Abstract
Variants in seven genes (LRRK2, GBA1, PRKN, SNCA, PINK1, PARK7 and VPS35) have been formally adjudicated as causal contributors to Parkinson’s disease; however, individuals with Parkinson’s disease are often unaware of their genetic status since clinical testing is infrequently offered. As a result, genetic information is not incorporated into clinical care, and variant-targeted precision medicine trials struggle to enrol people with Parkinson’s disease. Understanding the yield of genetic testing using an established gene panel in a large, geographically diverse North American population would help patients, clinicians, clinical researchers, laboratories and insurers better understand the importance of genetics in approaching Parkinson’s disease.
PD GENEration is an ongoing multi-centre, observational study (NCT04057794, NCT04994015) offering genetic testing with results disclosure and genetic counselling to those in the US (including Puerto Rico), Canada and the Dominican Republic, through local clinical sites or remotely through self-enrolment. DNA samples are analysed by next-generation sequencing including deletion/duplication analysis (Fulgent Genetics) with targeted testing of seven major Parkinson’s disease-related genes. Variants classified as pathogenic/likely pathogenic/risk variants are disclosed to all tested participants by either neurologists or genetic counsellors. Demographic and clinical features are collected at baseline visits.
Between September 2019 and June 2023, the study enrolled 10 510 participants across >85 centres, with 8301 having received results. Participants were: 59% male; 86% White, 2% Asian, 4% Black/African American, 9% Hispanic/Latino; mean age 67.4 ± 10.8 years. Reportable genetic variants were observed in 13% of all participants, including 18% of participants with one or more ‘high risk factors’ for a genetic aetiology: early onset (<50 years), high-risk ancestry (Ashkenazi Jewish/Basque/North African Berber), an affected first-degree relative; and, importantly, in 9.1% of people with none of these risk factors. Reportable variants in GBA1 were identified in 7.7% of all participants; 2.4% in LRRK2; 2.1% in PRKN; 0.1% in SNCA; and 0.2% in PINK1, PARK7 or VPS35 combined. Variants in more than one of the seven genes were identified in 0.4% of participants.
Approximately 13% of study participants had a reportable genetic variant, with a 9% yield in people with no high-risk factors. This supports the promotion of universal access to genetic testing for Parkinson’s disease, as well as therapeutic trials for GBA1 and LRRK2-related Parkinson’s disease.
Keywords: Parkinson’s disease, genetic testing, genetic counselling, LRRK2, GBA1, clinical trials
Cook et al. present results from an ongoing multicentre observational study offering genetic testing with results disclosure and genetic counselling to individuals with Parkinson's disease across North America. Among the approximately 8000 participants who have received results to date, 13% have a reportable genetic variant.
Introduction
Parkinson’s disease is a neurodegenerative disease characterized by progressive motor disability and non-motor symptoms.1 To date, seven genes (LRRK2, GBA1, PRKN, SNCA, PINK1, PARK7, VPS35) have been curated by the ClinGen Parkinson’s Disease Gene Curation Expert Panel (PD GCEP) as having a causal relationship with Parkinson’s disease (https://search.clinicalgenome.org/kb/affiliate/10079). Prior studies have identified pathogenic variants in Parkinson’s disease-linked genes in about 5%–10% of people with Parkinson’s disease in the US and Europe.2-10 However, only a small fraction of people with Parkinson’s disease receive genetic testing due to lack of awareness among clinicians and patients, cost issues and lack of clinician confidence in providing results to patients.5,11 Previous studies have shown a greater likelihood of positive (abnormal) gene test results among those with an earlier age at onset (AAO), positive family history or certain ancestry such as Ashkenazi Jewish, Spanish Basque or North African Berber.12 However, recent research indicates that different gene variants may be found in individuals of diverse ancestries from various countries and global regions.13
There are several clinical reasons why individuals with Parkinson’s disease might wish to have genetic testing: to aid in diagnosis, to answer the question, ‘Why did I get Parkinson’s disease?’, to inform prognosis, to assist in treatment and life decisions and to clarify the Parkinson’s disease risk for other family members. In addition, there is growing interest within the pharmaceutical industry in genetically targeted precision medicine. Accordingly, in the research and direct-to-consumer (DTC) spaces, there has been a notable increase in genetic testing for Parkinson’s disease over the past decade.14,15 Over 10 000 people with Parkinson’s disease have ordered DTC testing from 23andMe that includes health risk assessment for Parkinson’s disease via limited, targeted testing of a single variant, each, within the genes LRRK2 and GBA1.16 Genetic information about Parkinson’s disease is being used to determine clinical trial eligibility for gene-specific trials.1,17
The genetics of Parkinson’s disease is complex. Counselling issues such as dominant and recessive inheritance, reduced penetrance, phase determination for carriers of two variants for recessive disorders, the uncertainty of the relationship between heterozygous forms of Parkinson’s disease thought to be recessively inherited and disease risk, the risk for both Gaucher disease and Parkinson’s disease in relation to some but not all GBA1 variants and the possibility of carrying disease-related variants in more than one Parkinson’s disease gene require thoughtful discussion with a clinician skilled in explaining genetic concepts (genetic counsellor or trained neurologist). We have shown that neurologists are uncomfortable with their knowledge of Parkinson’s disease genetics5 and access to genetic counsellors in North America is limited.18
With this background, the Parkinson’s Foundation, a non-profit Parkinson’s disease research and advocacy organization, launched PD GENEration (PD GENE) as a clinical study in 2019 with the goal of educating and empowering people with Parkinson’s disease by offering Clinical Laboratory Improvement Amendments (CLIA)-certified genetic testing with return of results in the context of genetic counselling at no cost to the participants.18 PD GENE addresses potential barriers to testing by providing genetic counselling in either English or Spanish, in local or remote settings. The study has had three phases thus far. In its pilot phase, we verified feasibility of the study and documented both a strong community interest and satisfaction with the testing process among both participants and providers with no significant adverse psychological sequelae in participants.18 Here, we report the genetic testing results along with clinical data from the first three phases of the study through June 2023.
Materials and methods
Study design and participants
PD GENE is a multi-centre, observational and registry, clinical study (NCT04057794, NCT04994015) offering CLIA-certified genetic testing and genetic counselling to people with Parkinson’s disease in North America,18 with an original enrolment goal of 15 000 by 2025. The study was approved by centralized and site institutional review boards (IRBs), as well as the Scientific Review and Executive Committees of the Parkinson Study Group. All participants signed informed consents. The study has had three phases: a pilot phase (600 tested participants) and a clinical phase (the pilot participants plus an additional 1354 tested participants) in which detailed clinical data were collected, as well as the current registry phase with a simplified protocol (6347 tested participants). We include here data from all phases, described in more detail in the Supplementary material. Participants may enrol through their local study site or, remotely, through designated, national enrolling sites with IRB approval to provide telemedicine genetic testing and counselling on a national level or via the Parkinson’s Foundation website https://www.parkinson.org/pdgeneration. Participants are eligible for the study if they meet the Movement Disorder Society (MDS) Clinical Diagnostic Criteria for probable Parkinson’s disease based on examination, chart review or self-report,19 are at least 18 years of age, able to provide informed consent in English or Spanish, complete study activities and willing to undergo genetic testing and be informed of their results. In the registry phase, clinical examinations and chart reviews are not performed. Race and ethnicity are obtained by self-report, using categories developed by the US Census Bureau and collected by the study to assess outreach to underserved communities. Self-reported ancestry is collected to evaluate trends in genetic testing yields related to geographic/genealogic descent. Participants are asked to report if a parent, child and/or sibling had a Parkinson’s disease diagnosis established by a physician or by autopsy. All study materials were created in or translated to Spanish and culturally adapted for the Hispanic/Latino community, involving input from the PD GENE Latino Advisory Committee and professional translators. Sites that have bilingual staff are eligible to recruit Spanish-speaking participants. Further details about the processes and protocol of the study can be found at the Parkinson’s Foundation website (https://parkinson.org/pdgeneration).
