Abstract
The nonstructural adeno-associated virus type 2 Rep proteins are known to control viral replication and thus provide the single-stranded DNA genomes required for packaging into preformed capsids. In addition, complexes between Rep proteins and capsids have previously been observed in the course of productive infections. Such complexes have been interpreted as genome-linked Rep molecules associated with the capsid upon successful DNA encapsidation. Here we demonstrate via coimmunoprecipitation, cosedimentation, and yeast two-hybrid analyses that the Rep-VP association also occurs in the absence of packageable genomes, suggesting that such complexes could be involved in the preparation of empty capsids for subsequent encapsidation steps. The Rep domain responsible for the observed Rep-VP interactions is situated within amino acids 322 to 482. In the presence of all Rep proteins, Rep52 and, to a lesser extent, Rep78 are most abundantly recovered with capsids, whereas Rep68 and Rep40 vary in association depending on their expression levels. Rep78 and Rep52 are bound to capsids to roughly the same extent as the minor capsid protein VP2. Complexes of Rep78 and Rep52 with capsids differ in their respective detergent stabilities, indicating that they result from different types of interactions. Rep-VP interaction studies suggest that Rep proteins become stably associated with the capsid during the assembly process. Rep-capsid complexes can reach even higher complexity through additional Rep-Rep interactions, which are particularly detergent labile. Coimmunoprecipitation and yeast two-hybrid data demonstrate the interaction of Rep78 with Rep68, of Rep68 with Rep52, and weak interactions of Rep40 with Rep52 and Rep78. We propose that the large complexes arising from these interactions represent intermediates in the DNA packaging pathway.
Adeno-associated virus type 2 (AAV-2) is a human parvovirus dependent on coinfection with a helper virus, such as adenovirus or herpesvirus, for efficient reproduction (for reviews, see references 2, 3, and 28). The 4.7-kb single-stranded DNA (ssDNA) genome is encapsidated in an icosahedral virion 20 to 24 nm in diameter. The genome consists of two open reading frames (ORFs) flanked by inverted terminal repeats (ITRs) of 145 nucleotides. The right ORF encodes the three capsid proteins VP1, VP2, and VP3, which are translated from alternative translation start sites and have molecular masses of 87, 72, and 62 kDa, respectively. The capsid proteins are present in the mature virion in a 1:1:10 ratio (4, 33). Capsid assembly occurs in the nucleus and requires only the expression of the capsid proteins (34, 45). In the absence of Rep proteins, capsids accumulate predominantly in the nuceoli (45). Encapsidation of the ssDNA takes place in the nucleoplasm. Immunofluorescence data indicate that the capsid proteins and the nonstructural Rep proteins colocalize in certain areas of the nucleus (18, 45) and that the Rep proteins are able to influence the subnuclear capsid distribution (45). Coimmunoprecipitation experiments have shown that Rep and capsid proteins can form complexes (31, 46). These complexes are at least partially stabilized by covalent linkage of Rep78 to single-stranded AAV-2 DNA in the virion (32), in analogy to findings for the autonomous parvovirus minute virus of mice (MVM) (8).
The left ORF encodes the four nonstructural Rep proteins, with molecular masses of 78, 68, 52, and 40 kDa, respectively. The two large Rep proteins are required for AAV-2 DNA replication (14, 40) and influence AAV-2 gene expression (1, 17, 24, 30, 41, 42). They have ATP-dependent helicase and endonuclease activities (19, 21). The two small Rep proteins strongly stimulate ssDNA accumulation (6), which may suggest a role in ssDNA encapsidation. In principle, Rep78 and Rep68 are sufficient for the production of infectious virions (15); however, coexpression of Rep52 and Rep40 increases the infectious titer by up to 1,000-fold. Recent experiments also revealed an ATP-dependent helicase activity associated with Rep52 (37).
The two ITRs of the AAV genome serve as origins of AAV DNA replication and are necessary and sufficient for packaging (26, 35). A specific packaging sequence within the ITR has so far not been identified. In vivo pulse-labeling experiments have shown that the number of empty particles decreases at the same rate as the number of DNA-containing mature virions increases over the course of a viral infection (29). Interpretation of these data led to the hypothesis that ssDNA is packaged into preformed empty capsids. This model is supported by findings relating to autonomous parvoviruses for which capsids with attached DNA as potential packaging intermediates have been visualized by electron microscopy (27). Recently, infectious AAV-2 has been produced in cell-free systems which might help to analyze the mechanism of DNA encapsidation in more detail (9, 47).
Encapsidation of the DNA is a crucial step in the production of wild-type and recombinant AAV-2. In both cases, the number of empty capsids exceeds the number of packaged infectious virions. The underlying mechanisms both of capsid assembly and DNA packaging are not yet understood. In particular, there is no explanation for the apparent specificity of the AAV-2 DNA packaging process. Here we describe complexes between Rep proteins and capsids as well as between different Rep proteins which provide a molecular basis for the specific association of AAV-2 DNA with preformed capsids prior to encapsidation. A model describing the formation of the initial encapsidation complexes is presented.
MATERIALS AND METHODS
Materials.
The polyclonal guinea pig Rep antisera αRep-S, αRep-M, and αRep-A against the Rep proteins derived from spliced mRNAs (Rep68 and Rep40) and against all Rep proteins, respectively, and the monoclonal antibodies (MAbs) 303.9 (directed against the Rep proteins) and B1 (directed against the capsid proteins) have been described previously (42, 46). A polyclonal guinea pig antiserum (αRep-US) specifically reacting with the Rep proteins derived from the nonspliced mRNAs (Rep78 and Rep52) was generated by immunization of guinea pigs with the peptide GKVPDACTACDLVNVDLDDCIFEQ according to conventional protocols (12). The rabbit polyclonal antisera 48, 51, and 87, reacting with the VP proteins, were prepared after immunization by using the VP1 capsid protein which was expressed in a baculovirus system and then gel purified.
The expression vectors pRep, including Rep78, Rep68, Rep52, Rep40, M225, M274, M324, Stop482, Stop515, and RepK340H are identical to the corresponding pKEXRep constructs previously described (17). The mutant Rep52ΔXS was generated via deletion of the XmnI-SalI region of Rep52. Through blunting of the SalI site and ligation to the blunt XmnI end, the original Rep reading frame was conserved. Constructs containing point mutations (E379K, E379Q, E391I, E391T, K404I, K404T, and P415H) in the Rep helicase domain have been previously described (25) and were kindly provided by N. Muzyczka. SalI-SwaI fragments were removed from these Rep/Cap expression constructs and inserted into a cytomegalovirus (CMV)-Rep52/40 expression construct (pRepM225). pDGΔVP was prepared by homologous recombination. For this, a 1.1-kb ApaI fragment which comprises nucleotides 2946 to 4049 of the VP ORF was deleted from pΔTR (42), resulting in plasmid pΔTRΔVP. The 1.8-kb XbaI/HindIII fragment from pΔTRΔVP was then cotransformed with pDG (11) linearized with SwaI into Escherichia coli BJ5183 for homologous recombination (5).
For quantitation of the sensitivity of MAbs 303.9 and B1, Rep68 and VP3 were expressed and purified as described earlier (16, 38).
Cell culture, virus infection, and plasmid transfection.
