression mediated by Xiang-lian pill: an integrated in silico exploration and experimental validation. J. Ethnopharmacol. 2022;298 doi: 10.1016/j.jep.2022.115586. [DOI] [PubMed] [Google Scholar]
- 75.Zhang M., Tao Z., Gao L., Chen F., Ye Y., Xu S., et al. Toosendanin inhibits colorectal cancer cell growth through the Hedgehog pathway by targeting Shh. Drug Dev. Res. 2022;83(5):1201–1211. doi: 10.1002/ddr.21951. [DOI] [PubMed] [Google Scholar]
- 76.Tu Y., Tan L., Lu T., Wang K., Wang H., Han B., et al. Glytabastan B, a coumestan isolated from Glycine tabacina, alleviated synovial inflammation, osteoclastogenesis and collagen-induced arthritis through inhibiting MAPK and PI3K/AKT pathways. Biochem. Pharmacol. 2022;197 doi: 10.1016/j.bcp.2022.114912. [DOI] [PubMed] [Google Scholar]
- 77.Zhu D., Chen C., Liu X., Wang S., Zhu J., Zhang H., et al. Osteosarcoma cell proliferation suppression via SHP-2-mediated inactivation of the JAK/STAT3 pathway by tubocapsenolide A. J. Adv. Res. 2021;34:79–91. doi: 10.1016/j.jare.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan J.H., Xu M.M., Zhou L.M., Gui Z.W., Huang L., Li X.G., et al. Integrating network pharmacology deciphers the action mechanism of Zuojin capsule in suppressing colorectal cancer. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2022;96 doi: 10.1016/j.phymed.2021.153881. [DOI] [PubMed] [Google Scholar]
- 79.Wang R., Hu X., Wang J., Zhou L., Hong Y., Zhang Y., et al. Proanthocyanidin A1 promotes the production of platelets to ameliorate chemotherapy-induced thrombocytopenia through activating JAK2/STAT3 pathway. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2022;95 doi: 10.1016/j.phymed.2021.153880. [DOI] [PubMed] [Google Scholar]
- 80.Yuan R., Zhao W., Wang Q.Q., He J., Han S., Gao H., et al. Cucurbitacin B inhibits non-small cell lung cancer in vivo and in vitro by triggering TLR4/NLRP3/GSDMD-dependent pyroptosis. Pharmacol. Res. 2021;170 doi: 10.1016/j.phrs.2021.105748. [DOI] [PubMed] [Google Scholar]
- 81.Liu X., Hu Q., Wang W., Ma H., Pu J., Cui J., et al. A protein-fragment complementation assay reveals that celastrol and gambogic acid suppress ERα mutants in breast cancer. Biochem. Pharmacol. 2021;188 doi: 10.1016/j.bcp.2021.114583. [DOI] [PubMed] [Google Scholar]
- 82.Wang W., Wu Y., Chen S., Liu X., He J., Wang S., et al. Shikonin is a novel and selective IMPDH2 inhibitor that target triple-negative breast cancer. Phytother Res. : PT. 2021;35(1):463–476. doi: 10.1002/ptr.6825. [DOI] [PubMed] [Google Scholar]
- 83.Callies O., Hernández Daranas A. Application of isothermal titration calorimetry as a tool to study natural product interactions. Nat. Prod. Rep. 2016;33(7):881–904. doi: 10.1039/c5np00094g. [DOI] [PubMed] [Google Scholar]
- 84.Feng G., Han K., Yang Q., Feng W., Guo J., Wang J., et al. Interaction of pyrogallol-containing polyphenols with mucin reinforces intestinal mucus barrier properties. J. Agric. Food Chem. 2022;70(30):9536–9546. doi: 10.1021/acs.jafc.2c03564. [DOI] [PubMed] [Google Scholar]
- 85.Xue B., DasGupta D., Alam M., Khan M.S., Wang S., Shamsi A., et al. Investigating binding mechanism of thymoquinone to human transferrin, targeting Alzheimer's disease therapy. J. Cell. Biochem. 2022;123(8):1381–1393. doi: 10.1002/jcb.30299. [DOI] [PubMed] [Google Scholar]
- 86.Kabe Y., Koike I., Yamamoto T., Hirai M., Kanai A., Furuhata R., et al. Glycyrrhizin derivatives suppress cancer chemoresistance by inhibiting progesterone receptor membrane component 1. Cancers. 2021;13(13) doi: 10.3390/cancers13133265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J., Liu Y., He X., Teng B., McRae J.M. Valonea tannin: tyrosinase inhibition activity, structural elucidation and insights into the inhibition mechanism. Molecules. 