Genetic testing and genetic counselling
Following participant consent and sample collection, testing of the genes LRRK2, GBA1, PRKN, SNCA, PINK1, PARK7 and VPS35 is performed at Fulgent Genetics, a CLIA-certified US laboratory. These genes are recognized as major causes of Parkinson’s disease and are included on most commercial Parkinson’s disease genetics panels.14 In addition, the seven genes on this panel were previously adjudicated by an independent panel of Parkinson’s disease experts using a recently developed framework supported by the National Institutes of Health (NIH)-funded Clinical Genome Resource (ClinGen)20 that found sufficient evidence to support a gene-disease relationship for all of them.
As previously reported,21 next-generation sequencing (NGS) and data analysis are performed on genomic DNA. Genomic DNA isolated from accessioned samples (blood or buccal saliva) is prepared into libraries using a customized hybrid capture enrichment protocol, targeting key coding exons and splicing junctions based on IDT xGen Lockdown probe chemistry (Integrated DNA Technologies, Inc., Coralville, IA, USA). Paired-end sequencing is then performed on DNA libraries on the Illumina platform 2500 HiSeq or NovaSeq 6000, using 300 bp reads [size of genomic fragments are 400–500 bp peak size (range 150–900 bp)]. Following alignment to the human genome reference sequence (assembly GRCh37/hg19), variants are detected in regions with at least 10× coverage. For specimens, 99% and 98% of coding regions and splicing junctions (±20 bp of canonical exon splice donor) of the genes listed are sequenced with coverage of at least 10× and 20×, respectively, or by Sanger sequencing. However, the average coverage is usually >100 from this assay, and we achieve a typical coverage >50× (except for GBA1 exons 9–11). For germline variants, 20× is typically considered sufficient to call a heterozygous variant with an allele fraction of about 50%. In addition, all the variants with quality scores less than 500 (roughly 40× coverage for a heterozygous variant) in the NGS-based panel sequencing are confirmed by targeted Sanger sequencing. The genes are also evaluated for large deletions and/or duplications, and putative deletions or duplications identified are confirmed by quantitative PCR (qPCR) or multiplex ligation-dependent probe amplification (MLPA) by MRC-Holland (Amsterdam, The Netherlands). The analysis of single exon deletions and duplications is performed on the PRKN gene. The NGS misalignment analysis is performed on the GBA1 gene to avoid pseudogene interference. When a potential variant misalignment is identified, long-range PCR is performed to confirm variants and gene conversion.22 To verify homozygosity for variants in the GBA1 gene, for each exon, the lab utilizes two sets of primers to account for possible allele dropout. This method in combination with NGS data and misalignment analyses, greatly reduces the likelihood of allele dropout that would be categorized incorrectly. When a single pathogenic or likely pathogenic variant is identified in a gene with autosomal recessive inheritance, e.g. PRKN, PINK1 or PARK7, 100% of coding sequences of that gene are covered either through NGS or Sanger sequencing technologies. For bioinformatics, the Fulgent Germline v2019.2 pipeline is used for analysis.
Variants are initially curated using automated ranking rules and further interpreted manually using locus-specific databases, literature searches and other molecular biological principles. All variants detected in the reportable region (i.e. coding exons and 20 bp flanking introns) are assessed based on the American College of Medical Genetics and Genomics (ACMG) guideline for sequence variant interpretation.23 Variants are classified into five-tier categories: pathogenic (P), likely pathogenic (LP), variants of uncertain significance (VUS), likely benign and benign. GBA1 variants are classified based on their pathogenicity to cause Gaucher disease, except for the GBA1 c.1093G>A (p.Glu365Lys)/E365K/E326K variant, which is associated with Parkinson’s disease risk and reduced glucocerebrosidase enzymatic activity but not with Gaucher disease.24,25 Details regarding ranking rules used for curation and additional methodology applied to the GBA1 gene are provided in the Supplementary material, Methods section.
Variants deemed reportable in this study include those classified as P/LP, whereas VUS are not reported to the clinician or the participant. There is some controversy about the relationship between certain GBA1 variants and the development of Parkinson’s disease. We chose to report the GBA1 c.1093G>A (p.Glu365Lys)/E365K/E326K variant, which is referred to as a ‘risk variant’ with a low penetrance for Parkinson’s disease (about twice the population risk) and no risk for Gaucher disease,25 because at the time the study began, individuals with Parkinson’s disease who carried this variant were eligible to participate in a GBA1-Parkinson’s disease therapeutic trial. For this reason, this risk variant was felt to be clinically actionable. However, another GBA1 risk variant, c.1223C>T (p.Thr408Met)/T408 M/T369M, was not reported back to participants as its level of pathogenicity was controversial at study onset but is included as a ‘disease-relevant result’.24 Monoallelic P/LP heterozygous variants of autosomal recessive Parkinson’s disease genes such as PRKN were considered as reportable, due to the potential implications for reproductive risks in relatives, despite lack of consensus about causation in Parkinson’s disease.26
VUS are not disclosed to participants but are catalogued for research use and shared with a global consortium of Parkinson’s disease geneticists and clinicians to centralize and harmonize discussions of VUS identified across multiple studies (ClinGen Parkinson’s Disease Variant Curation Expert Panel: https://clinicalgenome.org/affiliation/50079/). Participants in PD GENE are asked if they wish to give permission to be recontacted for further research based on their Parkinson’s disease gene status and can opt in to receive additional genetic information from the study, such as VUS that are later classified as P/LP. Curated gene variants will be deposited in the NIH ClinGen and ClinVar repositories. Coded, de-identified DNA samples and raw sequence data are stored and are available to researchers upon request.
All participants receive genetic test results (negative or positive) via an in-person or remote genetic counselling session provided by neurologists or certified genetic counselors, at local or nationally enrolling sites. Test results are interpreted by clinicians for participants in the context of their clinical findings and medical and family histories. All participants receive a copy of their gene test laboratory report and a summary of the genetic counselling session. Those with biallelic GBA1 P/LP variants are referred to local Gaucher centres for further evaluation and counselling.
Statistical analysis
All statistical analyses were performed using R 4.2.2. For continuous variables, mean, standard deviation (SD), medians and interquartile range (IQR) are provided. For categorical data, the percentage and counts for each category over total number of available participants are provided. To compare differences in clinical measures of interests that are continuous, such as AAO between groups, we used Mann–Whitney test to reduce the type I error due to non-normality of the outcome variables. For the categorical outcomes, such as sex, we used Fisher’s exact test to reduce the type I error due to sparse cell counts (less than five in any cell) or chi-squared test with two-sided test. All P-values are reported for two-sided tests.
To provide the results in Tables 1 and 3, our primary analyses, we conducted a total of 33 discovery tests comparing Parkinson’s disease disease-related items. Using Bonferroni correction for multiple testing, our statistical significance threshold was 0.05/33 or 0.0015.
Table 1.
