Abstract
Background
Joint destruction is a hallmark of autoantibody-positive rheumatoid arthritis (RA), though the severity is highly variable between patients. The processes underlying these interindividual differences are incompletely understood.
Methods
We performed a genome-wide association study on the radiological progression rate in 384 autoantibody-positive patients with RA. In stage-II 1557 X-rays of 301 Dutch autoantibody-positive patients with RA were studied and in stage-III 861 X-rays of 742 North American autoantibody-positive patients with RA. Sperm-Associated Antigen 16 (SPAG16) expression in RA synovium and fibroblast-like synoviocytes (FLS) was examined using Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) and immunohistochemistry. FLS secrete metalloproteinases that degrade cartilage and bone. SPAG16 genotypes were related to matrix metalloproteinase (MMP)-3 and MMP-1 expression by FLS in vitro and MMP-3 production ex vivo.
Results
A cluster of single nucleotide polymorphisms (SNPs) at 2q34, located at SPAG16, associated with the radiological progression rate; rs7607479 reached genome-wide significance. A protective role of rs7607479 was replicated in European and North American patients with RA. Per minor allele, patients had a 0.78-fold (95% CI 0.67 to 0.91) progression rate over 7 years. mRNA and protein expression of SPAG16 in RA synovium and FLS was verified. FLS carrying the minor allele secreted less MMP-3 (p=1.60×10−2). Furthermore, patients with RA carrying the minor allele had lower serum levels of MMP-3 (p=4.28×10−2). In a multivariate analysis on rs7607479 and MMP-3, only MMP-3 associated with progression (p=2.77×10−4), suggesting that the association between SPAG16-rs7607479 and joint damage is mediated via an effect on MMP-3 secretion.
Conclusions
Genetic and functional analyses indicate that SPAG16 influences MMP-3 regulation and protects against joint destruction in autoantibody-positive RA. These findings could enhance risk stratification in autoantibody-positive RA.
Rheumatoid arthritis (RA) is characterised by persistent joint inflammation and joint destruction. The severity of radiographic destruction can be measured objectively and reflects the cumulative burden of inflammation. Up-to-date treatment strategies have improved the outcome of patients with RA, though the chronic and destructive character of the disease is not prevented. Anticitrullinated peptide antibody (ACPA)-positive patients with RA in particular are characterised by progressive joint destruction.1 The severity is highly variable between ACPA-positive patients, some develop almost no joint destruction, whereas others have a severely progressive disease (see online supplementary figure 1). Current treatment is not individualised; we are neither able to identify the patients that will have the most severe disease course nor do we understand the processes underlying these interindividual differences. To improve this, risk factor studies are required.
A genome-wide association analysis (GWAs) is a hypothesis-free approach to identify common genetic risk factors. Performed GWAs in RA consisted of a case-control design, mainly yielding genetic variants involved in disease susceptibility. However, therapeutic strategies are targeted to affect pathways of disease progression. For drug discovery and to achieve personalised medicine, interrogation of the genome with disease progression is attractive. As it requires high-quality longitudinal outcome data of a considerable number of patients, such GWAs are scarce. We present the first GWAs on long-term disease progression. The notion that the severity of joint destruction in RA is influenced by genetic factors is supported by our previous work on twins and the Icelandic RA-population, estimating the heritability of joint destruction at 45–58%.2 3 We evaluated high-quality radiological data of cross-sectional and longitudinal studies (2812 X-rays in total) of 1427 ACPA-positive patients with RA.
METHODS
Patients
All patients with RA fulfilled the 1987-ACR criteria4 and were ACPA positive. Approval was given by the ethics committees of all cohorts. Written informed consent was obtained from all participants. First, 396 ACPA-positive patients with RA from the North American Rheumatoid Arthritis Consortium (only one of the sib-pair) with hand X-rays, information on disease onset and genomewide single nucleotide polymorphism (SNP) data5 were assessed; after quality control 384 patients were studied (figure 1). These patients, diagnosed between 1953–2002, were treated conservatively; the mean follow-up was 13.9±10.5 years. Stage-II included 301 ACPA-positive patients with RA included in the Leiden Early Arthritis Clinic between 1993–2006.6 Hands and feet X-rays were made with yearly intervals during 7 years of follow-up. Treatment regimes changed over time as described elsewhere.6–8 In short, patients diagnosed in 1993–1995 were initially treated with NSAIDs, patients diagnosed in 1996–1998 were mainly treated with hydroxychloroquine or sulphasalazine and patients included from 1999 onwards were treated with methotrexate. In stage-III, 742 North American and Canadian ACPA-positive patients with RA were studied. In the National Databank for Rheumatic diseases (NDB)9 629 patients were included and 113 patients with RA in the Wichita cohort (Kansas, USA) in 1973–1993.10 The NDB-patients had one hand X-ray per person; the Wichita-patients had serial X-rays. Also these patients were recruited and followed in eras when biological therapies were uncommon.
Figure 1.