293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and 100 μg/ml of penicillin and streptomycin at 37°C in 5% CO2. Cells were coinfected with AAV-2 and adenovirus type 5 (Ad5) at 80% confluency. The medium was removed, and the cells were then incubated with AAV-2 (multiplicity of infection [MOI] of 20) and Ad5 (MOI of 2) in a total volume of 500 μl per 6-cm-diameter petri dish for 2 h. DMEM was added, and the cells were incubated at 37°C in 5% CO2 for 72 h.
Cells were transfected as described elsewhere (7). 293T cells at approximately 60% confluency were transfected with 10 μg of plasmid DNA per 6-cm petri dish.
Preparation of cell extracts.
The cells were harvested at 72 h postinfection or posttransfection and washed with 5 ml of phosphate-buffered saline (PBS) and then with 1 ml of PBS per dish. Cells from a 6-cm dish were resuspended in a final volume of 0.5 ml or in 1 ml of buffer A (10 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM EDTA) or RIPA buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). To both buffers the complete proteinase inhibitor cocktail (Roche, Mannheim, Germany) was added as suggested by the manufacturer. The cell suspensions in buffer A were sonicated twice for 15 s. All extracts were cleared by centrifugation at an average of 17,600 × g for 5 min (4°C).
Sucrose gradients.
Nuclear extracts of 293T cells transfected with pDG from 10 10-cm petri dishes were prepared as previously described (46). Samples of 0.5 ml were loaded onto 10-ml sucrose gradients (5 to 30% sucrose in TNEM buffer [10 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 2 mM MgCl2, 5 mM β-mercaptoethanol]) and centrifuged for 2 h at 160,000 × g at 4°C in a swing-out rotor. Fractions of 1 ml were collected from the bottoms of the tubes. Rep and VP concentrations were controlled by precipitation in trichloroacetic acid (final concentration of 20%) of 200 μl of the fractions, followed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blot analysis. The remaining 800 μl of the fractions was used for immunoprecipitation experiments.
Western blot analysis.
Protein samples were electrophoresed on 15% polyacrylamide gels in the presence of SDS (39) and transferred to nitrocellulose membranes using semidry blotting equipment. The Rep and capsid proteins were detected with MAbs and peroxidase-coupled secondary antibodies using the enhanced chemoluminescence (ECL) detection kit (Amersham, Braunschweig, Germany) according to standard methods (12).
Immunoprecipitation experiments.
Three hundred to 500 μl of cell extract or 800 μl of sucrose gradient fractions was incubated overnight at 4°C with either 3 μl of polyclonal guinea pig antiserum or polyclonal rabbit antiserum. After this incubation, the samples were cleared of nonspecific protein precipitates by centrifugation at 17,600 × g for 5 min. The immunocomplexes were precipitated via the addition of 30 μl of protein A-Sepharose (Amersham-Pharmacia, Freiburg, Germany) (10% [wt/vol] in NETN [20 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40]). After overnight incubation at 4°C, the immunocomplex-protein A-Sepharose beads were washed three times with 1 ml of NETN buffer in which guinea pig antisera had been used. Immunoprecipitations carried out using rabbit antisera in RIPA buffer or buffer A were washed three times with 1 ml of the respective lysis buffer instead of NETN. The samples were boiled in protein loading buffer and analyzed by SDS-PAGE and Western blotting.
Two-hybrid vectors and interaction assay.
The Rep two-hybrid constructs were cloned as follows. Plasmids pGBT9 and pGAD424 (Clontech Laboratories, Inc., Palo Alto, Calif.) encoding the GAL4 DNA-binding and the GAL4 transactivation domain, respectively, were cut with SalI and PstI and ligated to a SalI/PstI fragment from HIV-LTR-OVEC (16) containing the EcoRV site derived from pBluescript SK2 adjacent to the PstI site. The resulting plasmids were digested with SalI and partially with EcoRV to excise only the HIV-LTR-derived inserts. The vector fragments were ligated to Rep78- and Rep52-encoding XhoI/SmaI fragments from pRep78 and pRep52, respectively, to obtain pGBT9-Rep78, pGBT-Rep52, pGAD424-Rep78, and pGAD424-Rep52. To generate the Rep68- and Rep40-encoding two-hybrid constructs, the NotI/XbaI-Rep78 fragments of pGBT9-Rep78 and pGAD424-Rep78, respectively, were replaced by the corresponding fragments from pRep68 and pRep40. The vector for the expression of the AAV-2 VP proteins in fusion with the yeast GAL4 activation domain (pGAD-VP) was generated by inserting the DraI/SnaBI fragment of pTAV2-0 into the blunted BamHI site of pGAD424. The corresponding pGBT-VP construct containing the AAV-2 cap gene fused to the gene for the yeast GAL4-binding domain was prepared by ligating the VP-containing EcoRI/SalI fragment of pGAD-VP into pGBT9 that had been digested with EcoRI and SalI.
The constructs were transformed according to a basic lithium acetate protocol (10) in every possible pairwise combination into Saccharomyces cerevisiae PJ69-4 (22), which contains three reporter genes, ADE2, HIS3, and lacZ, under the control of three independent promoters, Gal2, Gal1, and Gal7. The resulting 36 double transformants were selected on synthetic complete medium lacking leucine and tryptophan.
For the in vivo assays, fresh transformants were streaked onto tryptophan- and leucine-deficient medium lacking adenine or histidine or supplemented with a final concentration of 100 mM sodium phosphate (pH 7.0) and 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml, respectively, and incubated for 72 h at 30°C. Cotransformants displaying a positive reaction on at least one of the selective media were subjected to a liquid culture assay by quantitative colorimetric measurement of β-galactosidase activity using chlorophenol red β-d-galactopyranoside (CPRG) (36). The liquid culture assay was performed at least twice with three independent cotransformants and triple measurements.
RESULTS
Complex formation between Rep and VP proteins in the absence of packageable DNA.