Demographic characteristics of participants tested
| Variable | Tested (n = 8301) | Negative (n = 7231) | Positive (n = 1070) | P-value, positive versus negative |
|---|---|---|---|---|
| Age at enrolment, years, mean (SD), (IQR) | 67.4 (10.8), (61.0–74.3) | 67.7 (10.7), (61.7–75.0) | 65.2a (11.7), (58.0–73.0) | 3.63 × 10−10 |
| Reported sex, % male (count/total) | 59% (4902/8300) | 60% (4339/7230) | 53%a (563/1070) | 4.94 × 10−6 |
| Self-reported race % (count/total) | – | – | – | 0.239 |
| White | 86% (6978/8159) | 85% (6059/7111) | 88% (919/1048) | – |
| Alaska Native/American Indian | <1% (16/8159) | <1% (15/7111) | <1% (1/1048) | – |
| Asian | 2% (195/8159) | 2% (175/7111) | 2% (20/1048) | – |
| Native Hawaiian/Pacific Islander | <1% (7/8159) | <1% (6/7111) | <1% (1/1048) | – |
| Black/African American | 4% (350/8159) | 4% (312/7111) | 4% (38/1048) | – |
| Multiple | 3% (225/8159) | 3% (204/7111) | 2% (21/1048) | – |
| Self-reported ethnicity % (count/total) | – | – | – | 0.114 |
| Hispanic/Latino | 9% (707/7925) | 9% (601/6895) | 10% (106/1030) | – |
| AAO of PD, years, mean (SD), (IQR) | 61.1 (11.1), (54.0–69.0) | 61.5 (10.9), (55.0–69.0) | 58.3a (12.3), (51.0–67.0) | 3.53 × 10−15 |
| PD duration, years, mean (SD), (IQR) | 5.7 (5.8), (1.0–8.0) | 5.6 (5.7), (1.0–8.0) | 6.5a (6.6), (2.0–9.0) | 2.12 × 10−5 |
| Early onset (AAO of PD < 50 years), % (count/total) | 16% (1276/8049) | 15% (1040/7004) | 23%a (236/1045) | 9.79 × 10−10 |
| High-risk ancestryb, % (count/total) | 14% (1130/8301) | 12% (860/7231) | 25%a (270/1070) | 4.13 × 10−28 |
| First degree relative with PD, % (counts/total) | 20% (1579/7994) | 19% (1295/6969) | 28%a (284/1025) | 4.55 × 10−11 |
AAO = age at onset; IQR = interquartile range; PD = Parkinson’s disease; SD = standard deviation.
aStatistically significant after multiple test correction (P < 0.0015).
bAshkenazi Jewish, Spanish Basque, North African Berber.
Table 3.
Participant characteristics by gene variant subgroups
| P-values | |||||||
|---|---|---|---|---|---|---|---|
| Variable | Negative (n = 7231) | GBA1 (n = 638) | LRRK2 (n = 196) | PRKN, bi (n = 56) | GBA1 versus neg | LRRK2 versus neg | PRKN, bi versus neg |
| Male: female ratio, (n male/n female) | 1.50 (4339/2890) | 1.19 (347/291) | 0.81a (88/108) | 0.65 (22/34) | 0.006 | 3.19 × 10−05 | 0.002 |
| Self-reported race, count/total (%) | 0.617 | 0.003 | 0.191 | ||||
| White | 85% (6059/7111) | 88% (553/628) | 91% (172/190) | 73% (40/55) | – | – | – |
| Asian | 2% (175/7111) | 2% (10/628) | 0% (0/190) | 9% (5/55) | – | – | – |
| Black/African American | 4% (312/7111) | 4% (25/628) | 3% (6/190) | 5% (3/55) | – | – | – |
| Multiple | 3% (204/7111) | 2% (13/628) | 2% (3/190 | 5% (3/55) | – | – | – |
| Self-reported ethnicity %(count/total) | – | – | – | – | 0.260 | 0.013 | 6.00 × 10−5 |
| Hispanic/Latino | 9% (601/6895) | 7% (45/612) | 14% (27/188) | 27%a (15/55) | – | – | – |
| AAO of PD, years, mean (SD), (IQR) | 61.5 (10.8), (55.0–69.0) | 58.6a (10.8), (52.0–66.0) | 62.9 (9.56), (57.0–69.0) | 38.6a (14.8), (27.8–49.0) | 2.19 × 10−11 | 0.159 | 2.37 × 10−23 |
| PD duration, years, mean (SD), (IQR) | 5.60 (5.70), (1.00–8.00) | 5.72 (5.13), (2.00–8.00) | 6.04 (5.72), (2.00–8.00) | 15.0a (13.2), (4.00–23.0) | 0.052 | 0.173 | 1.98 × 10−8 |
| Early onset (AAO of PD <50 years), %(count/total) | 15% (1040/7004) | 21%a (130/620) | 9% (17/194) | 79%a (44/56) | 9.41 × 10−5 | 0.018 | 6.29 × 10−29 |
| High-risk ancestryb, %(count/total) | 12% (860/7231) | 19%a (123/638) | 62%a (122/196) | 4% (2/56) | 3.50 × 10−7 | 1.01 × 10−59 | 0.059 |
| First degree relative with PD %(counts/total) | 19% (1295/6969) | 21% (131/610) | 40%a (76/188) | 47%a (26/55) | 0.084 | 1.11 × 10−11 | 1.36 × 10−6 |
AAO = age at onset; bi = biallelic (homozygous or compound heterozygous); IQR = interquartile range; PD = Parkinson’s disease; SD = standard deviation.
aStatistically significant after multiple test correction (P < 0.0015).
bAshkenazi Jewish, Spanish Basque, North African Berber.
Results
Participant demographic characteristics
More than 10 510 people with Parkinson’s disease across North America have enrolled in the study (Fig. 1), of whom 8301 have completed genetic testing and received results as of 1 June 2023. Population descriptions are detailed in Table 1 and approximate expected frequencies among those who typically participate in research in the US.27 We observed that 16% of tested participants had early-onset disease (onset at <age 50). Fourteen per cent were of self-reported high-risk ancestry (Ashkenazi Jewish, North African Berber or Spanish Basque) and 20% reported having a first-degree relative with Parkinson’s disease (Table 1).
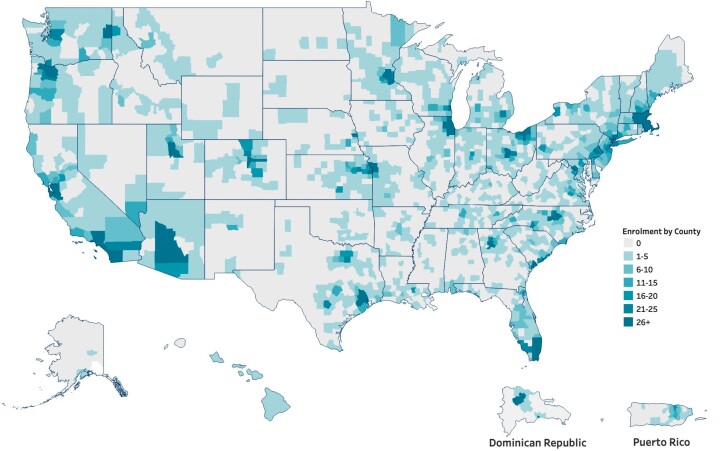
Figure 1.
PD GENEration heat map. Heat map showing broad enrolment of participants into PD GENEration by county in the US, Puerto Rico and the Dominican Republic. Use of telemedicine allowed for enrolment of participants from all 50 states, representing both urban and rural areas.
Genetic findings
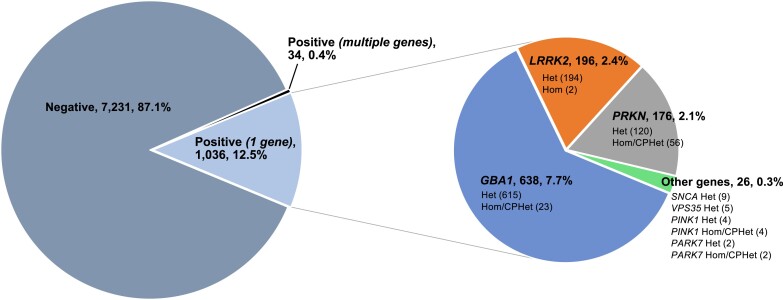
Reportable variants were found in 1070 (12.9%) of the 8301 participants (Table 2). GBA1 variants were the most frequently identified, followed by LRRK2 and PRKN (7.7%, 2.4% and 2.1%, respectively, and including both single and biallelic variants) (0.4% were multigene variant carriers not included) (Fig. 2). The yield for disease-relevant results, e.g. eliminating those who were heterozygous for PRKN, PINK1 and PARK7 P/LP variants and adding those individuals who had a c.1223C>T (p.Thr408Met)/T408M//T369M GBA1 risk variant, was overall 13.4% (1111/8301) and specifically, 10.1% (838/8301) for GBA1, 2.4% (196/8301) for LRRK2 and (56/8301) 0.67% for biallelic PRKN.