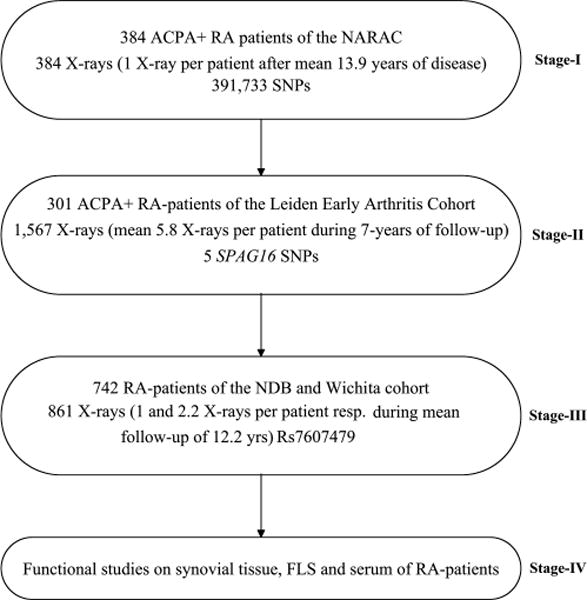
Study design. The mean (±SD) ages at disease onset of the patients in the three stages were: 40.8±11.9 years, 55.8±14.8 years and 45.8±13.3 years, respectively. The percentage of female patients was 73%, 86% and 76%, respectively. The years of diagnosis (min-max) of the patients were 1953–2002, 1993–2006 and 1963–1999, for the three stages respectively. The mean (min-max) follow-up durations were 13.9 (0.5–46) years, 5.8 (1–7) years and 12.2 (1–56) years. All patients were positive for the 1987-ACR criteria for rheumatoid arthritis (RA) and had ACPA. ACPA was measured on stored serum samples using second-generation ELISAs (anti-CCP2, Immunoscan RA Mark 2, Euro-Diagnostica, Arnhem, the Netherlands). The minor allele frequency of rs7607479 in the different cohorts were 0.36 (NARAC), 0.37 (Leiden Early Arthritis Clinic), 0.37 (NDB) and 0.35 (Wichita). ACPA, anticitrullinated peptide antibodies; FLS, fibroblast-like synoviocytes; NARAC, North-American Rheumatoid Arthritis Consortium; NDB, National Databank of rheumatic Diseases; RA, rheumatoid arthritis.
Radiological joint destruction
All X-rays were scored by trained readers at the Leiden University Medical Centre (JABvN in stage-I, MPMvdL in stage-II and DPCdR in stage-III) using the Sharp–van der Heijde method (SHS)11 blinded to clinical and genetic data. Serial X-rays were scored chronologically. In every stage ≥10% randomly selected X-rays were rescored. The intraobserver reliability (ICC) was 0.99, 0.91 and 0.98 in the three stages.
Genotyping
In stage-I GWAs genotyping was conducted with Illumina BeadChips (HumanHap 550 k).5 The dataset was filtered for SNPs on genotyping success rate>98%, minor allele frequency>0.1 and Hardy-Weinberg equilibrium (HWE) p<1×10−5. From chromosome 1–22, 391 733 SNPs passed quality control. Patients were excluded based on success rates<10% (n=2), evidence of identity-by-decent (n=0) or population substructures (n=10). Since after quality control the genomic inflation factor was 1.009 (see online supplementary figure 2), we did not further adjust for substructures. In total 384 patients were studied. In stage-II, a set of SNPs in SPAG16 were chosen based on the strongest association in stage-1 (p<9.5×10−5) and a pairwise R2 <0.8 were typed using Sequenom iPLEX. Doubtful calls were manually checked. Error and success rates (percentage of incorrect duplicates and of successful calls per SNP respectively) were <1% and >97%. None of the SNPs was out of HWE (p<0.001). In stage-III and functional studies, rs7607479 was typed using TaqMan platforms. The error rate and genotyping success rate were respectively <1% and >98%.
RA synovial tissue and fibroblast-like synoviocytes
Presence of SPAG16 protein in synovial tissue from ACPA-positive patients with RA that underwent joint replacement surgery (Hasselt University, Belgium) was studied using immunohistochemistry (see online supplementary methods). Fibroblast-like synoviocytes (FLS) were derived from synovial tissue specimens obtained from 26 patients with RA during joint replacement surgery (Department of Orthopaedic Surgery, Schulthess Clinic Zurich, Switzerland). Cells were cultured as described elsewhere12 and used between passages 4–8 for all experiments.
To isolate DNA 5×104 cells were cultured in 6-well plates 24 h after splitting cells were lysed in tissue lysis buffer and DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen, Hombrechtikon, Switzerland).
Total RNA was isolated using the miRneasy Kit (Qiagen, Hombrechtikon, Switzerland) including on-column DNaseI (Qiagen) digest, reversed transcribed and used for relative quantification of mRNA expression using human glyceraldehyde-3-phosphate dehydrogenase for internal standard sample normalisation as described previously (see online supplementary table 1 for primer sequences).12 Relative mRNA expression levels were calculated by the comparative threshold cycle method (ΔCt). The specificity of the SPAG16 transcript was also confirmed by loading the Real-Time Quantitative Polymerase Chain Reaction-products on a 1.5% agarose gel.
Matrix metalloproteinase (MMP)-1 and MMP-3 were measured using the human total MMP-1 ELISA-kit and the human total MMP-3 ELISA-kit (BD Biosciences, Allschwil, Switzerland) in cell culture supernatants from unstimulated cells according to the manufacturer’s instructions.
Subsequently, the MMP-3 mRNA and protein expression studies were repeated in tumor necrosis factorα and interleukin (IL)-1β stimulated FLS as described in the online supplementary methods.
Serum MMP-3 levels
Serum samples of patients with RA included in the replication cohort used in stage-II were studied. Samples were taken at mean disease duration of 4.2±2.2 years (range 1–9 years) and before 2000, an era when the antirheumatic drugs were mild and tight control strategies not implemented. Standard sandwich ELISA for mature MMP-3 was performed (BD Biosciences).
