Abstract
IKZF1 plays an essential role in lymphopoiesis, and somatic IKZF1 variants in acute lymphoblastic leukemia (ALL) are associated with poor prognosis. In this issue of Cancer Cell, Churchman et al. add to the list of leukemia predisposition genes with the identification and characterization of germline IKZF1 variants in childhood ALL.
Acute lymphoblastic leukemia (ALL), the most common childhood malignancy, has traditionally been regarded as a non-hereditary disease. With the exception of rare familial cases, initial genetic studies in ALL, as in other cancers, have largely focused on somatically acquired mutations in malignant cells. However, given that pediatric cancers occur early in life, it is perhaps not surprising that germline mutations predisposing to childhood cancers have increasingly been described, even among cases initially presumed to be sporadic (Kratz et al., 2016). Germline mutations inform medical management and advance our understanding of the role of these genes in development and tumorigenesis.
Germline TP53 mutations, for example, cause Li-Fraumeni syndrome, with increased risks of a variety of cancers, including ALL. Genomic profiling has identified somatic mutations in approximately 90% of low-hypodiploid ALL, with about 40% concurrently present in nontumor cells, suggesting likely germline origin (Holmfeldt et al., 2013). Another growing category of genes implicated in predisposition to lymphoid malignancy are those encoding transcription factors that orchestrate lymphoid development. Somatic variants of PAX5, critical for the maturation of B cells from pro/pre-B cells, are seen in about 30% of B-ALL, and germline variants have also been described in familial cases (Auer et al., 2014; Shah et al., 2013). Germline alterations in ETV6, encoding an ETS family transcriptional repressor important in the maintenance and differentiation of hematopoietic stem cells and megakaryopoiesis, lead to thrombocytopenia, ALL, and myeloid malignancies (Hock and Shimamura, 2017).
In this issue of Cancer Cell, Churchman, Mullighan, and colleagues add IKZF1 to the growing number of genes involved in lymphoid leukemia predisposition (Churchman et al., 2018). IKZF1 encodes IKAROS, a member of a zinc-finger transcription factor family. IKAROS has an N-terminal DNA binding domain composed of four zinc fingers and a C-terminal dimerization domain with two zinc fingers required for homo- or heterodimerization (Churchman and Mullighan, 2017; Marke et al., 2018) (Figure 1A). Dominant-negative isoforms result when DNA binding (N-terminal) is disrupted but dimerization (C-terminal) remains intact. The multifaceted role of IKAROS in cancer is also driven by interactions with an array of epigenetic regulator complexes. IKAROS interacts with the SWItch/Sucrose Non-Fermentable (SWI/SNF) related complex, the nucleosome remodeling and deacetylase (NuRD) complex, and the polycomb repressive complex 2 (PRC2) that regulates both transcriptional repression and activation across a large set of genes. These global epigenetic interactions and the role of IKAROS in transcriptional regulation are context-dependent on cell type and developmental timing. Murine models demonstrate an essential role for Ikzf1 in lymphopoiesis of all lymphoid lineages, as well as a role in hematopoietic differentiation. Mice with mutant Ikzf1 also develop lymphoid malignancies, either spontaneously or in combination with oncogenic transgenes (Marke et al., 2018).
Figure 1. IKZF1 Germline and Somatic Mutations.