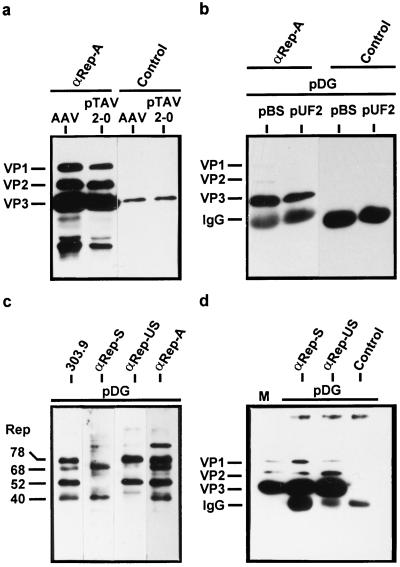
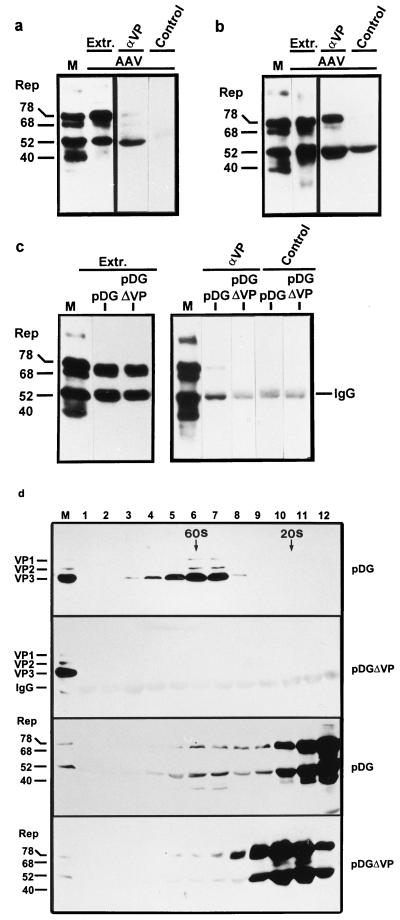
In the course of a productive AAV-2 infection, complexes between Rep and capsid proteins (VP proteins) can be demonstrated by coimmunoprecipitation with antibodies against the Rep proteins (31, 46). These complexes have been interpreted as being the result of ssDNA packaging in which Rep is covalently linked to the viral DNA and thereby indirectly associated with the capsid (31, 32). To determine whether the association of Rep proteins with capsid proteins or capsids requires AAV-2 DNA, Rep-VP complex formation was studied following transfection of Rep and VP protein expression constructs in the presence and absence of replicatable and packageable DNA. 293T cells were cotransfected either with the AAV-2 vector plasmid pUF2 (Fig. 1) (48) and the helper plasmid pDG (Fig. 1) (11), which provides AAV-2 and all adenovirus helper functions, or with pBluescript SK+ (pBS), a plasmid which can neither be replicated nor packaged into AAV-2 capsids, and the pDG helper plasmid. As a positive control, 293T cells were infected with AAV-2 (MOI, 20) and Ad5 (MOI, 2) or transfected with pTAV2-0 (Fig. 1), an infectious AAV-2 clone, and infected with Ad5 (MOI, 2). The Rep proteins were immunoprecipitated from extracts of cells harvested 72 h postinfection or posttransfection with polyclonal guinea pig antisera recognizing all four Rep proteins (αRep-A or αRep-M). Coprecipitated capsid proteins were detected by SDS-PAGE and Western blot analysis by using the monoclonal antibody B1 (45). It is evident from Fig. 2a that capsid proteins can be coprecipitated with the Rep proteins to the same extent following transfection of the pTAV2-0 infectious clone as after infection. In addition, coimmunoprecipitation of VP proteins occurred not only in the presence of replication- and packaging-competent DNA (pUF2) but also in the presence of replication- and packaging-deficient DNA (pBS) (Fig. 2b). Control precipitations with an unrelated guinea pig antiserum showed no precipitation of capsid proteins. To further validate these results, coimmunoprecipitations using two additional polyclonal antisera were performed from extracts of cells transfected with pDG alone. The specificities of the various guinea pig antisera were demonstrated by Western blot analysis (Fig. 2c). Whereas the αRep-S antiserum reacts only with Rep40 and Rep68 (see also reference 42), αRep-US specifically detects Rep78 and Rep52 in Western blot analysis. Both αRep-S and αRep-US coprecipitated the capsid proteins, indicating that Rep proteins derived from spliced and unspliced mRNA form complexes with the VP proteins (Fig. 2d). In addition, this result confirms that packageable DNA is not required for Rep-VP complex formation.
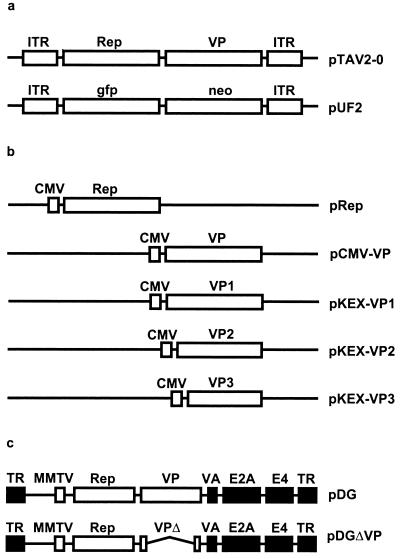
FIG. 1.
Diagrams of (a) packageable AAV plasmids, (b) expression constructs containing the rep and cap genes under control of the CMV immediate-early promoter, and (c) helper plasmids providing AAV-2 and all adenovirus helper functions. (a) pTAV2-0 (13) contains the complete AAV-2 genome coding for the Rep proteins (Rep) and the capsid proteins (VP); pUF2 (48) is a recombinant AAV-2 plasmid and contains the gene for a green fluorescent protein (gfp) under control of the CMV promoter and the neomycin-resistance gene (neo) under control of the herpes simplex virus thymidine kinase promoter. (b) pRep contains the respective rep genes shown in Fig. 3 and described in Materials and Methods. pCMV-VP (45) contains the complete VP gene and allows the expression of all three capsid proteins in the correct stoichiometry, whereas pKEX-VP1, pKEX-VP2, and pKEX-VP3 (34) harbor point mutations allowing expression of the individual VP proteins. (c) pDG (11) contains the Rep and VP expression cassette of AAV-2. The p5 promoter was exchanged for the mouse mammary tumor virus (MMTV) promoter. The adenovirus genes necessary for AAV production are indicated by black boxes. TR, Ad5 terminal repeat; VA, virus-associated genes I and II; E2A and E4, early region 2A and 4 of Ad5. pDGΔVP contains a deletion within the coding region for the capsid proteins (VPΔ). Both pDG and pDGΔVP lack the AAV-2 ITRs.
FIG. 2.
Coimmunoprecipitation of VP proteins with anti-Rep antisera in the presence and absence of AAV-2 DNA packaging. Extracts of 293T cells were prepared as described in Materials and Methods using RIPA buffer after coinfection with AAV-2 (AAV) and Ad5 or after transfection with pTAV2-0 and infection with Ad5 (a), after cotransfection with pDG and pBluescript SK+ (pBS) or pUF2, respectively (b), or after transfection with pDG (c and d). Immunoprecipitation was accomplished by incubating the extracts with a guinea pig anti-Rep polyclonal antiserum (αRep-A) or an unrelated guinea pig polyclonal antiserum (Control) (a and b) or guinea pig polyclonal antisera which recognize Rep78 and Rep52 (αRep-US) or Rep68 and Rep40 (αRep-S), respectively (d). The immunoprecipitates were analyzed by Western blotting using the anti-VP MAb B1 (46) and ECL detection. IgG, position of the IgG heavy chain fraction precipitated with protein A-Sepharose and detected by the peroxidase-coupled anti-mouse secondary antibody. M, marker extract derived from HeLa cells coinfected with AAV-2 and Ad5. (c) Western blot analysis with the different Rep antisera used for immunoprecipitation compared to MAb 303.9 (46).
Mapping of the VP interaction domain of the Rep proteins.