Statistical analysis
In all genetic analyses an additive model (1-df. test) was used. Since SHS are non-normally distributed, all radiological data were log transformed before analyses. The residuals of the regression analysis were randomly distributed around the zero-line, indicating a good fit of the model. Obtained effect sizes were back transformed to the original scale, indicating how many fold the joint destruction increased per year in the presence of the minor allele compared with the reference common genotype. The difference in joint damage increases per follow-up year by the power of the years of follow-up.
For datasets with one X-ray per patient (North American Rheumatoid Arthritis Consortium and NDB), the estimated yearly progression rate was calculated (total SHS/disease years at time of X-ray).13 This method assumes a linear progression rate over time and the absence of joint damage at disease onset. Here a linear regression with loge(estimated progression+1) as outcome variable performed. For the analyses in datasets with multiple measurements per patient (Leiden Early Arthritis Clinic and Wichita) a multivariate normal regression analysis was used to model loge(SHS+1) over time.14 15 This method takes advantage of within-patient correlation between the measurements. All genetic analyses were adjusted for age and sex. In stage-I and stage-III no influence of treatment on joint destruction was observed. In stage-II an effect of treatment on the radiographic progression was observed and analyses were adjusted for the applied treatment strategy as described previously.8 An inverse variance weighting meta-analysis testing for random effects16 was done on the Wichita and NDB data and on stage-II and stage-III.
In stage-I, 545K SNPs were genotyped, after quality control 391.733 SNPs were analysed. For GWA-studies the family wise error rate depends on linkage disequilibrium, the SNP density, the chosen test statistic, the population and the sample size.17 Large sample sizes decrease the per test significance level, because increased sample sizes change the distribution of the test statistic at rare variants to allow smaller p values. Hoggart et al showed that for the European population, an additive model, 660K SNPs and a sample size of 100 cases and 100 controls the per test significance level is 2.9×10−7, while for 1000 cases and 1000 controls the level decreases to 1.5×10−7.17 Interpolating these results to a sample size of 400 subjects resulted in a threshold for genome-wide significance for the present study of p≤2.7×10−7, with 2.7×10−7 <p<9.5×10−5 considered as highly suggestive. These cut-offs served to find variants of interest, which were proceeded to stage-II. In stage-II, stage-III and the functional analyses, p values <0.05 were considered statistically significant.
The associations of SNPs with MMPs’ expression and production were analysed using the non-parametric data for linear trend (1-df.) of Cuzick. Linear regression with loge(MMP-3) as outcome variable and SNP as test variable was used for analyses on serum levels. Similar as the Cuzick test a linear association was assumed (1-df. test). All analyses on the serum samples were corrected for sex, age, treatment and disease duration at time of serum collection. The relation between loge(MMP3) levels and progression of joint destruction in the next year (loge(SHS2-SHS1+1)) was analysed using linear regression.
PLINK V.1.0.6, Stata V.10.1 and SPSS V.18.0 were used.
RESULTS
Genetic analyses
Evaluating joint damage progression in stage-I revealed the strongest association for a cluster of SNPs located at 2q34, in the region of Sperm-Associated AntiGen16 (SPAG16) (figure 2, see online supplementary table 2). Of these, rs7607479 reached the predefined level of genome-wide significance (p=1.59×10−7). SPAG16 is located 132 kb downstream of IKFZ2 and there is no linkage disequilibrium (LD) (R2<0.05) between rs7607479 and any of the variants in IKZF2. Compared with the patients with the common genotype, patients with one minor rs7607479 allele had a 0.77-fold (95% CI 0.70 to 0.85) progression rate per year and patients carrying two minor alleles a 0.59-fold (95% CI 0.49 to 0.72) progression rate per year. The interindividual variance in joint destruction explained by rs7607479 was 6.6%.
Figure 2.
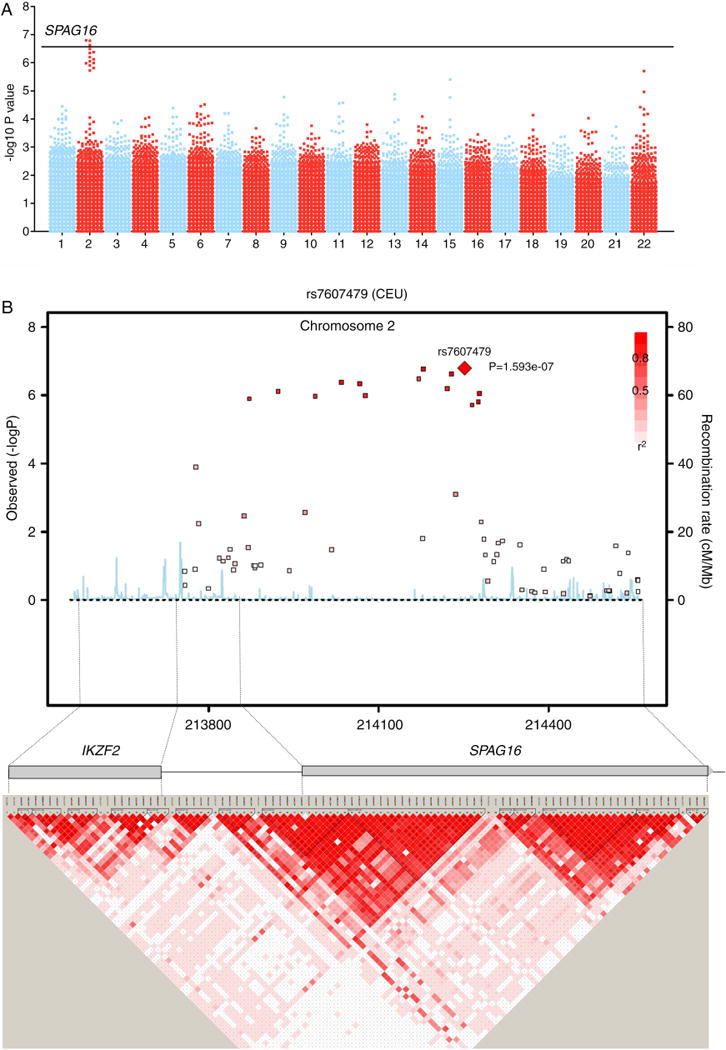
Results of the discovery stage. (A) Manhattan-plot showing the −log10 (p value) of the trend-test. p values are plotted against position on each chromosome. (B) Genomic structure (upper) and −log10 of SNPs in the GWAs around SPAG16 and linkage disequilibrium map (lower) in NARAC. GWAs, genome-wide association analysis; NARAC, North-American Rheumatoid Arthritis Consortium.