Table 2.
Genetic yield by population
| Population | Total tested | Reportable variant identified | Yield |
|---|---|---|---|
| All | 8301 | 1070 | 12.9% |
| High-risk factorsa | 3453 | 625 | 18.1% |
| No risk factorsb | 4406 | 399 | 9.10% |
| Early onset (AAO of PD<50 years) | 1276 | 236 | 18.4% |
| Late onset (AAO of PD ≥50 years) | 6773 | 809 | 11.9% |
| First degree relative with PD | 1579 | 284 | 18.0% |
| High-risk ancestryc | 1130 | 270 | 23.9% |
AAO = age at onset; PD = Parkinson’s disease.
aEarly-onset PD, high-risk ancestry or first degree relative with PD.
bWithout early-onset PD, high-risk ancestry and first degree relative with PD.
cAshkenazi Jewish, Spanish Basque, North African Berber.
Figure 2.
Reportable results. Pie chart (left) depicting overall positivity rate for reportable variants in a single gene or multiple genes among 8301 participants. Right: Categorization of single gene variants for the PD GENEration seven-gene panel. Values are number of participants followed by percentage of total. CPHet = compound heterozygous; Hom = homozygous; Het = heterozygous.
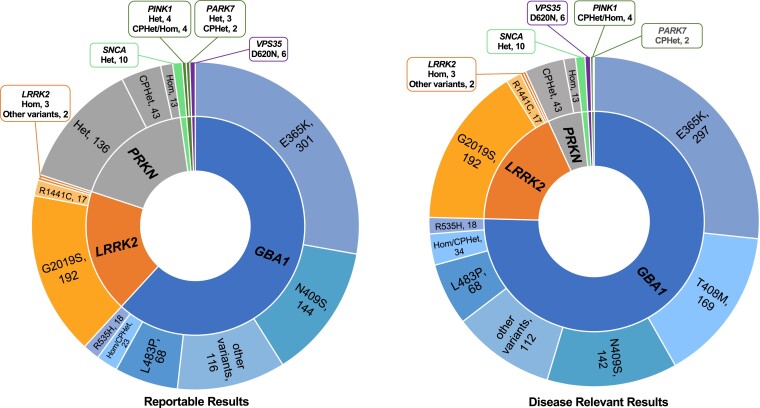
The most common reportable GBA1 variants identified were the c.1093G>A (p.Glu365Lys)/E365K/E326K risk variant (3.7%, 307), the c.1226A>G (p.Asn409Ser)/N409S/N370S variant (1.6%, 133) and the c.1448T>C (p.Leu483Pro)/L444P variant (0.8%, 65). Fourteen participants had two variants (phase not determined), including nine with apparent homozygous variants in the GBA1 gene. Six of these cases involved Gaucher disease-associated variants including c.1226A>G (p.Asn409Ser)/N409S/N370S, representing individuals with likely genotypic Gaucher disease, which was not an exclusion to the study. The most common LRRK2 variant identified was the c.6055G>A (p.Gly2019Ser) variant (177, 2.1%), including two participants who had homozygous c.6055G>A (p.Gly2019Ser) variants. Heterozygous PRKN variants were found in 120 participants: 43 were potentially compound heterozygous (phase not determined) and 13 had homozygous variants. Detected variants in the PRKN gene encompassed copy number variants (CNVs) (n = 32 unique types) and single nucleotide variants (SNVs)/insertions/deletions (indels) (n = 21 unique types). Additional details regarding gene variants observed are described in Figs 2 and 3 and the Supplementary Table 5. Of note, 34 participants (0.4%) had complex results encompassing variants in more than one gene; 32 cases involved the GBA1 gene, two cases were double heterozygous for the PRKN and LRRK2 genes, and one case had variants in GBA1, LRRK2 and PRKN (Supplementary Table 2). VUS data are catalogued and will be reported later.
Figure 3.
Variant level results. Variant level representation of results reportable to participants (left), compared to disease relevant results (right), which included GBA1 T408M and excluded heterozygous (Het) carriers of recessive genes. Individuals positive for multiple genes are counted in both gene categories resulting in adjusted totals between graphs. Variant counts are in the heterozygous state unless otherwise specified. Different variant inclusions between graphs resulted in shifts of some individuals from heterozygous to compound heterozygous (CPhet) categories (e.g. two individuals with GBA1 N409S/T369M represented as N370S on the left and compound heterozygous on the right). Hom = homozygous.
For those with high-risk ancestry (Ashkenazi Jewish/Basque/North African Berber), early AAO (<50 years) or an affected first-degree relative, the yield for reportable findings was 24%, 18% and 18%, respectively. In those with one or more of these genetic risk factors, 18% received a positive finding (Table 2). When excluding individuals with these predefined genetic risk factors, 9.1% received a positive report.
Genetic subgroup comparisons
Participant characteristics by genotype are provided in Table 3 and the Supplementary material. Additional subgroup data including data regarding double heterozygotes are provided in Supplementary Tables 2–4. Numbers were too small for some subgroup statistical analyses.
Self-reported race/ethnicity
Between individuals with positive and negative results, we did not see differences across self-reported race and ethnicity. However, individuals who were Hispanic/Latino were 3.6 times more likely to carry biallelic PRKN variants (P = 2.07 × 10−4) compared with non-Hispanic/non-Latino individuals, among those who tested positive.
Age at onset and Parkinson’s disease duration
Those with GBA1 variants or presumed compound heterozygous or homozygous PRKN variants had earlier AAO compared to those with negative results (58.6 and 38.6 versus 61.5 years; P = 2.19 × 10−11 and 2.37 × 10−23), whereas those with LRRK2 variants did not (Table 3). Carriers of SNCA variants had an earlier AAO compared with those with negative results (49.2 versus 61.5 years), though the numbers were too small for statistical comparison. The Parkinson’s disease duration was significantly greater among those with presumed compound heterozygous or homozygous PRKN variants.
Male to female ratio
The male to female ratio among those with negative results was 1.50, while both the LRRK2 and the biallelic PRKN groups had more females than males. Significance was reached in the LRRK2 group compared to non-carriers after correction for multiple comparisons (ratio = 0.81, P = 3.19 × 10−5) (Table 3).
High-risk ancestry and positive family history
There was a greater number of participants with high-risk ancestry among the LRRK2 and GBA1 subgroups compared with the negative group (62%, 19% versus 12%; P = 1.01 × 10−59 and 3.50 × 10−7). Those in the LRRK2 and PRKN compound heterozygous/homozygous groups were more likely to have a first-degree relative with Parkinson’s disease (40%, 47% versus 19%; P = 1.11 × 10−11 and 1.36 × 10−6). This was not observed for GBA1 variants (Table 3).
MDS-Unified Parkinson’s Disease Rating Scale, Hoehn and Yahr and Montreal Cognitive Assessment
The results of the clinical [MDS Unified Parkinson’s Disease Rating Scale (MDS-UPDRS), Hoehn and Yahr scale] and cognitive assessments [Montreal Cognitive Assessment (MoCA)] and genetic subgroups are reported in the 1954 individuals from the clinical phase who had available results (Table 4). There were no significant differences between scores of those with negative results versus those in the genetic subgroups (all P-values > 0.001). Individuals with SNCA variants had lower MoCA scores (mean 22.3), although numbers in this subgroup (n = 7) were too small for meaningful analysis.