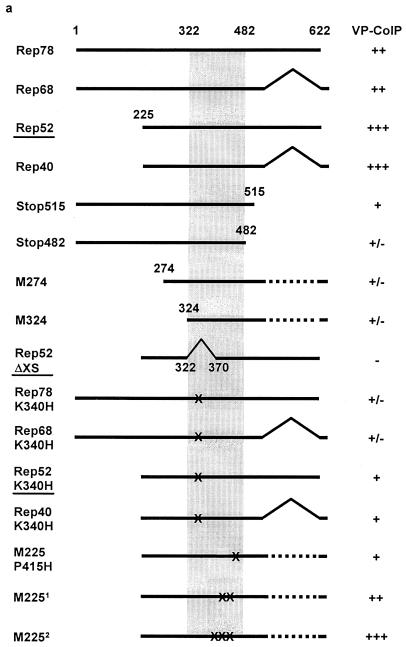
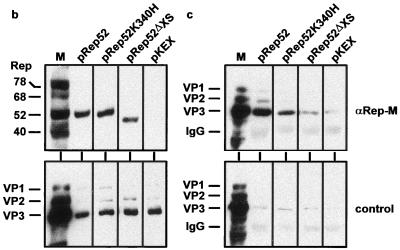
The ability of individual Rep proteins and Rep protein mutants to form complexes with VP proteins was tested by cotransfecting increasing amounts of the pCMV-VP expression construct (2, 4, or 8 μg) (Fig. 1) with fixed amounts of various Rep expression constructs (2 μg) (Fig. 1). The Rep ORF depicted in Fig. 1b is representative of the individual Rep proteins and Rep protein mutants shown in Fig. 3a. Rep and VP protein expression was controlled by Western analysis, and Rep-VP complex formation was analyzed by immunoprecipitation with αRep-A or αRep-M antiserum and Western blot analysis as described above. Figures 3b and c show examples of strong, weak, and noninteracting Rep proteins. All four Rep proteins showed a detectable interaction with VP proteins in this assay. Larger amounts of VP proteins were coprecipitated using the small Rep proteins than with the large Rep proteins. C- and N-terminal deletions as well as point mutations in the ATP-binding site of the Rep proteins and in proline 415 further reduced the interaction with the capsid proteins. An internal deletion between amino acids 322 and 370 (Rep52ΔXS) showed no detectable VP coprecipitation (Fig. 3b and c). This mutant turned out to be less soluble within the cell and therefore could only be tested at a reduced expression level. We concluded from these results that each of the Rep proteins is able to form complexes with the capsid proteins and that the most important interaction site(s) is contained within the domain spanning amino acids 322 to 482.
FIG. 3.
Mapping of the Rep protein domain critical for Rep-VP interactions. Extracts of 293T cells transfected with a fixed amount of either Rep expression plasmid (indicated in panel a) (2 μg/6-cm petri dish) and increasing concentrations of pCMV-VP (2, 4, or 8 μg/6-cm petri dish) were prepared in RIPA buffer and subjected to immunoprecipitation with the αRep-A or αRep-M polyclonal antiserum or an unrelated guinea pig antiserum (control) as described in Materials and Methods. Controls also included immunoprecipitation from cell extracts after cotransfection of the empty Rep expression vector (pKEX) and increasing amounts of pCMV-VP with the αRep antiserum or with the unrelated guinea pig antiserum. Detection of the VP proteins was accomplished as described in the legend to Fig. 2. For comparability, expression of the Rep and VP proteins was controlled in the extracts before immunoprecipitation. (a) Compilation of VP proteins coprecipitated with a number of Rep mutants. The amount of coprecipitated capsid proteins is given on a semiquantitative scale between +++ and −. (b) Rep and VP expression controls of the examples underlined in panel a. (c) The corresponding capsid proteins coprecipitated with the αRep-M serum or the control serum. M2251, constructs expressing small Rep proteins with mutations at K391T or K404T; M2252, small Rep proteins with mutations at E379K, E379Q, K391I, or K404I.
Characterization of Rep-VP complexes.
Since Rep-VP complex formation was not restricted to a subset of the four Rep proteins, the Rep content of the Rep-VP complexes was analyzed. Extracts of 293T cells that had been infected with AAV-2 (MOI, 20) and Ad5 (MOI, 2) were immunoprecipitated in the presence of RIPA buffer by using either a polyclonal VP antiserum (αVP#87) or the preimmune serum. The precipitates thus obtained were analyzed using a MAb (303.9) that reacts with all four Rep proteins. It becomes obvious from Fig. 4a that Rep52 was overrepresented in the precipitate, whereas Rep78 and Rep68 were present in lower amounts, although comparison with total Rep protein concentration present in the cell extract (Fig. 4a, extract lanes) shows that Rep78 and Rep68 were abundantly expressed. Rep40 was generally not detected in the immunoprecipitates except in low amounts in experiments in which Rep40 was more highly expressed. The same result was obtained from immunoprecipitations using two other anti-VP polyclonal rabbit antisera (#48 and #51) or after transfection of 293T cells with pTAV2-0 and overinfection with Ad5 (MOI, 5) (data not shown). No Rep proteins were detected following immunoprecipitation with the preimmune serum (control lanes). Immunoprecipitations shown so far were performed using RIPA cell extracts that contain considerable amounts of detergents. In order to analyze the composition of Rep-VP complexes under less stringent conditions, cell extracts prepared using detergent-free buffer A were used for immunoprecipitation with the VP antiserum (αVP#87). Figure 4b shows that under these conditions, considerably more Rep78 was recovered in the immunoprecipitate (lane αVP) than in the precipitation in RIPA buffer compared to the expression level of the Rep proteins in the cell extract (extract lane). However, such precipitations always showed a higher background with the preimmune serum (control lane).
FIG. 4.
Rep content of Rep-VP complexes in the presence and absence of packageable DNA and in the presence and absence of detergents. Extracts were prepared from 293T cells after coinfection with AAV-2 and Ad5 (a and b), after transfection of pDG or pDGΔVP (5 μg/6-cm petri dish) (c), or after transfection of pDG or pDGΔVP (15 μg/10-cm petri dish, 10 petri dishes each) (d). They were subjected to immunoprecipitation with a VP antiserum (VP#87; αVP) in RIPA buffer (a and c) or in buffer A (b). The precipitations in panel d were performed in buffer A using a Rep antiserum (αRep-M [44]) after fractionation of nuclear extracts by sucrose gradient centrifugation. The immunoprecipitates were analyzed by SDS-PAGE and Western blotting using the 303.9 anti-Rep MAb or the B1 anti-VP antibody. Five microliters of the extract was directly immunoblotted to determine the relative concentration of the Rep proteins present in the extract (Extr. in panels a, b, and c). M, marker extract derived from HeLa cells coinfected with AAV-2 and Ad5. IgG, position of the rabbit IgG immunoprecipitated with protein A-Sepharose cross-reacting with the peroxidase-coupled anti-mouse secondary antibody. Control, immunoprecipitations with the preimmune serum.
The Rep content of Rep-VP complexes in the absence of packageable DNA was analyzed by immunoprecipitation of the VP proteins in RIPA buffer from extracts of 293T cells transfected either with pDG or pDGΔVP. The pattern of Rep coprecipitation was similar to that obtained following infection of 293T cells with AAV-2 and Ad5 (Fig. 4c, αVP). Rep52 was present in higher amounts than Rep78, although the relative amount of Rep78 in the cell extract was almost equal to that of Rep52 (compare extract and αVP lanes). Rep68 and Rep40 were neither detected in the extract nor in the immunoprecipitate. In the immunoprecipitate obtained from 293T cells transfected with pDGΔVP (Fig. 1) in which the cap gene was deleted, no Rep proteins could be detected (Fig. 4c), demonstrating the specificity of the immunoprecipitates. In addition, control precipitations with the preimmune serum did not coprecipitate significant amounts of Rep proteins. The broad faint band at the position of Rep52 represents the immunoglobulin G (IgG) heavy chain fraction of the antiserum weakly detected by the anti-mouse secondary antibody used in the Western blot analysis.