In stage-II, all five tested SPAG16 SNPs were significantly associated with the rate of joint destruction; rs7607479 had the strongest association (see online supplementary table 3). However, none remained significant when conditioned on rs7607479 (see online supplementary table 4), indicating that the five significant SNPs reflect one signal. Over the 7-years studied, patients with one rs7607479 minor allele had a 0.81-fold progression rate and patients with two minor alleles a 0.65-fold progression rate compared with patients with the common genotype (95% CIs 0.68 to 0.96 and 0.45–0.93, p=2.16×10−2 figure 3A), indicating that 19% and 35% less joint destruction was observed.
Figure 3.
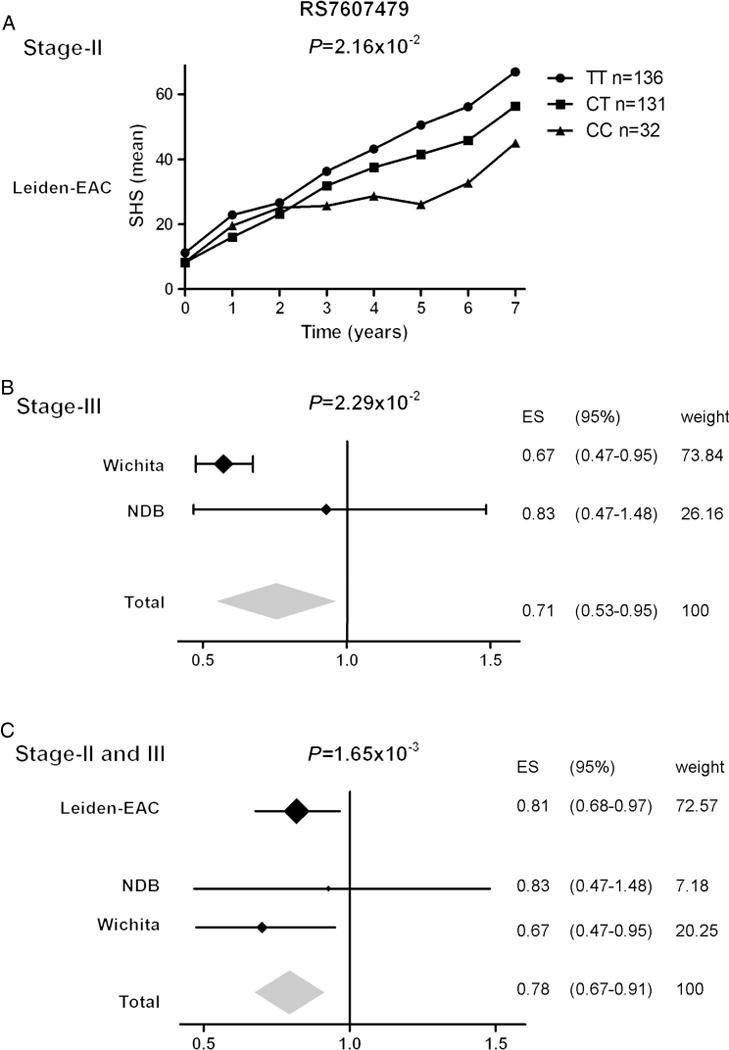
Results of the two replication stages and a meta-analysis on stage-II and stage-III. (A) rs7607479 and radiological progression rates in stage-II where patients with RA had serial measurements over 7 years. (B) rs7607479 and radiological progression rates in stage-III where patients with RA from the NDB had single X-rays and from the Wichita cohort serial X-rays; data of both cohorts were combined in inverse variance weighted meta-analysis. (C) The data of both replication stages were summarised an inverse variance weighted meta-analysis testing for random effects. The I2 was 0%. ES=effect size. For reasons of comparability all data were depicted for progression over a similar disease period (7 years). An effect size <1 indicated a protective effect. NDB, National Databank of rheumatic Diseases; RA, rheumatoid arthritis.
In stage-III, again a protective effect of the rs7607479 minor allele on progression of joint destruction was observed (p=2.29×10−2, 0.71-fold (95% CI 0.53 to 0.95) progression rate over 7 years in the presence of one minor allele, figure 3B).