2021;26(9) doi: 10.3390/molecules26092747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grzelczyk J., Budryn G., Peña-García J., Szwajgier D., Gałązka-Czarnecka I., Oracz J., et al. Evaluation of the inhibition of monoamine oxidase A by bioactive coffee compounds protecting serotonin degradation. Food Chem. 2021;348 doi: 10.1016/j.foodchem.2021.129108. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J., Sun Y., Zhong L.Y., Yu N.N., Ouyang L., Fang R.D., et al. Structure-based discovery of neoandrographolide as a novel inhibitor of Rab5 to suppress cancer growth. Comput. Struct. Biotechnol. J. 2020;18:3936–3946. doi: 10.1016/j.csbj.2020.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Z., Hu Q., Wang W., Lu S., Wu D., Ze S., et al. Natural product piperine alleviates experimental allergic encephalomyelitis in mice by targeting dihydroorotate dehydrogenase. Biochem. Pharmacol. 2020;177 doi: 10.1016/j.bcp.2020.114000. [DOI] [PubMed] [Google Scholar]
- 91.Zhou Y., Huang L., Ji S., Hou S., Luo L., Li C., et al. Structural basis for the inhibition of the autophosphorylation activity of HK853 by luteolin. Molecules. 2019;24(5) doi: 10.3390/molecules24050933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Budryn G., Grzelczyk J., Pérez-Sánchez H. Binding of red clover isoflavones to actin as A potential mechanism of anti-metastatic activity restricting the migration of cancer cells. Molecules. 2018;23(10) doi: 10.3390/molecules23102471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li F.J., Liu Y., Yuan Y., Yang B., Liu Z.M., Huang L.Q. Molecular interaction studies of acetylcholinesterase with potential acetylcholinesterase inhibitors from the root of Rhodiola crenulata using molecular docking and isothermal titration calorimetry methods. Int. J. Biol. Macromol. 2017;104(Pt A):527–532. doi: 10.1016/j.ijbiomac.2017.06.066. [DOI] [PubMed] [Google Scholar]
- 94.Suresh P.S., Kumari S., Sahal D., Sharma U. Innate functions of natural products: a promising path for the identification of novel therapeutics. Eur. J. Med. Chem. 2023;260 doi: 10.1016/j.ejmech.2023.115748. [DOI] [PubMed] [Google Scholar]
- 95.Thomas M.D., Langston-Unkefer P.J., Uchytil T.F., Durbin R.D. Inhibition of glutamine synthetase from pea by tabtoxinine-beta-lactam. Plant Physiol. 1983;71(4):912–915. doi: 10.1104/pp.71.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamashita M.M., Almassy R.J., Janson C.A., Cascio D., Eisenberg D. Refined atomic model of glutamine synthetase at 3.5 A resolution. J. Biol. Chem. 1989;264(30):17681–17690. doi: 10.2210/pdb2gls/pdb. [DOI] [PubMed] [Google Scholar]
- 97.Kreuzer J., Bach N.C., Forler D., Sieber S.A. Target discovery of acivicin in cancer cells elucidates its mechanism of growth inhibition†Electronic supplementary information (ESI) available: synthesis, cloning, protein expression, purification and biochemical assays. See DOI: 10.1039/c4sc02339k. Chem. Sci. 2014;6(1):237–245. doi: 10.1039/c4sc02339k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kindler H.L., Burris H.A., 3rd, Sandler A.B., Oliff I.A. A phase II multicenter study of L-alanosine, a potent inhibitor of adenine biosynthesis, in patients with MTAP-deficient cancer. Invest. N. Drugs. 2009;27(1):75–81. doi: 10.1007/s10637-008-9160-1. [DOI] [PubMed] [Google Scholar]
- 99.Riddiford L.M., Cherbas P., Truman J.W. Ecdysone receptors and their biological actions. Vitam. Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- 100.Fessler M.B. The challenges and promise of targeting the Liver X Receptors for treatment of inflammatory disease. Pharmacol. Therapeut. 2018;181:1–12. doi: 10.1016/j.pharmthera.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin C.Y., Gustafsson J. Targeting liver X receptors in cancer therapeutics. Nat. Rev. Cancer. 2015;15(4):216–224. doi: 10.1038/nrc3912. [DOI] [PubMed] [Google Scholar]
- 102.Bełtowski J. Liver X receptors (LXR) as therapeutic targets in dyslipidemia. Cardiovascular therapeutics. 2008;26(4):297–316. doi: 10.1111/j.1755-5922.2008.00062.x. [DOI] [PubMed] [Google Scholar]