Table 4.
Clinical measures and demographics of clinical cohort (n = 1954)
| Variable | Total (n = 1954) | Negative (n = 1665) | GBA1 (n = 169) | LRRK2 (n = 51) | PRKN, het (n = 27) | PRKN, bi (n = 18) | SNCA (n = 7)a |
|---|---|---|---|---|---|---|---|
| Clinical measurement, mean (SD), (IQR) | |||||||
| MoCA | 26.5 (3.00), (25.0–29.0) | 26.5 (2.90), (25.0–29.0) | 26.1 (3.40), (25.0–28.0) | 27.4 (2.30), (27.0–29.0) | 26.2 (2.90), (25.0–28.0) | 26.8 (2.20), (26.0–28.0) | 22.3 (2.50), (20.5–24.0) |
| MDS-UPDRS | 48.3 (23.1), (31.0–62.0) | 47.7 (22.3), (31.0–61.0) | 52.5 (25.8), (34.2–66.0) | 40.6 (25.6), (28.2–42.0) | 51.2 (26.2), (32.0–59.2) | 48.6 (33.3), (27.0–48.0) | 81.2 (19.0), (68.0–95.0) |
| Hoehn and Yahr | 2.0 (0.7), (2.0–2.0) | 2.0 (0.7), (2.0–2.0) | 2.0 (0.6), (2.0–2.0) | 2.0 (0.7), (2.0–2.0) | 1.9 (0.7), (2.0–2.0) | 2.3 (1.0), (2.0–2.5) | 2.3 (0.5), (2.0–2.5) |
| Demographic, mean (SD), (IQR) | |||||||
| Age, years | 64.7 (10.0), (59.0–72.0) | 65.1 (9.80), (59.0–72.0) | 62.4b(9.70), (56.0–69.0) | 66.2 (8.00), (61.0–71.0) | 64.8 (9.90), (60.0–71.0) | 52.3b (15.2), (37.5–62.5) | 54.4 (15.6), (43.0–60.0) |
| Sex, % males (n/total) | 57% (1110/1954) | 58% (960/1665) | 51% (87/169) | 47% (24/51)b | 63% (17/27) | 33% (6/18) | 57% (4/7) |
| AAO of PD, years | 59.3 (10.7), (53.0–67.0) | 59.8 (10.5), (53.0–67.0) | 57.2b (10.2), (50.0–65.0) | 60.9 (8.0), (55.0–66.5) | 57.9 (10.7), (50.0–65.0) | 41.2b (15.0), (30.8–49.5) | 50.0 (16.7), (39.0–56.5) |
| Disease duration | 5.40 (5.20), (2.00–8.00) | 5.30 (5.10), (2.00–8.00) | 5.20 (4.80), (2.00–8.00) | 5.30 (4.70), (1.50–8.00) | 7.00 (6.90), (1.00–11.5) | 11.2 (12.2), (2.20–15.0) | 4.40 (2.90), (2.00–5.50) |
AAO = age at onset; bi = biallelic (i.e. homozygous or compound heterozygous); het = heterozygous (i.e. single variant detected); IQR = interquartile range; MDS-UPDRS = Movement Disorder Society-Unified Parkinson’s Disease Rating Scale; MoCA = Montreal Cognitive Assessment; PD = Parkinson’s disease; SD = standard deviation.
aInsufficient count for statistical comparison.
b P < 0.001.
Discussion
In this study we counselled and tested 8301 participants for seven established Parkinson’s disease-related genes, focusing initially on the feasibility and safety of counselling/return of results through the local neurologist versus centralized genetic counselors (pilot phase),18 then the expansion of the study to multiple clinical sites in the US (clinical phase; Table 4), followed by a simplified study protocol to permit further geographic, racial and ethnic diversity (registry phase; Tables 1–3 andSupplementary Table 1). In this largest and most geographically diverse North American cohort ever tested, we demonstrate an overall yield of reportable genetic variants in 13% of unselected enrolled participants.
There are only a few large-scale, multinational studies with similar attributes to ours as regards ancestry, recruitment, genotyping of multiple genes including GBA1 and data analyses, and their results are consistent with our observations, with approximately an overall 14% disease-relevant yield, even though larger gene panels were used.6,7 Like our study, variants were most often found in GBA1 and LRRK2 at similar rates, approaching 10% and 3%, respectively. The most common variant observed was the GBA1 c.1093G>A (p.Glu365Lys) variant, also known as the E365K/E326K risk allele.6,7 Sequencing data from the UK, as part of the Tracking Parkinson’s study, showed lower yields for pathogenic variants for certain genes such as LRRK2 (0.9%), whereas the yield for GBA1 variants was similar.8,9 The c.6055G>A (p.Gly2019Ser) variant was the predominant abnormal finding in LRRK2, similar to reports of Parkinson’s disease genetic testing results in other European-based cohorts.6,8 As expected from published literature,3,6,8,28 Parkinson’s disease gene variants were found more frequently in those with earlier AAO < 50 (18%, 236/1276), with a first-degree family history of Parkinson’s disease (18%, 284/1579) or high-risk ancestries (24%, 270/1130) compared with our total population rate (Table 2).
Specifically, variants in GBA1 and PRKN (biallelic) were associated with earlier AAO, and those with LRRK2 variants were more likely to be self-reported as female, confirming prior observations.6,12 Female predominance among LRRK2 carriers with Parkinson’s disease is not yet fully explained, and our study does not shed further light on this question. Individuals with either LRRK2 or biallelic PRKN variants were more likely to report a first-degree relative with Parkinson’s disease, likely attributed to higher penetrance of these gene variants. Although self-reported race did not emerge as a significant variable, individuals who were Hispanic/Latino were more likely to have PRKN variants (biallelic) consistent with observations in the CORE-PD study in which those who were Hispanic were more likely to have PRKN variants than non-Hispanic individuals.12 Since we did not report back VUS by design, as additional variants are adjudicated in ClinGen/ClinVar using patient materials from this and other large studies, we anticipate that some VUS will be elevated to the status of P/LP and then the positivity rate in our cohort will increase.
The absolute number of people with Parkinson’s disease who tested positive for PINK1, PARK7, VPS35 and SNCA was extremely low, precluding any detailed comments about genotype-phenotypes or participant characteristics for these groups. In addition, results regarding clinical measures by genetic subgroups should be interpreted with caution due to various limitations in study design. They include the cross-sectional nature of the study, a potential referral bias, presentation in earlier phases of disease, examinations by telemedicine for some participants and informed consent requirements for participants to have the cognitive capacity to provide consent, which had the practical effect of excluding individuals with dementia or a low MoCA score.
We highlight that although we observed a yield of 18% for reportable results in participants with one of three identified genetic risk factors (early AAO, affected first degree relative, high-risk ancestry), notably, the positivity rate was 9% among participants in our cohort with none of these risk factors. As the study expands with plans to query more Parkinson’s disease-related genes, yields in both groups will even be greater. Our results provide compelling data to suggest that genetic testing should not be restricted to high-risk individuals, but rather should be offered to all people with Parkinson’s disease.3,6,7 For example, while as expected, GBA1 and LRRK2 variants were more common among people of Ashkenazi Jewish ancestry compared with those without any high-risk ancestries, most who possessed these variants were non-Ashkenazi Jewish (Supplementary Table 4). However, where resources are limited, testing can be prioritized according to perceived impact and patient interest. Specifically, in cases of limited resources, we propose prioritizing testing for those who would be interested in acting on positive results, e.g. for clinical trial participation or life/family planning.