Combining the results of rs7607479 on progression of joint destruction of stage-II and stage-III in a meta-analysis testing for random effects (I2=0%) demonstrated a 0.78-fold (95% CI 0.67 to 0.91) progression rate over 7 years per minor allele (p=1.65×10−3, figure 3C). A meta-analysis on all three cohorts is presented in online supplementary figure 3.
SPAG16 expression in the joint
We identified an association with SPAG16, a region with a largely unknown function. SPAG16 is expressed in sperm, testis,18–21 and in many other tissues like haematopoietic cells in the bone marrow, macrophages in the lung, glandular cells of the tractus digestivus and mammae (http://www.proteinatlas.org)). Information on the expression in synovium was lacking. Several isoforms are described; isoforms 1 and 2 are the best validated. Considering how SPAG16 could influence joint damage progression, we first evaluated whether it is expressed in the joint. Presence of SPAG16 protein was observed in synovial tissue of patients with RA using immunohistochemical staining (figure 4). Fluorescent double staining showed SPAG16 expression in FLS. The presence of SPAG16 transcripts was detected by Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) and subsequent agarose gel electrophoresis on cDNA derived from FLS of RA synovium (figure 5).
Figure 4.

Immunohistochemical expression of SPAG16 in synovial tissue and FLS. (A) Immunohistochemistry for the expression of SPAG16 in synovial tissue of a patient with RA (B) Isotype control on consecutive synovial tissue section. Staining was performed with diaminobenzidine (brown) with haematoxylin counterstaining of nuclei (blue). Scale bars represent 10 μm. (C) Fluorescent double staining of SPAG16 (green) and a FLS marker prolyl-4-hydroxylase-β (red) in the synovial tissue lining; cell nuclei are stained in blue. Representative photos of one of the five ACPA +RA patients tested. (D) Control staining on consecutive synovial tissue section. Scale bars represent 50 μm. ACPA, anticitrullinated peptide antibodies; FLS, fibroblast-like synoviocytes; RA, rheumatoid arthritis.
Figure 5.

Expression of SPAG16, studied using cDNA derived from FLS of RA tissue and using quantitative Real-time PCR and agarose gel electrophoresis for visualisation of bands. (A) Location of the primers used to amplify different SPAG16 transcripts are indicated by the arrows above the exons. Transcripts in bold have been experimentally validated. (B) PCR products were visualised on a 1.5% agarose gel; a 100 bp ladder was used and the expected amplicon length is 52 bp as indicated by the arrow. Sample 1 concerns testis cDNA as positive control, samples 2–7 are FLS of RA patients with genotypes TC, TC, TT (common genotype), CC (homozygous for the minor allele), TT and CC respectively. Sample 8 concerns the negative control. FLS, fibroblast-like synoviocytes; RA, rheumatoid arthritis.
SPAG16 expression in vitro in relation to genotypes
FLSs are key effector cells in the RA synovium and contribute actively to joint destruction by the release of MMPs.22–24 MMP-3 and MMP-1 are involved in the breakdown of extracellular matrix during tissue remodelling in normal physiological processes and disease processes, such as tumour metastasis and arthritis.25 Rs7607479 is located at an intronic region of SPAG16. SPAG16 expression did not correlate with rs7607479 genotypes (see online supplementary figure 4). The capacity of FLS to express or secrete MMPs can be measured in vitro and we studied this in relation to rs7607479 genotypes. In unstimulated FLS a tendency towards a lower mRNA expression of MMP-3 in FLS carrying the minor alleles of rs7607479 was observed. Furthermore, MMP-3 protein levels were significantly lower in supernatants of FLS carrying the minor allele compared with that of FLS with the common genotype (p=1.60×10−2, figure 6A). Since the fold induction after cytokine stimulation was similar between genotypes, these data show that even in inflammatory conditions rs7607479 genotypes are predictive for MMP-3 expression levels (see online supplementary figure 5). Rs7607479 was not associated with MMP-1 expression or production.
Figure 6.
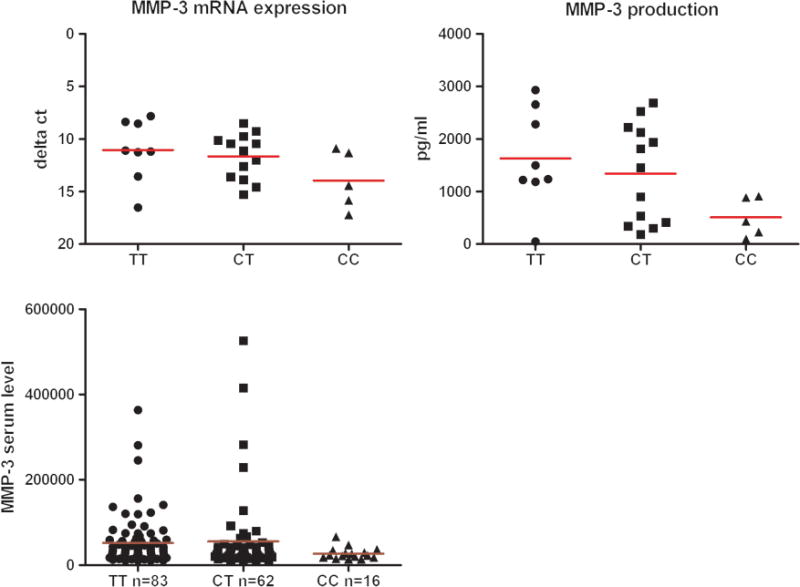
rs7607479-genotypes in relation to MMP-3 expression and production by FLS (A) and MMP-3 serum levels in patients with RA (B). (A) Presented are the mRNA expression (RT-qPCR) and protein production levels (ELISA on cell culture supernatants) of MMP-3 in relation to SPAG16-rs7607479 genotypes in FLS of 26 patients with RA. In FLS carrying the minor alleles of SPAG16-rs7607479 a tendency towards a lower expression of MMP-3 was observed (p=0.08) and a significantly lower protein production compared with FLS with the common genotype (p=1.60×10−2). (B) In patients with RA, presence of the minor allele of rs7607479 was associated with lower serum MMP-3 levels (p=4.28×10−2). FLS, fibroblast-like synoviocytes; MMP, matrix metalloproteinase; RA, rheumatoid arthritis.