The composition of the Rep-VP complexes formed in the absence of packageable AAV-2 DNA was also analyzed by cosedimentation of Rep proteins with capsids at 60S. In these experiments, nuclear extracts of cells transfected with pDG or pDGΔVP were fractionated by sucrose gradient centrifugation and subjected to Western blotting using VP antibodies (B1) or Rep antibodies (303.9). In some experiments, the Rep and VP proteins were concentrated by immunoprecipitation with anti-Rep antibodies prior to Western blot analysis (Fig. 4d); however, similar results were obtained without this concentration step (data not shown). It is evident that Rep78 and Rep52 show a peak in the capsid-containing fractions around 60S which does not appear in the absence of capsids (Fig. 4d, lanes pDGΔVP). This result was obtained in a detergent-free buffer system. It also shows that the DNA-free complexes contain predominantly Rep78 and Rep52 (at slightly higher levels). The repeatedly observed protein band that migrates slightly below Rep40 may represent a modified form of Rep40 or a degradation product of one of the four Rep proteins. The quantitation of the Rep and VP proteins in the 60S fractions from two representative sucrose gradients suggests that the Rep proteins are present in the 60S complexes in amounts comparable to or slightly higher than VP2 (Table 1).
TABLE 1.
Estimated Rep/VP ratios in 60S Rep-capsid complexesa
| Gradientb | Fraction | Rep52-VP2 | Rep78-VP2 |
|---|---|---|---|
| A | 6/7 | 3.9 | 1.4 |
| B | 7/8 | 2.6 | 0.7 |
Western blot analysis of the Rep and VP proteins in each sample chosen was done in parallel using MAbs 303.9 and B1, respectively. Exposure times were identical. Small amounts of Rep proteins detected in the gradients without VP proteins were subtracted. Both MAbs have comparable sensitivities as determined by Western blot analysis of serial dilutions of purified Rep68 and VP3 (data not shown). Ratios were estimated using the program Image Quant (Molecular Dynamics).
Rep and VP proteins in gradient A were concentrated by immunoprecipitation with an anti-Rep antiserum (αRep-A). Rep and VP proteins in gradient B were not concentrated.
Association of Rep proteins with free and assembled capsid proteins.
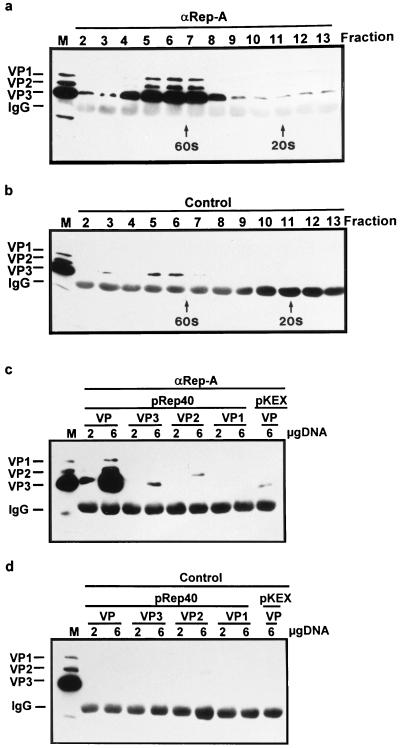
In order to determine whether the Rep-VP interaction occurs prior or subsequent to capsid assembly, the interactions of Rep proteins with nonassembled capsid proteins were analyzed. Nuclear extracts of pDG-transfected 293T cells were subjected to sucrose gradient centrifugation, and the resulting fractions were used in coimmunoprecipitation experiments. Using the αRep-A antiserum, capsid proteins were predominantly coprecipitated from fractions containing the fully assembled capsids, around 60S (Fig. 5a). However, some capsid proteins were also specifically coprecipitated in fractions corresponding to S values of less than 20S, suggesting that Rep proteins are not only associated with the assembled capsids but also with nonassembled VP proteins. To confirm the association of Rep proteins with free capsid proteins, coimmunoprecipitation experiments were performed via cotransfection of 293T with the various Rep expression constructs (pRep40, pRep52, pRep68, or pRep78) and constructs from which the individual capsid proteins VP1, VP2, or VP3 (Fig. 1, pKEX-VP1, pKEX-VP2, and pKEX-VP3) are expressed (34), but they show very poor or, in the case of VP3, no capsid assembly (38). To monitor Rep association with capsids, the three capsid proteins were coexpressed from plasmid pCMV-VP (45). Using the αRep-A antiserum, only small amounts of the separately expressed VP3 or VP2 and no VP1 could be detected, whereas the assembled capsids (lane VP) in the control experiment were abundantly recovered (Fig. 5c and d; only the experiment with pRep40 is shown; similar results were obtained for the other Rep proteins). These results demonstrate that VP3 and probably also VP2 are able to interact with the Rep proteins also in their nonassembled states. The specificity of these precipitations is shown by the two controls (Fig. 5c, lane pKEX/VP, and 5d). The relatively weak VP signals obtained following coprecipitation of individual VP proteins with the Rep proteins can be explained by the small amount of VP molecules present in these complexes, compared to complexes involving capsids. Finally, data from two-hybrid interaction assays also suggest that Rep78 and Rep52 can interact with nonassembled VP proteins (see Fig. 8D and E). Such interactions were not observed in this assay for the other Rep proteins.
FIG. 5.
Association of Rep proteins with free and assembled capsid proteins. (a and b) Sucrose gradient fractions obtained from nuclear extracts of 293T cells transfected with pDG (10 10-cm petri dishes; 15 μg of pDG each) were subjected to immunoprecipitation with the αRep-A or control polyclonal antisera in RIPA buffer as described in Materials and Methods. The positions of 60S and 20S were determined by using empty AAV-2 capsids (60S) and α-macroglobulin (20S) in parallel gradients. (c and d) 293T cells were cotransfected with the expression constructs pRep40 (4 μg/6-cm petri dish) and pCMV-VP (VP), pKEX-VP3 (VP3), pKEX-VP2 (VP2), and pKEX-VP1 (VP1) (2 or 6 μg/6-cm petri dish) or with the empty vector pKEX (4 μg/6-cm petri dish) and pCMV-VP (VP) (6 μg/6-cm petri dish). Whole cell extracts were immunoprecipitated with the αRep-A and control antisera in RIPA buffer as described in Materials and Methods. VP proteins were detected by Western blotting using MAb B1 and ECL. M, marker extract derived from HeLa cells coinfected with AAV-2 and Ad5. IgG, position of guinea pig IgG immunoprecipitated with protein A-Sepharose cross-reacting with the peroxidase-coupled anti-mouse secondary antibody.
FIG. 8.