- 103.Hazra S., Rasheed A., Bhatwadekar A., Wang X., Shaw L.C., Patel M., et al. Liver X receptor modulates diabetic retinopathy outcome in a mouse model of streptozotocin-induced diabetes. Diabetes. 2012;61(12):3270–3279. doi: 10.2337/db11-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sommer S., Fuqua S.A. Estrogen receptor and breast cancer. Semin. Cancer Biol. 2001;11(5):339–352. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 105.Malíková J., Swaczynová J., Kolár Z., Strnad M. Anticancer and antiproliferative activity of natural brassinosteroids. Phytochemistry. 2008;69(2):418–426. doi: 10.1016/j.phytochem.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 106.Sundaram R., Naresh R., Shanthi P., Sachdanandam P. Efficacy of 20-OH-ecdysone on hepatic key enzymes of carbohydrate metabolism in streptozotocin induced diabetic rats. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2012;19(8–9):725–729. doi: 10.1016/j.phymed.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 107.Giovanelli J., Owens L.D., Mudd S.H. Mechanism of inhibition of spinach beta-cystathionase by rhizobitoxine. Biochim. Biophys. Acta. 1971;227(3):671–684. doi: 10.1016/0005-2744(71)90016-7. [DOI] [PubMed] [Google Scholar]
- 108.Yasuta T., Satoh S., Minamisawa K. New assay for rhizobitoxine based on inhibition of 1-aminocyclopropane-1-carboxylate synthase. Appl. Environ. Microbiol. 1999;65(2):849–852. doi: 10.1128/aem.65.2.849-852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mok Y.Y., Atan M.S., Yoke Ping C., Zhong Jing W., Bhatia M., Moochhala S., et al. Role of hydrogen sulphide in haemorrhagic shock in the rat: protective effect of inhibitors of hydrogen sulphide biosynthesis. Br. J. Pharmacol. 2004;143(7):881–889. doi: 10.1038/sj.bjp.0706014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nilsson L.K., Linderholm K.R., Engberg G., Paulson L., Blennow K., Lindström L.H., et al. Elevated levels of kynurenic acid in the cerebrospinal fluid of male patients with schizophrenia. Schizophr. Res. 2005;80(2–3):315–322. doi: 10.1016/j.schres.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 111.Asimakopoulou A., Panopoulos P., Chasapis C.T., Coletta C., Zhou Z., Cirino G., et al. Selectivity of commonly used pharmacological inhibitors for cystathionine β synthase (CBS) and cystathionine γ lyase (CSE) Br. J. Pharmacol. 2013;169(4):922–932. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yun S., Lee Y.J., Choi J., Kim N.D., Han D.C., Kwon B.M. Acacetin inhibits the growth of STAT3-activated DU145 prostate cancer cells by directly binding to signal transducer and activator of transcription 3 (STAT3) Molecules. 2021;26(20) doi: 10.3390/molecules26206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Y.C., Feng N., Li W.W., Tu P.F., Chen J.P., Han J.Y., et al. Costunolide plays an anti-neuroinflammation role in lipopolysaccharide-induced BV2 microglial activation by targeting cyclin-dependent kinase 2. Molecules. 2020;25(12) doi: 10.3390/molecules25122840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cassiano C., Esposito R., Tosco A., Casapullo A., Mozzicafreddo M., Tringali C., et al. Chemical proteomics-guided identification of a novel biological target of the bioactive neolignan magnolol. Front. Chem. 2019;7:53. doi: 10.3389/fchem.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lu F., Wang D., Li R.L., He L.Y., Ai L., Wu C.J. Current strategies and technologies for finding drug targets of active components from traditional Chinese medicine. Front. Biosci. 2021;26(9):572–589. doi: 10.52586/4968. [DOI] [PubMed] [Google Scholar]
- 116.Zhu Y., Ouyang Z., Du H., Wang M., Wang J., Sun H., et al. New opportunities and challenges of natural products research: when target identification meets single-cell multiomics. Acta Pharm. Sin. B. 2022;12(11):4011–4039. doi: 10.1016/j.apsb.2022.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.