SPAG16 expression ex vivo in relation to genotypes
Given these in vitro data, we studied whether SPAG16-rs7607479 was associated with MMP-3 production in vivo, reflected by the serum level. Patients with the minor rs7607479 allele had lower MMP-3 serum levels. For each minor allele of rs7607479 patients displayed 0.83-fold MMP-3 levels compared with patients with the common genotype (95% CI 0.70 to 0.996, p=4.28×10−2, figure 6B). Additionally, lower MMP-3 levels were associated with less radiological progression during the next year (p=1.76×10−4, see online supplementary figure 6). When rs7607479 and MMP-3 serum levels were analysed together in one analysis, only MMP-3 serum levels were significantly associated with progression of joint destruction (p=2.77×10−4, see online supplementary table 5), suggesting that the association between SPAG16-rs7607479 and joint destruction is mediated by a mechanism affecting MMP-3 levels.
DISCUSSION
RA is composed of an ACPA-positive and ACPA-negative subset, which have different genetic susceptibility factors.1 26 Although ACPA-positive RA is more severe, the severity varies between patients and the mechanisms responsible are unknown. Elucidation of these mechanisms allows developing targeted interventions in patients most at risk for a severe destructive disease course. We identified a common variant in SPAG16 of which the minor allele is predictive for less severe joint damage progression and associated with less secretion of the matrix degrading enzyme MMP-3, that is known25 27 and also here observed to be associated with radiological progression.
While we describe an association at this locus with RA severity, we do not at this point know the exact causal SNP. We mined 1000Genomes pilot1 CEU population to identify proxies that could be of functional relevance. We extracted all SNPs in strong LD (R2>0.8) with rs7607479. Of the 159 SNPs identified, none were in the protein-coding region (see online supplementary table 6). The regulatory potential was subsequently studied in Haploreg, a tool for exploring annotations of the non-coding genome (see online supplementary table 7). While there are potential candidates, further fine-mapping studies are needed to investigate functional causal alleles at this locus.
Despite the gene name, SPAG16 expression is not confined to sperm and the association observed with joint damage was not confined to men and equally present in women (data not shown). The gene is located on chromosome 2 and expressed in various tissues. We found that SPAG16 is expressed in the synovium and FLS. FLS are key effector cells because of their capacity to degrade cartilage and bone.24 25 In severe combined immunodeficiency mouse models and in vitro studies this is reflected by the invasiveness of RA FLS.24 We have previously shown that the in vitro invasive capacity of FLS is associated with the expression of MMP-3.28 Moreover we observed that the invasiveness of FLS is an individual characteristic that is associated with the severity of joint damage progression.29 The present data revealed that rs7607479 genotypes were associated with MMP-3 production by FLS and the multivariate analyses suggested that the effect of SPAG16-rs7607479 was partly mediated via an effect on MMP-3. Altogether these data imply that SPAG16-rs7607479 influences the expression of the matrix degrading enzyme MMP-3, thereby influencing joint damage progression. Up to now little is known about the function of SPAG16. Since SPAG16 is predicted to have a role in the basal structure of the primary cilium, we excluded malformation of the primary cilium in FLS with different genotypes. MicroRNA-4438 is the only known miR that is encoded within the SPAG16 gene and is located in the same intron as rs7607479. However, this microRNA was not expressed in FLS (data not shown). Further studies are required to unravel the pathway with which rs7607479 regulates MMP-3 production. Additionally, rs7607479 may affect pathways in other cell types as well, which were not studied here.
Presence of high-quality phenotypic outcome data of populations with RA from different continents is a strength of this study. High-quality data were obtained by accurate X-ray scoring and by evaluation of cohorts with serial X-rays within patients. Having repeated measurements within patients yields more precise estimations of the progression rate than single measurements which facilitates differentiating between true effects and noise.30 Another strength is studying patients treated in an era with less intense treatment strategies than today; presumably their natural disease course was relatively less affected.
This study is the first GWAs on radiological progression in ACPA-positive RA. HLA-DRB1 shared epitopes are known to associate with a severe joint damage; though these alleles actually predispose to ACPA-development, that associate with severe RA.31 This explains why in this ACPA-positive population the HLA-region was not associated with RA severity. Two recent candidate gene studies identified variants (among others in IL-15) as risk factors for radiological progression in the total population of ACPA-positive and ACPA-negative patients. 8 32 These variants did not appear in the top list of this study where a slightly different patient population was evaluated (we confined to ACPA-positive RA). Furthermore, the effect size of SPAG16-rs7607479 observed in stage-I (0.77-fold progression rate/year) was considerably larger than the effect size of IL-15 observed in stage-I of the present study (1.06-fold progression rate/year, p<0.05) and previously published studies (1.03-fold progression rate/year; p<0.01).8 31
Even though our GWA contained relatively low numbers of patients, the power in the present study was largely the result of the preciseness of the outcome data. Furthermore, as mentioned above, the effect observed was considerable. This is in contrast to the results of many GWAs in complex disorders where effect sizes are generally small.