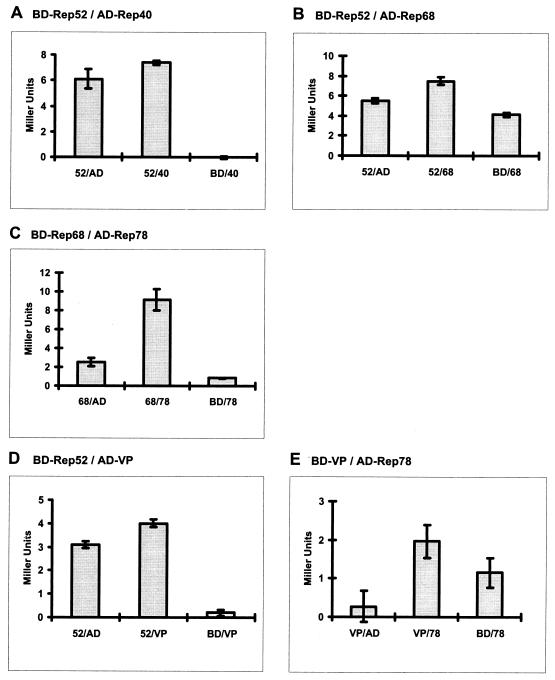
Rep-Rep and Rep-VP interactions detected in the yeast two-hybrid system. The Rep and VP proteins were fused to the GAL4 activation domain and GAL4-binding domain, respectively, as indicated. The pairs of fusion proteins were coexpressed in S. cerevisiae PJ69-4 and tested for interaction in a liquid culture assay. A positive interaction resulted in a higher expression of β-galactosidase. Enzyme activity (shown in Miller units) was measured as a function of chlorophenol red cleaved from CPRG (chlorophenol-red-β-d-galactopyranoside). For each protein pair, three independent cotransformants were measured in triplicate. Standard deviations based on differences between the three cotransformants are indicated.
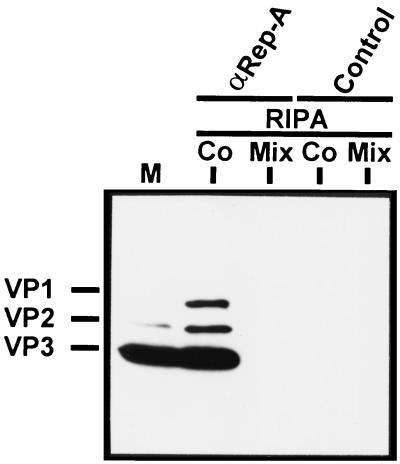
From the data so far obtained, Rep-capsid complexes could either be formed by the association of Rep proteins with assembled capsids or with VP proteins during the assembly process. To distinguish between the two mechanisms, we compared Rep-capsid complexes obtained by coexpression of Rep and capsid proteins with complexes obtained upon separate expression and coincubation of the extracts in vitro. Very little capsid assembly occurs in vitro (38), so that any resulting interactions of Rep proteins with capsids or VP proteins can be said to be assembly independent. Whereas coexpression of Rep40 with the VP proteins led to the formation of a stable complex, no VP proteins were coprecipitated with Rep40 when the Rep and VP proteins were separately expressed and then mixed in vitro (Fig. 6). Similar results were obtained using Rep52 (data not shown). This result also emphasizes that the abundance of the capsid proteins in the cell extract does not lead to a nonspecific coprecipitation with the Rep proteins.
FIG. 6.
Immunoprecipitation of Rep-VP complexes after Rep-VP coexpression or separate expression of Rep and VP proteins and subsequent mixing of extracts. Extracts from 293T cells cotransfected (Co) or separately transfected with pRep40 and pCMV-VP were prepared in RIPA buffer. The extracts of the separately transfected cells were combined (Mix), and all extracts (with equal capsid and Rep protein concentrations) were subjected to immunoprecipitation with the αRep-A and control antisera. The precipitated proteins were analyzed by Western blotting using the antibody B1 for detection of the VP proteins as described in the legend to Fig. 2. M, marker extract derived from HeLa cells coinfected with AAV-2 and Ad5.
Taken together, these data suggest that complex formation involving the VP and Rep proteins occurs during capsid assembly and results in the formation of stable Rep-capsid complexes that cannot be formed by mixing cellular extracts in vitro.
Rep-Rep complex formation.
Since all Rep proteins are able to form complexes with VP proteins, we were interested in determining whether Rep-Rep protein interactions could contribute to a capsid-associated Rep protein complex possibly involved in AAV-2 DNA packaging. Rep-Rep protein interactions were investigated by coimmunoprecipitation and two-hybrid analyses.
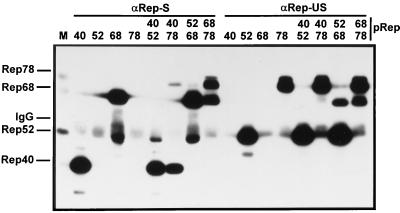
The individual Rep proteins were either expressed separately or in combination via cotransfection and immunoprecipitated using αRep-S or αRep-US polyclonal antisera. Extracts were prepared in buffer A, because the detergents present in RIPA buffer destabilize Rep-Rep interactions. The immunoprecipitated proteins were visualized by Western blot analysis. Small amounts of Rep52 and Rep78 could be coimmunoprecipitated with Rep40 (Fig. 7). With Rep68, however, Rep78 was coprecipitated in a 1:1 ratio. An interaction between Rep68 and Rep52 was difficult to detect using αRep-S, since a polypeptide of about the same molecular weight as Rep52, which also reacted with the 303.9 antibody, was precipitated after expression of Rep68 alone (Fig. 7, αRep-S, lane 68). This protein most likely represents a degradation product of Rep68 that was still recognized by the αRep-S serum. Immunoprecipitation using the αRep-US antiserum, however, showed a clear coprecipitation of Rep68 with Rep52 as well as with Rep78, confirming and specifying a previously observed interaction of Rep52 with one of the large Rep proteins (30). In contrast to the experiment with αRep-S, the weak interactions of Rep40 with Rep52 and Rep78 could not be detected.
FIG. 7.
Rep-Rep interactions in the presence and absence of DNA replication. Extracts from 293T cells that had been transfected with pRep40, pRep52, pRep68, or pRep78 or cotransfected with the different Rep expression vectors in various combinations, as indicated, were prepared and subjected to immunoprecipitation in buffer A with αRep-S, αRep-US, or control antiserum as described in Materials and Methods. Precipitated Rep proteins were detected by Western blotting using MAb 303.9. M, marker extract derived from HeLa cells coinfected with AAV-2 and Ad5. IgG, position of guinea pig IgG immunoprecipitated with protein A-Sepharose cross-reacting with the peroxidase-coupled anti-mouse secondary antibody.
In the two-hybrid system, the Rep as well as the VP proteins were fused to the GAL4 activation (pGAD424) or binding (pGBT9) domains, respectively and cotransformed into the yeast strain PJ69-4 in every possible pairwise combination. The cotransformants were analyzed for the expression of three reporter genes under the control of the GAL4 activatable promoters Gal1, Gal2, and Gal7, respectively. Cotransformants able to activate the least stringent promoter (Gal7) were subjected to a quantitative β-galactosidase liquid culture assay. The β-galactosidase activities are indicated as Miller units (Fig. 8). Rep52 and Rep78 showed a background activation of all three promoters when fused to the GAL4 DNA-binding domain. Based on these assays, clear interactions could be demonstrated for Rep52 and Rep68 as well as for Rep68 and Rep78 fused to either of the GAL4 domains (Fig. 8B and C), thus supporting the results from the coimmunoprecipitation experiments. Rep52 and Rep40 showed a slight activation of the Gal7 promoter only when Rep40 was fused to the GAL4 activation domain (Fig. 8A).