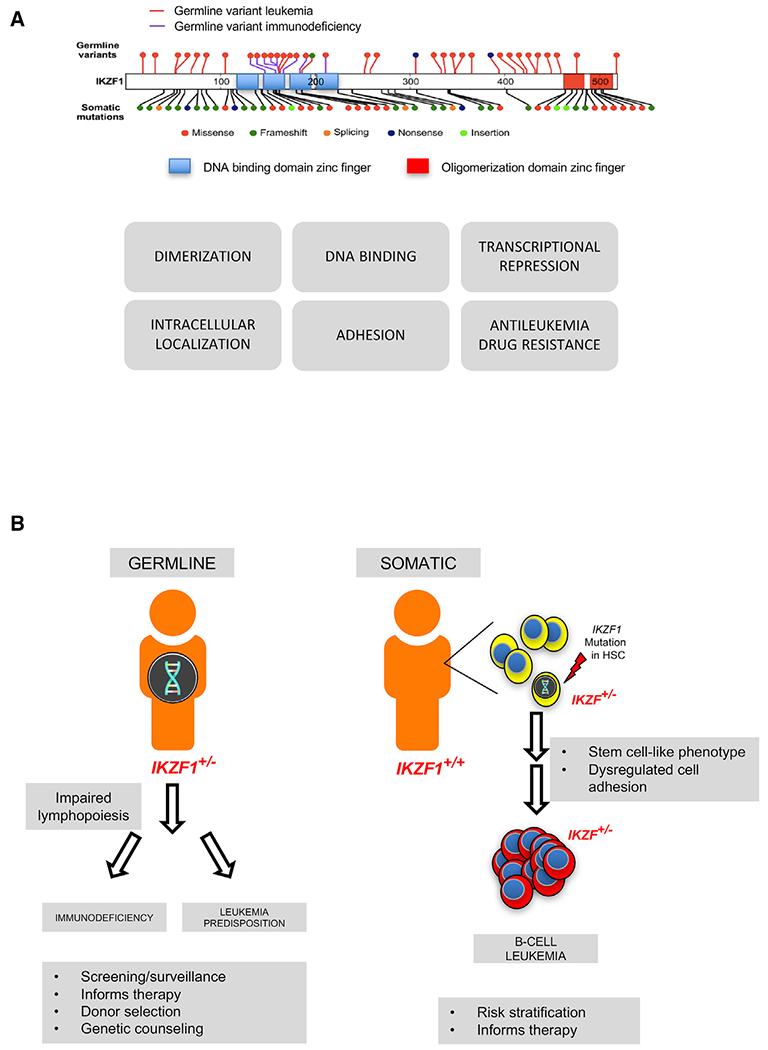
(A) IKZF1 structural domains and mutations (top), modified from Churchman et al. (2018), and functional assays used by Churchman et al. to assess pathogenicity of identified IKZF1 variants (bottom). (B) Germline IKZF1 mutations are associated with immunodeficiency and predisposition to B cell leukemia. Acquisition of somatic mutations in IZKF1 also promote the development of B cell leukemia. HSC, hematopoietic stem cell. Figure 1B modified from https://pixabay.com/en/man-user-profile-person-icon-42934/.
In human leukemia, somatic alterations in IKZF1 are seen in about 15% of B-ALL overall. IKZF1 mutations are found in over 70% of BCR-ABL1 (Ph+) ALL and in Ph-like ALL and are associated with adverse outcomes. Poor outcomes are also associated with alterations of IKZF1 in standard-risk B-ALL subtypes, including those with high hyperdiploidy. Somatic deletions or sequence alterations in IKZF1 lead to loss of function or dominant-negative isoforms, resulting in acquisition of a hematopoietic stem-cell-like phenotype with impaired leukemia cell maturation and dysregulated cell adhesion (Churchman and Mullighan, 2017; Marke et al., 2018).
Previously reported germline mutations in IKZF1 have been studied in the context of inherited immunodeficiency, with mutations in the DNA-binding zinc finger domain leading to common variable immunodeficiency (CVID) and reduced numbers of B cells. Interestingly, 2 of 29 such individuals in one series were noted to also develop B-ALL (Kuehn et al., 2016), suggesting a role for IKZF1 in human leukemia.