In conclusion, in three data-sets of ACPA-positive patients with RA we identified a common genetic risk factor (44% of patients with RA have one allele, 11% two alleles) that is associated with less severe joint damage progression and found a connection to a disease-relevant biological pathway, namely the production of MMP-3. These findings provide insight into the processes underlying the interindividual differences in progression of joint destruction in RA and could enhance risk stratification in ACPA-positive RA.
Acknowledgments
The authors thank Dr F Wolfe for kindly providing the X-rays and clinical data of his patients. G Cavet from Crescendo Bioscience is acknowledged for the help with the MMP-3 ELISA on the serum samples. P Künzler is acknowledged for help with the cell culture.
Funding The work of RK is supported by a grant of the Dutch Arthritis Foundation. The work of AHMvdH-vM is supported by a grant of the Dutch Organisation of Health care and development. VS, KS and LdB are supported by grants from Research Foundation Flanders (FWO-Vlaanderen), Hasselt University and the Transnationale Universiteit Limburg. The work of CO and SG was supported by a grant of IAR. SG was supported by FP-7 TEAM. PKG is supported by grants from the National Institutes of Health (RO1-AR-44422; RO1-AI-68759; RC2AR059092-01) and the Alliance for Lupus Research, in addition to support from Eileen Ludwig Greenland and the Boas family. The research has also been funded by The European Community Seventh Framework Program FP7 Health-F2-2008-223404 (Masterswitch), IMI-BTCure, BBMRI-NL a Research Infrastructure financed by the Dutch government (NWO 184.021.007), and the Netherlands Organization for Scientific Research (917.66.344).
Footnotes
Contributors
RK performed the analyses, QH and RK performed the analysis of stage-I, JJH-D gave statistical advice, JABvN, DPCdR and MPMvdL scored all X-rays, JS, GS-R, NM-O, LR-R and PKG were involved in genotyping, KK, KS, CO and LdB performed the functional experiments, PS, CO, SG and VS supervised the functional experiments, REMT and FASK provided advice, PKG, TWJH and AHMvdH-vM were involved in the design and collection of the patient cohorts, AHMvdH-vM designed and supervised the study, AHMvdH-vM and RK wrote the first draft, all authors contributed to the final manuscript.
Competing interests A patent application has been filed on the base of the results presented in this article (‘Means and methods for the determination of the joint destruction progression rate in rheumatoid arthritis patients’, patent pending, US application No. 61/611.494).
Ethics approval Local ethical committee.
Provenance and peer review Not commissioned; externally peer reviewed.
Handling editor Tore K Kvien
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/annrheumdis-2013-204050).
References
- 1.van der Helm-van Mil AHM, Huizinga TW, de Vries RR, et al. Emerging patterns of risk factor make-up enable subclassification of rheumatoid arthritis. Arthritis Rheum. 2007;56:1728–35. doi: 10.1002/art.22716. [DOI] [PubMed] [Google Scholar]
- 2.Van der Helm-van Mil AHM, Kern M, Gregersen PK, et al. Variation in radiologic joint destruction in rheumatoid arthritis differs between monozygotic and dizygotic twins and pairs of unrelated patients. Arthritis Rheum. 2006;54:2028–30. doi: 10.1002/art.21872. [DOI] [PubMed] [Google Scholar]
- 3.Knevel R, Gröndal G, Huizinga TW, et al. Genetic predisposition of the severity of joint destruction in rheumatoid arthritis: a population-based study. Ann Rheum Dis. 2012;71:707–9. doi: 10.1136/annrheumdis-2011-200627. [DOI] [PubMed] [Google Scholar]
- 4.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 5.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N Engl J Med. 2007;20:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Rooy DP, van der Linden MP, Knevel R, et al. Predicting arthritis outcomes–what can be learned from the Leiden Early Arthritis Clinic? Rheumatology (Oxford) 2011;50:93–100. doi: 10.1093/rheumatology/keq230. [DOI] [PubMed] [Google Scholar]
- 7.Lard LR, Edworthy SM, Bloch DA, et al. Early and aggressive treatment of rheumatoid arthritis patients affects the association of HLA class II antigens with progression of joint damage. Arthritis Rheum. 2002;46:899–905. doi: 10.1002/art.10151. [DOI] [PubMed] [Google Scholar]
- 8.Knevel R, Krabben A, Brouwer E. Genetic variants in IL-15 Associate with Progression of Joint Destruction in Rheumatoid Arthritis, a Multi Cohort Study. Ann Rheum Dis. 2012;71:1651–7. doi: 10.1136/annrheumdis-2011-200724. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe F, Michaud K. The National Data Bank for rheumatic diseases: a multi-registry rheumatic disease data bank. Rheumatology (Oxford) 2011;50:16–24. doi: 10.1093/rheumatology/keq155. [DOI] [PubMed] [Google Scholar]
- 10.Choi HK, Hernán MA, Seeger JD, et al. Methotrexate and mortality in patients with rheumatoid arthritis: a prospective study. Lancet. 2002;6:1173–7. doi: 10.1016/S0140-6736(02)08213-2. [DOI] [PubMed] [Google Scholar]