Taken together, these data demonstrate binding of Rep52 to Rep68 as well as Rep68 to Rep78 and also indicate that Rep40 can weakly bind both Rep52 and Rep78.
DISCUSSION
In this report we have described the protein-protein interactions of AAV-2-encoded polypeptides which result in complexes between Rep proteins and AAV-2 capsids. The same type of complex was formed regardless of whether packageable DNA was present, suggesting that it is not formed as the result of DNA packaging but rather constitutes an intermediate required for encapsidation.
Several reports have previously described complexes of Rep proteins with AAV-2 capsids (23, 31, 32, 46). However, they have been primarily interpreted based on the MVMp paradigm described by Cotmore and Tattersall (8), as a covalent linkage of Rep78 or Rep68 to the 5′ terminus of the genome formed during the terminal resolution reaction and preserved at the outside of the capsid after DNA packaging has been completed (31, 32). A functional role for this type of complex has been discussed and linked to the nuclear uptake of parvovirus genomes during an infection or to the release of assembled virus from the cell (8, 31, 32). Our interpretation suggests a role for Rep-VP complexes in the DNA packaging process, in particular at the initiation step during which the viral DNA must interact with the parvovirus capsid. A role in DNA packaging has also been favored by Cotmore and Tattersall (8); however, such a role would involve a DNA-independent protein-protein interaction between structural and nonstructural proteins, which the authors did not observe. Here, we have presented several lines of evidence demonstrating that such protein-protein interactions do indeed occur and that they result in capsids displaying Rep proteins on the outside of the viral shell. We have demonstrated this by coimmunoprecipitating capsids with four different anti-Rep antisera. Similarly, Rep proteins could be coprecipitated using three different anti-VP antisera but only when capsid proteins were coexpressed. We have also isolated Rep proteins by immunoaffinity chromatography on a matrix bound by MAb A20, which recognizes assembled capsids (data not shown). In addition, the small Rep proteins, which do not show sequence-specific DNA binding (21), can form complexes with VP, further suggesting that DNA is not involved in the initial complex formation. Analysis of Rep mutants showed that sequences near or within the ATP-binding domain are required for this interaction. Several Rep mutants showed strongly reduced or no interaction with capsids, emphasizing the specificity of the Rep-capsid associations. Finally, we could also detect a weak interaction of Rep78 and Rep52 with VP proteins by using the yeast two-hybrid system. A DNase-resistant complex of Rep proteins with recombinant and wild-type AAV-2 virions has also been observed by Kube et al. (23), who suggested a relationship between the AAV-2-mediated influence on cell proliferation and the capsid-associated Rep proteins. The authors observed that the association of Rep proteins with the capsid surface is sensitive to repeated CsCl centrifugation, i.e., to high salt concentrations, which is in line with our own experience (data not shown). Additionally, an indirect piece of evidence supporting a DNA-independent interaction of Rep proteins with AAV-2 capsids is suggested by the strong influence that the Rep proteins have on the intranuclear localization of assembled empty capsids in HeLa cells (45).
At this stage the mechanism by which protein-protein interactions can result in stable Rep-virion complexes is still unclear. Rep-VP interactions can occur prior to assembly of the capsid proteins into capsids. This is demonstrated by coimmunoprecipitation of VP proteins with anti-Rep antibodies from sucrose gradient fractions sedimenting below 20S, by coimmunoprecipitation of individually expressed capsid proteins which have a strongly reduced capsid forming capacity (38), and by the two-hybrid system in which the capsid proteins are fused to protein domains and are therefore prevented from forming capsids. That this interaction with nonassembled VP proteins plays an important role in Rep-capsid complex formation is demonstrated by the fact that coexpression of Rep and VP proteins leads to a much more efficient complex formation than the mere association of Rep proteins with capsids in vitro. Complex formation in vitro may be less efficient for several reasons. The actual protein concentrations in the nucleus and the availability of chaperones would certainly be different during coexpression in vivo compared to coincubation in vitro, although the amounts of Rep and VP proteins in the extracts were the same. Nonetheless, the strikingly higher stability of complexes formed after coexpression, compared to that after coincubation, strongly suggests that protein-protein interactions during the assembly process are important for the formation of this type of Rep-capsid complex.
The composition of the Rep-capsid complexes, as determined by immunoprecipitation with different VP antibodies in the presence and absence of AAV-2 DNA, suggests an abundance of Rep52, slightly less Rep78, and a small amount of Rep68 associated with the capsid. In some experiments, we could also detect traces of Rep40. Since all Rep proteins are able to form such complexes when individually expressed with capsid proteins, one would expect that they are represented in the capsids according to their expression levels. Rep78 is normally expressed at equal or higher levels than Rep52 in coinfection experiments, yet the capsids precipitated in RIPA buffer always showed a higher content of Rep52 than of Rep78. That these ratios are not immunoprecipitation artefacts is supported by the quantitation of Rep and VP proteins in sucrose gradient fractions without any precipitation (Table 1). The determined ratios suggest a Rep78/VP2 ratio of about 1 and a slightly higher ratio of 2 to 4 for Rep52/VP2. However, an exact determination of the Rep/VP stoichiometry in these complexes would require a purification of the complexes. Precipitations performed in buffer A (without detergents) gave much higher recoveries of Rep78, suggesting a difference in the binding stabilities of Rep78 and Rep52 to the capsid. Since Rep-Rep interactions are unstable in RIPA buffer, the additional amounts of Rep78 (and Rep68) recovered by immunoprecipitation in buffer A could thus result from Rep-Rep interactions in addition to the detergent-stable complexes of Rep52, Rep78, and Rep68 with the capsid. The low levels of Rep40 in the Rep-capsid complexes is difficult to interpret, especially as capsids were very efficiently coprecipitated with Rep antisera when Rep40 was overexpressed together with the VP proteins. In infection experiments and after transfection of pDG, however, expression levels of Rep40 were variable, but altogether low, making it difficult to judge whether the low recovery in these experiments was due to the low protein concentration or rather to a lower stability of Rep40 in the complex. In several cosedimentation experiments, a polypeptide migrating slightly faster than Rep40 was observed at 60S. It remains to be determined whether this protein represents a modified form of Rep40 or a degradation product of one of the Rep proteins.