Churchman and colleagues now identify IKZF1 as a new leukemia predisposition gene (Churchman et al., 2018). They investigated an index family, with B-ALL arising in two members who were found to share a heterozygous germline deletion in IKZF1, c.del556 (D186fs), leading to a protein truncated at residue 192. Other carriers of the truncating IKZF1 mutation in the pedigree were found to be lymphopenic without frank clinical immunodeficiency. The authors investigate the prevalence of IKZF1 mutations with targeted sequencing of remission samples from an impressive 4,963 cases of childhood leukemia. They identified 43 cases from these cohorts, with a total of 28 unique IKZF1 variants, the majority from patients with B-ALL. To confirm the germline origin of a subset of these variants, the authors analyzed bone marrow-derived mesenchymal stromal cells from four patients and identified the same IKZF1 genetic aberrations. No deletions were identified with germline single-nucleotide polymorphism array or with whole-genome sequencing on a subset of 697 cases.
Churchman and colleagues next assessed the functional consequences of each of the 28 IKZF1 variants, testing their ability to repress transcription of a known IKAROS target, DNA binding, dimerization, subcellular localization, and effects on cell adhesion. They also assessed sensitivity to antineoplastic drugs, including the tyrosine kinase inhibitor dasatinib and dexamethasone. The authors observed functional consequences for 22 of the 28 IKZF1 variants. The authors note only a 65% concordance of pathogenicity between in silico prediction algorithms and the outcome of these functional assays. The observation that different assays identified different patterns of functional impairments across the spectrum of IKZF1 variants highlights the complexity of assessing variant pathogenicity. The current work uses both genomics and functional studies to define the relevance of identified germline variants as a benchmark for future studies.
Taken together, Churchman and colleagues identify a new leukemia predisposition gene, IKZF1, first shown within a family and then expanded to include almost 1% of B-ALL cases otherwise presumed to be sporadic. The identification of a germline cancer risk carries profound clinical impact (Godley and Shimamura, 2017). For patients requiring a bone marrow transplant, germline genetic diagnosis allows the screening of familial transplant donors to avoid choosing an affected but clinically silent donor. Some germline cancer predisposition disorders require tailored therapies to avoid excessive treatment-related toxicities or to minimize relapse risk. Diagnosis of genetic cancer predisposition may also inform surveillance strategies for cancer and other associated medical co-morbidities. It is of critical importance, therefore, to distinguish germline from somatic mutations.
This study highlights the insights gained from studying rare families with cancer predisposition to advance our overall understanding of these genes in cancer biology and in normal development (Figure 1B). Although each cancer predisposition gene individually accounts for a relatively small number of patients, taken together as a group, germline cancer predisposition constitutes an increasingly significant subset of malignancies previously believed to be sporadic. As we continue to characterize the genomic landscape of leukemias, the list of cancer predisposition genes is likely to expand.
REFERENCES
- Auer F, Rüschendorf F, Gombert M, Husemann P, Ginzel S, Izraeli S, Harit M, Weintraub M, Weinstein OY, Lerer I, et al. (2014). Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia 28, 1136–1138. [DOI] [PubMed] [Google Scholar]
- Churchman ML, and Mullighan CG (2017). Ikaros: exploiting and targeting the hematopoietic stem cell niche in B-progenitor acute lymphoblastic leukemia. Exp. Hematol 46, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Qian M, Te Kronnie G, Zhang R, Yang W, Zhang H, Lana T, Tedrick P, Baskin R, Verbist K, et al. (2018). Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell 33, this issue, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godley LA, and Shimamura A (2017). Genetic predisposition to hematologic malignancies: management and surveillance. Blood 130, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, and Shimamura A (2017). ETV6 in hematopoiesis and leukemia predisposition. Semin. Hematol 54, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. (2013). The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet 45, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz CP, Stanulla M, and Cavé H (2016). Genetic predisposition to acute lymphoblastic leukemia: Overview on behalf of the I-BFM ALL Host Genetic Variation Working Group. Eur. J. Med. Genet 59, 111–115. [DOI] [PubMed] [Google Scholar]
- Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, Maffucci P, Pierce KR, Abbott JK, Voelkerding KV, et al. (2016). Loss of B cells in patients with heterozygous mutations in IKAROS. N. Engl. J. Med 374, 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marke R, van Leeuwen FN, and Scheijen B (2018). The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica 103, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, et al. (2013). A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat. Genet 45, 1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]