Motivations to return genetic results to people with Parkinson’s disease include: (i) people are interested in knowing their genetic status11,18; (ii) different genetic forms of Parkinson’s disease can have strikingly different prognoses4; (iii) emerging evidence that genotype may be relevant for aspects of clinical care28; and (iv) genetic status can determine eligibility for precision medicine clinical trials. To date, there are no US Food and Drug Administration (FDA)-approved interventions to modify disease progression in Parkinson’s disease, so approved treatments are all symptomatic. A potential explanation for the failure of multiple clinical trials to demonstrate disease-modifying effect is that Parkinson’s disease pathogenesis is heterogenous. There is reason to hope that therapies targeting Parkinson’s disease with specific associated genetic variants, such as GBA1 or LRRK2 variants, might demonstrate disease modification in a subset of people with Parkinson’s disease.17 Our findings show that there are many previously unidentified people with Parkinson’s disease who could qualify for precision medicine trials, especially ones focused on GBA1 (approaching 10% of all participants) and, to a lesser extent, LRRK2.
The PD GENE study has attempted to reduce previously identified barriers to genetic testing in Parkinson’s disease, including cost, physician knowledge and comfort with genetics (through training materials and coursework), the perceived low yield of genetic testing and patient awareness.5 Strengths of the study include its large and rapidly growing cohort, streamlining of the protocol and use of telehealth strategies to facilitate enrolment from outside the usual geographic (Fig. 1) and cultural reach of academic centres and inclusion of bilingual clinicians and Spanish language materials. The study is also able to contribute patient materials, genetic test results and expertise to several national and global Parkinson’s disease genetics programs, including ClinGen/ClinVar (described above), the Global Parkinson’s Genetics Project (GP2), the Black and African American Connections to Parkinson’s Disease (BLAAC-PD) and Latin American Research consortium on the GEnetics of Parkinson’s disease (LARGE-PD), each targeting Black/African American and Hispanic/Latino Parkinson’s disease populations, respectively.
Despite early efforts, a weakness of this study remains its relative lack of racial and ethnic diversity. Therefore, our results may not be directly applicable to non-European populations. In addition, we acknowledge that in the data analyses stratified by race that these results are less informative as it is increasingly recognized that race is only a social construct based on physical attributes, not accurately representing genetic differences. We recognize that both self-enrolment and physicians who recruit for this study may be biased towards people with early age at onset, positive family history or high-risk ancestry, resulting in some degree of enrichment. However, the study continues to encourage all people with Parkinson’s disease who are interested in testing to participate and to encourage study sites to enrol all-comers, removing these historic barriers to testing. We further acknowledge that the vast majority of study participants received a negative result and that additional genetic research is required to identify risk factors not investigated here (e.g. polygenic risk score). As noted earlier, the interpretation of reported clinical measures by genetic subgroups of the smaller PD GENE cohort were limited by the study design.
In the future, we plan to enhance our ongoing efforts to enrol underrepresented populations and to help improve the understanding of Parkinson’s disease gene variants in these groups. To address this, the PD GENE study has since added the site Morehouse College of Medicine/Grady Hospital in Atlanta, Georgia (traditionally Black/African American) and sites in Puerto Rico and the Dominican Republic. More recently, the study has expanded access to native Hawaiian populations by partnering with Queen’s Medical Center in Honolulu, Hawaii. In addition, PD GENE samples will be included as part of the Aligning Science Across Parkinson’s Global Parkinson’s Genetic Program (ASAP-GP2), in which individuals will be genotyped with the Global Diversity Array (plus Neurobooster)29 for almost 2 million variants genome wide. GP2 will use this information to address both global and local ancestry, and this will shed some light on the admixture present in our cohorts, as well as determining ancestral origins of the pathogenic variants identified in PD GENE. This will also allow for a greater ability to characterize unique variants across populations looking for any enrichment.
In summary, this large study of genetic testing for Parkinson’s disease in North America and Caribbean sites confirms a relatively high rate of positive (abnormal) results, approximately 13% for reportable or disease-relevant variants. Positive results were observed in up to 18% in people with genetic risk factors such as early AAO, high-risk ancestry or an affected first degree relative, but also 9% in people lacking any of these risk factors. As trials of gene-specific potentially disease-modifying treatments have begun, and genetic results may impact disease prognosis, possibly for management, and with certainty and clarity related to familial risks, we believe that clinical genetic testing should be offered to all people with Parkinson’s disease to empower them to act upon their genetic findings. Several strategies are underway to enhance the recruitment of other underserved populations into the last third of the study’s planned cohort.
Supplementary Material
Acknowledgements
This study was funded by the Parkinson’s Foundation, which contributed to study design and execution and was made possible by the generous support of the Parkinson’s community. The authors would like to thank the participants who have willingly given their time to advance research by participating in this study. This work was supported, in part, by the Intramural Research Program of the National Institute on Aging (NIA). We would like to thank our collaborators: Global Parkinson’s Genetics Program (GP2); GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). For a complete list of GP2 members see https://gp2.org.
Appendix 1.
PD GENEration Collaborators
Study Core Collaborators
Rebeca De Leon, Amasi Kumeh, Zachary Meyer, Anny Coral-Zambrano, Min-Jae Kim, Camilla Ruiz, Yun Lu, Elisa Ahmanson, Andra Matthews, Sarah Lawrence, Yun Lu, Susan Li, Uma Ragunathan, Deborah Baker, Betty Lyda, Ken Eaton, Olga Pikul, Ett Muneath, Karen Hodgeman, Yan Meng, Rachel Blake, Laura Heathers, Michelle Totten, Amanda Miller, Malia Rumbaugh.
Site Investigators
Rajesh Pahwa, William De Jesus, Josh Shulman, David Simon, Hemant Pandey, Okeanis Eleni Vaou, Anna Hohler, Ihtsham ul Haq, Henry Moore, Stuart Isaacson, Jason Aldred, Ariane Park, Steven Gunzler, Jill Ostrem, Carlie Tanner, Deborah Hall, Rachel Saunders-Pullman, Matthew Barrett, Chantale Branson, Karen Blindauer, Tsao-Wei Liang, Gonzalo Revuelta, Elizabeth Zauber, Julie Schwartzbard, Tritia Yamasaki, Rohit Dhall, Danielle Englert, Giulietta Riboldi, Annie Killoran, Kathleen McKee, Nina Browner, Connie Marras, Peter Lin, Irene Richard, Amy Hellman, Gian Pal, Kelly Mills, John Morgan, Mustafa Siddiqui, Jeanne Feuerstein, Jeffrey Cooney, Joseph Savitt, Tarannum Khan, Houman Homayounh, Stephen Lee, Harini Sarva, David Hinkle, Luis Forastieri, Frances Velez, Angel Vinuela, Thomas Beach, Charles Adler.
Coordinators
Karen Williams, Max Galarce, Lucas George, Rachel Lewandowski, Evalyn Mackenzie, Amanda Chan, Alia Neibaur, Kellie Keith, Emily Leonard, Gita Golonzka, April Langhammer, Sherry Neisen, Hiba Saade, Jamie Fong, Ruby Rendon, MaryBeth Serrano, Mansi Sharma, Arjun Laud, Isabella Montanaro, Tyler Tribble, Lisa Damron, Neda Almassi, Whitney Hartstone, Aine Russell, Hannah Babcock, Jerrod Cook, Giulia Andrews, Sean Guardado, Kaitlyn Ramsey, Savannah Reis, Claudia Cano, Claire Hennum, Amanda Kiefer, Katherine Ambrogi, Victoria Klee, Victoria Miller, Aaron Daley, Marc Rosenbaum, Deborah Raymond, Maya Rawal, Virginia Norris, Tazrin Rahman, Sherline Sauveur, LaShawn Baker, Paula Phabia-Millbrook, Marie Mejaki, Michelle Rochman, Sandra Wilson, Conner Mroz, Lauren Perrey-Moore, Jonathan Castano, Renee Wagner, Michael Nsoesie, Kimberly Gamble, Kelly Astudillo, Heena Olalde, Min-Jae Kim, Geidy Serrano.