- 11.Van der Heijde D. How to read X-rays according to the Sharp/van der Heijde method. J Rheumatol. 2000;27:261–3. [PubMed] [Google Scholar]
- 12.Ospelt C, Brentano F, Rengel Y, et al. Overexpression of Toll-like Receptors 3 and 4 in Synovial Tissue from patients with Early Rheumatoid Arthritis. Arthritis Rheum. 2008;58:3684–92. doi: 10.1002/art.24140. [DOI] [PubMed] [Google Scholar]
- 13.Strand V, Landéwé R, van der Heijde D. Using estimated yearly progression rates to compare radiographic data across recent randomised controlled trials in rheumatoid arthritis. Ann Rheum Dis. 2002;61(Suppl 2):ii64–6. doi: 10.1136/ard.61.suppl_2.ii64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. New York: John Wiley and Sons; 2004. Longitudinal data: basic concepts; pp. 19–29. [Google Scholar]
- 15.Van der Helm-van Mil AHM, Knevel R, van der Heijde DM, et al. How to avoid phenotypic misclassification in using joint destruction as an outcome measure for rheumatoid arthritis? Rheumatology (Oxford) 2010;49:1429–35. doi: 10.1093/rheumatology/keq013. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Hoggart CJ, Clark TG, De Iorio M, et al. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–85. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Zariwala MA, Mahadevan MM, et al. A heterozygous mutation disrupting the SPAG16 gene results in biochemical instability of central apparatus components of the human sperm axoneme. Biol Reprod. 2007;77:864–71. doi: 10.1095/biolreprod.107.063206. [DOI] [PubMed] [Google Scholar]
- 19.Nagarkatti-Gude DR, Collodel G, Hill LD, et al. Spag16, an axonemal central apparatus gene, encodes a male germ cell nuclear speckle protein that regulates SPAG16 mRNA expression. PLoS One. 2011;6:e20625. doi: 10.1371/journal.pone.0020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somers V, Govarts C, Somers K, et al. Autoantibody profiling in Multiple Sclerosis reveals novel antigenic candidates. J Immunol. 2008;180:3957–63. doi: 10.4049/jimmunol.180.6.3957. [DOI] [PubMed] [Google Scholar]
- 21.Pennarun G, Bridoux AM, Escudier E, et al. Isolation and expression of the human hPF20 gene orthologous to Chlamydomonas PF20: evaluation as a candidate for axonemal defects of respiratory cilia and sperm flagella. Am J Respir Cell Mol Biol. 2002;26:362–70. doi: 10.1165/ajrcmb.26.3.4738. [DOI] [PubMed] [Google Scholar]
- 22.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–55. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firestein GS. In: Kelly’s Textbook of Rheumatology. Firestein GS, editor. Vol. 8. Philadelphia, PA: Saunders Elsevier; 2009. pp. 1035–86. [Google Scholar]
- 24.Pap T, Meinecke I, Müller-Ladner U, et al. Are fibroblasts involved in joint destruction? Ann Rheum Dis. 2005;64(Suppl 4):iv52–4. doi: 10.1136/ard.2005.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niedermeier M, Pap T, Korb A. Therapeutic opportunities in fibroblasts in inflammatory arthritis. Best Pract Res Clin Rheumatol. 2010;24:527–40. doi: 10.1016/j.berh.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Plenge RM. Recent progress in rheumatoid arthritis genetics: one step towards improved patient care. Curr Opin Rheumatol. 2009;21:262–71. doi: 10.1097/BOR.0b013e32832a2e2d. [DOI] [PubMed] [Google Scholar]
- 27.Houseman M, Potter C, Marshall N, et al. Baseline serum MMP-3 levels in patients with Rheumatoid Arthritis are still independently predictive of radiographic progression in a longitudinal observational cohort at 8 years follow up. Arthritis Res Ther. 2012;14:R30. doi: 10.1186/ar3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolboom TC, Pieterman E, van der Laan W, et al. Invasive properties of fibroblast-like synoviocytes: correlation with growth characteristics and expression of MMP-1, MMP-3, and MMP-10. Ann Rheum Dis. 2002;61:975–80. doi: 10.1136/ard.61.11.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolboom TC, van der Helm-Van Mil AH, Nelissen RG, et al. Invasiveness of fibroblast-like synoviocytes is an individual patient characteristic associated with the rate of joint destruction in patients with rheumatoid arthritis. Arthritis Rheum. 2005;52:1999–2002. doi: 10.1002/art.21118. [DOI] [PubMed] [Google Scholar]
- 30.Van der Helm-van Mil AH, Knevel R, van der Heijde D, et al. How to avoid phenotypic misclassification in using joint destruction as an outcome measure for rheumatoid arthritis? Rheumatology (Oxford) 2010;49:1429–35. doi: 10.1093/rheumatology/keq013. [DOI] [PubMed] [Google Scholar]
- 31.van der Helm-van Mil AH, Verpoort KN, Breedveld FC, et al. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anti-cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54:1117–21. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 32.de Rooy DP, Yeremenko NG, Wilson AG, et al. Genetic studies on components of the Wnt signalling pathway and the severity of joint destruction in rheumatoid arthritis. Ann Rheum Dis. 2013;72:769–75. doi: 10.1136/annrheumdis-2012-202184. [DOI] [PubMed] [Google Scholar]