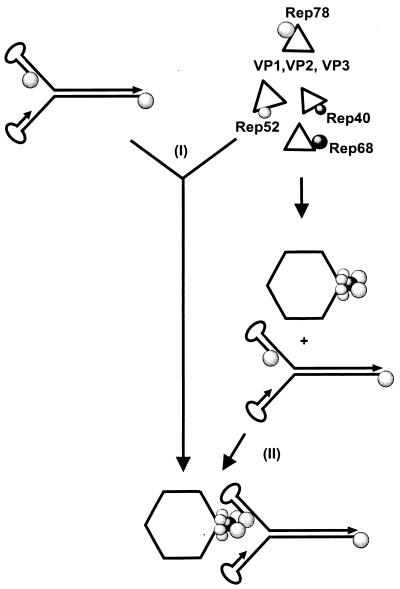
A problem that all viruses must solve is how to specifically package their genome. Current concepts for the solution of this problem postulate a specific interaction of a part of the genome, i.e., the packaging sequence, with components of the packaging apparatus and capsid. For the autonomous parvoviruses, a specific interaction of the 3′ hairpin structure of MVM with the capsid (43) and an interaction of a 3′-terminal fragment of the Aleutian mink disease virus with VP1 (44) have been demonstrated. In contrast, a direct interaction of AAV-2 ITRs, which are necessary and sufficient for AAV-2 genome encapsidation, with free capsid proteins or capsids produced in the absence of Rep proteins could not be demonstrated. Alternatively, the required specific association of AAV-2 DNA with the capsids could be achieved through Rep78 or Rep68 binding to the AAV-2 ITRs (20), followed by interaction with the capsid. We can imagine two mechanisms by which this could be accomplished (Fig. 9). The large Rep proteins bound to the AAV-2 DNA could (i) become directly incorporated into the capsid during capsid assembly or (ii) associate with capsids via interactions with the small and large Rep proteins already incorporated into the capsids. The fact that AAV-2 DNA packaging can be reconstituted in cell-free extracts (9, 47) tends to favor the second mechanism, because capsid assembly in such cell-free systems is inefficient (38). A preencapsidation complex, as postulated by this model, could also explain how the packaging of AAV-2 DNA genomes engaged in DNA replication or mRNA synthesis is avoided. Specific interactions of the Rep proteins covalently linked to the 5′ end of the genome with the preencapsidation complex could selectively sequester these molecules for DNA packaging. A question of future interest will concern the mechanism by which the virus avoids the initiation of packaging of multiple genomes and whether this is regulated by the number of Rep molecules anchored to the capsid surface. The results and the model presented here provide a basis for the analysis of these questions and a better understanding of the parvovirus DNA packaging process.
FIG. 9.
Working hypothesis for the specific binding of AAV-2 DNA to capsids based on Rep-capsid complexes as described in this report. (I) The association of Rep proteins with nonassembled capsid proteins would allow the binding of a Rep-bound single-stranded AAV genome to the capsid in a synchronous process. (II) In an alternative two-step process, the Rep-capsid complexes are formed first and are subsequently bound by the Rep-tagged packageable DNA to initiate the packaging process. Both processes involve Rep-VP, Rep-capsid, Rep-Rep, and Rep-DNA interactions.
ACKNOWLEDGMENTS
We thank Dirk Grimm for excellent assistance in the preparation of pDGΔVP and for critical reading of the manuscript. We are also grateful to Jean-Claude Jauniaux and Celina Cziepluch for technical and theoretical assistance in use of the yeast two-hybrid system.
REFERENCES
- 1.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the Rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Bohensky R A. Adeno-associated viruses: an update. Advances virus res. 1987;32:243–305. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 4.Buller R M, Rose J A. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore S F, Tattersall P. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J Virol. 1989;63:3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Lu S, Munshi N C. In vitro packaging of an infectious recombinant adeno-associated virus 2. Gene Ther. 1997;4:1167–1172. doi: 10.1038/sj.gt.3300514. [DOI] [PubMed] [Google Scholar]
- 10.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 11.Grimm D, Kern A, Rittner K, Kleinschmidt J A. Novel tools for the production and purification of recombinant AAV vectors. Hum Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Heilbronn R, Burkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–3018. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holscher C, Kleinschmidt J A, Burkle A. High-level expression of adeno-associated virus (AAV) Rep78 or Rep68 protein is sufficient for infectious-particle formation by a Rep-negative AAV mutant. J Virol. 1995;69:6880–6885. doi: 10.1128/jvi.69.11.6880-6885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horer M. Ph.D Thesis. HeidelbergHeidelberg, Germany: Ruprecht-Karls-Universität; 1996. [Google Scholar]
- 17.Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter L A, Samulski R J. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J Virol. 1992;66:317–324. doi: 10.1128/jvi.66.1.317-324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 20.Im D S, Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im D S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kube D M, Ponnazhagan S, Srivastava A. Encapsidation of adeno-associated virus type 2 Rep proteins in wild-type and recombinant progeny virions: Rep-mediated growth inhibition of primary human cells. J Virol. 1997;71:7361–7371. doi: 10.1128/jvi.71.10.7361-7371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyostio S R, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarty D M, Ni T H, Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992;66:4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller D E, Siegl G. Maturation of parvovirus LuIII in a subcellular system. II. Isolation and characterization of nucleoprotein intermediates. J Gen Virol. 1983;64:1055–1067. doi: 10.1099/0022-1317-64-5-1055. [DOI] [PubMed] [Google Scholar]
- 28.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 29.Myers M W, Carter B J. Assembly of adeno-associated virus. Virology. 1980;102:71–82. doi: 10.1016/0042-6822(80)90071-9. [DOI] [PubMed] [Google Scholar]
- 30.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad K M, Trempe J P. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology. 1995;214:360–370. doi: 10.1006/viro.1995.0045. [DOI] [PubMed] [Google Scholar]
- 32.Prasad K M, Zhou C, Trempe J P. Characterization of the Rep78/adeno-associated virus complex. Virology. 1997;229:183–192. doi: 10.1006/viro.1996.8431. [DOI] [PubMed] [Google Scholar]
- 33.Rose J A, Maizel J K, Inman J K, Shatkin A J. Structural proteins of adeno-associated viruses. J Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruffing M, Zentgraf H, Kleinschmidt J A. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol. 1992;66:6922–6930. doi: 10.1128/jvi.66.12.6922-6930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider S, Buchert M, Hovens C M. An in vitro assay of beta-galactosidase from yeast. Biotechniques. 1996;20:960–962. doi: 10.2144/96206bm03. [DOI] [PubMed] [Google Scholar]
- 37.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinbach S, Wistuba A, Bock T, Kleinschmidt J A. Assembly of adeno-associated virus type 2 capsids in vitro. J Gen Virol. 1997;78:1453–1462. doi: 10.1099/0022-1317-78-6-1453. [DOI] [PubMed] [Google Scholar]
- 39.Thomas J O, Kornberg R D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tratschin J D, Tal J, Carter B J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral rep gene product. Mol Cell Biol. 1986;6:2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weger S, Wistuba A, Grimm D, Kleinschmidt J A. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willwand K, Hirt B. The minute virus of mice capsid specifically recognizes the 3′ hairpin structure of the viral replicative-form DNA: mapping of the binding site by hydroxyl radical footprinting. J Virol. 1991;65:4629–4635. doi: 10.1128/jvi.65.9.4629-4635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willwand K, Kaaden O R. Capsid protein VP1 (p85) of Aleutian disease virus is a major DNA-binding protein. Virology. 1988;166:52–57. doi: 10.1016/0042-6822(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 45.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, Muzyczka N. In vitro packaging of adeno-associated virus DNA. J Virol. 1998;72:3241–3247. doi: 10.1128/jvi.72.4.3241-3247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]