Study Partners
Julia Shirvan, Judith Peterschmitt, Pablo Sardi, Cornelis Blauwendraat, Christine Klein, Parkinson Study Group.
Contributor Information
Lola Cook, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Jennifer Verbrugge, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Tae-Hwi Schwantes-An, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Jeanine Schulze, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Tatiana Foroud, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Anne Hall, Parkinson’s Foundation, NewYork, NY 10018, USA.
Karen S Marder, Neurology, Columbia University Irving Medical Center, New York, NY 10032, USA.
Ignacio F Mata, Genomic Medicine, Lerner Research Institute, Cleveland Clinic, Cleveland OH 44106, USA.
Niccolò E Mencacci, The Ken & Ruth Davee Department of Neurology, Northwestern University, Chicago, IL 60611, USA.
Martha A Nance, Struthers Parkinson’s Center, Golden Valley, MN 55427, USA.
Michael A Schwarzschild, Department of Neurology, Massachusetts General Hospital, Boston, MA 02114, USA.
Tanya Simuni, The Ken & Ruth Davee Department of Neurology, Northwestern University, Chicago, IL 60611, USA.
Susan Bressman, Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
Anne-Marie Wills, Department of Neurology, Massachusetts General Hospital, Boston, MA 02114, USA.
Hubert H Fernandez, Genomic Medicine, Lerner Research Institute, Cleveland Clinic, Cleveland OH 44106, USA.
Irene Litvan, Department of Neurosciences, University of California San Diego, San Diego, CA 92093, USA.
Kelly E Lyons, Department of Neurology, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Holly A Shill, The Muhammad Ali Parkinson’s Center, Barrow Neurological Institute, Phoenix, AZ 85013, USA.
Carlos Singer, Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Thomas F Tropea, Department of Neurology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Nora Vanegas Arroyave, Department of Neurology, Baylor College of Medicine, Houston, TX 77030, USA.
Janfreisy Carbonell, Centro Cardioneuro Oftalmológico y Trasplante, Santo Domingo 10306, República Dominicana.
Rossy Cruz Vicioso, Medicina Interna, Clínica Unión Médica del Norte, Santiago de los Caballeros 51000, República Dominicana.
Linn Katus, Neurology, Columbia University Irving Medical Center, New York, NY 10032, USA.
Joseph F Quinn, Brain Institute, Oregon Health & Sciences University, Portland, OR 97239, USA.
Priscila D Hodges, Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA.
Yan Meng, Fulgent Genetics, Temple City, CA 91780, USA.
Samuel P Strom, Illumina Inc., San Diego, CA 92122, USA.
Cornelis Blauwendraat, Laboratory of Neurogenetics, National Institute on Aging, National Institute of Health, Bethesda, MD 20892, USA.
Katja Lohmann, Institute of Neurogenetics, University of Lübeck, 23538 Lübeck, Germany.
Cynthia Casaceli, Clinical Trials Coordination Center, University of Rochester Medical Center, Rochester, NY 14627, USA.
Shilpa C Rao, Parkinson’s Foundation, NewYork, NY 10018, USA.
Kamalini Ghosh Galvelis, Parkinson’s Foundation, NewYork, NY 10018, USA.
Anna Naito, Parkinson’s Foundation, NewYork, NY 10018, USA.
James C Beck, Parkinson’s Foundation, NewYork, NY 10018, USA.
Roy N Alcalay, Parkinson’s Foundation, NewYork, NY 10018, USA; Neurology, Columbia University Irving Medical Center, New York, NY 10032, USA; Movement Disorders Division, Tel Aviv Sourasky Medical Center, Tel Aviv 6423906, Israel.
Data availability
Deidentified data will be made available to qualified researchers who submit and provide a valid research question. Enquiries should be directed to R.N.A.
Funding
Parkinson’s Foundation.
Competing interests
L.C. and J.S. received partial funding of salaries from the Michael J. Fox Foundation (MJFF) and the Parkinson’s Foundation; J.V. received partial funding of salaries from MJFF and the Parkinson’s Foundation and received travel funding for the Movement Disorders Society International Congress, Madrid, Spain, September 2022, abstract and poster presentation ‘PD GENEration Clinical Phase: Genetic Diagnostic Yield and Clinical Characteristics’ from the Parkinson’s Foundation and paid to Indiana University School of Medicine; T.F. received funding of grants from Parkinson’s Foundation for acting as a Steering Committee Member and received funding for grants/contracts from Parkinson’s Foundation; A.H. is a research advocate and received funding of grants from Parkinson’s Foundation for acting as a Steering Committee Member; K.S.M. received funding of grants from Parkinson’s Foundation for acting as a Steering Committee Member, receives funding of grants from NIH, MJFF, CHDI, HSG, HDSA, Prilenia, Novartis, Roche and Springer and receives consulting fees from Novartis and funding for leadership on the Enroll HD Oversight Committee (CHDI) and HSG steering committee; I.F.M. received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member and receives funding of grants/conference support from MJFF, ASAP, NIH and Cleveland Clinic Foundation; N.E.M. received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member, receives funding of grants from GP2 Monogenic Network, receives payment/honoraria for MDS lectures and received support for attending the MDS congress in Madrid, September 2022; M.A.N. received compensation for work as a Steering Committee member for studies conducted by Bial and Neurocrine, for consulting work from Roche, Novartis and Uniqure, grant funding from CHDI and HDSA and received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member, received payment for presentation at Parkinson’s Foundation and Parkinson Study Group conferences, received support for attending Parkinson Study Group conference and is the Chair of Genetics and environment Working Group for the Parkinson Study Group; M.A.S. received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member, receives funding of grants or conference support from NIH, MJFF, Farmer Family Foundation, Sergey Brin Family Foundation, American Parkinson’s Disease Association, Cure Parkinson’s, Parkinson’s Foundation, Harvard School of Public Health (NIH, DoD), GSK and GE Healthcare, receives consulting fees through the Parkinson Study Group for PSG advisory services [including to Bial, Biogen (LUMA/LIGHTHOUSE trials global SC), UCB (ORHCESTRA trial PSG SC)] and through Sutter Health (NIA), Northwestern University (NINDS), Cure Parkinson’s for Steering Committee services (for TOPAZ and SPARX3), International Linked Clinical Trial Committee (CP), participates on an Alzheimer’s drug trial Data Monitoring Committee, provides services for Eli Lilly & Co. and is Chair, Executive Committee for Parkinson Study Group; T.S. received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member, receives funding of grants from Amneal, Biogen, Roche, Neuroderm, Sanofi, Prevail and UCB, is an investigator for NINDS, MJFF, Parkinson’s Foundation studies, receives consulting fees from 4D Pharma, Acadia, AcureX, AskBio, Amneal, Blue Rock Therapeutics, Caraway Therapeutics, Critical Path for Parkinson's Consortium (CPP), Denali, MJFF, Neuroderm, Sanofi, Sinopia, Sunovion, Roche, Takeda, UCB, Vanqua Bio and Voyager and is on the advisory board for Acadia, AcureX, AskBio, Amneal, Denali, Sunovion and Roche and the Scientific Advisory Board for 4D Pharma, Neuroderm, Sanofi and UCB; A.M.W. received funding of grants from the Parkinson’s Foundation for acting as a Steering Committee Member and receives funding of grants from NIA/NIH, Roche/Genentech, Biogen and their institution, participates on Data Safety Monitoring Board or Advisory Board for Ono Pharmaceuticals and Amylyx Pharmaceuticals; S.B. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration; H.H.F. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration, receives funding of grants from Biogen, MJFF, NINDS and Roche, receives consulting fees for the Parkinson Study Group, Cerevel, Amneal, AbbVie, receives royalties as a book author from Springer Publishing, is Co-Chair of the PSG Executive Committee, is a member of the Scientific Advisory Board for CCXDP and receives payment as Editor-in-Chief of Parkinsonism and Related Disorders from Elsevier; I.L. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration, receives funding from the National Institutes of Health grants: 2R01AG038791-06A, U01NS100610, U01NS80818, R25NS098999; U19 AG063911-1 and 1R21NS114764-01A1, MJFF, the Parkinson Foundation, Lewy Body Association, CurePSP, Roche, Abbvie, Biogen, Centogene. EIP-Pharma, Biohaven Pharmaceuticals, Novartis and United Biopharma SRL and UCB and is on the Scientific Advisory Board for Amydis and on the Scientific Advisory Board of the Rossy PSP Program at the University of Toronto; H.A.S. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration, receives grants from UCB Pharma, Transposon Therapeutics, Jazz Pharmaceuticals, Barrow Neurological Foundation, MJFF and NINDS/NIH and receives consulting fees from Sage/Biogen, AbbVie and Parkinson Study Group/NQ; C.S. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration, receives grants from Pharma Two B, TEVA Pharmaceuticals, Amneal Pharmaceuticals, Revance Therapeutics and Sunovion Pharmaceuticals; T.F.T. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration and receives grants from MJFF and NIH; N.V.A. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration and receives grants from MJFF and NIH; R.C.V. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration; L.K. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration; J.F.Q. receives funding of a grant through institution for acting as a site principal investigator for PD GENEration, receives funding for acting as support to their institution as an LBDA RCOE and serves on DSMB for NIH and for Travere Pharmaceuticals; P.D.H. received partial funding of salary from the Parkinson’s Foundation and receives funding for acting on the PD GENE Latino Advisory Committee; Y.M. is an employee of Fulgent Genetics; S.P.S. received support for attending meetings from Illumina, Inc; S.C.R. is employed by the Parkinson’s Foundation; K.G.G. is employed by the Parkinson’s Foundation; A.N. is employed by the Parkinson’s Foundation; J.C.B. is employed by the Parkinson’s Foundation and receives funding for grants through their institution from NIH; R.N.A. received grants from the Parkinson’s Foundation for acting as a Steering Committee Member, receives funding from NIH, Department of Defense, MJFF and the Silverstein Foundation for GBA/Parkinson’s disease. He received consulting fees from Biogen, Biohaven, Capsida, Gain Therapeutics, Sanofi, Servier, Takeda and Vanqua bio. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Bloem BR, Okun MS, Klein C. Parkinson’s disease. Lancet. 2021;397:2284–2293. [DOI] [PubMed] [Google Scholar]
- 2. Lesage S, Brice A. Parkinson’s disease: From monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18(R1):R48–R59. [DOI] [PubMed] [Google Scholar]
- 3. Benitez BA, Davis AA, SC J, et al. . Resequencing analysis of five Mendelian genes and the top genes from genome-wide association studies in Parkinson’s disease. Mol Neurodegener. 2016;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim CY, Alcalay RN. Genetic forms of Parkinson’s disease. Semin Neurol. 2017;37:135–146. [DOI] [PubMed] [Google Scholar]
- 5. Alcalay RN, Kehoe C, Shorr E, et al. . Genetic testing for Parkinson disease: Current practice, knowledge, and attitudes among US and Canadian movement disorders specialists [published correction appears in genet med. 2019. Genet Med. 2020; 22:574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skrahina V, Gaber H, Vollstedt EJ, et al. . The Rostock international Parkinson's disease (ROPAD) study: Protocol and initial findings. Mov Disord. 2021;36:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hill EJ, Robak LA, Al-Ouran R, et al. . Genome sequencing in the Parkinson disease clinic. Neurol Genet. 2022;8:e200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan MMX, Malek N, Lawton MA, et al. . Genetic analysis of Mendelian mutations in a large UK population-based Parkinson's disease study. Brain. 2019;142:2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malek N, Weil RS, Bresner C, et al. . Features of GBA-associated Parkinson's disease at presentation in the UK tracking Parkinson's study. J Neurol Neurosurg Psychiatry. 2018;89:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Lau LML, Breteler MMB. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5:525–535. [DOI] [PubMed] [Google Scholar]
- 11. Maloney KA, Alaeddin DS, von Coelln R, et al. . Parkinson’s disease: Patients’ knowledge, attitudes, and interest in genetic counseling. J Genet Couns. 2018;27:1200–1209. [DOI] [PubMed] [Google Scholar]
- 12. Alcalay RN, Caccappolo E, Mejia-Santana H, et al. . Frequency of known mutations in early-onset Parkinson disease: Implication for genetic counseling: The consortium on risk for early onset Parkinson disease study. Arch Neurol. 2010;67:1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simpson C, Vinikoor-Imler L, Nassan FL, et al. . Prevalence of ten LRRK2 variants in Parkinson’s disease: A comprehensive review. Parkinsonism Relat Disord. 2022;98:103–113. [DOI] [PubMed] [Google Scholar]
- 14. Cook L, Schulze J, Verbrugge J, et al. . The commercial genetic testing landscape for Parkinson’s disease. Parkinsonism Relat Disord. 2021;92:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richards S, Mu W, Nusbaum R, Lincoln K, Solimine J. The genetic testing experience of individuals with Parkinson's disease. Mov Disord Clin Pract. 2023;10:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneider SA, Hizli B, Alcalay RN. Emerging targeted therapeutics for genetic subtypes of parkinsonism. Neurotherapeutics. 2020;17:1378–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook L, Verbrugge J, Schwantes-An TH, et al. . Providing genetic testing and genetic counseling for Parkinson's disease to the community. Genet Med. 2023;25:100907. [DOI] [PubMed] [Google Scholar]
- 19. Postuma RB, Berg D, Stern M, et al. . MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2015;30:1591–1601. [DOI] [PubMed] [Google Scholar]
- 20. Strande NT, Riggs ER, Buchanan AH, et al. . Evaluating the clinical validity of gene-disease associations: An evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bleyer AJ, Westemeyer M, Xie J, et al. . Genetic etiologies for chronic kidney disease revealed through next-generation renal gene panel. Am J Nephrol. 2022;53:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee CY, Yen HY, Zhong AW, Gao H. Resolving misalignment interference for NGS-based clinical diagnostics. Hum Genet. 2021;140:477–492. [DOI] [PubMed] [Google Scholar]
- 23. Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mallett V, Ross JP, Alcalay RN, et al. . GBA p.T369M substitution in Parkinson disease: Polymorphism or association? A meta-analysis. Neurol Genet. 2016;2:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duran R, Mencacci NE, Angeli AV, et al. . The glucocerobrosidase E326 K variant predisposes to Parkinson's disease, but does not cause Gaucher’s disease. Mov Disord. 2013;28:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lubbe SJ, Bustos BI, Hu J, et al. . Assessing the relationship between monoallelic PRKN mutations and Parkinson’s risk. Hum Mol Genet. 2021;30:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Di Luca DG, Macklin EA, Hodgeman K, et al. . Enrollment of participants from marginalized racial and ethnic groups. Neurol Clin Pract. 2023;13:e200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pal G, Mangone G, Hill EJ, et al. . Parkinson disease and subthalamic nucleus deep brain stimulation: Cognitive effects in GBA mutation carriers. Ann Neurol. 2022;91:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bandres-Ciga S, Faghri F, Majounie E, et al. NeuroBooster Array: A genome-wide genotyping platform to study neurological disorders across diverse populations. [Preprint] medRxiv. doi: 10.1101/2023.11.06.23298176 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified data will be made available to qualified researchers who submit and provide a valid research question. Enquiries should be directed to R.N.A.